Abstract
Identification of Mycobacterium tuberculosis antigens inducing cellular immune responses is required to improve the diagnosis of and vaccine development against tuberculosis. To identify the antigens of M. tuberculosis that differentiated between tuberculosis (TB) patients and healthy contacts based on T cell reactivity, the culture filtrate of in vitro grown M. tuberculosis was fractionated by two-dimensional liquid phase electrophoresis and tested for the ability to stimulate T cells in a whole blood assay. This approach separated the culture filtrate into 350 fractions with sufficient protein quantity (at least 200 μg of protein) for mass spectrometry and immunological analyses. High levels of interferon-γ (IFN-γ) secretion were induced by 105 fractions in healthy contacts compared with TB patients (p < 0.05). Most interesting was the identification of 10 fractions that specifically induced strong IFN-γ production in the healthy contact population but not in TB patients. Other immunological measurements showed 42 fractions that induced significant lymphocyte proliferative responses in the healthy contact group compared with the TB patients. The tumor necrosis factor-α response for most of the fractions did not significantly differ in the tested groups, and the interleukin-4 response was below the detectable range for all fractions and both study groups. Proteomic characterization of the 105 fractions that induced a significant IFN-γ response in the healthy contacts compared with the TB patients led to the identification of 59 proteins of which 24 represented potentially novel T cell antigens. Likewise, the protein identification in the 10 healthy “contact-specific fractions” revealed 16 proteins that are key candidates as vaccine or diagnostic targets.
Tuberculosis (TB)1 is a major health problem throughout the world. A recent World Health Organization report shows that TB has been increasing at a rate of 1% per year, and an estimated 9.2 million new cases arise each year (1). Although TB is preventable, there has been an increase in its incidence in recent years. Re-emergence of TB is mainly due to its association with human immunodeficiency virus infection (2) and also due to the occurrence of multidrug-resistant strains of the causative agent, Mycobacterium tuberculosis (3).
Vaccination of general population is cost effective and represents one of the best biological measures for disease control. The current vaccine against tuberculosis, Bacille Calmette-Guérin (BCG), has been administered to more people than any other vaccine. The side effects of BCG are tolerable, and it prevents miliary and meningeal tuberculosis in young children. In striking contrast, it affords limited and highly variable protection (0–80%) against pulmonary TB (4). Thus, BCG does not seem to be a satisfactory vaccine (5, 6) and necessitates exploration of newer strategies to improve BCG or to develop a more effective vaccine.
One of the potential strategies for the development of an improved TB vaccine involves the use of the proteins secreted by M. tuberculosis during growth. There is evidence that proteins actively secreted by M. tuberculosis during growth induce cell-mediated immune responses by causing expansion of specific interferon-γ (IFN-γ)-producing T lymphocytes that are capable of recognizing and exerting antimicrobial effects against infected macrophages (7). The importance of IFN-γ pathways in host defense against M. tuberculosis was clarified by experimental studies on IFN-γ knock-out mice as well as the identification and characterization of humans with mutations in IFN-γ receptor (8, 9).
Several studies have been carried out to define the secreted proteome of M. tuberculosis. The earliest study aimed at the identification of mycobacterial culture filtrate proteins, using chromatography and N-terminal sequencing to identify eight culture filtrate proteins (10). Later, many studies used two-dimensional (2D) PAGE combined with sensitive mass spectrometric methods for identification of proteins. The above mentioned approaches have identified nearly 300 culture filtrate proteins (11–13).
Identification of T cell antigens in a complex mixture was first done by a T cell Western blot method (14). Later, two-dimensional separation methods were used that involved protein separation by either IEF (15) or chromatography (16) in the first dimension and preparative SDS-PAGE followed by whole gel elution (17) in the second dimension. Mouse T cell antigens of M. tuberculosis were identified using this method (15). Mycobacterial antigens that induce an immune response in healthy household contacts and treated TB patients were also mapped using this approach (16).
In the present study, 2D liquid phase electrophoresis (LPE) along with an in vitro IFN-γ assay and LC-MS/MS were used to identify potential human T cell antigens. Systematic screening of the M. tuberculosis culture filtrate (CF) proteome and comparative evaluation of cellular immune responses between TB patients and healthy contacts led to the identification of 59 proteins in the most immunogenic 2D LPE fractions. Twenty-four potentially novel T cell antigens were identified, and 16 proteins were identified in 10 2D LPE fractions that differentiated healthy contacts from TB patients based on IFN-γ responses.
MATERIALS AND METHODS
Growth of M. tuberculosis and Preparation of Culture Filtrate Protein (CFP)
M. tuberculosis H37Rv (ATCC 27294) colonies were transferred from Lowenstein-Jensen slants to 2 ml of Sauton's medium (Himedia Laboratories), and the cells were dispersed using glass beads under sterile conditions. The bacterial cell suspension was transferred to 10 ml of Sauton's medium in a McCartney bottle for incubation at 37 °C for 2 weeks. The bacilli were then transferred to 200 ml of Sauton's liquid medium, grown in shaker culture for 4 weeks, transferred to a 4-liter culture flask containing 2 liters of Sauton's medium, and grown as stationary culture for 4 weeks at 37 °C. The bacilli in the culture were harvested by centrifugation at 3000 rpm for 30 min. The culture supernatant was filter-sterilized using a 0.20-μm filter (Pal Gelman Laboratory, Saint Germain en Laye, France), and the filtrate was concentrated using the Quixstand benchtop hollow fiber system (GE Healthcare). The protein content in the CF was estimated using a BCA assay (Pierce). The proteins were then distributed into smaller volumes along with sodium azide at a final concentration of 0.2% and stored at −80 °C for later use.
Separtation of CFP by 2D LPE
CFP (300–350 mg) was solubilized in 60 ml of a buffer containing 8 m urea, 1 mm DTT, 5% glycerol, 2% digitonin, and 2% ampholytes (pH 3.0–10.0 and pH 4.0–6.0 at a ratio of 1:4) (Bio-Rad). The sample was loaded onto a liquid IEF system (Rotofor, Bio-Rad) maintained at 4 °C. Separation was achieved by applying constant power (12 watts) until the voltage stabilized at ∼1400 V. Focusing was continued for an additional 30 min and terminated. The individual IEF fractions were harvested, and their pH value was determined. Each aliquot was subjected to SDS-PAGE, and the proteins were visualized by staining with silver nitrate. A total of three IEF runs was performed, and fractions having a similar pH from each run were pooled.
Preparative SDS-PAGE was performed with individual IEF fractions. Briefly, each IEF fraction (protein quantities ranged from 0.08 to 43 mg) was prepared for SDS-PAGE by the addition of 6× SDS-PAGE sample buffer (18) and heating at 95 °C for 5 min. The reduced samples were loaded on 16 × 20-cm polyacrylamide gels comprising a 4% stack over a 12.5% resolving gel. The stacking gel contained a single 13-cm-long sample well. Electrophoresis was performed at a constant current of 50 mA/gel until the dye front was ∼2 cm from the bottom of the gel. The gel was equilibrated in elution buffer (60 mm Tris (pH 9.4), 40 mm CAPS) for 10 min and transferred to a Whole Gel Eluter (Bio-Rad) according to the manufacturer's instructions. The proteins were eluted from the gel using a constant current of 250 mA for 1 h. Thirty protein fractions (∼2.5 ml each) were harvested, and the protein concentration of each fraction was determined using the BCA assay (Pierce). Prior to testing for T cell stimulation, the 2D LPE fractions were filter-sterilized using a 0.2-μm filter. An aliquot (10 μg) of each eluted fraction was analyzed by SDS-PAGE and silver-stained (19).
