Abstract
CooA is a heme-dependent CO-sensing transcription factor that has three observable heme coordination states. There is some evidence that each CooA heme state has a distinct protein conformation; the goal of this study was to characterize these conformations by measurement of their structural stabilities through guanidine hydrochloride (GuHCl) denaturation. By studying the denaturation processes of the Fe(III) state of WT CooA and several variants, it was possible characterize independent unfolding processes for each domain of CooA. This information was used to compare the unfolding profiles of various CooA heme activation states (Fe(III), Fe(II), and Fe(II)-CO) to show that the heme coordination state changes the stability of the effector binding domain. A mechanism consistent with the data predicts that all CooA coordination states and variants undergo unfolding of the DNA-binding domain between 2 and 3 M GuHCl with a free energy of unfolding of ~17 kJ/mol, while unfolding of the heme domain is variable and dependent on the heme coordination state. The findings support a model in which changes in heme ligation alter the structural stability of the heme domain and dimer interface, but do not alter the stability of the DNA-binding domain. These studies provide evidence that the domains of transcription factors are modular, and that allosteric signaling occurs through changes in the relative positions of the protein domains without affecting the structure of the DNA-binding region.
Keywords: allostery, heme sensor, transcription factor, equilibrium unfolding, protein conformation, conformational states
Allosteric activation of proteins is a process by which a change in protein activity is triggered by binding of an effector molecule. The induced-fit (or end-state) theory of allostery postulates that changes at the regulatory site of an allosteric protein induce a conformational change such that the inactive and active states of the protein are structurally distinct (1, 2). Ideally, structural information collected in the presence and absence of effector molecule would allow for characterization of the structural requirements for protein activation. In reality, however, it can be difficult to acquire structures for all conformational states of a protein. Furthermore, a thorough understanding of protein allostery requires information about the thermodynamics of the transitions between these conformations, not just images of their static endpoints.
Heme dependent gas-sensors are a burgeoning group of allosteric proteins with a wide range of sizes, domain architecture and function (3); yet, it has been difficult to characterize the global structural changes that accompany effector binding among members of this group. In these proteins, the activation process is initiated by binding a gas molecule at the heme cofactor, and the structural and coordination rearrangements in the heme domain are transmitted to a domain that is responsible for activity of the protein. Studies of truncated heme domains have shed light on structural changes that occur in and around the heme pocket (4–8), but global conformational changes in heme-containing sensor proteins are largely uncharacterized due to the absence of crystal structures of full-length sensor proteins. Our best understanding of structure function relationships in heme-containing gas sensors is based on studies of CooA1, the only gas-sensor to be crystallized as a full-length polypeptide (9). Although several structures of CooA proteins exist (10, 11), only one corresponds to the wild-type (WT) form of the protein in a physiologically relevant state (9). Many questions about allosteric regulation in CooA and heme-dependent sensors still remain to be answered, most notably: how does the signaling molecule cooperate with protein structural elements to transmit the allosteric signal and alter global protein structure?
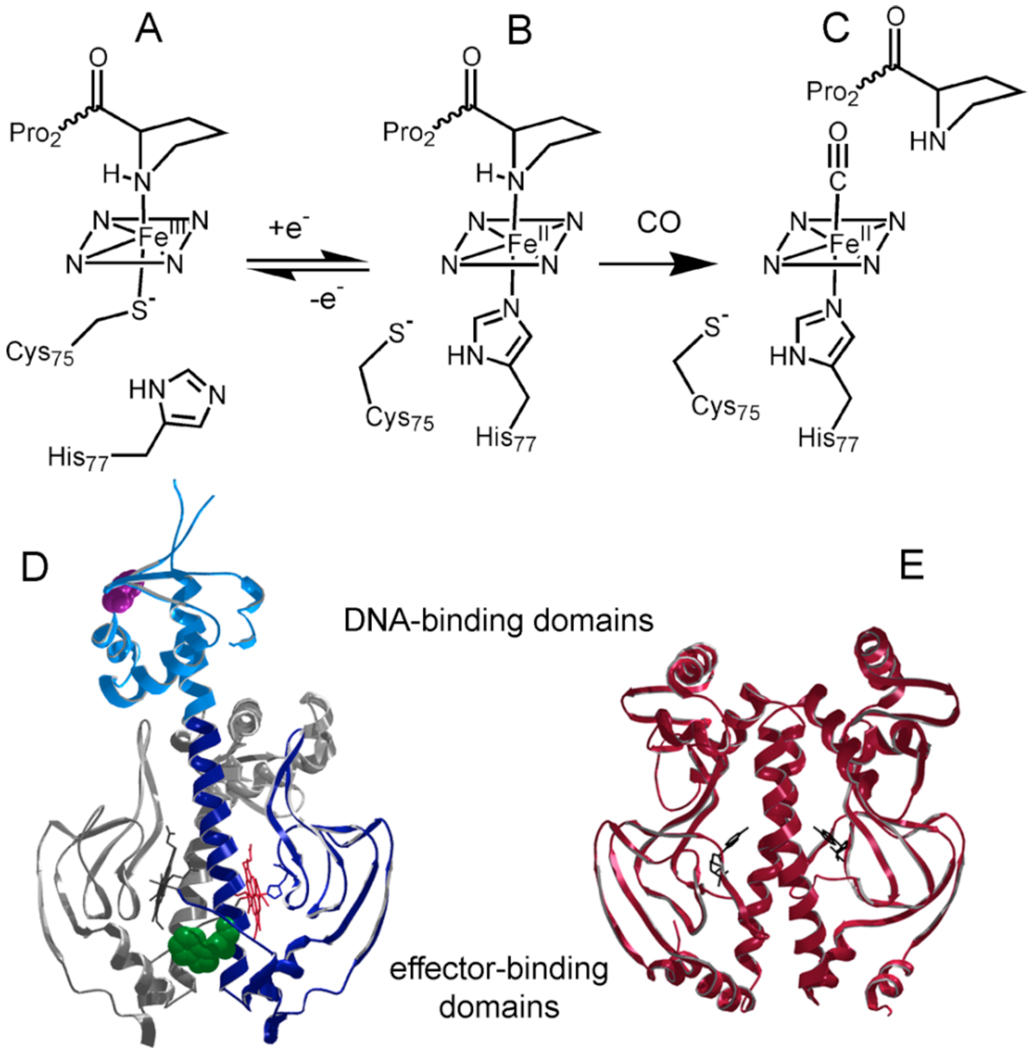
Rhodospirillum rubrum CooA is a CO-sensing transcription factor (12). In the presence of CO, CooA induces production of a carbon-monoxide dehydrogenase (CODH) system that allows the organism to survive on CO as a sole energy source (12, 13). CooA is a 49 kDa homodimer with an N-terminal effector-binding domain and a C-terminal DNA-binding domain that contains a helix-turn-helix motif (14). The effector domain of CooA contains two hemes (one in each monomer), and three heme activation states are known for Rr CooA (Figure 1). Under aerobic conditions, Rr CooA has a Fe(III) heme that is six-coordinate with a proximal cysteine ligand and a distal N-terminal proline ligand (15–17). This state is incapable of binding CO and is inactive for DNA-binding. Upon reduction, CooA undergoes a proximal ligand switch from cysteine to histidine, but retains the distal proline ligand (9, 15, 18, 19). This state is poised for activation. Exposure of the reduced state to CO results in replacement of the distal proline to give a heme with histidine/CO coordination (20). The CO-bound state is active for binding to DNA (12, 21, 22). It has been suggested that CooA has three distinct conformations corresponding to the three heme coordination states (Figure 1) (18).
Figure 1. Heme activation states and structure of CooA.
(A) In the Fe(III) state, the heme is ligated by Cys75 and Pro2 (proline is from the opposite monomer) (15–17). (B) When the heme is reduced, the Cys75 ligand is replaced by His77 (9, 15, 18, 19). (C) CooA is activated when CO binds to the reduced heme, displacing the Pro2 ligand and enabling DNA binding (20). (D & E) Ribbon diagrams of the Fe(II) state of CooA (D) and CRP (E). Both proteins are dimers, with similar structure in the effector-binding and DNA-binding domains. CRP is in the active state with cAMP effector bound and DNA-binding helices exposed for contact with DNA. CooA (one monomer in color) is in an inactive state, with the DNA-binding domains rotated such that the helices that contact DNA are solvent inaccessible. The effector domain of CooA is dark blue, while the DNA-binding domain is light blue. Trp110 (green) and Phe164 (purple) of CooA are shown with space-filled side-chains; in F164W CooA, Phe164 is replaced by tryptophan. The truncated variant contains residues 1–132, which is the dark blue domain in this representation. This figure was prepared using PDB viewer and PDB files 1FT9 and 1G6N (9, 24).
CooA is a member of the cAMP receptor protein (CRP)/fumarate nitrite reductase (FNR) family of homodimeric transcriptional regulators (9). Members of this family appear to have a common set of structural elements that are critical for allosteric activation. One of these elements is a coiled-coil dimer interface that has some degree of leucine zipper character (9, 23, 24). Evidence suggests that effector binding has a direct influence on residues at this dimer interface. Mutations in CooA that strengthen the leucine zipper interactions result in effector-independent DNA-binding activity (25), and cAMP appears to regulate CRP through specific contacts with the residues at the dimer interface (26). The significance of the subunit interface is most obvious in FNR, which is regulated through a monomer-dimer equilibrium (27). Another common element is a “hinge” sequence that exists between the dimer interface and the DNA-binding domain. Mutagenesis of hinge residues has shown that flexibility in this region is essential for proper communication of signal in both CooA and CRP (9, 28–32).
It has been suggested that activation in CooA and CRP is dependent on repositioning of the DNA-binding domain through the hinge to allow exposure of a critical helix (the F helix) for contact with DNA. All studies of the inactive structures of CooA (9) and CRP (33, 34) show the F-helix to be buried and unable to make contacts with DNA. Conversely, in the studies of the active states of CooA and CRP, the F-helix is on the surface of the protein and exposed for interactions with DNA (24, 33, 35). The structure of active CRP in the presence of DNA clearly shows hydrogen bonding between three residues in each F-helix and DNA (36, 37), and mutations in the Rr CooA F-helix at suspected H-bonding positions disrupted CooA-DNA interactions (38). In addition, many other studies of CooA and CRP activation have implicated an effector-dependent change in the position of the DNA-binding domain (10, 11, 35) (reviewed in (35)).
In the current study, we sought to further characterize conformational differences among the activation states of a single member of the CRP family, Rr CooA. If global conformational changes occur in CooA, they might be expected to impart changes in overall structural stability of the protein. A method that allows for measurement and comparison of protein stabilities is chemical denaturation (39), wherein differences in the slope, transition midpoints, and overall shape of the unfolding curves signify structural differences amongst proteins. Ligand binding has been shown to alter protein stability (40–45). Furthermore, heme proteins such as hemoglobin (46, 47) and cytochrome c (48) have been unfolded in multiple heme coordination states, and stability differences have been observed. The current studies were designed to measure possible variations in stability for the conformations that correspond to the three heme coordination states of Rr CooA.
