Abstract
The chondrogenic potential of multipotent mesenchymal stem cells (MSCs) makes them a promising source for cell-based therapy of cartilage defects; however, the exact intracellular molecular mechanisms of chondrogenesis as well as self-renewal of MSCs remain largely unknown. To gain more insight into the underlying molecular mechanisms, we applied isobaric tag for relative and absolute quantitation (iTRAQ) labeling coupled with on-line two-dimensional LC/MS/MS technology to identify proteins differentially expressed in an in vitro model for chondrogenesis: chondrogenic differentiation of C3H10T1/2 cells, a murine embryonic mesenchymal cell line, was induced by micromass culture and 100 ng/ml bone morphogenetic protein 2 treatment for 6 days. A total of 1756 proteins were identified with an average false discovery rate <0.21%. Linear regression analysis of the quantitative data gave strong correlation coefficients: 0.948 and 0.923 for two replicate two-dimensional LC/MS/MS analyses and 0.881, 0.869, and 0.927 for three independent iTRAQ experiments, respectively (p < 0.0001). Among 1753 quantified proteins, 100 were significantly altered (95% confidence interval), and six of them were further validated by Western blotting. Functional categorization revealed that the 17 up-regulated proteins mainly comprised hallmarks of mature chondrocytes and enzymes participating in cartilage extracellular matrix synthesis, whereas the 83 down-regulated were predominantly involved in energy metabolism, chromatin organization, transcription, mRNA processing, signaling transduction, and cytoskeleton; except for a number of well documented proteins, the majority of these altered proteins were novel for chondrogenesis. Finally, the biological roles of BTF3l4 and fibulin-5, two novel chondrogenesis-related proteins identified in the present study, were verified in the context of chondrogenic differentiation. These data will provide valuable clues for our better understanding of the underlying mechanisms that modulate these complex biological processes and assist in the application of MSCs in cell-based therapy for cartilage regeneration.
Mesenchymal stem cells (MSCs)1 are multipotent cells found in several adult tissues that can be expanded in vitro and differentiate into multiple mesoderm-type cells, including chondrocytes, thus representing a promising source for cell-based therapy of cartilage defects (1). Chondrogenic differentiation of MSCs in vitro closely resembles in vivo chondrogenesis, including mesenchymal cell condensation, chondrocyte differentiation, and maturation, which is elaborately modulated by signals initiated by cell-cell and cell-matrix interactions as well as a variety of growth and differentiation factors (2–4). Despite the enhanced interest and accumulating reports on making use of MSCs in cartilage repair and regeneration, the exact molecular events that occur in chondrogenic differentiation of MSCs remain largely unclear (5).
MSCs are routinely isolated from bone marrow according to their property of adhesion to plastic, which results in a morphologically, phenotypically, and functionally heterogeneous population of cells (6, 7). Because of the absence of defined markers, it is hard to obtain a morphologically and functionally homogeneous population, especially given the biological complexity derived from different ages and genetic backgrounds of the donors. To facilitate the molecular mechanism investigation and further in-depth biological function analysis, we chose a well established in vitro chondrogenic model in our primary proteomics study. C3H10T1/2, a murine embryonic mesenchymal cell line (8), has been demonstrated to differentiate into multiple mesenchymal lineages such as chondrocytes, osteoblasts, and adipocytes (9). Therefore, this cell line is regarded as a model for MSCs, representing a homogeneous population of multipotential cells that do not spontaneously differentiate under normal culture conditions, and hence is an ideal vehicle for in vitro study of chondrogenesis (10). Additionally, a recent report by Zhao et al. (11) revealed that under the same inductive conditions C3H10T1/2 cells showed chondrogenic differentiation potentials comparable to primary murine bone marrow-derived MSCs, suggesting that this cell line is a good alternative cell source for investigating chondrogenic differentiation. Consistent with a previous report (10), chondrogenic differentiation of C3H10T1/2 cells was induced by a high density micromass environment and BMP-2 treatment in the present study. This in vitro model has also been used in gene expression profiling of mesenchymal chondrogenesis (12).