Identification of Proteins
Proteins were digested in gel or in solution with modified trypsin (Roche Applied Science). For in-gel digestions, 10 μg of selected 2D LPE fractions was resolved by SDS-PAGE using 4–12% NuPAGE bis-Tris polyacrylamide gels and MES-SDS running buffer (Invitrogen). The gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad), and the resulting bands were excised from the gel. Digestion of proteins with modified trypsin was performed as described previously (20, 21). For in-solution digestions, 5 μg of each fraction was digested with 0.8 μg of modified trypsin in 10% acetonitrile, 0.2 m ammonium bicarbonate and incubated overnight at 37 °C. The reactions were terminated by the addition of 3 μl of 10% TFA. The resulting peptides from the tryptic digestions were dried; suspended in 15 μl of 5% acetonitrile, 0.1% acetic acid; and applied to a 0.2 × 50-mm C18 reversed phase HPLC column (Agilent Technologies, Santa Clara, CA). The peptides were eluted with an increasing gradient of acetonitrile at a flow rate of 5 μl/min using an Agilent 1100 capillary HPLC solvent delivery system. The effluent was introduced directly into an linear trap quadrupole electrospray ion trap mass spectrometer (Thermo-Finnigan, San Jose, CA). The electrospray needle of the mass spectrometer was operated at 4 kV with a sheath gas flow of nitrogen at 30 p.s.i. and a heated capillary temperature of 200 °C. Data-dependent tandem MS was used to generate fragment ions of individual peptides. The five most intense ions from the full MS scan were selected for fragmentation. The precursor ion was placed on the dynamic exclusion list for 1 min and Tandem MS was performed for each precursor ion, a maximum of two times.
MS/MS data were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA; Version 27, Revision 12) and X! Tandem (Global Proteome Machine Organization; Version 2006.04.01.2) software packages with data interrogation against the annotated M. tuberculosis genome (3912 entries). Fragment ion mass tolerance of 0.50 Da, parent ion tolerance of 1.5 Da, oxidation of methionine, and acrylamide adduct of cysteine were specified in Sequest and X! Tandem as part of the search criteria and variable modifications. The allowance for missed cleavages was 2. Scaffold (Version Scaffold-01-06-05; Proteome Software Inc., Portland, OR) was used to validate peptide and protein identifications using a two-peptide minimum and 95.0% peptide and protein probabilities as specified by the Peptide and Protein Prophet algorithms (22, 23).The Universal Protein Resource (UniProt) database was searched to get the subcellular localization information of identified proteins.
Study Population
The study was approved by the Institutional Ethics Committee of the Tuberculosis Research Centre, and informed consent was obtained from all persons who were enrolled in this study.
Ten patients with pulmonary TB were enrolled at the Tuberculosis Research Centre clinic. The subjects of this group had not undergone antituberculosis treatment when recruited for the study. Their age ranged from 26 to 52 years, and the male to female ratio was 7:3. Two spot and one overnight sputum specimens were collected from each patient. Sputum specimens were examined for acid-fast bacteria by fluorescence microscopy (auramine O phenol staining), and the samples were examined microscopically. All the TB patients were positive by sputum smear microscopy. For culture, the sputum specimens were processed by modified Petroff's method, inoculated onto Lowenstein-Jensen medium, and incubated for up to 8 weeks at 37 °C (24). Blood was collected from these subjects before treatment.
Seven individuals who shared living quarters with the tuberculosis patient agreed to join the study as healthy contacts whose age ranged from 28 to 55 years. The male to female ratio was 5:2. Three heavily exposed health care workers, working closely with pulmonary tuberculosis patients for at least 2 years at the Tuberculosis Research Centre, were also included in the study. Their age ranged from 28 to 35 years, and all were male. These individuals had no history of tuberculosis on the basis of personal history, physical examination, chest x-ray, and negative acid-fast bacilli sputum smear microscopy.
Tuberculin skin test using 2 tuberculin units of purified protein derivative (PPD) (Statens Serum Institut, Copenhagen, Denmark) was performed on all subjects, and an induration of 15 mm or more after 48 h was considered positive. All 10 healthy contacts enrolled in this study were PPD skin test-positive. Of the 10 TB patients, seven were skin test-negative, and three were positive. All subjects were human immunodeficiency virus-negative as determined by Tridot (J. Mitra and Co.) and Retroquic (Qualprodiagnostics) assays with serum. Heparinized blood (21 ml) was collected from all the subjects for the immunological assays presented in this study.
Lymphocyte Proliferation Assay (LPA)
An LPA was performed by diluting whole blood 1:10 in RPMI 1640 medium (Sigma) supplemented with glutamine (0.29 g/liter), penicillin (100 IU/ml), streptomycin (0.1 mg/ml), and amphotericin B (5 mg/ml) and culturing in 96-well flat bottom tissue culture plates. These cultures were stimulated with the 2D LPE fractions and with PPD and phytohemagglutinin as Positive controls. Cells cultured under similar conditions without any stimulation served as the negative control. Each antigen or fraction was added in triplicate wells to a final concentration of 5 μg/ml. The antigen-stimulated cells were cultured for 6 days at 37 °C in a 5% CO2 atmosphere (Hera Cell, Kendro Laboratories). Sixteen hours before the termination of the cultures, 1 μCi of tritiated thymidine ([3H]thymidine) (Board of Radiation and Isotope Technology, Mumbai, India) was added to each well. The cells were harvested onto glass fiber filters using a cell harvester (PHD, Cambridge Technology Ltd., Watertown, MA), and the filter discs were dried overnight. An aliquot (2 ml) of scintillation fluid (0.05 mg/ml 1,4-bis[2-(5-phenyloxazolyl)]benzene and 4 mg/ml 2,5-diphenyloxazole in toluene) was added to each filter disc and counted using a liquid scintillation β counter (Wallac oy, Torku, Finland). The proliferation was measured as uptake of [3H]thymidine by the cells and expressed as cpm. The mean cpm value of the triplicate culture was calculated. Proliferation was expressed as the stimulation index (SI): SI = mean cpm with antigen/mean cpm without antigen. Two analyses were performed with the lymphocyte proliferation data. In the first analysis, the mean cpm value obtained for the 10 healthy contacts was compared with that of the 10 TB patients for each of the fractions P values were calculated using the Mann-Whitney U test (Graphpad Software, San Diego, CA), and p values <0.05 were considered significant. Based on the p value, the fractions were grouped as “very highly significant” (p < 0.0005), “highly significant” (p < 0.005), “significant” (p < 0.05), and “non-significant” (p > 0.05). In the second analysis, a subject showing a cutoff value of ≥3 SI was classified as a responder, and the number of responders in each group was counted.
Cytokine Measurements
For quantification of cytokines (IFN-γ, TNF-α, and IL-4), cell-free culture supernatants were harvested after 6 days of in vitro stimulation by fractions and stored immediately (at −80 °C) until assayed. Cytokine production was determined by a standard ELISA technique using commercially available BD OptEIA kits (BD Biosciences) according to the manufacturer's instructions. The OD values were read at 450 nm using an ELISA reader (Molecular Devices, Sunnyvale, CA).
Two types of analyses were performed with the IFN-γ results. In the first analysis, the actual amount of IFN-γ secreted (pg/ml) in response to each fraction was calculated. The levels induced by each fraction were compared in the TB patient and healthy contact groups using the Mann-Whitney U test (Graphpad Software), and p values <0.05 were considered significant. Based on the p value, fractions were grouped as very highly significant (p < 0.0005), highly significant (p < 0.005), significant (p < 0.05) IFN-γ-inducing, and non-significant (p > 0.05).
In the second analysis, the cutoff value for IFN-γ was fixed for each fraction by using the mean + 2 S.D. of IFN-γ levels in TB patients (susceptible population). Any subject with an IFN-γ value above the cutoff point was classified as a responder. The number of responders to each fraction in each study group was calculated.
RESULTS
2D LPE Separation of CFP
The yield of M. tuberculosis CFP after 4 weeks of culture was 50 ± 5 mg/liter. A large quantity of culture filtrate protein (1 g) was used as the starting material to ensure sufficient protein for immunological analysis and molecular identification of fractions. Initial experiments showed that when quantities of CFP greater than 350 mg were applied to preparative IEF excessive precipitation resulted. Therefore, three technical replicates of preparative IEF runs were performed using 300–350 mg of protein per run. The 20 fractions collected from each replicate were analyzed by SDS-PAGE (data not shown). Fractions from each replicate were pooled based on their pH values, resulting in a total of 20 pooled fractions, each corresponding to a specific pH range. The pH of the separated IEF fractions ranged from 2.5 to 12.9, and the protein content varied from 0.08 to 43 mg.