We report herein GuHCl unfolding studies of Fe(III) CooA monitored by visible, fluorescence, and CD spectroscopy. We used two CooA variants to explore the mechanism of unfolding in the WT protein: a truncated form of CooA that lacks the DNA-binding domain provided information specific to the heme domain; and F164W CooA, a variant containing an additional tryptophan fluorescence signal, provided information specific to the DNA-binding domain. We found that the domains of Fe(III) WT CooA and variants unfold independently, with distinct transitions that are easily visible in the unfolding curves. Based on the results from the variants, we were able to interpret the differences in the Fe(III), Fe(II), and Fe(II)-CO WT CooA denaturation curves. We fit these unfolding curves for multiple unfolding intermediates to generate free energy values that describe the stabilities of the CooA domains. Models describing the denaturation process are presented which have implications for the function of both gas-sensing heme proteins and allosterically regulated transcription factors. Our findings demonstrate that domain modularity plays a role in signal transduction and activation in these proteins.
MATERIALS AND METHODS
Materials
MOPS was purchased from Sigma. Guanidine hydrochloride (ultrapure) was from ICN. Sodium dithionite was purchased from Fluka. Unless otherwise specified, all manipulations were carried out in 25 mM MOPS buffer (pH 7.4) with 100 mM NaCl.
Strains and plasmids
The construction of strains overexpressing WT CooA in E. coli backgrounds has been previously described (18, 25). F164W CooA was overexpressed in strain UQ2412, which is a derivative of strain UQ1639 (18). F164W CooA was constructed in a pEXT20-based expression plasmid as previously described (49). Site directed mutagenesis involved PCR amplification of CooA with primers designed to incorporate the desired nucleotide changes as described elsewhere (50).
WT, F164W, and truncated CooA purification
WT CooA was purified to greater than 95% as described (18) and stored at −80°C in 25 mM MOPS buffer (pH 7.4) with 100 mM NaCl until use. F164W CooA was purified and stored using the same procedures. The CooA truncate was purified as described (51) and stored at −80°C in 25 mM MOPS buffer (pH 7.4) with 750 mM NaCl, 10% glycerol, and 2 mM DTT. WT and F164W CooA had the same visible absorption spectrum, and were assumed to have the same ε424 for the Fe(III) state. Protein concentration was determined by extinction coefficient of the heme cofactor (ε424 = 105 mM−1cm−1) (52), or by bicinchoninic acid assays (53).
In vitro protein activity assays
Activity of WT and F164W CooA was measured at room temperature using a fluorescence polarization method (49). Briefly, a 26-base pair target DNA containing pCooF was labeled with Texas Red on the 5’ end of one strand of the DNA. The labeled oligonucleotide was titrated with protein, and the protein-DNA complex was detected by an increase of fluorescence anisotropy. Dissociation constants were calculated by non-linear least squares fit (54).
Visible absorption spectroscopy
CooA unfolding was measured by loss of intensity at strongest band in the absorption spectrum (the Soret peak) with increasing concentration of GuHCl. The Soret wavelengths used were: 424 nm for Fe(III) WT, Fe(III) F164W, and Fe(III) truncate CooA; 425 nm for Fe(II) WT CooA; and 423 nm for Fe(II)-CO WT CooA) (51, 52). Visible absorption spectra were recorded at room temperature on a Varian Cary 4 Bio spectrophotometer with a bandwidth of 0.5 nm. Samples of protein (2 µM) or hemin (4 µM) were monitored over the full wavelength range (350 to 800 nm) to investigate heme changes. Intensity was measured using a cuvette with a 1 cm pathlength.
CD spectroscopy
CD experiments were performed at room temperature with an Aviv Model 202SF Stopped Flow CD Spectrometer or an MOS-450 spectropolarimeter from BioLogic (BioLogic Science Instruments, Claix, France). CD spectra were measured over a range of 200 nm to 400 nm with a path length of 0.1 or 0.2 cm. Spectra were recorded in 1 nm increments with at least a 1 s averaging time. Single wavelength measurements were taken with a 10 s averaging time. The protein concentration was 2 µM for all samples. CD data for WT CooA and F164W CooA were recorded in 25 mM MOPS, 100 mM NaCl, pH 7.4 buffer. The Fe(II) state for WT CooA was generated by the addition of a minimal amount of buffer containing dithionite to Ar purged samples of the Fe(III) protein. The Fe(II)-CO state for WT CooA was prepared by addition of CO gas to the headspace of the corresponding Fe(II) sample. Conversion to the appropriate state (as well as minimal residual concentration of dithionite) was confirmed by visible absorption spectroscopy.
Fluorescence spectroscopy
All fluorescence measurements were made on an ISS PC1 photon-counting fluorometer (ISS Instruments Inc., Champaign, IL) at room temperature. Data were recorded in a cuvette with a 2 mm excitation and a 1 cm emission pathlength. All protein samples were 2 µM in concentration. Emission spectra were recorded from 290 to 400 nm with excitation and emission slits of 1.0 mm. Corrections were made by subtracting the spectrum of the appropriate buffer. For titrations, an excitation wavelength of 296 nm was used to select for tryptophan fluorescence with excitation slits of 0.5 mm and emission slits of 2.0 mm. Samples were monitored at an emission wavelength of 352 nm (the wavelength of maximum intensity changes between 0 and 6.25 M GuHCl) to measure unfolding.
Titrations with guanidine hydrochloride
GuHCl stocks were prepared by dissolving solid denaturant in the buffer appropriate for storage of each protein, followed by filtration. Concentrations of denaturant were determined by refractive index (55). In each GuHCl denaturation experiment, 26 samples of CooA were titrated with GuHCl from 0 to 6.25 M, at a protein concentration of 2 µM, unless otherwise specified. Unfolding was intiated by dilution of a concentrated protein stock into the appropriate GuHCl buffer. All samples were incubated at room temperature for 2 h before measurement using the procedures outlined above. The data shown in Figure 2–Figure 4 are one representative data set, while Figure 5 shows raw intensity data with error bars representing the standard deviation from three measurements.
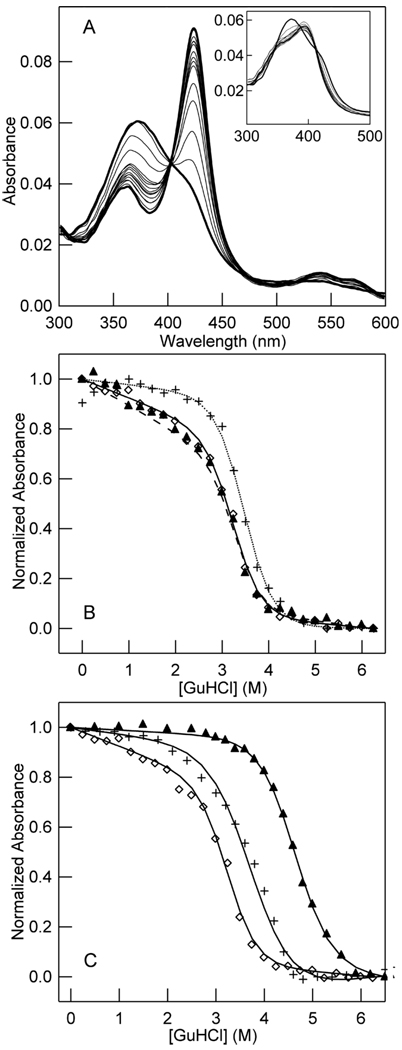
Figure 2. Denaturation of CooA monitored by visible spectroscopy of the heme for WT and variants of CooA in the Fe(III) state (A, B) and the Fe(II) and Fe(II)-CO states of WT CooA (C).
Samples of a fixed protein concentration (2 µM) were titrated with GuHCl from 0–6.25 M and allowed to unfold for 2 h at 25 °C before the full absorption spectrum was measured. Lines represent fits to a two-state unfolding model for all proteins and variants. (A) Visible absorption spectra show isosbestic changes between 0 and 3.75 M GuHCl. Samples between 3.75 and 6 M GuHCl show an additional isosbestic transition (A-inset). (B) Changes in Soret intensity at 424 nm as a function of increasing GuHCl for Fe(III) WT (◇), Fe(III) F164W (▲), and Fe(III) truncated CooA (+). (C) Changes in the absorbance intensity of Fe(III) WT (◇), Fe(II) WT (+), and Fe(II)-CO WT (▲) CooA at the Soret peak for each species (424, 425, and 423 nm, respectively) as a function of GuHCl concentration.
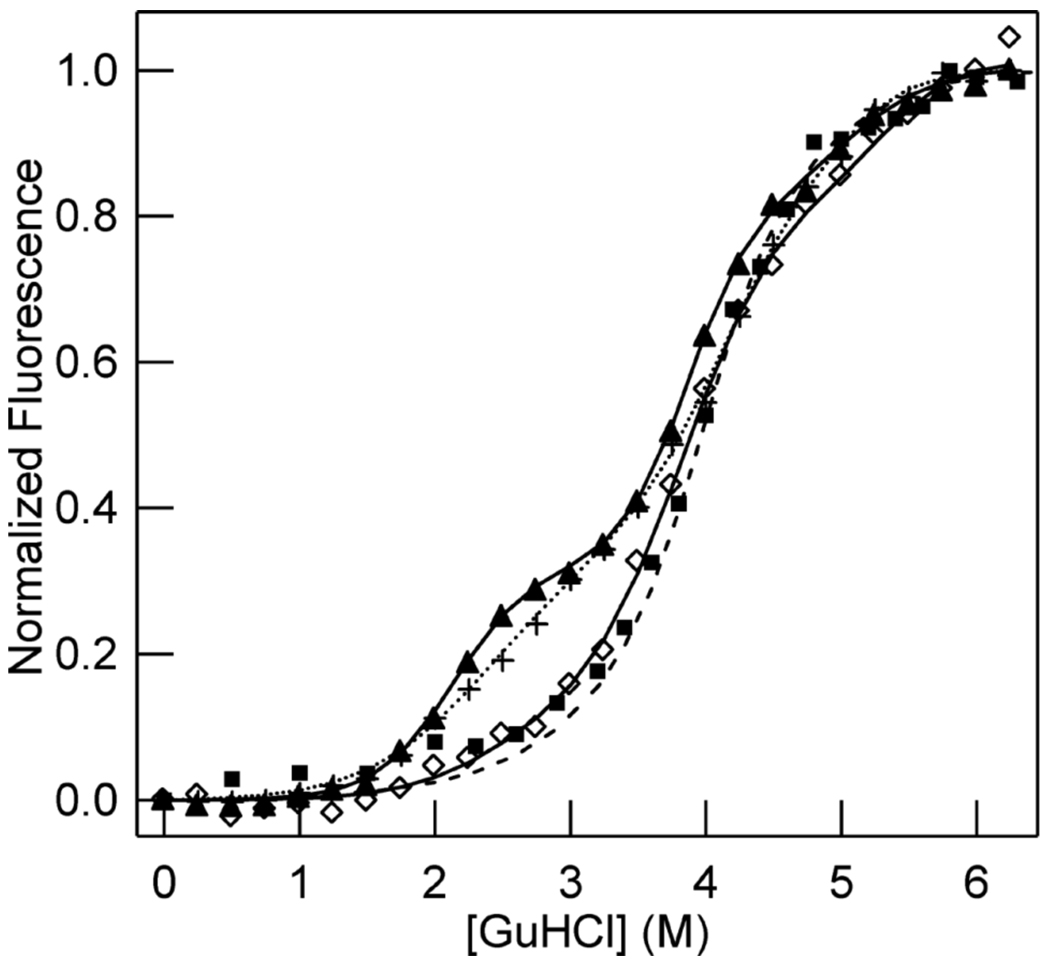
Figure 4. Denaturation of WT and F164W CooA monitored by tryptophan fluorescence intensity.