Recently, proteomics approaches have been applied to the studies of MSCs, such as the secretome of embryonic stem (ES) cell-derived MSCs (13), the global effects of transforming growth factor-β on MSCs (14), differential expression profiling of membrane proteins of MSCs undergoing osteoblast differentiation (15), and proteome analysis of rat MSC subcultures (16). However, the comprehensive expression profiling of MSCs undergoing chondrogenic differentiation has not been reported yet (17). To gain further understanding of the molecular mechanism underlying this stringently modulated process, we applied an iTRAQ labeling coupled with on-line 2D LC/MS/MS proteomics technology to quantitatively assess the protein expression profile of the in vitro chondrogenesis model. The iTRAQ labeling coupled with LC/MS/MS is a gel-free quantitative proteomics technology that uses amine-specific isobaric tags to compare the intensity of reporter ions of labeled peptides and infer quantitative values for corresponding proteins (18).
In this study, we compared the protein profile of chondrogenically differentiated C3H10T1/2 cells with those of undifferentiated cells by using iTRAQ labeling coupled with on-line 2D LC/MS/MS. A total of 1756 proteins were identified. Of them, 100 significantly altered proteins were identified (95% confidence interval), and six of them were validated by Western blotting. Further functional categorization revealed that these altered proteins could play essential roles in lineage-specific differentiation, promoting a nonspecific stem cell state or the commitment to other lineages. Finally, biological functions of a number of these intriguing proteins were preliminarily validated in the context of chondrogenesis. Such findings will advance our understanding of the underlying intracellular mechanisms that modulate chondrogenic differentiation as well as self-renewal of MSCs and in turn promote their application in cartilage defect therapy.
EXPERIMENTAL PROCEDURES
Cell Culture
Cell culture media and media supplements were purchased from Invitrogen. C3H10T1/2 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Monolayer cultures were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 50 units/ml penicillin G, and 50 μg/ml streptomycin and incubated at 37 °C in 5% (v/v) CO2 in a humidified incubator. Media were changed every 3 days.
Chondrogenesis Induction
The micromass culture technique modified from Ahrens et al. (19) was adopted for chondrogenic differentiation experiments. Briefly, subconfluent C3H10T1/2 cells were trypsinized and resuspended at a concentration of 107 cells/ml in Ham's F-12 medium with 10% (v/v) fetal bovine serum. A 10-μl drop of cell suspension was placed in a well of a 24-well culture plate and allowed to adhere for 2–3 h at 37 °C and 5% (v/v) CO2, then 1 ml of medium containing 100 ng/ml human recombinant BMP-2 purchased from R&D Systems (Minneapolis, MN) was added to the cultures, and controls were cultured in the same manner but without supplementation of human recombinant BMP-2. The cultures were fed every 3 days with fresh media with or without BMP-2, and day 6 cultures were used for the proteomics analysis.
Alcian Blue Staining and Quantification
For Alcian blue staining, cultures were rinsed twice with PBS, fixed in 4% (w/v) paraformaldehyde for 15 min, and incubated in 1% (w/v) Alcian blue 8-GX (Sigma) in 0.1 n HCl (pH 1.0) overnight. For quantitative analysis, Alcian blue-stained cultures were extracted with 6 m guanidine HCl for 2 h at room temperature. The absorption of the extracted dye was measured at 650 nm in a microplate reader (Bio-Rad).