The second dimension preparative SDS-PAGE performed on each IEF fraction resulted in 30 subfractions for a cumulative total of 600. All the fractions were numbered by IEF fraction number first followed by whole gel elution fraction number (for e.g. 1_1, 3_4, etc.). Initially, the fractions were analyzed by analytical SDS-PAGE. On analysis, it was observed that each fraction showed one to three bands. The protein quantity of each fraction ranged from 50 μg to 4 mg. A minimum of 200 μg of protein was required for immunological as well as proteomic characterization. Of the 600 2D LPE fractions, 350 possessed 200 μg or more of protein, and these were selected for further analyses.
Definition of Immunodominant Fractions for TB Healthy Contacts
IFN-γ, an important effector cytokine in tuberculosis infection (7), was used as a marker of antigen-specific T cell activation in whole blood assays with the 2D LPE fractions. In general, the antigen-induced IFN-γ levels were higher in the healthy contact group when compared with the TB patients. The level of IFN-γ secreted by TB patients for all 2D LPE fractions ranged from 0 to 2000 pg/ml, whereas for healthy contacts, it ranged from 0 to 8000 pg/ml. Of the 350 2D LPE fractions screened, 105 induced significant IFN-γ levels in the healthy contact group compared with the TB patient group (supplemental Table S1). Based on the IFN-γ levels and the significance in the difference between the two study populations, the reactive fractions were subdivided into three groups as follows: (i) 32 fractions induced very highly significant levels of IFN-γ (p < 0.0005), (ii) 34 fractions stimulated highly significant levels of IFN-γ (p < 0.005), and (iii) 39 fractions produced “significant levels” of IFN-γ (p < 0.05) in the healthy contacts as compared with the TB patients. There were no fractions at all that induced significantly higher levels of IFN-γ in TB patients as compared with the healthy contacts.
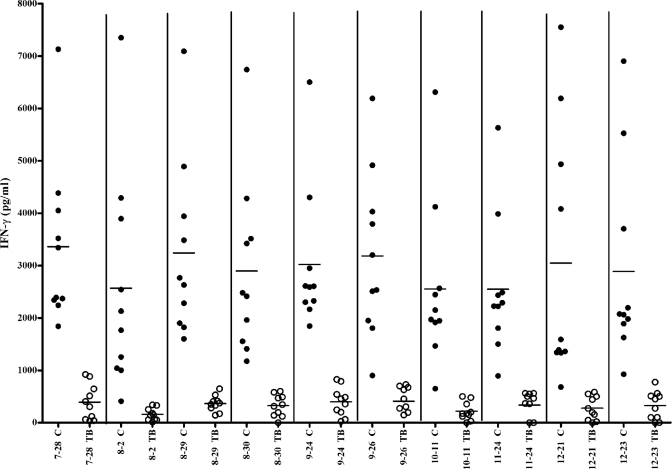
Interestingly, nine of the 2D LPE fractions designated very highly significant and one designated highly significant for IFN-γ production in the healthy contact population showed a positive IFN-γ response in all healthy contacts (n = 10) but a negative response in all TB patients (n = 10) based on the mean IFN-γ response +2 SD cutoff value (Fig. 1). These “contact-specific fractions” are listed in Table I.
Fig. 1.
IFN-γ response of 10 contact-specific fractions. “C” represents the response of healthy contacts, and “TB” represents the response of TB patients. Filled circles represent the Individual contact response for each fraction. Open circles represent the Individual patient response for each fraction. Horizontal bars represent the median value of the cytokine response (Patient or contact group).
Table I. Immune parameters studied in contact-specific fractions.
Significance was calculated using the Mann-Whitney U test.
| Protein fraction | IFN-γ |
LPA |
||
|---|---|---|---|---|
| Significance | p value | Significance | p value | |
| 7_28 | a | 0.0003 | NSb | 0.1230 |
| 8_2 | a | 0.0005 | c | 0.0185 |
| 8_29 | d | 0.0011 | d | 0.0089 |
| 8_30 | a | 0.0003 | d | 0.0022 |
| 9_24 | a | 0.0001 | NS | 0.0524 |
| 9_26 | a | 0.0001 | c | 0.0355 |
| 10_11 | a | 0.0001 | NS | 0.0892 |
| 11_24 | a | 0.0001 | NS | 0.1927 |
| 12_21 | a | 0.0001 | c | 0.0378 |
| 12_23 | a | 0.0003 | NS | 0.0753 |
a Statistically very highly significant value (p < 0.0005).
b NS, statistically non-significant.
c Significant value (p < 0.05).
d Statistically highly significant value (p < 0.005).
Other cytokines such as TNF-α and IL-4 are known to influence the outcome of TB (7, 25). Thus, these two cytokines were also measured in the whole blood assays with the 2D LPE fractions to determine whether there were other immunological markers that could differentiate the healthy contacts and TB patients. All the 2D LPE fractions induced a TNF-α response, but this response was not significantly different between the two study populations. It was noted that the spontaneous as well as antigen-induced TNF-α level was found to be higher in TB patients when compared with healthy contacts for all the fractions of which three fractions (6_7, 6_8, and 9_9) showed a statistically significant difference (data not shown). In contrast to the TNF-α response, the IL-4 response was found to be below the detectable range in all the cell culture supernatants from both groups (data not shown).
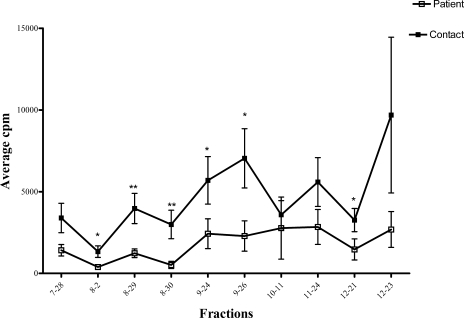
A fourth immunological parameter evaluated was induction by the 2D LPE fractions of lymphocyte proliferation (LP) in the whole blood. Of the 350 fractions screened, 42 (12%) induced a significant proliferative response in the healthy contacts as compared with the TB patients (supplemental table S2). Eight of these fractions were designated highly significant (p < 0.005), and 34 were designated significant (p < 0.05). All of the highly significant LP-inducing fractions except 8_30 showed positive LP responses in all 10 healthy contacts, and LP positivity in TB patients ranged from four to eight individuals depending on the 2D LPE fraction (supplemental Table S2). The significant LPA fractions gave a positive LPA stimulation index for seven to 10 individuals in the healthy contact group, and in the TB patient group, four to 10 individuals were LPA-positive for the same fractions. Thus, unlike the IFN-γ assay, no 2D LPE fractions were identified that could differentiate all healthy contacts from TB patients by the LPA. There was also variability in the correlation between the IFN-γ and LP responses. All but three of the 2D LPE fractions inducing significant LP responses also induced significant IFN-γ secretion (supplemental table S2). However, several of the fractions not inducing a significant LP response did induce significant IFN-γ responses (data not shown). The variability in these two immunological measurements carried over to the 10 contact-specific fractions defined through the IFN-γ response. Specifically, five of the contact-specific fractions (8_2, 8_29, 8_30, 9_26, and 12_21) induced highly significant or significant LP responses in healthy contacts as compared with the TB patients (Fig. 2), However, the remaining five contact-specific fractions did not result in significant differences in the LP responses between healthy contacts and TB patients (Table I).
Fig. 2.
LPA response of 10 contact-specific fractions. Statistical analysis was performed with the Mann-Whitney U test. *, significant value (p < 0.05); **, highly significant value (p < 0.005). This error bar represents median ± standard error.
Molecular Characterization of Immunodominant 2D LPE Fractions
Given that current in vitro T cell-based diagnostic assays for TB are based on the measurement of IFN-γ responses (26) and the majority of anti-TB vaccine candidates were selected based on their ability to induce an antigen-specific IFN-γ response (27), we focused protein identification efforts on those 2D LPE fractions that resulted in a significant IFN-γ response in the healthy contact population. Specifically, the IFN-γ-inducing 2D LPE fractions were analyzed by in-gel and in-solution proteolytic digestions followed by LC-MS/MS, and data interrogation via Sequest and collation via Scaffold were used to identify the dominant proteins in these immunologically reactive fractions (supplemental Tables S3, S4, and S5).