Samples of a fixed concentration of CooA (2 µM) were titrated with GuHCl from 0–6.25 M and allowed to unfold for 2 h at 25 °C before the fluorescence was measured. The normalized, background-corrected fluorescence intensity at 352 nM at each concentration of GuHCl are plotted. The lines represent fits using a four-state model (see below). Changes in fluorescence of Fe(III) WT (◇), Fe(III) F164W (▲), Fe(II)-CO F164W (+), and Fe(II)-CO WT (■) CooA as a function of GuHCl are shown.
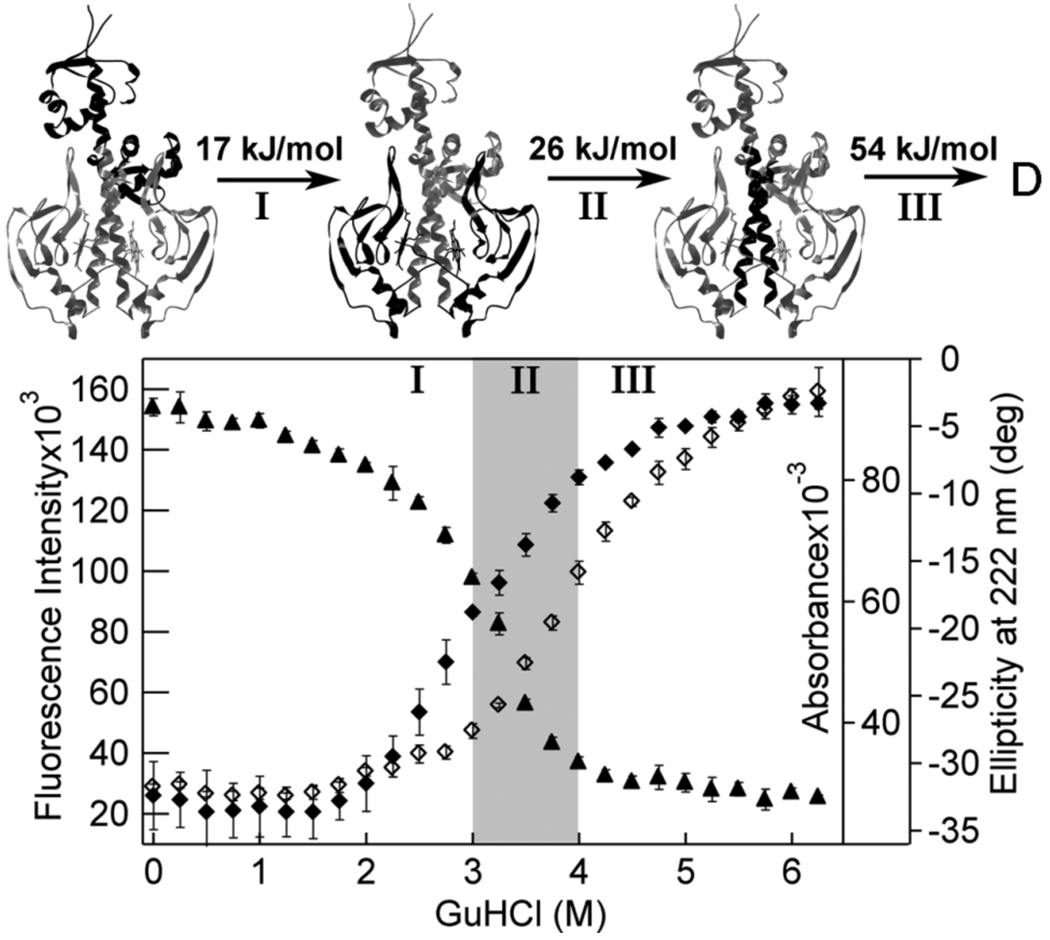
Figure 5. Model for denaturation of Fe(III) WT CooA.
The averaged raw data as a function of GuHCl concentration collected for Fe(III) WT CooA using fluorescence intensity at 352 nm (◇), absorbance intensity at 424 nm (▲), and CD intensity at 222 nm (♦) are plotted for comparison. The error bars represent the standard deviation for three measurements. Above the chart, the CooA crystal structure is presented with the area unfolded in each step darkened for reference (the protein is presented in the native state for clarity). In Step I, between 0 and 3 M GuHCl, the visible absorbance intensity shows a modest decrease. The CD and fluorescence intensities show more significant changes between 2 and 3 M GuHCl. In this step, the protein unfolds from native to an intermediate that has an unfolded DNA-binding domain and a mostly native effector domain. The free energy associated with unfolding between Step I and II has been fitted to 17 kJ/mol. In Step II, between 3 and 4 M GuHCl, all methods show dramatic changes, indicating that the heme domain unfolds and the protein reaches a second intermediate. The free energy of this step has been fitted to ~26 kJ/mol. In Step III, above 4 M GuHCl, the fluorescence continues to increase, which is due to final unfolding of the effector domain and dissociation of the monomers, with a fitted free energy of ~54 kJ/mol. The free energy values are taken from the fits to visible, fluorescence, and CD, as shown in Figure 2–Figure 4, and are an approximation of the values derived from all methods and variants of Fe(III) CooA.
Fe(III) WT CooA titrations were carried out at different concentrations of protein. The samples were prepared using the titration conditions given above, but at 4, 2, or 0.5 µM protein. Changes in CooA were measured using visible absorbance, CD, and fluorescence as described.
Preparation and handling of the Fe(II) CooA and Fe(II)-CO CooA samples and titrations required special techniques to maintain the proper oxidation state. All buffers and GuHCl stock solutions were degassed by a first step of filtration through a 0.2-µm filter or centrifugation at 10,000 × g, followed by a second step of purging with Ar gas. For Fe(II) CooA and Fe(II)-CO CooA titrations, the protein stock was reduced with solid Na2(S2O4) (dithionite) before it was titrated. For Fe(II)-CO CooA titrations, the Fe(II) CooA stock solution was exposed to CO gas in the headspace to form the CO adduct. Samples for unfolding titrations were prepared in a nitrogen-filled glovebox, using the methods described above. After incubating for 2 h in the glovebox, visible absorbance or CD spectra were measured for each sample. The presence of the appropriate heme coordination state was confirmed by visible absorption spectroscopy both immediately before addition of GuHCl to the samples and immediately before measurement of unfolding by the requisite spectroscopic method.
Measurement of fluorescence unfolding curves for Fe(II)-CO WT and Fe(II)-CO F164W CooA required removal of excess dithionite reductant. Dithionite quenches tryptophan fluorescence, however quenching does not occur when the reductant is fully oxidized. Fluorescence spectroscopy is possible for the Fe(II)-CO state of CooA due to the slow CO off-rates for Fe(II)-CO CooA and CO-bound samples may be exposed to an aerobic environment without a change in heme coordination. To measure fluorescence of the CO-adduct, the samples were exposed to air to allow for oxidation of the dithionite immediately before measurement of tryptophan fluorescence. The presence of the appropriate heme coordination state and complete removal of dithionite were confirmed by visible absorption spectroscopy immediately prior to measurement of fluorescence. The same techniques are not possible for detection of fluorescence in Fe(II) WT CooA because the protein reoxidizes readily in the absence of reductant.
Reversibility
Fe(III) WT CooA cannot be diluted from 6 M to 0 M GuHCl without precipitation. In an exemplary experiment, a sample of 50 µM Fe(III) WT CooA and 6 M GuHCl was allowed to unfold for two hours at room temperature before being placed in a Float-A-Lyzer (Spectrum Laboratories) regenerated cellulose dialysis cartridge with a molecular weight cutoff of 10,000 Da. The sample was then dialyzed against two changes of 2 M GuHCl before being dialyzed against 0 M GuHCl buffer. Precipitation was observed when the protein was dialyzed against GuHCl concentrations below 0.5 M, and visible absorption spectra indicated that the heme environment of the refolded Fe(III) WT CooA was severely perturbed.
A titration-dilution method was pursued to investigate reversibility in the CD and fluorescence spectra. WT Fe(III) CooA (50 µM) was denatured for 2 h at a concentration of 5 M GuHCl at room temperature. The presence of unfolded CooA was confirmed through visible spectroscopy. The protein was then diluted to 2 µM and the final GuHCl concentration desired, and allowed to incubate for 2 h at room temperature. An elevated fluorescence signal was observed for dilution from 5 M GuHCl to below 3 M GuHCl; therefore, a separate dilution was carried out from 3 M to 0.5 M GuHCl. Only in CD spectroscopy did we observe full recovery of signal, indicating the refolding of the secondary structure between 5 M to 0.5 M GuHCl. Persistent precipitation below 0.5 M GuHCl prevented investigation of the activity of the renatured protein, as high concentrations of salt interfere with the CooA activity assay
Quaternary structure measurements
GuHCl-induced unfolding of WT Fe(III) CooA was analyzed by size exclusion chromatography using a Superdex 75 prepacked column (16 mm× 600 mm; gel volume 120 mL; GE). The high GuHCl concentrations greatly perturbed the elution times for CooA, and calibration using molecular weight standards confirmed that protein separation was not adequate for analysis of CooA quaternary structure at 5 M GuHCl. Furthermore, size exclusion chromatography was not possible for Fe(II) or Fe(II)-CO CooA, which require reducing conditions to maintain the heme states. Other methods, such as analytical ultracentrifugation, were unsuccessful for analysis of CooA quaternary state due to the formation of aggregates.
Analysis of denaturation data
The analysis of two-state unfolding behavior using linear extrapolation has been described (56), and we used the standard two-state methods and equations to analyze visible absorbance data for all protein variants and states, and CD unfolding data for Fe(III) truncated CooA. Briefly, the pre- and post-transition baseline slopes and intercepts were fitted using a linear least squares regression, and these parameters were fixed while the free energy (ΔGH2O) and slope (m) for each curve were fitted to the equations described using linear extrapolation (57).
CD data for Fe(III) WT, Fe(III) F164W, Fe(II) WT and Fe(II)-CO CooA were fitted for a three state model with a single intermediate, and all fluorescence data were fitted to a four state model with two intermediates (see below). The choice of a model was based on i) evidence for multiple states in the full-wavelength visible absorption spectra; ii) the presence of an intermediate indicated by irreversibility of unfolding; and iii) use of the fewest number of states that could reasonably fit the obvious transitions in the unfolding curves. Several models are possible for the unfolding process of CooA, but we chose the simplest model that would allow for comparison of unfolding curves and determination of relative values for free energy of unfolding among CooA states and variants.