Western Blotting
Cultures were washed with ice-cold phosphate buffered saline and lysed in a buffer containing 50 mm Tris-HCl (pH 7.6), 5 mm EDTA, 50 mm NaCl, 30 mm sodium pyrophosphate, 50 mm NaF, 0.1 mm Na3VO4, 1% (v/v) Triton X-100, 1 mm PMSF, and a protease inhibitor mixture tablet (Roche Applied Science). Lysates were clarified by centrifugation at 15,000 × g for 10 min at 4 °C, and the protein concentration of the supernatant was measured by the Bradford protein assay (Bio-Rad). Samples containing 30 μg of total protein were separated by 12% (w/v) SDS-PAGE and transferred onto a PVDF membrane (Millipore, Bedford, MA). After incubating for 1 h with blocking buffer (5% (w/v) nonfat milk in TBS-T (0.05% (v/v) Tween 20 in Tris-buffered saline)), the membrane was probed with the indicated primary antibodies diluted in blocking buffer overnight at 4 °C. After being extensively washed with TBS-T, the membrane was incubated with horseradish peroxidase-conjugated antibody to mouse (Kirkegaard and Perry Laboratories, Inc.) or rabbit (Cell Signaling Technology) diluted in blocking buffer (1:2000) for 1 h at room temperature. Bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and recorded on x-ray films (Fuji Medical, Tokyo, Japan). Finally, the visualized bands were quantified by QUANTITY ONE software on a GS-800 densitometer (Bio-Rad). The following antibodies were used: monoclonal anti-collagen type II (Lab Vision Corp.) (1:1000), monoclonal anti-β-tubulin (Sigma-Aldrich) (1:5000), rabbit polyclonal antibody to protein-lysine 6-oxidase (LOX) (Santa Cruz Biotechnology, Inc.) (1:500), rabbit polyclonal antibody to Histone H2A.X (Abcam) (1:2000), and monoclonal antibodies to MARCKS and hnRNPM (Santa Cruz Biotechnology, Inc.) (1:1000).
Flow Cytometry
For flow cytometric analysis, the cultures were detached by 0.1% (w/v) trypsin and collagenase type II (Sigma-Aldrich) digestion followed by washing and fixation. The resulting pellets were resuspended in 1% (w/v) bovine serum albumin (Roche Applied Science) for 30 min in room temperature to block the nonspecific binding sites. Afterward, the samples were incubated with anti-collagen type II monoclonal antibody (Clone 2B1.5, Neomarker) at 4 °C for 8 h and then stained with a FITC-conjugated goat anti-mouse secondary antibody (Molecular Probes, Inc.) at room temperature for 1 h. Flow cytometric acquisition and data analysis were performed with an Epics ALTRA flow cytometer and EXPO32TM software (Beckman Coulter, Inc., Miami, FL). As a negative control, the cells were incubated only with the FITC-conjugated secondary antibody. Three independent flow cytometric experiments were performed.
iTRAQ Labeling, Sample Cleaning, and Desalting
All the reagents and buffers needed for iTRAQ labeling and cleaning were obtained from Applied Biosystems (Foster City, CA). iTRAQ labeling was performed according to the manufacturer's instructions. Briefly, at day 6, total cell lysates were collected as described for Western blotting, and protein concentration was determined by the Bradford assay. Subsequently, 100 μg of protein was precipitated with ice-cold acetone overnight at −20 °C. Protein pellets were dissolved, denatured, alkylated, and digested with trypsin (Sigma; 1:20, w/w, 37 °C for 18 h). To label peptides with iTRAQ reagent, 1 unit of label (defined as the amount of reagent required to label 100 μg of protein) was thawed and reconstituted in 70 μl of ethanol; digestions from BMP-2-treated and untreated C3H10T1/2 cells were labeled with 117and 114 iTRAQ reagents, respectively; and then the samples were incubated at room temperature for 1 h and pooled. In the present study, three independent chondrogenic induction and iTRAQ labeling experiments were carried out.