When all of the data were combined, a total of 59 proteins were identified for the 105 2D LPE fractions that had significant IFN-γ responses for the healthy contact population (Table II). For those proteins in the 10–30-kDa molecular mass range, most were identified with one to seven peptides with a maximum of seven unique peptide sequences found for the protein Rv2626c followed by Cfp-17 (Rv1827) with six unique peptides. Proteins in the molecular mass range of 31–120 kDa were identified by matching 1–12 distinct peptides with 12 distinct peptide sequences found to match KatG (Rv1908c) in fraction 13_12 (supplemental Tables S6, S7, S8, and S9 contain the detailed proteomics data for each LPE fraction inducing a significant IFN-γ response). Among the 59 identified novel T cell antigens, the UniPort database showed that 12 were secretory, four proteins localized to the cell membrane, 15 proteins localized to the cytoplasm, and one protein had an integral membrane location. Subcellular localization information for 27 proteins was not annotated in the UniPort database (Table II).
Table II. Biological and immunological roles of proteins identified.
Proteins in bold are novel T cell antigens identified in this study. NA, subcellular localization information was not annotated in the UniPort database.
| Protein name | Gene numbera | Biological function | Immunological studies | Subcellular location |
|---|---|---|---|---|
| Acn | Rv1475c | Tricarboxylic acid cycle aconitase enzyme | No report | NA |
| AcpM | Rv2244 | Involved in fatty acid biosynthesis | Human B cell antigen (38) | Cytoplasm |
| Adk | Rv0733 | ATP AMP transphosphorylase (adenylate kinase) | Mouse T cell antigen (15) | Cytoplasm |
| Ald | Rv2780 | Secreted l-alanine dehydrogenase | No report | Secreted |
| BfrB | Rv3841 | Possible bacterioferritin | Mouse T cell and human B cell antigen (15) | NA |
| Cfp-2 | Rv2376c | Function unknown | Human T cell antigen (32) | Secreted |
| Cfp-10 | Rv3874 | Conserved hypothetical protein | Human T cell antigen (37) | Secreted |
| Cfp-17 | Rv1827 | A substrate for PknB | Human T cell antigen (28) | NA |
| DnaK | Rv0350 | Probable chaperone | Human T cell antigen (64) | NA |
| Esat-6 | Rv3875 | Function unknown | Human T cell antigen (34) | Secreted |
| FabG4 | Rv0242c | Probable 3-oxoacyl-(acyl-carrier protein) reductase | No report | NA |
| Fba | Rv0363c | Involved in glycolysis (fructose-bisphosphate aldolase) | No report | NA |
| FbpA | Rv3804c | Possesses mycolyltransferase activity | Human T cell antigen (36) | Secreted |
| FbpB | Rv1886c | Possesses mycolyltransferase activity | Human T cell antigen (65) | Secreted |
| FbpC | Rv0129c | Mycolyltransferase 85C | Human T cell antigen (66) | Secreted |
| Frr | Rv2882c | Ribosomal recycling factor | No report | Cytoplasm |
| GgtB | Rv2394 | Probable γ-glutamyltranspeptidase precursor | No report | NA |
| GlcB | Rv1837c | Probable malate synthase | Human B cell antigen (67) | Cytoplasm |
| GlnA1 | Rv2220 | Involved in glutamine synthesis | Human T cell antigen (68) | Cytoplasm |
| GroEL | Rv0440 | Molecular chaperone | Human T cell antigen (69) | Cytoplasm |
| GroES | Rv3418c | Molecular chaperone | Human T cell antigen (69) | Cytoplasm |
| HspX | Rv2031c | Heat shock protein (hsp 16.3) | Human T cell antigen (35) | Secreted |
| Icd2 | Rv0066c | Probable isocitrate dehydrogenase | Human B cell antigen (70) | NA |
| KatG | Rv1908c | Multifunction enzyme exhibiting catalase, peroxidase, and peroxynitrase activity | Human T cell antigen (71) | NA |
| LppX | Rv2945c | Function unknown | Human T cell antigen (73) | Cell membrane |
| LppZ | Rv3006 | Function unknown | Human B cell response in TB (74) | NA |
| LprA | Rv1270c | Function unknown | Human T cell antigen (72) | Cell membrane |
| MmsA | Rv0753c | Probable methylmalonate-semialdehyde dehydrogenase | No report | NA |
| ModD | Rv1860 | Function unknown | Human T cell antigen (75) | Secreted |
| Mpt63 | Rv1926c | Function unknown | Human T cell antigen (76) | Secreted |
| Mpt64 | Rv1980c | Function unknown | Human T cell antigen (77) | Secreted |
| NdkA | Rv2445c | Probable nucleoside-diphosphate kinase | Human T cell antigen (16) | Cytoplasm |
| Pgi | Rv0946c | Probable glucose-6-phosphate isomerase | No report | Cytoplasm |
| PhoS1 | Rv0934 | Involved in inorganic phosphate transport | Human T cell antigen (78) | Cell membrane |
| Pks13 | Rv3800c | Polyketide synthase | No report | NA |
| PpiA | Rv0009 | Probable iron-regulated peptidyl-prolyl cis-trans isomerase | Human B cell antigen (38) | Cytoplasm |
| ProA | Rv2427c | Probable γ-glutamyl phosphate reductase protein | No report | Cytoplasm |
| pstS2 | Rv0932c | Involved in inorganic phosphate transport | Mouse T cell and human B cell antigen (74) | Cell membrane |
| Rpi | Rv2465c | Ribose-5-phosphate isomerase | Human T cell antigen (79) | NA |
| Rv0577 | Rv0577 | Function unknown | Human T cell antigen (15) | NA |
| Rv1324 | Rv1324 | Possible thioredoxin | No report | NA |
| Rv1558 | Rv1558 | Function unknown | No report | NA |
| Rv1910c | Rv1910c | Function unknown | No report | NA |
| Rv2204c | Rv2204c | Function unknown | No report | NA |
| Rv2251 | Rv2251 | Function unknown | No report | NA |
| Rv2626c | Rv2626c | Function unknown | Human T cell antigen (15) | NA |
| Rv2721c | Rv2721c | Function unknown | No report | Integral to membrane |
| Rv3169 | Rv3169 | Function unknown | No report | NA |
| Rv3716c | Rv3716c | Function unknown | No report | NA |
| SahH | Rv3248c | S-Adenosyl l-homocysteine hydrolase | No report | Cytoplasm |
| SodA | Rv3846 | Superoxide dismutase | Human T cell antigen (80) | Secreted |
| Tal | Rv1448c | Probable transaldolase | No report | Cytoplasm |
| TB16.3 | Rv2185c | Function unknown | Human T cell antigen (16) | NA |
| TB18.6 | Rv2140c | Function unknown | Mouse T cell antigen (81) | NA |
| TB49.2 or CIP50 | Rv0462 | Probable protein involved in phagosomal maturation arrest | No report | Cytoplasm |
| TB8.4 | Rv1174c | Function unknown | Human T cell antigen (16) | NA |
| Tpx or Cfp-20 | Rv1932 | Probable thioperoxidase | Human T cell antigen (15) | NA |
| Trxc | Rv3914 | Thioredoxin | Human T cell antigen (16) | NA |
| Wag 31 | Rv2145c | Function unknown | Human T cell antigen (82) | Cytoplasm |
a Gene number represents the designation of the corresponding open reading frame based on the information from the M. tuberculosis genome project (83).
The most relevant group of 2D LPE fractions are those designated contact-specific fractions. Proteomic characterization of these 10 fractions demonstrated 16 proteins (Table III), 13 of which (Cfp-10, DnaK, FpbB, Esat-6, GroEL2, GroES, HspX, KatG, PhoS1, PstS1, Rv2626c, TB8.4, and TrxC) were previously reported as human T cell antigens. Three proteins (AcpM, Adk, and Rv3716c) identified in these fractions are potentially new human T cell antigens. Proteins could not be identified in two contact-specific fractions (8_2 and 8_30) even though these fractions induced notable IFN-γ responses. Among the 16 proteins identified in contact-specific fractions, four proteins were secretory, two proteins were cell membrane-associated, and four proteins have a cytoplasmic location. Subcellular localization of the remaining six proteins was not annotated in the UniPort database (Table III).