Although the four-state model is presented below, the three-state fits follow the same method but include only one intermediate. The four-state model assumed for the unfolding of Fe(III) WT CooA is shown in equation 1.
| (1) |
Using this model, equilibrium constants are defined for the folding processes, in which the denatured state D folds to each successive species, as shown in equation 2– equation 4.
| (2) |
| (3) |
| (4) |
The observed signal is proportional to the signal intensity of the species (Y), corrected for the fractional population of each state (f) (equation 5)
| (5) |
and the fraction of each state is related to the concentration of the state over the sum of all species (equation 6–equation 9).
| (6) |
| (7) |
| (8) |
| (9) |
Taking the fraction of folded protein as an example, these fractions are related to the equilibrium constants by equation 10.
| (10) |
The equilibrium constant at each guanidinium hydrochloride concentration for each folding process, (D→F), (D→I), (D→J), is related to the free energy of this process. Using the linear regression method for analysis of denaturation data (56), equation 5 may be rearranged to give an expression for total spectral intensity in terms of ΔGH2O and slope of each folding process. The data for total intensity were fitted by fixing the intensity of each intermediate and varying ΔGH2O and slope for each folding process. Error between the calculated intensity and the measured data was minimized using non-linear least squares regression. The values reported for the free energy change between the folded protein and the intermediates ΔGFI, ΔGIJ, ΔGJD were acquired by calculating the difference between the free energy of each folding process and the free energy of the conversion from folded to denatured protein (e.g. ΔGFI = ΔGDI-ΔGDF). Typically, the free energy values collected from these fits were correlated, but starting free energy values and spectral intensities for intermediates were used that were derived from the two-state fits for domain stability and the observed spectra of the above protein variants. The data were fitted to give the free energy of folding in the absence of denaturant, but are presented as free energy of unfolding (ΔGH2O) in Table 1 for clarity.
Table 1.
Comparison of calculated free energies for the various steps in the denaturation of CooA
| State/Protein | Probe | ΔG1H2O(kJ/mol) (~2–3 M GuHCl) |
m1 | ΔG2H2O (kJ/mol) (~3–4 M GuHCl) |
m2 | ΔG3H2O(kJ/mol) (>4 M GuHCl) |
m3 |
|---|---|---|---|---|---|---|---|
| Fe(III) WT | visa | 26 (1.1) | 8.1 (0.08) | ||||
| CDa | 18 (0.1) | 6.3 (0.8) | 28 (1.7) | 6.2 (0.4) | |||
| fluora | 16 (3.3) | 6.8 (1.6) | 24 (3.1) | 6.4 (0.8) | 54 (7.8) | 9.9 (1.1) | |
| Fe(III) F164W | visa | 27 (2.5) | 7.9 (0.6) | ||||
| CDb | 12 (0.4) | 4.7 (0.1) | 29 (2.3) | 6.3 (0.8) | |||
| fluora | 13 (5.6) | 6.4 (2.4) | 28 (8.6) | 7.4 (2.2) | 51 (9.7) | 8.7 (1.3) | |
| Fe(III) truncate | visa | 28 (2.7) | 7.7 (0.5) | ||||
| CDc | 24 | 5.3 | |||||
| Fe(II)-CO WT | visa | 29 (1.3) | 7.3 (0.5) | ||||
| CDa | 17 (3.7) | 7.0 (1.5) | 21 (1.6) | 4.7 (0.4) | |||
| fluorb | 12 (1.7) | 5.6 (1.1) | 22 (3.2) | 6.8 (1.3) | 29 (1.1) | 6.2 (0.7) | |
| Fe(II)-CO F164W | fluora | 13 | 6.4 | 20 | 5.9 | 29 | 6.7 |
| Fe(II) WT | visa | 22 (0.1) | 5.6 (0.4) | ||||
| CDb | 16 (0.3) | 5.5 (0.2) | 46 (0.9) | 10.1 (0.9) |
Individual titration curves collected at room temperature were fitted to the appropriate model to get values for free energy of unfolding in the absence of denaturant (ΔGH2O), and the values from three fits were averaged for each unfolding transition with the standard deviation for each transition shown as error.
Free energies are averages of values from two curves with the standard deviation shown as error.
Values represent a single measurement.
RESULTS
Visible absorption spectroscopy allows for observation of unfolding of the effector domain in Fe(III) WT and Fe(III) variant CooA proteins
The heme of CooA presents an opportunity to follow protein unfolding using visible absorption spectroscopy to monitor changes in protein environment around the cofactor. Visible absorbance data for the unfolding of Fe(III) WT CooA show the presence of at least one intermediate heme state during the unfolding process (Figure 2A). Fe(III) WT CooA has a Soret peak in the visible spectrum at 424 nm that disappears with increasing concentrations of GuHCl. From 0 to 3.75 M GuHCl the heme spectra show an isosbestic change in absorbance marked by the disappearance of the strong Soret band at 424 nm and the growth of a broad band at roughly 360 nm. These spectra then show a second isosbestic change between 3.75 and 6.25 M GuHCl. The final spectrum of unfolded Fe(III) WT CooA is similar to that of Fe(III) PPIX in 6.25 M GuHCl (data not shown). The absorbance unfolding profile of Fe(III) WT CooA shows an unfolding transition between 3 and 4 M GuHCl (Figure 2B). Visible absorbance unfolding curves for titrations of Fe(III) WT CooA at 2 and 4 µM were the same; however, at protein concentrations of 0.5 µM, the unfolding curves show transition regions that occur at lower concentrations of GuHCl than those at 2 and 4 µM. At 4 µM protein, the unfolding curve shows a decrease in intensity in the pre-transition region (Figure 2B). We attribute these pre-transition changes to alterations in the heme environment at low concentrations of GuHCl (<3 M) due to unfolding of the DNA binding region (vide infra).
We unfolded two protein variants to aid in characterization of the CooA unfolding process. The first variant that was unfolded is a truncated version of Fe(III) CooA that contains residues 1–132 of the WT protein. The truncated protein consists of only the heme domain of CooA and has been crystallized as a dimer with almost identical structure to the heme domain of the WT protein (51). The Fe(III) truncated version of CooA shows a visible absorbance unfolding transition that occurs in the same GuHCl concentration range as that of Fe(III) WT CooA, signifying that removal of the DNA-binding domain has virtually no effect on the stability of the heme domain as observed through visible spectroscopy. Unlike Fe(III) WT CooA, the Fe(III) truncate shows minimal intensity changes between 0–3 M GuHCl, indicating that that the decrease exhibited by WT Fe(III) CooA at these concentrations may be related to an unfolding process in the DNA-binding domain.
The second variant, Fe(III) F164W CooA, has a substitution to provide an additional tryptophan fluorescence signal in the DNA-binding domain and was selected to help in interpretation of the fluorescence unfolding data (see below). The mutated residue is far from the heme domain (Figure 1D), and has no effect on the native absorbance, fluorescence, or CD spectrum of the protein (see Supporting Information Figure S1 for CD spectra). As is expected, the Fe(III) F164W variant shows the same visible absorbance unfolding profile as the Fe(III) WT protein (Figure 2B). Taken together, these results suggest that: i) the heme domain of Fe(III) CooA is a structural unit that unfolds between 3 and 4 M GuHCl regardless of the absence or presence of a DNA-binding domain, and ii) visible absorbance data primarily reflect changes in the heme domain, and iii) the slope in the pre-transition region of Fe(III) WT CooA curve appears to be correlated with changes in the DNA-binding domain.
Changes in heme ligation and activation state of CooA affect the unfolding of the heme domain
To detect possible differences in the structural stability of the various heme coordination states of CooA, we compared visible absorbance changes that accompany GuHCl denaturation of Fe(II)-CO and Fe(II), and Fe(III) WT CooA (Figure 2C). Upon titration of Fe(II)-CO WT CooA with increasing concentrations of GuHCl, the Soret peak at 423 nm in the visible absorption spectrum converts isosbestically to a peak at 410 nm, and the final state (6.25 M GuHCl) has a spectrum that appears to be the same as that of Fe(II)-CO protoporphyrin IX (PPIX) in 6.25 M GuHCl solution (data not shown). The visible absorbance unfolding profile of Fe(II)-CO WT CooA differs from that of Fe(III) WT CooA in that the Fe(II)-CO Soret peak shows a large decrease in absorbance intensity between 4 and 5 M GuHCl (Fig. 2C).
The Fe(II) WT CooA visible unfolding profile differs from both the Fe(II)-CO WT and Fe(III) WT unfolding curves. Exposure of Fe(II) WT CooA to increasing concentrations of GuHCl causes a decrease in intensity of the Soret peak at 425 nm, and the final state appears to be the same as free Fe(II) PPIX in 6.25 M GuHCl (data not shown). The Soret absorbance intensity of Fe(II) WT CooA starts to show significant decreases at 2 M GuHCl that continue until 4.75 M GuHCl (Figure 2C). These data would suggest that the Fe(II) WT CooA heme domain has stability to GuHCl denaturation that is intermediate between the lower stability of Fe(III) WT CooA and the higher stability of the Fe(II)-CO WT CooA. The increased stability of the Fe(II)-CO heme domain as compared to the Fe(III) state is not derived from changes in the DNA-binding domain. We titrated the Fe(II)-CO state of truncated CooA (residues 1–132) with GuHCl (data not shown), and the absorbance intensity of the CO-bound form of the variant showed no change between 0 and 4.5 M denaturant. Fe(II)-CO truncate CooA showed a folding transition between 4.5 and 6 M denaturant, which is within a concentration range much higher than the transition observed for the Fe(III) truncate CooA (3–4 M). This result suggests that the visible absorbance unfolding curves for the WT CooA protein correlate to the relative stability of the heme domain in the three states of CooA, and are unrelated to the presence of the DNA binding domain.
The visible absorbance data for unfolding of the various states and variants of CooA were fitted to a two-state model according to published methods (Table 1) (56). These fits give a relatively consistent ΔGH2O (free energy of unfolding in the absence of denaturant) of ~27 kJ/mol for Fe(III) WT, Fe(III) truncated, and Fe(III) F164W CooA. Fits for the Fe(II)-CO WT CooA and Fe(II) WT CooA absorbance unfolding curves give a ΔGH2O of 29 kJ/mol and 22 kJ/mol, respectively.
CD spectroscopy allows for observation of unfolding of both the heme and DNA-binding domains of Fe(III) WT CooA
Monitoring the CD spectrum of CooA as a function of GuHCl concentration is a logical way to examine unfolding of both domains of the WT protein. The DNA-binding domain of CooA is rich in α-helix, and the protein dimer interface in the effector-binding domain is a long α-helical leucine-zipper motif. Both of these domains are expected to contribute to the native CD signal at 222 nm. A comparison of the native CD spectrum of the Fe(III) WT and Fe(III) truncate CooA (Supporting Information Figure S1) shows the variant to have a much weaker intensity at 222 nm, presumably because the variant lacks the signal component that is derived from the helix-turn-helix motif.
The CD spectrum of Fe(III) WT CooA shows a decrease in intensity with increasing GuHCl concentration, consistent with unfolding of α-helix in the protein. The greatest change in CD signal occurs in a range of GuHCl concentrations (2–4 M GuHCl, Figure 3A) that differs from that observed for the Fe(III) WT visible absorbance unfolding curves (>3 M, Figure 2B). The CD intensity shows changes over a broad range of GuHCl concentrations, possibly because CD data reflect the unfolding of both CooA domains. The CD unfolding profile showed little concentration dependence for all studied protein concentrations. CD profiles for unfolding of Fe(III) F164W CooA are comparable to the profiles for Fe(III) WT CooA (Figure 3A), which suggests similar unfolding processes.