Prior to on-line 2D nanoscale LC (nano-LC)/MS/MS analysis, iTRAQ-labeled samples were cleaned up and desalted. A cation exchange cartridge system (Applied Biosystems) was used to remove the reducing reagent, SDS, excess iTRAQ reagents, undigested proteins, and trypsin in the labeled sample mixture that would interfere with the LC/MS/MS analysis. Before cation exchange, the concentration of buffer salts in the labeled samples was reduced below 10 mm by diluting 10-fold with cation exchange buffer-load (10 mm K2HPO4 in 25% (v/v) acetonitrile at pH 3.0). The pH of the diluted sample was checked: it should be between 2.5 and 3.3; otherwise it was adjusted to 3.0 with phosphoric acid. The diluted sample mixture was loaded onto the cation exchange cartridge, after washing with 10 column volumes of buffer-load, peptides were eluted with 500 μl of buffer-elute (10 mm K2HPO4 in 25% (v/v) acetonitrile, 350 mm KCl at pH 3.0). Afterward, the eluate of the cation exchange was desalted using an Agilent 1100 series HPLC system equipped with an autosampler, 2/6 valve, and diode array detector (220 nm) (Agilent, Waldbronn, Germany). Eluates of cation exchange were diluted with 0.1% (v/v) FA (v/v in H2O) to reduce the concentration of acetonitrile to 5% (v/v), afterward the dilution was injected onto a 4.6-mm-inner diameter × 150-mm C18 reversed-phase column (5 μm, 80 Å; Agilent, Waldbronn, Germany), flushed with phase A (5% (v/v) acetonitrile, 0.1% (v/v) FA in H2O) at 1 ml/min for 10 min, and finally peptides were eluted with 65% (v/v) phase B (90% (v/v) acetonitrile, 0.1% (v/v) FA in H2O). Absorbance at 220 nm was monitored, and the maximal absorption peak was collected and divided into two aliquots. Each aliquot was dried in a Heto vacuum concentrator (Heto-Holten A/S, Allerod, Denmark).
On-line 2D LC/MS/MS
2D nano-LC/MS/MS analyses were conducted on a nano-HPLC system (Agilent, Waldbronn, Germany) coupled to a hybrid Q-TOF mass spectrometer (QSTAR XL, Applied Biosystems) equipped with a nano-ESI source (Applied Biosystems) and a nano-ESI needle (Picotip, FS360-50-20; New Objective Inc., Woburn, MA). AnalystTM 1.1 software was used to control QSTAR XL mass spectrometry and the nano-HPLC system and to acquire mass spectra. Vacuum-dried iTRAQ-labeled peptides were reconstituted in phase A and injected at a flow rate of 10 μl/min onto a high resolution strong cation exchange (SCX) column (Bio-SCX, 300-μm inner diameter × 35 mm; Agilent, Wilmington, DE), which was on line with a C18 precolumn (PepMap, 300-μm inner diameter × 5 mm; LC Packings). After loading, the SCX column and C18 precolumn were flushed with a 16-step gradient sodium chloride solution (0, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 125, 150, 200, 300, and 400 mm) for 5 min and phase A for 10 min at a flow rate of 15 μl/min. Afterward, the precolumn was switched on line with a nanoflow reversed-phase column (VYDAC 218MS, 75-μm inner diameter × 100 mm; Grace, Hesperia, CA), and the peptides concentrated and desalted on the precolumn were separated using a 120-min linear gradient from 12 to 30% (v/v) phase B (0.1% (v/v) FA in ACN) at a flow rate of 400 nl/min.
The Q-TOF instrument was operated in positive ion mode with ion spray voltage typically maintained at 2.0 kV. Mass spectra of iTRAQ-labeled samples were acquired in an information-dependent acquisition mode. The analytical cycle consisted of a 0.7-s MS survey scan (400–1600 m/z) followed by three 2-s MS/MS scans (100–2000 m/z) of the three most abundant peaks (i.e. precursor ions), which were selected from the MS survey scan. Precursor ion selection was based upon ion intensity (peptide signal intensity above 25 counts/s) and charge state (2+ to 5+), and once the ions were fragmented in the MS/MS scan, they were allowed one repetition before a dynamic exclusion for a period of 120 s. Because of the iTRAQ tags, the parameters for rolling collision energy (automatically set according to the precursor m/z and charge state) were manually optimized. Under CID, iTRAQ-labeled peptides fragmented to produce reporter ions at 114.1 and 117.1, and fragment ions of the peptides were simultaneously produced, resulting in sequencing of the labeled peptides and identification of the corresponding proteins. The ratios of the peak areas of the two iTRAQ reporter ions reflected the relative abundances of the peptides and the proteins in the samples. External calibration of mass spectrometer was carried out using reserpine and trypsinized BSA routinely.