Table III. Proteins identified in contact-specific fractions.
Proteins in bold represent the novel proteins identified in this study. NA, subcellular localization information was not annotated in UniPort database.
| Protein fraction | Protein name | Gene numbera | Subcellular location |
|---|---|---|---|
| 7_28 | GroES | Rv3418c | Cytoplasm |
| 8_29 | AcpM | Rv2244 | Cytoplasm |
| 8_29 | Esat-6 | Rv3875 | Secreted |
| 8_29 | TB8.4 | Rv1174c | NA |
| 9_24 | Cfp-10 | Rv3874 | Secreted |
| 9_24 | Rv3716c | Rv3716c | NA |
| 9_24 | TrxC | Rv3914 | NA |
| 9_26 | HspX | Rv2031 | Secreted |
| 10_11 | DnaK | Rv0350 | NA |
| 10_11 | PhoS1 | Rv0934 | Cell membrane |
| 10_11 | PstS2 | Rv0932c | Cell membrane |
| 11_24 | FbpB | Rv1886c | Secreted |
| 11_24 | Rv2626c | Rv2626c | NA |
| 12_21 | Adk | Rv0733 | Cytoplasm |
| 12_21 | KatG | Rv1908c | NA |
| 12_23 | GroEL | Rv0440 | Cytoplasm |
a Gene number represents the designation of the corresponding open reading frame based on the information from the M. tuberculosis genome project (83).
The other 22 very highly significant IFN-γ-inducing fractions that were not “contact-specific” demonstrated 20 proteins (supplemental Table S3). Of these, 10 were already reported as T cell antigens (Table II), and 10 (BfrB, GgtB, LpdC, MmsA, Pks13, Rv1910c, Rv1558, Rv2204c, Rv2251, and Rv2721c) are reported here as potentially novel human T cell antigens. Mass spectrometric analyses identified 15 proteins in the 34 highly significant IFN-γ inducing fractions and 24 proteins in the 39 significant fractions. In the highly significant and significant IFN-γ-inducing fractions, an additional 11 potentially new human T cell antigens were identified: Ald, FabG4, ProA, Tal, Rv1324c, Rv3169, aconitase (Acn), Fba, Frr, Pgi, and SahH (supplemental Tables S4 and S5).
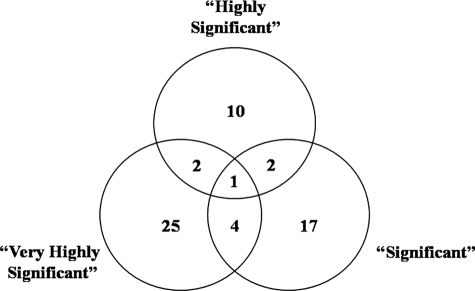
A comparison of identified proteins across all of the IFN-γ-inducing fractions (very highly significant, highly significant, and significant) revealed only modest overlap in the proteins identified for each group. Specifically, only one protein (Rv2465c) was found in a 2D LPE fraction of each group, and a total of eight proteins was shared between two of the groups (Fig. 3). These data demonstrate that immunological responses are stratified with respect to different antigens, and it is this stratification that has allowed for the identification of the 10 contact-specific fractions and their corresponding proteins.
Fig. 3.
Venn diagram of proteins identified in immunodominant fractions. Very Highly Significant represents the proteins identified in fractions that induced very highly significant levels of IFN-γ (p < 0. 0005) in the healthy contacts as compared with the TB patients. Highly Significant represents the proteins identified in fractions that induced highly significant levels of IFN-γ (p < 0.005) in the healthy contacts as compared with the TB patients. Significant represents the proteins identified in fractions that produced significant levels of IFN-γ (p < 0.05) in the healthy contacts as compared with the TB patients.
DISCUSSION
This present study was developed to identify the antigens of M. tuberculosis that have a potential use in a TB vaccine or in diagnostic development. Like many previous T cell antigen discovery efforts (15, 16, 28), we focused on the secreted proteins of M. tuberculosis. However, our current efforts differ from earlier studies, which used experimental animals (15), whereas our study aims at immunological responses to human TB. Moreover, the present study used the differential immune response of TB patients and healthy contacts to identify those proteins of M. tuberculosis with the greatest potential as protective or diagnostic antigens.
Although the immunological mechanisms of protection against tuberculosis are not fully understood, consistent evidence shows a dependence on antigen-specific T lymphocytes and the ability to stimulate the antimycobacterial activity of macrophages through the release of IFN-γ. The central role of IFN-γ in the control of TB is clearly demonstrated by experiments showing that disruption of the IFN-γ gene in mice and mutation of IFN-γ receptor gene in humans result in increased susceptibility to TB infection (8, 9). Therefore, the ability to stimulate T cell release of IFN-γ has been used as a critical criterion for the identification of protective antigens for tuberculosis.
In our current studies, medical, paramedical, and laboratory staff and healthy household members that are in close contact with TB patients but remain healthy with no evidence of disease are viewed as the “protected” population (contacts). Multiple studies provide evidence that antigens recognized by the protected group, but not active TB patients, can be considered for vaccine development strategies by using IFN-γ response as a protective correlate (29–31). Although donor to donor variation exists in antigen recognition and magnitude of response, this approach is considered a highly viable method to identify the protective antigens (28). In agreement with a previous study (32), healthy contacts in our study displayed a strong IFN-γ response to the 2D LPE fractions as compared with that of TB patients. Additionally, the stratification of the IFN-γ response allowed for the identification of 10 2D LPE fractions possessing at least 16 proteins that are specific for the protected (contact) population versus the susceptible (TB patient) population.
The Quantiferon TB Gold assay is a well established T cell-based in vitro diagnostic assay for infection with M. tuberculosis and is a significant improvement to PPD skin testing in a non-endemic population for tuberculosis surveillance (33). However, in endemic settings, its ability to distinguish between infected and diseased individuals is severely limited (33). In fact our unpublished data with healthy controls and healthy household contacts of TB patients in Chennai, India reveal that 36 and 79% of these two populations, respectively, were positive to the Quantiferon TB Gold assay. This is compared with a 96% positive response to the same assay in smear-positive TB patients for the same area. The 10 contact-specific fractions identified in this study and the corresponding proteins show a positive IFN-γ response for 100% (10 of 10) healthy contacts and no reactivity for TB patients. These data provide evidence of proteins that if used in conjunction with the current Quantiferon TB Gold assay would allow for differentiation of infected and diseased individuals. Furthermore, the proteins of the contact-specific fractions are also potential tools to monitor infected, healthy individuals for the development of active tuberculosis.
Many proteins in the CFP have been identified as potent T cell antigens including Esat-6 (34), TB8.4 (16), HspX (35), Ag85 complex (36), and Cfp-10 (37). However, there is no direct correlation between the relative concentration of these antigens in CF and their immunological relevance. For instance, Esat-6, which is present in low amounts in culture filtrate, acts as one of the most potent T cell antigens (34). Of the 350 2D LPE fractions screened, we defined 105 fractions that induced significant levels of IFN-γ in healthy contacts as compared with the TB patients. In these fractions, 59 different proteins were identified. Among these proteins, 35 observed as immunodominant antigens in our study were reported in earlier studies (Table II) either as T cell antigens in animal experiments or human studies or as B cell-stimulating antigens. Such concordance of our results with the pre-existing reports shows the reliability of the method used. Equally exciting is the fact that this methodology identified 24 proteins as potentially novel T cell antigens.
Among the 105 fractions inducing IFN-γ in healthy contacts, 10 fractions containing 16 proteins were considered contact-specific. Of these, three proteins were identified as novel human T cell antigens (Adk, AcpM, and Rv3716c) along with the 13 already reported T cell antigens. Adk, reported as a mouse T cell antigen (15), was identified as a human T cell antigen in the present study. AcpM, previously identified as a weak B cell antigen, is reported here as strong T cell antigen (38). The presence of Rv3716c protein in culture supernatants was reported earlier in proteomics studies (11, 39), and it was identified as an immunodominant molecule for the first time in the present study.