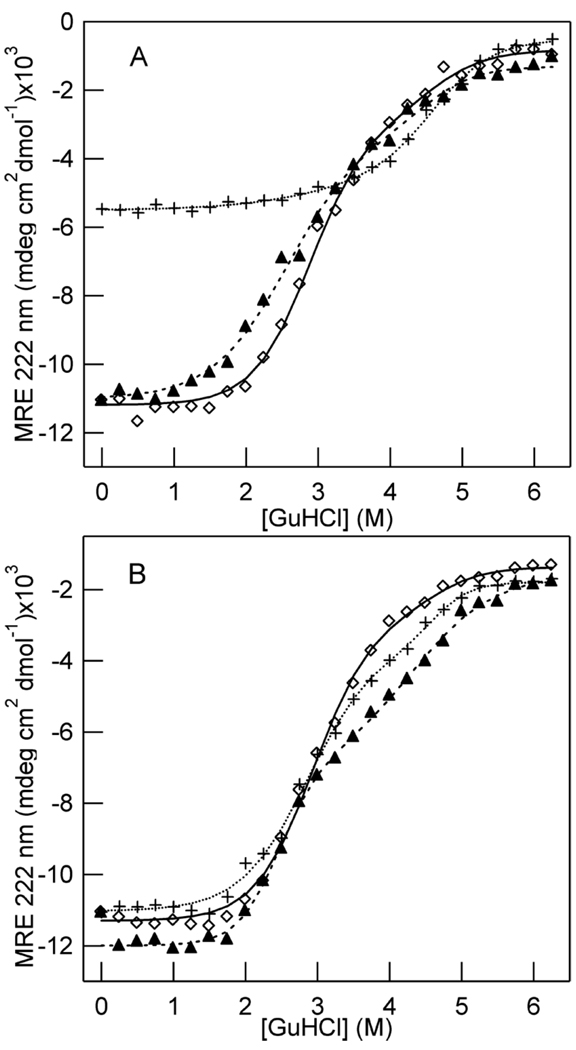
Figure 3. Denaturation of CooA monitored by CD spectroscopy for Fe(III) WT, Fe(III) F164W, Fe(III) truncated CooA (A) and Fe(II) WT and Fe(II)-CO WT CooA (B).
Samples of a fixed concentration of CooA (2 µM) were titrated with GuHCl from 0–6.25 M and were allowed to unfold for 2 h at 25 °C before measurement of CD intensity. The CD intensity at 222 nm is plotted as a function of GuHCl concentration. The solid curves represent fits to a three-state model in the case of WT and F164W CooA, and a two-state model in the case of Fe(III) truncated CooA. (A) Unfolding curves for Fe(III) WT (◇), Fe(III) F164W (▲), and Fe(III) CooA truncation variant (+) consisting only of the heme domain. (B) Unfolding curves for Fe(III) (◇), Fe(II) (+), and Fe(II)-CO (▲) WT CooA.
To account for the contribution of each domain to the unfolding of Fe(III) WT CooA, we unfolded the Fe(III) truncated variant of CooA, which lacks the helix-turn-helix DNA-binding domain. The CD unfolding curve of the truncated variant is substantially different from that of the full-length Fe(III) WT CooA protein (Figure 3A); changes for Fe(III) truncate CooA do not occur until 4 M GuHCl, and the profile shows a single unfolding transition. No changes are observed between 2 and 3 M GuHCl in the truncate CooA CD unfolding profile, indicating that the effector domain is stable in this range of GuHCl. These results support the proposal that the Fe(III) WT CooA DNA-binding domain unfolds between 2 and 3 M GuHCl.
Heme dependent changes in CooA structure result in differences in CD unfolding curves at high concentrations of GuHCl
To detect possible differences in stability of the α-helix in the various heme coordination states of CooA, we carried out GuHCl denaturation studies of Fe(II) and Fe(II)-CO WT CooA and monitored the CD signal at 222 nm. CD spectra of three heme coordination states of CooA show minimal differences in α-helical secondary structure (Supporting Information Figure S1). Upon titration with GuHCl, the CD spectra of Fe(II) and Fe(II)-CO WT CooA both show a decrease in intensity, indicating a loss of α-helical secondary structure. A comparison of the CD unfolding profiles at 222 nm of the three heme states of WT CooA (Figure 3B) indicates that the states show similar changes in CD intensity at concentrations of GuHCl below 3 M. All states show an unfolding transition in the CD data that covers a broad range of GuHCl concentrations compared to transitions observed in the visible absorption spectra. Above 3 M GuHCl, the curves diverge, and the Fe(II)-CO WT CooA protein shows less change in ellipticity (greater stability) with increasing GuHCl than the Fe(II) and Fe(III) states. If, as we have supposed, the CD unfolding curve for concentrations greater than 3 M GuHCl primarily reflects signal from the heme domain, the CD data suggest that changes in the coordination state or stability of the heme affect the unfolding and stability of α-helix in this domain.
The CD data were fitted for the presence of a folding intermediate, and energies were calculated for the process of going from native state to intermediate, and intermediate to unfolded state. The three-state fits for the CD curves of Fe(III) WT, Fe(III) F164W, Fe(II), and Fe(II)-CO CooA gave free energy values for the second unfolding step (intermediate→unfolded) that were consistent with those from two-state fits of the visible absorbance unfolding data (Table 1). These results indicate that the second step in the CD unfolding process might be the same as is detected in the visible absorbance unfolding data. The Fe(III) truncated CooA CD unfolding curve was fitted to a two-state model to give a ΔGH2O of 24 kJ/mol, also consistent with the visible absorbance data for this variant. The stability values for the first step in the CD unfolding process range from 12 to 18 kJ/mol, values that are significantly lower than the energies of the second step. The fits support the conclusion that visible absorbance data report on unfolding of the heme domain, while CD data report on unfolding of both the DNA-binding and heme domains.
Fluorescence spectra of the denaturation of Fe(III) WT CooA and Fe(III) F164W CooA reveal independent unfolding of the DNA-binding domain
Fe(III) WT CooA fluorescence spectra show an increase in tryptophan fluorescence intensity with increasing concentrations of GuHCl. The native Fe(III) WT protein has a weak tryptophan fluorescence signal, likely due to heme-induced quenching of fluorescence emission. A quenching effect is not surprising in light of the Fe(II) WT CooA crystal structure, which shows close proximity between the tryptophan residues (Trp110) and the heme cofactors (Figure 1) (9). As the protein is exposed to increasing concentrations of GuHCl, the fluorescence intensity increases and the emission maximum shifts from 340 nm to 352 nm (Supporting Information, Figure S2). Similar to observations in the visible absorbance unfolding data of Fe(III) WT CooA, the fluorescence data show a significant change in intensity corresponding to the unfolding transition at GuHCl concentrations >3 M (Figure 4). As was observed in the visible absorbance unfolding curves, fluorescence unfolding curves for titrations of Fe(III) WT CooA at 2 and 4 µM protein were superimposable, while fluorescence unfolding curves collected at 0.5 µM protein showed a shift such that the transition was at lower GuHCl concentrations. The fluorescence unfolding data for Fe(III) WT CooA are similar to data collected by visible absorbance, signifying that fluorescence reports on the folding state of the heme domain.
Since the fluorescence spectra and location of the tryptophan residues of Fe(III) WT CooA suggest that their emission is fully dependent on their proximity to the heme cofactor, fluorescence spectra of unfolding of the WT protein do not provide any information beyond what is available through visible absorption spectroscopy. To better understand the unfolding behavior of the DNA-binding domain of Fe(III) WT CooA, we unfolded a CooA variant, Fe(III) F164W CooA, that has a substitution to provide an additional tryptophan fluorescence signal in the DNA-binding domain. If the Fe(III) F164W CooA domains unfold independently, the fluorescence denaturation profile would be expected show two transitions: one for denaturation of the DNA-binding domain due to Trp164, and one for unfolding of the heme domain due to Trp110. The fluorescence intensity increases during an unfolding titration of Fe(III) F164W CooA, indicating that in native Fe(III) F164W CooA protein, both Trp164 and Trp110 are quenched. The fluorescence unfolding profile of Fe(III) F164W CooA shows two significant unfolding transitions (Figure 4). The first unfolding transition of Fe(III) F164W CooA occurs at concentrations between 2 and 3 M GuHCl. We interpret the first transition to be due to unfolding of the DNA-binding domain of Fe(III) F164W CooA, made apparent by the additional tryptophan signal in the DNA-binding domain. The second transition occurs at concentrations >3 M GuHCl, and is fully coincident with the curve for unfolding of Fe(III) WT CooA. The clear separation of the two transitions suggests that the two domains of Fe(III) F164W CooA are unfolding independently. Furthermore, the coincidence of the unfolding curves above 3 M GuHCl, both in fluorescence and visible absorbance data, further reinforces the interpretation that the heme domain of CooA is unfolding independently at concentrations >3M GuHCl.
Changes in heme ligation and activation state have little effect on the fluorescence unfolding curves of WT and F164W CooA
A comparison of the fluorescence unfolding profile of Fe(II)-CO WT CooA with that of Fe(III) WT CooA shows that the curves nearly overlap completely (Figure 4). The difference between the two profiles occurs at high GuHCl concentrations (>4 M GuHCl). Most notably, Fe(III) WT CooA shows a continual fluorescence increase above 5 M GuHCl, while the Fe(II)-CO protein appears to reach a final fluorescence equilibrium and has a post-transition baseline. These results are contrary to the significant differences in unfolding curves that is observed in the visible absorbance unfolding data, but the fluorescence results may be complicated by the fact that in CooA, tryptophan fluorescence emission is a reporter of the environments of both the heme cofactor and the dimer interface (Figure 1D), as discussed in more detail below.
Direct comparison of the fluorescence intensity changes, as represented in Figure 4, may not fully reveal differences in Fe(II)-CO WT and Fe(III) WT CooA unfolding. In order to interpret the fluorescence unfolding data, it is useful to examine the changes in fluorescence intensity peak maximum wavelength (λmax,F). Dissociation of CooA into monomers would cause both i) a significant change in the λmax,F as the tryptophans move from a region of low polarity to one that is exposed to solvent (58), and ii) and a moderate increase in tryptophan fluorescence intensity due to an increase in distance from one heme, even if the protein monomers are fully native. Thus, increases in fluorescence intensity that occur upon CooA denaturation may reflect both the unfolding of the protein heme domain and dissociation of the protein from dimers to monomers. A significant increase in fluorescence λmax,F for Fe(II)-CO WT CooA occurs between 3 and 4 M GuHCl (Supporting Information, Figure S2). Since this change is not coincident with the unfolding transition in the visible absorbance unfolding curve, it is possible that this increase in Fe(II)-CO CooA λmax,F is not due to unfolding of the heme domain, but to the dissociation of the protein into monomers. The changes in λmax,F observed for Fe(III) WT CooA differ considerably from the data for Fe(II)-CO WT CooA, indicating possible differences in GuHCl-dependent dissociation of the dimers into monomers for the two protein activation states. Experiments were conducted to examine the dependence of CooA quaternary structure on GuHCl concentration, but results for these experiments were equivocal (see Materials and Methods).
We investigated the fluorescence unfolding behavior of Fe(II)-CO F164W CooA for comparison to the Fe(III) F164W CooA and Fe(II)-CO WT CooA data. The fluorescence unfolding curve shows two transitions, as was observed for unfolding of Fe(III) F164W CooA. Similar to the Fe(III) fluorescence unfolding curve, the first transition for unfolding of Fe(II)-CO F164W CooA is apparent between 2 and 3 M GuHCl. The second transition appears at GuHCl >3 M, and the fluorescence unfolding curves for both states of the WT and F164W proteins are coincident between 3 and 6.25 M GuHCl. These results indicate that the DNA-binding domain of Fe(II)-CO CooA unfolds independently of the heme domain, and with the same stability as is observed for the Fe(III) state.