Protein Identification and False Discovery Rate (FDR) Analysis
For protein identification, the complete set of raw data files (*.wiff) of each run was analyzed together using ProteinPilot software 2.0.1 (revision 67476), which consisted of the Paragon and Pro GroupTM algorithms. The Paragon search algorithm used a sequence tag algorithm to calculate sequence temperature values for identification of peptides from a database (20). The parameters for searching were as follows: iTRAQ fourplex peptide labeled; trypsin digestion; methyl methane thiosulfate alkylation of cysteine residue; instrument, QSTAR ESI; identification focus, biological modifications; non-redundant International Protein Index (IPI) mouse sequence database version 3.62 (date of release, August 2009; with a total of 56,727 protein entries) from the European Bioinformatics Institute selected; and software defaults used for other parameters needed for searching.
Afterward, the Pro Group algorithm assembled the peptide evidence from the Paragon algorithm to find the smallest number of proteins that could explain all the fragmentation spectral evidence. The core philosophy of the Pro Group algorithm was that the spectral evidence used to prove the detection of one protein could not be used again to prove the detection of a second protein; therefore, two types of scores for each protein were calculated: Total ProtScore and Unused ProtScore. The Total ProtScore was based on all peptides pointing to a protein and was analogous to protein scores reported by other protein identification softwares, whereas the Unused ProtScore was calculated from the peptide evidence that had not already been used to explain more confident proteins. The Pro Group algorithm greatly reduced the redundancy and suppressed the false positives. In addition, the Pro Group algorithm also could distinguish protein isoforms; a specific protein isoform would be reported only if unique evidence (peptide) existed for this isoform (from Understanding the Pro Group Algorithm, Applied Biosystems). To obtain an even more reliable protein list, in the present study, proteins were identified on the basis of having at least two distinct peptides with 99% confidence and 2.0 contribution to Unused ProtScore. In other words, only proteins with Unused ProtScore greater than or equal to 4.0 were included in the final protein list.
To evaluate the rate of erroneously identified proteins, the same set of MS spectral data was searched as above but against a decoy database, a randomized version of the database generated by DecoyDBB software (21). The following formula was used to calculate the FDR of protein identification: FDR = (FP/FP + TP) × 100% (FP, false positives, number of random hits; TP, true positives, number of normal hits).
Protein Quantification and Identification of Differentially Expressed Proteins
Protein quantification was also performed by ProteinPilot software, which automatically calculated the relative abundance of iTRAQ-labeled peptides and the corresponding proteins. Corrections were made for impurity of iTRAQ reagents based on the data provided by the manufacturer. For pipetting and other similar errors in analyses, iTRAQ ratios were normalized by autobias, which used all data obtained from a 2D LC/MS/MS analysis to calculate the bias correction factor.
In the present study, the thresholds for protein differential expression were mean ± 1.96S.D. (95% confidence interval). Proteins whose 117/114 ratio met the differential expression threshold in at least two of the three biological experiments are shown in the main text, whereas those that appeared in only one of the three biological experiments are listed in the supplemental material.