The novel proteins identified in this study can be classified into five groups: (i) the metabolic enzymes Acn (13), GgtB (40), Pgi (41), Tal, and FabG4; (ii) proteins with a reduced presence or that are absent in BCG such as Ald (42) and Rv1558 (43); (iii) proteins whose genes are up-regulated during various stress conditions including Fba (44), Rv1324 (45), MmsA (46), ProA (47), Rv2204c (48), and Rv2721c (49); (iv) proteins that are absent in hypervirulent Beijing isolates, Rv1910c (50) and Rv3179 (51); and (v) proteins identified as virulence factors such as LpdC/Rv0462 (52), SahH (53), Pks13 (54, 55), Frr (56), and Fba (53).
The presence of metabolic enzymes has been previously reported in the CF or extracellular milieu of M. tuberculosis and various pathogens (57). The extracellular localization of these proteins may be due to autolysis given the prolonged culture time (4 weeks) of M. tuberculosis. However, it is also possible they are present because of specialized secretion systems (58). The extracellular metabolic enzymes of M. tuberculosis also have been identified as virulence factors (53, 59). A recent study (53) identified a list of proteins having plasminogen binding activity in M. tuberculosis. Interestingly, this included the immunodominant antigens Ag85 complex, DnaK, Mpt51, and GlnA1 as well as the potentially novel T cell antigens (LpdC, SahH, and Fba) identified in the immunodominant fractions of the current study.
Interaction between virulence factors and IFN-γ has been clearly documented in the case of LpdC, a protein identified in this study. One of the activities of this protein is participating in the inhibition of phagolysosomal fusion by binding with coronin (a protein involved in phagolysosomal fusion). Experiments showed that IFN-γ was able to inhibit LpdC protein association with coronin (60). In the present study, the Lpd-containing fraction induced increased IFN-γ secretion in the protected population compared with TB patients, thus corroborating that Lpd is produced in vivo.
It is generally believed that low molecular mass proteins of M. tuberculosis are predominantly recognized by human T cells (61, 62), and most of the studies on human T cell antigens have focused on low molecular mass or selective pooling of fractions (16, 28) and reported that antigens <15 kDa were highly immunogenic. In the present study, only 14.8% of the immunodominant T cell antigens were represented by antigens with a predicted mass of less than 15 kDa. A stronger immune response to the proteins below 25 kDa was observed in the present study (48.6%) in agreement with previous reports (16, 63). However, a sizable number of recognized proteins (∼31%) were also found to be greater than 40 kDa. Earlier studies have shown that secreted and membrane-associated mycobacterial proteins were immunogenic (16, 79) and can be tested as vaccine targets. In this study, we found 12 secretory and five cell membrane-associated proteins to be immunogenic (Table II). These proteins can be tested as potential vaccine targets.
Further animal and human studies are needed to explore the vaccine and diagnostic potential of the candidate antigens identified in this study. Experiments are underway to test the endemic normal population and TB patients who have successfully completed chemotherapy and remained quiescent for 1 year. These studies will help define the utility of the 16 contact-specific antigens in distinguishing between protected and “susceptible” populations and whether IFN-γ responses to these antigens return after successful chemotherapeutic treatment of TB.
Supplementary Material
Acknowledgments
We thank the patients and healthy contacts for voluntarily participating in the study.
* This work was supported, in whole or in part, by the National Institutes of Health through NIAID under the International Center for Excellence in Research/Tuberculosis Research Center program and NIAID Contract HHSN266200400091c.
 The on-line version of this article (available at http://www.mcponline.org) contains Supplemental data-1 contain immunological and proteomic information of analyzed fraction (Tables S1–S9). Supplemental data-1 contain MS/MS spectra of single peptide based identification.
The on-line version of this article (available at http://www.mcponline.org) contains Supplemental data-1 contain immunological and proteomic information of analyzed fraction (Tables S1–S9). Supplemental data-1 contain MS/MS spectra of single peptide based identification.
1 The abbreviations used are:
- TB
- tuberculosis
- CF
- culture filtrate
- IFN-γ
- interferon-γ
- LPA
- lymphocyte proliferation assay
- CFP
- culture filtrate protein
- IL-4
- interleukin-4
- BCG
- Bacille Calmette-Guérin
- 2D
- two-dimensional
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PPD
- purified protein derivative
- SI
- stimulation index
- TNF-α
- tumor necrosis factor-α
- LP
- lymphocyte proliferation
- Acn
- aconitase.
REFERENCES
- 1.World Health Organization (2008) Global Tuberculosis Control—Surveillance, Planning, Financing, WHO Report 2008, World Health Organization, Geneva [Google Scholar]
- 2.Kaufmann S. H., McMichael A. J. (2005) Annulling a dangerous liaison: vaccination strategies against AIDS and tuberculosis. Nat. Med 11, S33–S44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz M. A., Wright A., Laszlo A., De Muynck A., Portaels F., Van Deun A., Wells C., Nunn P., Blanc L., Raviglione M. (2006) Epidemiology of antituberculosis drug resistance (the Global Project on Anti-tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet 368, 2142–2154 [DOI] [PubMed] [Google Scholar]
- 4.Lienhardt C., Zumla A. (2005) BCG: the story continues. Lancet 366, 1414–1416 [DOI] [PubMed] [Google Scholar]
- 5.Fine P. E. (1989) The BCG story: lessons from the past and implications for the future. Rev. Infect Dis 11, Suppl. 2, S353–359 [DOI] [PubMed] [Google Scholar]
- 6.Orme I. M. (1999) New vaccines against tuberculosis. The status of current research. Infect. Dis. Clin. North Am 13, 169–185, vii–viii [DOI] [PubMed] [Google Scholar]
- 7.Nicod L. P. (2007) Immunology of tuberculosis. Swiss Med. Wkly 137, 357–362 [DOI] [PubMed] [Google Scholar]
- 8.Flynn J. L., Chan J., Triebold K. J., Dalton D. K., Stewart T. A., Bloom B. R. (1993) An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med 178, 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jouanguy E., Altare F., Lamhamedi S., Revy P., Emile J. F., Newport M., Levin M., Blanche S., Seboun E., Fischer A., Casanova J. L. (1996) Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N. Engl. J. Med 335, 1956–1961 [DOI] [PubMed] [Google Scholar]
- 10.Nagai S., Wiker H. G., Harboe M., Kinomoto M. (1991) Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect. Immun 59, 372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungblut P. R., Schaible U. E., Mollenkopf H. J., Zimny-Arndt U., Raupach B., Mattow J., Halada P., Lamer S., Hagens K., Kaufmann S. H. (1999) Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol 33, 1103–1117 [DOI] [PubMed] [Google Scholar]
- 12.Rosenkrands I., King A., Weldingh K., Moniatte M., Moertz E., Andersen P. (2000) Towards the proteome of Mycobacterium tuberculosis. Electrophoresis 21, 3740–3756 [DOI] [PubMed] [Google Scholar]
- 13.Målen H., Berven F. S., Fladmark K. E., Wiker H. G. (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7, 1702–1718 [DOI] [PubMed] [Google Scholar]
- 14.Abou-Zeid C., Filley E., Steele J., Rook G. A. (1987) A simple new method for using antigens separated by polyacrylamide gel electrophoresis to stimulate lymphocytes in vitro after converting bands cut from Western blots into antigen-bearing particles. J. Immunol. Methods 98, 5–10 [DOI] [PubMed] [Google Scholar]
- 15.Covert B. A., Spencer J. S., Orme I. M., Belisle J. T. (2001) The application of proteomics in defining the T cell antigens of Mycobacterium tuberculosis. Proteomics 1, 574–586 [DOI] [PubMed] [Google Scholar]