The fluorescence data for unfolding of Fe(III) WT, Fe(III) F164W, Fe(II)-CO WT and Fe(II)-CO F164W CooA were fitted according to a four-state model with two intermediates based on the information gained from all methods of observation. (Fluorescence spectroscopy is not possible for Fe(II) WT CooA due to buffer requirements for a stable Fe(II) state, see Materials and Methods). In the fits, the presence of two unfolding intermediates was assumed, and the energies correspond to transitions from native folded protein to a first intermediate, from the first intermediate to a second intermediate, and from the second intermediate to unfolded protein. The four-state fits give stabilities for the first and second transition that are in the range of those from the absorbance and CD fitted curves (Table 1). Most notably, the fluorescence and CD fits give similar energies for the initial step (range for all fits is 13–18 kJ/mol), which is undoubtedly a measure of the stability of the DNA-binding domain. The second and third steps have a range of stability values for the different heme ligation states, suggesting that either: i) they correspond to different processes the in the various states, or ii) they correspond to processes that are associated with different free energy changes for each protein state.
The use of variants for analysis of CooA unfolding data
It is possible that the variants unfold in a way that is qualitatively different from the WT protein, but this is unlikely for several reasons. First, F164W CooA has comparable CO-dependent activation behavior to the WT protein (data not shown), indicating that the variant has the same structure as WT CooA. Also, the Fe(III) F164W CooA visible and CD unfolding data are almost coincident with those for Fe(III) WT for all methods, supporting the conclusions based on the activity data. A crystal structure of the Fe(II) truncate is nearly identical to the structure of the heme domain of the Fe(II) full-length protein (51). Furthermore, the similarity of visible unfolding curves for the full-length Fe(III) WT and Fe(III) truncated proteins confirms that the truncate is a good model for the heme domain of the full-length protein.
A mechanism for CooA unfolding includes multiple steps
By integrating data from WT, F164W, and truncated CooA, we propose a plausible mechanism for CooA unfolding. A scheme describing this mechanism is shown in Figure 5, and the key conclusions are summarized below. Because CooA is a dimer of monomer units, each with two domains and a non-covalently associated cofactor, the unfolding process is undoubtedly complex. The most salient indicator of multi-state unfolding in CooA is the lack of overlap observed in the visible absorbance, fluorescence, and CD unfolding curves for the Fe(III) WT protein (Figure 5). These curves indicate that the unfolding process is not simultaneous for the tryptophan, heme, and α-helix in the protein. Multistate unfolding of CooA is also supported by the apparent presence of non-equilibrium behavior in reversibility experiments (described in the Materials and Methods and Supporting Information, Figure S3). In these experiments, the fluorescence refolding behavior of Fe(III) WT CooA is complicated, which may be related to irreversible changes at the heme cofactor. Independent unfolding of individual domains has been reported for several proteins (41, 59–62), and the ability of a helix-turn-helix motif to unfold independently has also been studied in some detail (63). For CooA, changes in protein secondary structure that occur during the unfolding process between 0 and 3 M are fully reversible, and appear to be an independent unfolding event within one domain.
We propose a model in which unfolding of CooA is a four-state process with three distinguishable steps. The first step occurs between 0 and 3 M GuHCl and is due to unfolding of the DNA-binding domain. The visible intensity, CD intensity, and fluorescence intensity of Fe(III) WT CooA show that structural changes affect a region of the protein that is rich in α-helix , but they do not affect the heme (Figure 5). The Fe(III) truncate CooA does not show any change in structure between 0 and 3 M GuHCl (Figures 2B, 3A), confirming that the changes in α-helical content of the protein are not due to unfolding of the heme domain. Both Fe(III) F164W and Fe(II)-CO F164W CooA show an additional unfolding transition at GuHCl between 2 and 3 M GuHCl in the fluorescence profile (Figure 4), likely due to unfolding of the DNA-binding domain made apparent by the tryptophan fluorophore in that domain of the variant. Together, the variant unfolding data confirm that the DNA-binding domain of CooA unfolds at concentrations of GuHCl less than 3 M, and the unfolding process of this domain is independent of the activation state of the heme domain.
A second step in CooA denaturation is unfolding of the heme domain. In all states of CooA, the major visible absorbance unfolding transition occurs at concentrations greater than 3 M GuHCl (Figure 2C), indicating that unfolding of the heme domain occurs within this range of denaturant concentrations. This conclusion is also supported by the observation that the Fe(III) truncate shows minimal change in CD intensity at concentrations of GuHCl lower than 3 M (Figure 3A). Changes in ligation state of the heme appear to affect the unfolding stability of the heme domain of CooA. The inactive Fe(III) CooA state shows a visible absorbance unfolding transition between 3 and 4 M GuHCl (Figure 2B), but the Fe(II) and Fe(II)-CO WT CooA states do not show major structural changes until 4 and 5 M GuHCl, respectively (Figure 2C).
A third step likely involves changes in CooA quaternary structure. At concentrations greater than 4 M GuHCl, after the visible absorbance has reached a post-transition baseline, the Fe(III) WT CooA fluorescence and CD intensities continue to change (Figure 5). These changes are accompanied by an increase in the fluorescence peak maximum at GuHCl concentrations greater than 4 M (Supporting Information Figure S2), which may be attributable to the dissociation of Fe(III) WT CooA subunits (58). Comparison of unfolding profiles at varying protein concentrations (4, 2, and 0.5 µM) confirmed that a dissociation process occurs during denaturation of Fe(III) CooA. At 0.5 µM protein, the major visible and fluorescence transition shifted such that the transition midpoints were at lower GuHCl concentrations (data not shown). These results confirm that a dissociation reaction is influencing the unfolding process of the hemes and tryptophans, which reside at the dimer interface.
If the fluorescence peak maximum is an indicator of quaternary structure in CooA, then we may conclude that Fe(II)-CO WT CooA dissociates into monomers at much lower concentrations of GuHCl than is observed for Fe(III) WT CooA. Fe(II)-CO WT CooA shows a change in fluorescence peak maximum wavelength at GuHCl concentrations lower than 3 M (Supporting Information Figure S2), a denaturant concentration that is much less than is necessary to unfold the heme domain as suggested by visible absorbance unfolding curves (Figure 2C). This result has two important implications, which will be discussed below: i) although unfolding of CooA is a multistep process for all protein conformations, the order of the individual steps may vary, and ii) protein-protein interactions at the dimer interface of Fe(II)-CO CooA may be much more sensitive to disruption by denaturant than they are at the interface of Fe(III) CooA.
DISCUSSION
CooA is an exemplary member of two general classes of proteins and has played an important role in the understanding of structure-function relationships in each class. When first crystallized in 2000 (9), inactive Fe(II) CooA gave insight into the activation pathway of the CRP/FNR family of transcriptional regulators by representing the only known structure of an inactive state of the family. In addition, the Fe(II) CooA structure was also a major advancement for gas-sensing heme proteins in that it was the first full-length heme-dependent gas-sensor crystallized. For each class, gas-sensors and CRP-family transcription factors, the main structural features of CooA have been used to posit mechanisms that involve cofactor repositioning, movement at subunit interfaces, rotations of domains, and other detailed structural changes that may accompany activation (64). Yet, one issue remains: conclusive structural information exists for only one state of Rr CooA, making any proposals regarding allosteric mechanisms speculative. In the current work, we approach the question of CooA activation using a different technique that allows for physical comparison of the various activation states of the protein in solution: protein unfolding studies using GuHCl as a chemical denaturant. Our experiments allow us to conclude that activation of CooA occurs with a change in protein stability, but this change is not a reflection of a change in global stability of the entire protein. Instead, we show through a multi-step unfolding mechanism that structural changes may be localized to specific domains and regions of the protein.
Fits to the CooA unfolding data show consistent stability of the DNA-binding domain and variable unfolding of the heme domain
Based on the four-state model proposed above, we were able to fit the unfolding data for CooA to equilibrium expressions for zero, one or two intermediates using non-linear least squares analysis. We were selective about using a model that best described observed intermediates, multiple unfolding transitions, and reversibility for each protein variant and/or state and method of observation. A comparison of the free energy values obtained is given in Table 1.
The transition that occurs between 2 and 3 M GuHCl ((ΔG1H2O,CD and ΔG1H2O, fluor in Table 1) is attributed to unfolding of the DNA-binding domain of CooA. The free energy of unfolding associated with this transition is consistent and has a value of 16±3 kJ/mol when averaged over all samples and methods. This consistency was wholly unexpected for the three states of CooA. Models for CooA activation suggest that activation of the protein is dependent on a rotation of the DNA-binding domain to reposition recognition helices for proper contact with DNA (9). It was assumed that this rotation would require stabilizing contacts with the heme domain such that either the global conformational stability of the protein would change, or at the very least, the DNA-domain would be differentially stabilized in one conformation or the other. Instead, the DNA-domain appears to act as an independent structural unit with stability that is unchanged by heme coordination changes in the effector domain. Such independent unfolding behavior has been reported for other DNA-binding domains (59, 63). Furthermore, a truncated helix-turn-helix domain has been shown to have very low stability to denaturants (65), supporting our finding that the DNA-binding domain of CooA is the least stable structural element.
The effector domain of CooA appears to undergo changes in structural stability upon changes in ligation state, which is reflected in different free energy values for the unfolding process as monitored by visible absorption (Table 1, ΔG2H2O, vis for Fe(III) and Fe(II) CooA; ΔG3H2O, vis for Fe(II)-CO CooA). The values derived from fits of the visible absorption data give unfolding energies for the heme domain of the Fe(III), Fe(II), and Fe(II)-CO CooA states in the range of 20–29 kJ/mol. Changes in the stability of heme proteins with changes in heme state, ligation, or heme pocket have been well characterized in cytochrome proteins (48, 66–69), and also reported for hemoglobin (46, 47). The differences in stability of the heme domain amongst the three states of CooA are smaller than those observed in other heme proteins (ΔΔGH2O for Fe(III)/Fe(II) cytochrome c has been reported at ~30 kJ/mol) (48, 69), but unlike single-domain proteins, CooA may be able to compensate for changes in heme ligation by altering contacts at a subunit interface (vide infra). Although there is some inconsistency for CooA in the values calculated for the fluorescence and CD data corresponding to unfolding of the heme domain, these values are consistently significantly greater than any fitted value of ΔG1H2O, reinforcing the conclusion that the heme domain unfolding and DNA-binding domain unfolding are distinct processes. Limitations in the analysis may prohibit unequivocal determination of the relative energies of heme domain unfolding in the three states of CooA; however, two conclusions are clear. Changes in heme ligation and activation state affect the structural stability of the heme domain, and the unfolding of this domain occurs independently from and with a different change in free energy than the DNA-binding domain.
The nature of the third unfolding process of CooA is more difficult to interpret for the three states of CooA. For Fe(III) WT CooA, we assigned the free energy of the step (ΔG3H2O,fluor =~54 kJ/mol) to dissociation of the dimer subunits to monomers. In contrast, Fe(II)-CO CooA fluorescence data suggest that in this state of the protein, the subunit dissociation step may occur at lower concentrations of denaturant (Supporting Information Figure S2). For the Fe(II)-CO state, dissociation of subunits would correspond to the process in the fluorescence and CD data that is fitted with ΔG2H2O, fluor and ΔG2H2O, CD. Accordingly, dissociation of Fe(II)-CO CooA subunits would occur with a change in free energy of 22 kJ/mol, less than half of the magnitude that was calculated for dissociation of subunits of the Fe(III) protein! Interpretation of the Fe(II) CooA unfolding data is even more complicated. Fits for Fe(II) WT CooA give values for ΔG2H2O,CD that are much greater than ΔG2H2O,vis (46 and 22 kJ/mol, respectively). These results suggest that CD is able to detect a cooperative process in the Fe(II) state that includes both unfolding of the heme domain and dissociation at the subunit interface. Unfortunately, without fluorescence data to describe the dissociation process, it is impossible to conclusively attribute free energy values to unfolding steps in Fe(II) CooA.