RNA Extraction and Quantitative Real Time PCR
The methods for RNA extraction and quantitative real time PCR were the same as in our previous report (22). Briefly, total RNA was extracted and purified using the RNeasy kit (Qiagen, Valencia, CA). Gene products were analyzed by using SYBR Green PCR Master Mix (Applied Biosystems) and specific primers in a 7300 Real Time PCR System (Applied Biosystems). Relative gene expression levels were calculated as ratios of the mRNA levels normalized against those of 18 S rRNA. All the results were expressed as means ± S.D. of three independent experiments. Primer sequences are reported in supplemental Table 1.
Lactate Assay
Lactate concentration in the conditioned medium of day 6 cultures was measured by using a lactate assay kit (Biovision) according to the manufacture's instructions. The absorption at 570 nm was measured by a microplate reader (Bio-Rad).
Short Interfering RNA (siRNA) Transfection
ON-TARGETplus SMARTpool siRNA against Btf3l4 (L-045446-01-0005) or Fbln5 (M-059161-01-0005) was transfected into the C3H10T1/2 cells, respectively. Transfection was performed according to the manual (Thermo Science). Briefly, the cells were collected and subcultured in growth medium without antibiotics; the cells were 90–95% confluent at the time of transfection. siRNAs were incubated with Lipofectamine 2000 (Invitrogen), and then the mixture was added into the culture. Six hours after transfection, the cells were trypsinized, inoculated as micromass, and treated with 100 ng/ml BMP-2 for chondrogenesis induction as aforementioned.
Statistical Analysis
Data are presented as the mean ± S.D. Comparison between means was assessed by unpaired Student's t test or one-way analysis of variance using SPSS10.0 software. Statistical significance was set at p < 0.05. Unless otherwise specified, all assays were performed in triplicate.
RESULTS
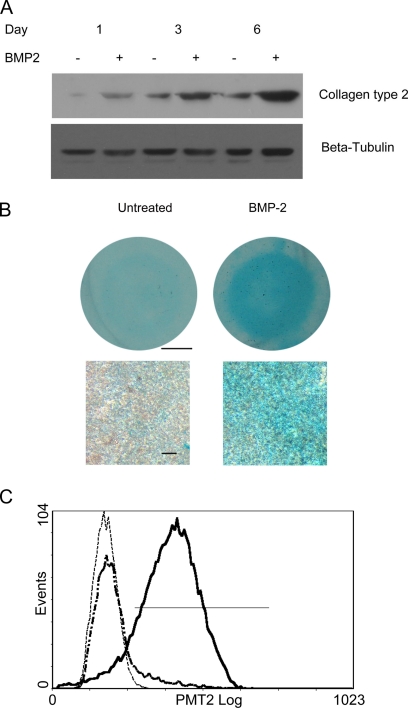
To gain more insight into the molecular mechanisms underlying chondrogenic differentiation as well as self-renewal of MSCs, we applied iTRAQ labeling coupled with on-line 2D nano-LC/MS/MS to quantitatively assess the global protein expression profile of a well established in vitro chondrogenesis model (10). To evaluate the chondrogenesis of C3H10T1/2 cells in micromass culture treated with BMP-2, glycosaminoglycans (GAGs), and collagen type II, two markers for mature chondrocytes, were detected by Alcian blue staining and Western blotting, respectively. In line with previous reports (12), increased expression of collagen type II and positive staining of Alcian blue were observed in BMP-2-treated cultures, demonstrating that the cultures underwent chondrogenesis beginning on day 3 and more overtly on day 6 (Fig. 1, A and B). Furthermore, to quantitatively evaluate the percentage of collagen type II-positive cells in the micromass cultures, cells from day 6 cultures were subject to flow cytometry analysis. As shown in Fig. 1C, the collagen type II-positive cells of BMP-2-treated and untreated C3H10T1/2 cells in micromass culture were 86.5 ± 5.5 and 10.9 ± 2.3%, respectively, showing that most of the cells in micromass cultures were chondrogenically differentiated by micromass culture and BMP-2 treatment. In addition, these results also revealed the existence of spontaneous differentiation of C3H10T1/2 cells when cultured in high density, although the rate of occurrence was limited. Taken together, these data demonstrated the chondrogenesis of the C3H10T1/2 cells in the present study.