- 16.Sable S. B., Kumar R., Kalra M., Verma I., Khuller G. K., Dobos K., Belisle J. T. (2005) Peripheral blood and pleural fluid mononuclear cell responses to low-molecular-mass secretory polypeptides of Mycobacterium tuberculosis in human models of immunity to tuberculosis. Infect. Immun 73, 3547–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen P., Heron I. (1993) Simultaneous electroelution of whole SDS-polyacrylamide gels for the direct cellular analysis of complex protein mixtures. J. Immunol. Methods 161, 29–39 [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld J., Capdevielle J., Guillemot J. C., Ferrera P. (1992) In gel digestion of proteins for internal sequence analysis after 1 or 2-D gel electrophoresis. Anal. Biochem 203, 173–179 [DOI] [PubMed] [Google Scholar]
- 21.Hellman U., Wernstedt C., Góñez J., Heldin C. H. (1995) Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem 224, 451–455 [DOI] [PubMed] [Google Scholar]
- 22.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 24.Selvakumar N., Sekar M. G., Ilampuranan K. J., Ponnuraja C., Narayanan P. R. (2005) Increased detection by restaining of acid-fast bacilli in sputum samples transported in cetylpyridinium chloride solution. Int. J. Tuberc. Lung Dis 9, 195–199 [PubMed] [Google Scholar]
- 25.Rook G. A., Dheda K., Zumla A. (2005) Immune responses to tuberculosis in developing countries: implications for new vaccines. Nat. Rev. Immunol 5, 661–667 [DOI] [PubMed] [Google Scholar]
- 26.Lalvani A. (2007) Diagnosing tuberculosis infection in the 21st century: new tools to tackle an old enemy. Chest 131, 1898–1906 [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann S. H. (2006) Envisioning future strategies for vaccination against tuberculosis. Nat. Rev. Immunol 6, 699–704 [DOI] [PubMed] [Google Scholar]
- 28.Lim J. H., Kim H. J., Lee K. S., Jo E. K., Song C. H., Jung S. B., Kim S. Y., Lee J. S., Paik T. H., Park J. K. (2004) Identification of the new T-cell-stimulating antigens from Mycobacterium tuberculosis culture filtrate. FEMS Microbiol. Lett 232, 51–59 [DOI] [PubMed] [Google Scholar]
- 29.Havlir D. V., Wallis R. S., Boom W. H., Daniel T. M., Chervenak K., Ellner J. J. (1991) Human immune response to Mycobacterium tuberculosis antigens. Infect. Immun 59, 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres M., Herrera T., Villareal H., Rich E. A., Sada E. (1998) Cytokine profiles for peripheral blood lymphocytes from patients with active pulmonary tuberculosis and healthy household contacts in response to the 30-kilodalton antigen of Mycobacterium tuberculosis. Infect. Immun 66, 176–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grotzke J. E., Lewinsohn D. M. (2005) Role of CD8+ T lymphocytes in control of Mycobacterium tuberculosis infection. Microbes Infect 7, 776–788 [DOI] [PubMed] [Google Scholar]
- 32.Vekemans J., Ota M. O., Sillah J., Fielding K., Alderson M. R., Skeiky Y. A., Dalemans W., McAdam K. P., Lienhardt C., Marchant A. (2004) Immune responses to mycobacterial antigens in the Gambian population: implications for vaccines and immunodiagnostic test design. Infect. Immun 72, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies P. D., Pai M. (2008) The diagnosis and misdiagnosis of tuberculosis. Int. J. Tuberc. Lung Dis 12, 1226–1234 [PubMed] [Google Scholar]
- 34.Doherty T. M., Demissie A., Olobo J., Wolday D., Britton S., Eguale T., Ravn P., Andersen P. (2002) Immune responses to the Mycobacterium tuberculosis-specific antigen ESAT-6 signal subclinical infection among contacts of tuberculosis patients. J. Clin. Microbiol 40, 704–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geluk A., Lin M. Y., van Meijgaarden K. E., Leyten E. M., Franken K. L., Ottenhoff T. H., Klein M. R. (2007) T-cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infect. Immun 75, 2914–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horwitz M. A., Harth G., Dillon B. J., Maslesa-Galic S. (2005) Enhancing the protective efficacy of Mycobacterium bovis BCG vaccination against tuberculosis by boosting with the Mycobacterium tuberculosis major secretory protein. Infect. Immun 73, 4676–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillon D. C., Alderson M. R., Day C. H., Bement T., Campos-Neto A., Skeiky Y. A., Vedvick T., Badaro R., Reed S. G., Houghton R. (2000) Molecular and immunological characterization of Mycobacterium tuberculosis CFP-10, an immunodiagnostic antigen missing in Mycobacterium bovis BCG. J. Clin. Microbiol 38, 3285–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weldingh K., Rosenkrands I., Okkels L. M., Doherty T. M., Andersen P. (2005) Assessing the serodiagnostic potential of 35 Mycobacterium tuberculosis proteins and identification of four novel serological antigens. J. Clin. Microbiol 43, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt F., Donahoe S., Hagens K., Mattow J., Schaible U. E., Kaufmann S. H., Aebersold R., Jungblut P. R. (2004) Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol. Cell. Proteomics 3, 24–42 [DOI] [PubMed] [Google Scholar]
- 40.Dayaram Y. K., Talaue M. T., Connell N. D., Venketaraman V. (2006) Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol 188, 1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu S., Chen J., Dobos K. M., Bradbury E. M., Belisle J. T., Chen X. (2003) Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 2, 1284–1296 [DOI] [PubMed] [Google Scholar]
- 42.Chen J. M., Alexander D. C., Behr M. A., Liu J. (2003) Mycobacterium bovis BCG vaccines exhibit defects in alanine and serine catabolism. Infect. Immun 71, 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollenkopf H. J., Grode L., Mattow J., Stein M., Mann P., Knapp B., Ulmer J., Kaufmann S. H. (2004) Application of mycobacterial proteomics to vaccine design: improved protection by Mycobacterium bovis BCG prime-Rv3407 DNA boost vaccination against tuberculosis. Infect. Immun 72, 6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenkrands I., Slayden R. A., Crawford J., Aagaard C., Barry C. E., 3rd, Andersen P. (2002) Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol 184, 3485–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidders B., Withers M., Kendall S. L., Bacon J., Waddell S. J., Hinds J., Golby P., Movahedzadeh F., Cox R. A., Frita R., Ten Bokum A. M., Wernisch L., Stoker N. G. (2007) Quantification of global transcription patterns in prokaryotes using spotted microarrays. Genome Biol 8, R265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tailleux L., Waddell S. J., Pelizzola M., Mortellaro A., Withers M., Tanne A., Castagnoli P. R., Gicquel B., Stoker N. G., Butcher P. D., Foti M., Neyrolles O. (2008) Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE 3, e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danelishvili L., Wu M., Young L. S., Bermudez L. E. (2005) Genomic approach to identifying the putative target of and mechanisms of resistance to mefloquine in mycobacteria. Antimicrob. Agents Chemother 49, 3707–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dosanjh N. S., Rawat M., Chung J. H., Av-Gay Y. (2005) Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol. Lett 249, 87–94 [DOI] [PubMed] [Google Scholar]
- 49.Betts J. C., McLaren A., Lennon M. G., Kelly F. M., Lukey P. T., Blakemore S. J., Duncan K. (2003) Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother 47, 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsolaki A. G., Gagneux S., Pym A. S., Goguet de la Salmoniere Y. O., Kreiswirth B. N., Van Soolingen D., Small P. M. (2005) Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol 43, 3185–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stavrum R., Valvatne H., Bø T. H., Jonassen I., Hinds J., Butcher P. D., Grewal H. M. (2008) Genomic diversity among Beijing and non-Beijing Mycobacterium tuberculosis isolates from Myanmar. PLoS ONE 3, e1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee K. Y., Erdjument-Bromage H., Tempst P., Nathan C. F. (2005) S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc. Natl. Acad. Sci. U.S.A 102, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xolalpa W., Vallecillo A. J., Lara M., Mendoza-Hernandez G., Comini M., Spallek R., Singh M., Espitia C. (2007) Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7, 3332–3341 [DOI] [PubMed] [Google Scholar]