The fluorescence results imply that the mechanism for unfolding of Fe(II)-CO CooA differs from that of the Fe(III) state; in the Fe(II)-CO state, the protein dissociates to monomers before the heme domain undergoes unfolding. The role of the dimer interface in the transmission of allosteric signal in the CRP/FNR family has been well established (9, 23–26, 70, 71), and our results support proposals that changes in heme ligation affect subunit interactions and stability at the dimer interface of CooA. Intriguingly, the inactive (effector-free) state of CRP appears to have an unfolding mechanism that is more similar to Fe(II)-CO active CooA than it is to Fe(III) inactive CooA. It has been shown that inactive CRP unfolds through a mechanism where dissociation at the dimer interface precedes subunit unfolding (71). In this study, the authors fit the inactive CRP unfolding data to get free energy values for both the dissociation and unfolding processes. The reported free energy values are comparable to those we obtain for inactive Fe(III) CooA, yet the order of denaturation steps for inactive CRP differs from the order for Fe(III) CooA. This finding may be due to structural differences between inactive Fe(III) CooA, which may be “locked” into an inactive state, and inactive Fe(II) CooA, which may be “poised” for activation and may be more similar inactive CRP.
The relationship of CooA structure to function
Our models for CooA unfolding have interesting implications for the structural differences among the activation states of CooA. We anticipated that that the structural rearrangements that accompany heme coordination changes would be global, and significant differences were anticipated for the stability of the entire protein in each state. Contrary to this expectation, the changes in structural stability that occur with changes in heme coordination are localized to specific regions of the protein. Given that we observe independent stability changes for the domains of CooA, what can be learned about the structural changes that accompany allosteric regulation?
The unfolding data illustrate how activation in CooA is related to changes in protein secondary, tertiary, and quaternary structure. The invariant stability of the DNA-binding domain and the variable stability of the effector-binding heme domain suggest modularity within the protein structure. The DNA-binding domain has a fixed architecture and acts as a unit amidst structural changes in the rest of the protein. In contrast, the heme domain experiences structural changes that affect local stability. From these conclusions, we may speculate about the role of CooA structure in the activation mechanism of the protein. CO binding to the heme of CooA causes significant change in the secondary structure of the heme domain, as is clear from the changes in stability of this domain. These changes then affect the quaternary structure of the protein by altering contacts at the dimer interface, making the Fe(II)-CO state more susceptible to denaturant-induced dissociation into monomers. The contacts at the dimer interface are then able to influence the protein tertiary structure by repositioning a structurally-intact DNA-binding domain. This mechanism is supported by recent CooA crystal structures: they show that the conversion from Fe(II) CooA to Fe(II)-CO CooA occurs with change in the position of the heme cofactor (11), and each structure has a different relative orientation of a structurally invariant DNA-binding domain (9–11).
These observations do not rule out a dynamic mechanism for allostery in CooA. Recently, dynamic models have been proposed to describe activation in allosteric proteins (72, 73). With respect to heme sensor proteins, this theory would predict that when in one heme coordination state, the protein would show a high level of flexibility and would sample multiple conformations on a relatively short timescale. Changes in heme coordination upon gas binding (or release) would cause a “stiffening” of the structure to select for one particular conformational activation state. For CooA, we envision that the inactive Fe(II) state may show dynamic flexibility in the position of the DNA-binding domain, which may sample conformations that are represented in the positions in the Fe(II) CooA crystal structures (9, 10). Binding of CO to CooA may stabilize the active conformation by decreasing the motion of the DNA-binding domain and stabilizing a position with exposed F-helices for contact with DNA, as is seen in the crystal structure of a constitutively active CO-bound CooA variant (11). Consistent with those proposals, studies have reported on ligand-induced changes in the dynamics of CRP (74, 75). While the studies herein give no direct evidence for a dynamic mechanism for CooA regulation, dynamic variation of the DNA-binding domain orientation would explain why we were unable to observe three distinct global conformations for CooA: the DNA-binding domain would be equally susceptible to denaturation in each coordination state because its movement would expose the entire domain to solvent.
Supplementary Material
Figure which shows comparison of far UV-CD spectra for the Fe(III), Fe(II), and Fe(II)-CO WT CooA and Fe(III) F164W and Fe(III) truncated CooA (Supporting Information Figure S1). Figure which shows changes in the peak maximum of fluorescence as a function of GuHCl concentration for Fe(III) and Fe(II)-CO WT and Fe(III) F164W and Fe(III) truncate CooA (Supporting Information Figure S2). The reversibility of the unfolding reaction is shown in Supporting Information Figure S3, which compares the fluorescence and CD intensities for forward and reverse titrations of Fe(III) WT CooA with GuHCl. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENTS
We thank Mary Conrad for constructing the truncated CooA variant, Jose Serate for the purification of F164W CooA, professors Silvia Cavagnero and Thomas Brunold for the use of their equipment. Finally, we would like to acknowledge Professor M. Thomas Record, Professor Gary Roberts, Dr. Catherine Royer, Professor Karen Fleming, and Jocelyn Pinkert for helpful discussions and critical reading of this manuscript.
Footnotes
This work was supported by NIH grant HL-66147 to J.N.B.
Abbreviations: CooA, a CO-sensing heme-containing transcription factor; GuHCl, guanidine hydrochloride; Fe(III) CooA, an inactive form of Rhodospirillum rubrum CooA that has a ferric heme; Fe(II) CooA, an inactive form of Rr CooA that has a ferrous heme; Fe(II)-CO CooA, the active form of Rr CooA that has CO bound at the ferrous heme; WT, wild-type; F164W CooA, a variant of WT CooA that has a substitution at position 164 (Phe→Trp); CODH, carbon monoxide dehydrogenase; CRP, c-AMP receptor protein; FNR, fumarate nitrite reductase; MOPS, 3-(N-morpholino)propanesulfonic acid; PCR, polymerase chain reaction; DTT, dithiothreitol; CD, circular dichroism; PPIX, protoporphyrin IX; ΔGH2O, free energy of unfolding in the absence of denaturant; λmax,F, wavelength of maximum intensity for a fluorescence spectrum; ΔΔGH2O, difference in unfolding free energy measured for two proteins or two protein states
REFERENCES
- 1.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 2.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 3.Gilles-Gonzalez M-A, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Nat. Acad. Sci. USA. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong W, Hao B, Chan MK. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 2000;39:3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 6.Key J, Moffat K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry. 2005;44:4627–4635. doi: 10.1021/bi047942r. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Phillips GN., Jr Crystallization and x-ray diffraction analysis of the sensor domain of the HemAT aerotactic receptor. Acta Crystallogr., Sect D: Biol. Crystallogr.D. 2003;59:749–751. doi: 10.1107/s0907444903003299. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Olson JS, Phillips GN., Jr Biophysical and kinetic characterization of HemAT, an aerotaxis receptor from Bacillus subtilis. Biophys. J. 2005;88:2801–2814. doi: 10.1529/biophysj.104.047936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanzilotta WN, Schuller DJ, Thorsteinsson MV, Kerby RL, Roberts GP, Poulos TL. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 10.Komori H, Inagaki S, Yoshioka S, Aono S, Higuchi Y. Crystal structure of CO-sensing transcription activator CooA bound to exogenous ligand imidazole. J. Mol. Biol. 2007;367:864–871. doi: 10.1016/j.jmb.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Borjigin M, Li H, Lanz ND, Kerby RL, Roberts GP, Poulos TL. Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA. Acta Crystallogr., Sect D: Biol. Crystallogr. D. 2007;63:282–287. doi: 10.1107/S0907444906051638. [DOI] [PubMed] [Google Scholar]
- 12.Shelver D, Kerby RL, He Y, Roberts GP. Carbon monoxide-induced activation of gene expression in Rhodospirillum rubrum requires the product of CooA, a member of the cyclic AMP receptor protein family of transcriptional regulators. J. Bacteriol. 1995;177:2157–2163. doi: 10.1128/jb.177.8.2157-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerby RL, Ludden PW, Roberts GP. Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 1995;177:2241–2244. doi: 10.1128/jb.177.8.2241-2244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 15.Aono S, Ohkubo K, Matsuo T, Nakajima H. Redox-controlled ligand exchange of the heme in the CO-sensing transcriptional activator CooA. J. Biol. Chem. 1998;273:25757–25764. doi: 10.1074/jbc.273.40.25757. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds MF, Shelver D, Kerby RL, Parks RB, Roberts GP, Burstyn JN. EPR and electronic absorption spectroscopies of the CO-sensing CooA protein reveal a cysteine-ligated low-spin ferric heme. J. Am. Chem. Soc. 1998;120:9080–9081. [Google Scholar]
- 17.Clark RW, Youn H, Parks RB, Cherney MM, Roberts GP, Burstyn JN. Investigation of the role of the N-terminal proline, the distal heme ligand in the CO sensor CooA. Biochemistry. 2004;43:14149–14160. doi: 10.1021/bi0487948. [DOI] [PubMed] [Google Scholar]
- 18.Shelver D, Thorsteinsson MV, Kerby RL, Chung S-Y, Roberts GP, Reynolds MF, Parks RB, Burstyn JN. Identification of two important heme site residues (cysteine 75 and histidine 77) in CooA, the CO-sensing transcription factor of Rhodospirillum rubrum. Biochemistry. 1999;38:2669–2678. doi: 10.1021/bi982658j. [DOI] [PubMed] [Google Scholar]
- 19.Dhawan IK, Shelver D, Thorsteinsson MV, Roberts GP, Johnson MK. Probing the heme axial ligation in the CO-sensing CooA protein with magnetic circular dichroism spectroscopy. Biochemistry. 1999;38:12805–12813. doi: 10.1021/bi991303c. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, Ishikawa H, Takahashi S, Ishimori K, Morishima I, Nakajima H, Aono S. Binding of CO at the Pro2 side is crucial for the activation of CO-sensing transcriptional activator CooA. 1H NMR spectroscopic studies. J. Biol. Chem. 2001;276:11473–11476. doi: 10.1074/jbc.C100047200. [DOI] [PubMed] [Google Scholar]
- 21.Aono S, Nakajima H, Saito K, Okada M. A novel heme protein that acts as a carbon monoxide-dependent transcriptional activator in Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 1996;228:752–756. doi: 10.1006/bbrc.1996.1727. [DOI] [PubMed] [Google Scholar]