Fig. 1.
Western blotting, histochemical, and flow cytometry analyses of chondrogenic differentiation of C3H10T1/2 cells in micromass culture treated with BMP-2. A, time course of BMP-2-induced collagen type II expression. Total protein was extracted at the indicated times, and collagen type II expression was determined by Western blotting. β-Tubulin was used to confirm equal loading. B, after 6 days, BMP-2-treated (right) and untreated (left) cultures were fixed and stained with Alcian blue. Scale bar, 2.5 mm (top) and 50 μm (bottom). C, representative flow cytometry histogram of three independent experiments illustrating that the majority of the cells in the micromass culture treated with BMP-2 were collagen type II-positive (86.5 ± 5.5%), whereas the percentage of these positive cells in the untreated control was smaller (10.9 ± 2.3%). The solid line represents the C3H10T1/2 cells treated with BMP-2 for 6 days, the dash-dotted line represents the untreated controls, and the dotted line represents the negative control.
For identifying proteins whose expression levels were either up- or down-regulated during chondrogenesis, total cell lysates from day 6 cultures were subjected to iTRAQ labeling and on-line 2D LC/MS/MS analyses as described under “Experimental Procedures.” Three biological replicates were performed to provide information about the reproducibility of the in vitro chondrogenesis model; in addition, iTRAQ-labeled samples from biological replications 2 and 3 were analyzed repeatedly to evaluate the analytical reproducibility of the on-line 2D LC/MS/MS system and enhance protein coverage (23).
Protein Identification and FDR Analyses
According to the criteria for protein identification mentioned under “Experimental Procedures,” more than 1000 proteins were identified in each of the five individual analyses (Table I). Unambiguous protein identification was critical for the following data processing and final conclusion; therefore, in the beginning of data analysis, we evaluated the certainty of the protein identification criteria using decoy database searches. As showed in supplemental Table 2, the stringent criteria achieved a very high confidence in protein identification as FDRs of the five individual analyses were from 0.08 to 0.40%, and the average FDR was 0.21%. Additionally, the relatively high number of identified peptides with 99% confidence in each of the five corresponding decoy analyses underlined the necessity of the stringent criteria for true protein identification (24). Detailed information on peptides and proteins identified by searching against normal and decoy databases for the five individual LC/MS/MS runs are provided in supplemental Tables 3–5.
Table I. Spectra, peptides, and proteins identified and quantified by searching against IPI mouse database of three independent iTRAQ experiments.
| Total spectra | Identified peptides (confidence = 99%) | Identified peptides (confidence = 99%, contribution = 2) | Proteins before grouping (unused >2.0) | Identified proteins (unused >2.0) | Identified proteins (unused ≥4.0) | Quantified proteins (unused ≥4.0) | Non- redundant proteinsa | Total identified proteinsb | |
|---|---|---|---|---|---|---|---|---|---|
| Sample 1 | 48,270 | 21,757 | 6,580 | 3,561 | 1,291 | 1,008 | 1,005 | 1,008 | 1,756 |
| Sample 2-1 | 45,851 | 26,386 | 7,045 | 4,035 | 1,425 | 1,138 | 1,136 | 1,298 | |
| Sample 2-2 | 46,550 | 27,126 | 6,684 | 3,668 | 1,262 | 1,011 | 1,011 | ||
| Sample 3-1 | 49,276 | 31,115 | 6,967 | 4,037 | 1,390 | 1,054 | 1,052 | 1,470 | |
| Sample 3-2 | 48,937 | 32,063 | 8,629 | 4,898 | 1,673 | 1,323 | 1,322 |
a The number of non-redundant proteins of samples 2 and 3 was derived from combining the spectral data from duplicated analyses.
b The number of proteins derived from merging the identified proteins under identical accession numbers and/or gene symbols in three independent biological samples.