- 54.Portevin D., De Sousa-D'Auria C., Houssin C., Grimaldi C., Chami M., Daffé M., Guilhot C. (2004) A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. U.S.A 101, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A., Mai D., Kumar A., Steyn A. J. (2006) Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U.S.A 103, 11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vizcaíno N., Cloeckaert A., Dubray G., Zygmunt M. S. (1996) Cloning, nucleotide sequence, and expression of the gene coding for a ribosome releasing factor-homologous protein of Brucella melitensis. Infect. Immun 64, 4834–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pancholi V., Chhatwal G. S. (2003) Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol 293, 391–401 [DOI] [PubMed] [Google Scholar]
- 58.Sibbald M. J., Ziebandt A. K., Engelmann S., Hecker M., de Jong A., Harmsen H. J., Raangs G. C., Stokroos I., Arends J. P., Dubois J. Y., van Dijl J. M. (2006) Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev 70, 755–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinhikar A. G., Vargas D., Li H., Mahaffey S. B., Hinds L., Belisle J. T., Laal S. (2006) Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol. Microbiol 60, 999–1013 [DOI] [PubMed] [Google Scholar]
- 60.Deghmane A. E., Soualhine H., Bach H., Sendide K., Itoh S., Tam A., Noubir S., Talal A., Lo R., Toyoshima S., Av-Gay Y., Hmama Z. (2007) Lipoamide dehydrogenase mediates retention of coronin-1 on BCG vacuoles, leading to arrest in phagosome maturation. J. Cell Sci 120, 2796–2806 [DOI] [PubMed] [Google Scholar]
- 61.Boesen H., Jensen B. N., Wilcke T., Andersen P. (1995) Human T-cell responses to secreted antigen fractions of Mycobacterium tuberculosis. Infect. Immun 63, 1491–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demissie A., Ravn P., Olobo J., Doherty T. M., Eguale T., Geletu M., Hailu W., Andersen P., Britton S. (1999) T-cell recognition of Mycobacterium tuberculosis culture filtrate fractions in tuberculosis patients and their household contacts. Infect. Immun 67, 5967–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gulle H., Fray L. M., Gormley E. P., Murray A., Moriarty K. M. (1995) Responses of bovine T cells to fractionated lysate and culture filtrate proteins of Mycobacterium bovis BCG. Vet. Immunol. Immunopathol 48, 183–190 [DOI] [PubMed] [Google Scholar]
- 64.Floto R. A., MacAry P. A., Boname J. M., Mien T. S., Kampmann B., Hair J. R., Huey O. S., Houben E. N., Pieters J., Day C., Oehlmann W., Singh M., Smith K. G., Lehner P. J. (2006) Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science 314, 454–458 [DOI] [PubMed] [Google Scholar]
- 65.Derrick S. C., Yang A. L., Morris S. L. (2004) A polyvalent DNA vaccine expressing an ESAT6-Ag85B fusion protein protects mice against a primary infection with Mycobacterium tuberculosis and boosts BCG-induced protective immunity. Vaccine 23, 780–788 [DOI] [PubMed] [Google Scholar]
- 66.D'Souza S., Rosseels V., Romano M., Tanghe A., Denis O., Jurion F., Castiglione N., Vanonckelen A., Palfliet K., Huygen K. (2003) Mapping of murine Th1 helper T-Cell epitopes of mycolyl transferases Ag85A, Ag85B, and Ag85C from Mycobacterium tuberculosis. Infect. Immun 71, 483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh K. K., Dong Y., Belisle J. T., Harder J., Arora V. K., Laal S. (2005) Antigens of Mycobacterium tuberculosis recognized by antibodies during incipient, subclinical tuberculosis. Clin. Diagn. Lab. Immunol 12, 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tullius M. V., Harth G., Horwitz M. A. (2003) Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun 71, 3927–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qamra R., Mande S. C., Coates A. R., Henderson B. (2005) The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis 85, 385–394 [DOI] [PubMed] [Google Scholar]
- 70.Banerjee S., Nandyala A., Podili R., Katoch V. M., Murthy K. J., Hasnain S. E. (2004) Mycobacterium tuberculosis (Mtb) isocitrate dehydrogenases show strong B cell response and distinguish vaccinated controls from TB patients. Proc. Natl. Acad. Sci. U.S.A 101, 12652–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drake W. P., Dhason M. S., Nadaf M., Shepherd B. E., Vadivelu S., Hajizadeh R., Newman L. S., Kalams S. A. (2007) Cellular recognition of Mycobacterium tuberculosis ESAT-6 and KatG peptides in systemic sarcoidosis. Infect. Immun 75, 527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pecora N. D., Gehring A. J., Canaday D. H., Boom W. H., Harding C. V. (2006) Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J. Immunol 177, 422–429 [DOI] [PubMed] [Google Scholar]
- 73.Al-Attiyah R., Mustafa A. S. (2004) Computer-assisted prediction of HLA-DR binding and experimental analysis for human promiscuous Th1-cell peptides in the 24 kDa secreted lipoprotein (LppX) of Mycobacterium tuberculosis. Scand. J. Immunol 59, 16–24 [DOI] [PubMed] [Google Scholar]
- 74.Målen H., Søfteland T., Wiker H. G. (2008) Antigen analysis of Mycobacterium tuberculosis H37Rv culture filtrate proteins. Scand. J. Immunol 67, 245–252 [DOI] [PubMed] [Google Scholar]
- 75.Kumar P., Amara R. R., Challu V. K., Chadda V. K., Satchidanandam V. (2003) The Apa protein of Mycobacterium tuberculosis stimulates gamma interferon-secreting CD4+ and CD8+ T cells from purified protein derivative-positive individuals and affords protection in a guinea pig model. Infect. Immun 71, 1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulrichs T., Munk M. E., Mollenkopf H., Behr-Perst S., Colangeli R., Gennaro M. L., Kaufmann S. H. (1998) Differential T cell responses to Mycobacterium tuberculosis ESAT6 in tuberculosis patients and healthy donors. Eur. J. Immunol 28, 3949–3958 [DOI] [PubMed] [Google Scholar]
- 77.Tavares R. C., Salgado J., Moreira V. B., Ferreira M. A., Mello F. C., Leung J. A., Singh M., Fonseca Lde S., Saad M. H. (2006) Cell proliferation and interferon-gamma response to recombinant MBP-3, NarL, MT-10.3, and 16 kDa Mycobacterium tuberculosis antigens in Brazilian tuberculosis patients. Mem. Inst. Oswaldo Cruz 101, 857–861 [DOI] [PubMed] [Google Scholar]
- 78.Fonseca D. P., Benaissa-Trouw B., van Engelen M., Kraaijeveld C. A., Snippe H., Verheul A. F. (2001) Induction of cell-mediated immunity against Mycobacterium tuberculosis using DNA vaccines encoding cytotoxic and helper T-cell epitopes of the 38-kilodalton protein. Infect. Immun 69, 4839–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sinha S., Kosalai K., Arora S., Namane A., Sharma P., Gaikwad A. N., Brodin P., Cole S. T. (2005) Immunogenic membrane-associated proteins of Mycobacterium tuberculosis revealed by proteomics. Microbiology 151, 2411–2419 [DOI] [PubMed] [Google Scholar]
- 80.Khera A., Singh R., Shakila H., Rao V., Dhar N., Narayanan P. R., Parmasivan C. N., Ramanathan V. D., Tyagi A. K. (2005) Elicitation of efficient, protective immune responses by using DNA vaccines against tuberculosis. Vaccine 23, 5655–5665 [DOI] [PubMed] [Google Scholar]
- 81.Weldingh K., Hansen A., Jacobsen S., Andersen P. (2000) High resolution electroelution of polyacrylamide gels for the purification of single proteins from Mycobacterium tuberculosis culture filtrate. Scand. J. Immunol 51, 79–86 [DOI] [PubMed] [Google Scholar]
- 82.Hermans P. W., Abebe F., Kuteyi V. I., Kolk A. H., Thole J. E., Harboe M. (1995) Molecular and immunological characterization of the highly conserved antigen 84 from Mycobacterium tuberculosis and Mycobacterium leprae. Infect. Immun 63, 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.