- 22.He Y, Shelver D, Kerby RL, Roberts GP. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 1996;271:120–123. doi: 10.1074/jbc.271.1.120. [DOI] [PubMed] [Google Scholar]
- 23.Moore LJ, Kiley PJ. Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 2001;276:45744–45750. doi: 10.1074/jbc.M106569200. [DOI] [PubMed] [Google Scholar]
- 24.Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J. Mol. Biol. 2000;304:847–859. doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 25.Kerby RL, Youn H, Thorsteinsson MV, Roberts GP. Repositioning about the dimer interface of the transcription regulator CooA: a major signal transduction pathway between the effector and DNA-binding domains. J. Mol. Biol. 2003;325:809–823. doi: 10.1016/s0022-2836(02)01203-2. [DOI] [PubMed] [Google Scholar]
- 26.Youn H, Kerby RL, Conrad M, Roberts GP. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 28.Garges S, Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985;41:745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Adhya S, Garges S. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc. Nat. Acad. Sci. USA. 1992;89:9700–9704. doi: 10.1073/pnas.89.20.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harman JG, McKenney K, Peterkofsky A. Structure-function analysis of three cAMP-independent forms of the cAMP receptor protein. J. Biol. Chem. 1986;261:16332–16339. [PubMed] [Google Scholar]
- 31.Ryu S, Kim J, Adhya S, Garges S. Pivotal role of amino acid at position 138 in the allosteric hinge reorientation of cAMP receptor protein. Proc. Nat. Acad. Sci. USA. 1993;90:75–79. doi: 10.1073/pnas.90.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aono S, Matsuo T, Shimono T, Ohkubo K, Takasaki H, Nakajima H. Single transduction in the transcriptional activator CooA containing a heme-based CO sensor: isolation of a dominant positive mutant which is active as the transcriptional activator even in the absence of CO. Biochem. Biophys. Res. Commun. 1997;240:783–786. doi: 10.1006/bbrc.1997.7746. [DOI] [PubMed] [Google Scholar]
- 33.Won H-S, Yamazaki T, Lee T-W, Yoon M-K, Park S-H, Kyogoku Y, Lee B-J. Structural understanding of the allosteric conformational change of cyclic AMP receptor protein by cyclic AMP binding. Biochemistry. 2000;39:13953–13962. doi: 10.1021/bi000012x. [DOI] [PubMed] [Google Scholar]
- 34.Baichoo N, Heyduk T. Mapping cyclic nucleotide-induced conformational changes in cyclic AMP receptor protein by a protein footprinting technique using different chemical proteases. Protein Sci. 1999;8:518–528. doi: 10.1110/ps.8.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harman JG. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 36.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 37.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc. Nat. Acad. Sci. USA. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aono S, Takasaki H, Unno H, Kamiya T, Nakajima H. Recognition of target DNA and transcription activation by the CO-sensing transcriptional activator CooA. Biochem. Biophys. Res. Commun. 1999;261:270–275. doi: 10.1006/bbrc.1999.1046. [DOI] [PubMed] [Google Scholar]
- 39.Pace CN, Grimsley GR, Scholtz JM. Denaturation of proteins by urea and guanidine hydrochloride. Protein Folding Handbook. 2005;1:45–69. [Google Scholar]
- 40.Vijayanathan V, Greenfield NJ, Thomas TJ, Ivanova MM, Tyulmenkov VV, Klinge CM, Gallo MA, Thomas T. Effects of estradiol and 4-hydroxytamoxifen on the conformation, thermal stability, and DNA recognition of estrogen receptor beta. Biochem Cell Biol. 2007;85:1–10. doi: 10.1139/o06-144. [DOI] [PubMed] [Google Scholar]
- 41.Kristl S, Zhao S, Falsone SF, Somerville RL, Kungl AJ. The influence of ATP on the association and unfolding of the tyrosine repressor ligand response domain of Haemophilus influenzae. Biochem. Biophys. Res. Commun. 2001;280:81–84. doi: 10.1006/bbrc.2000.4076. [DOI] [PubMed] [Google Scholar]
- 42.Masino L, Martin SR, Bayley PM. Ligand binding and thermodynamic stability of a multidomain protein, calmodulin. Protein Sci. 2000;9:1519–1529. doi: 10.1110/ps.9.8.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobson KF, Housley NA, Pedigo S. Ligand-linked stability of mutants of the C-domain of calmodulin. Biophys. Chem. 2005;114:43–52. doi: 10.1016/j.bpc.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Murugan R, Mazumdar S. Role of substrate on the conformational stability of the heme active site of cytochrome P450cam: effect of temperature and low concentrations of denaturants. J Biol Inorg Chem. 2004;9:477–488. doi: 10.1007/s00775-004-0544-1. [DOI] [PubMed] [Google Scholar]
- 45.Lynch SM, Boswell SA, Colon W. Kinetic stability of Cu/Zn superoxide dismutase is dependent on its metal ligands: implications for ALS. Biochemistry. 2004;43:16525–16531. doi: 10.1021/bi048831v. [DOI] [PubMed] [Google Scholar]
- 46.Puett D, Friebele E, Hammonds RG., Jr A comparison of the conformational stabilities of homologous hemoproteins. Myoglobin from several species, human hemoglobin and subunits. Biochim. Biophys. Acta. 1973;328:261–277. doi: 10.1016/0005-2795(73)90260-2. [DOI] [PubMed] [Google Scholar]
- 47.Elbaum D, Pandolfelli ER, Herskovits TT. Denaturation of human and Glycera dibranchiata hemoglobins by the urea and amide classes of denaturants. Biochemistry. 1974;13:1278–1284. doi: 10.1021/bi00703a034. [DOI] [PubMed] [Google Scholar]
- 48.Cohen DS, Pielak GJ. Entropic stabilization of cytochrome c upon reduction. J. Am. Chem. Soc. 1995;117:1675–1677. [Google Scholar]
- 49.Thorsteinsson MV, Kerby RL, Youn H, Conrad M, Serate J, Staples CR, Roberts GP. Redox-mediated transcriptional activation in a CooA variant. J. Biol. Chem. 2001;276:26807–26813. doi: 10.1074/jbc.M102758200. [DOI] [PubMed] [Google Scholar]
- 50.Chiang LW, Kovari I, Howe MM. Mutagenic oligonucleotide-directed PCR amplification (Mod-PCR): An efficient method for generating random base substitution mutations in a DNA sequence element. PCR Methods and Applications. 1993;2:210–217. doi: 10.1101/gr.2.3.210. [DOI] [PubMed] [Google Scholar]
- 51.Kuchinskas M, Li H, Conrad M, Roberts G, Poulos TL. The role of the DNA-binding domains in CooA activation. Biochemistry. 2006;45:7148–7153. doi: 10.1021/bi052609o. [DOI] [PubMed] [Google Scholar]
- 52.Shelver D, Kerby RL, He Y, Roberts GP. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Nat. Acad. Sci. USA. 1997;94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 54.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 55.Nozaki Y. Preparation of guanidine hydrochloride. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]
- 56.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 57.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl.alpha.-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 58.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
- 59.Vreuls C, Filee P, Van Melckebeke H, Aerts T, De Deyn P, Llabres G, Matagne A, Simorre JP, Frere JM, Joris B. Guanidinium chloride denaturation of the dimeric Bacillus licheniformis BlaI repressor highlights an independent domain unfolding pathway. Biochem J. 2004;384:179–190. doi: 10.1042/BJ20040658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conejero-Lara F, Parrado J, Azuaga AI, Smith RA, Ponting CP, Dobson CM. Thermal stability of the three domains of streptokinase studied by circular dichroism and nuclear magnetic resonance. Protein Sci. 1996;5:2583–2591. doi: 10.1002/pro.5560051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markovic-Housley Z, Cooper A, Lustig A, Flukiger K, Stolz B, Erni B. Independent folding of the domains in the hydrophilic subunit IIABman of the mannose transporter of Escherichia coli. Biochemistry. 1994;33:10977–10984. doi: 10.1021/bi00202a017. [DOI] [PubMed] [Google Scholar]
- 62.Bogusky MJ, Dobson CM, Smith RA. Reversible independent unfolding of the domains of urokinase monitored by 1H NMR. Biochemistry. 1989;28:6728–6735. doi: 10.1021/bi00442a028. [DOI] [PubMed] [Google Scholar]
- 63.Religa TL, Johnson CM, Vu DM, Brewer SH, Dyer RB, Fersht AR. The helix-turn-helix motif as an ultrafast independently folding domain: the pathway of folding of Engrailed homeodomain. Proc Natl Acad Sci U S A. 2007;104:9272–9277. doi: 10.1073/pnas.0703434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts GP, Kerby RL, Youn H, Conrad M. CooA, a paradigm for gas sensing regulatory proteins. J Inorg Biochem. 2005;99:280–292. doi: 10.1016/j.jinorgbio.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 65.Felitsky DJ, Record MT., Jr Thermal and urea-induced unfolding of the marginally stable lac repressor DNA-binding domain: a model system for analysis of solute effects on protein processes. Biochemistry. 2003;42:2202–2217. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- 66.Bartalesi I, Bertini I, Di Rocco G, Ranieri A, Rosato A, Vanarotti M, Vasos PR, Viezzoli MS. Protein stability and mutations in the axial methionine loop of a minimal cytochrome c. J Biol Inorg Chem. 2004;9:600–608. doi: 10.1007/s00775-004-0558-8. [DOI] [PubMed] [Google Scholar]
- 67.Sabahi A, Wittung-Stafshede P. Unfolding the unique c-type heme protein, Chlamydomonas reinhardtii cytochrome f. Biochim Biophys Acta. 2002;1596:163–171. doi: 10.1016/s0167-4838(02)00214-5. [DOI] [PubMed] [Google Scholar]
- 68.Wittung-Stafshede P, Lee JC, Winkler JR, Gray HB. Cytochrome b562 folding triggered by electron transfer: approaching the speed limit for formation of a four-helix-bundle protein. Proc Natl Acad Sci U S A. 1999;96:6587–6590. doi: 10.1073/pnas.96.12.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pascher T, Chesick JP, Winkler JR, Gray HB. Protein folding triggered by electron transfer. Science. 1996;271:1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 70.Polit A, Bonarek P, Kepys B, Kedracka-Krok S, Gorecki A, Wasylewski Z. Kinetic studies of cAMP-induced allosteric changes in mutants T127I, S128A, and T127I/S128A of the cAMP receptor protein from Escherichia coli. J. Biol. Chem. 2003;278:43020–43026. doi: 10.1074/jbc.M306398200. [DOI] [PubMed] [Google Scholar]
- 71.Cheng X, Gonzalez ML, Lee JC. Energetics of intersubunit and intrasubunit interactions of Escherichia coli adenosine cyclic 3',5'-phosphate receptor protein. Biochemistry. 1993;32:8130–8139. doi: 10.1021/bi00083a011. [DOI] [PubMed] [Google Scholar]
- 72.Kern D, Zuiderweg ERP. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 73.Cooper A, Dryden DTF. Allostery without conformational change. A plausible model. Eur. Biophys. J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 74.Dong A, Malecki JM, Lee L, Carpenter JF, Lee JC. Ligand-induced conformational and structural dynamics changes in Escherichia coli cyclic AMP receptor protein. Biochemistry. 2002;41:6660–6667. doi: 10.1021/bi020036z. [DOI] [PubMed] [Google Scholar]
- 75.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure which shows comparison of far UV-CD spectra for the Fe(III), Fe(II), and Fe(II)-CO WT CooA and Fe(III) F164W and Fe(III) truncated CooA (Supporting Information Figure S1). Figure which shows changes in the peak maximum of fluorescence as a function of GuHCl concentration for Fe(III) and Fe(II)-CO WT and Fe(III) F164W and Fe(III) truncate CooA (Supporting Information Figure S2). The reversibility of the unfolding reaction is shown in Supporting Information Figure S3, which compares the fluorescence and CD intensities for forward and reverse titrations of Fe(III) WT CooA with GuHCl. This material is available free of charge via the Internet at http://pubs.acs.org.