Relative Quantification, Analytical, and Biological Reproducibility Analyses
In the present quantitative proteomics study, almost all identified proteins were quantified (1753 versus 1756) (Table I and supplemental Table 4). To estimate the analytical reproducibility of our proteomics study, linear regression analyses were performed on ln-transformed 117/114 ratios of duplicate analyses of samples 2 and 3 (supplemental Fig. 1). Pearson correlation coefficients for samples 2 and 3 were 0.948 and 0.923, respectively (p < 0.0001). Thus, the ratios of the two duplicate analyses were significantly positively correlated, indicating the good analytical reproducibility of the on-line 2D LC/MS/MS system. Thereby, spectral data from two duplicate analyses were merged and searched again to enhance the coverage of protein identification and to “average” the expression ratios of proteins identified in samples 2 and 3 (Table I and supplemental Table 6). Linear regression analyses were also performed on the ratios of three independent iTRAQ labeling experiments to evaluate the biological reproducibility and reliability. Pearson correlation coefficients between samples 1 and 2, samples 1 and 3, and samples 2 and 3 were 0.881, 0.869, and 0.927, respectively (p < 0.0001) (supplemental Fig. 2), showing the good biological reproducibility of the in vitro chondrogenesis model.
Data Normalization
For normalization of iTRAQ ratios derived from five individual analyses, the 117/114 ratios of a set of the most widely used internal controls detected in the present iTRAQ experiments, including tubulin, G6pdh, and MAPK1/3 (ERK1/2), were critically assessed. As shown in supplemental Table 7, the 117/114 ratios of these proteins were mainly around 1.0; for example, the average 117/114 ratio of 11 tubulin isoforms was 1.052, and the corresponding R.S.D. was 7.4%. The expression ratios of β-tubulin and MAPK1/3 were further verified by Western blotting (Fig. 3A). So we chose autobias in the present study, and the ratios of each analysis were automatically normalized by the ProteinPilot software.
Fig. 3.
Validation of differentially expressed proteins detected by iTRAQ labeling and mass spectrometry. A, differential expression of HNRNPM, Histone H2A.x, MARCKS, LOX, ERK1/2, and β-tubulin was verified by Western blotting. Total cell lysates were extracted from three independent experiments. B, histogram depicting the similar changing tendency between the protein expression ratios measured by iTRAQ and Western blotting (WB). Bars showed the S.D.
Non-redundant Protein Grouping and Homology Search
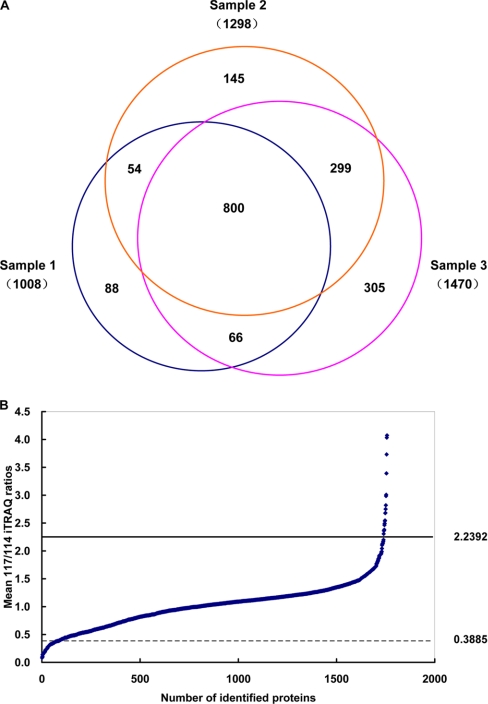
Subsequently, proteins identified from three independent experiments (supplemental Table 6) were grouped, and proteins under identical accession number and/or gene symbol were merged. In addition, by using Wu-BLAST2 and UniProt Knowledgebase, a homology search was performed for those proteins that had no name or were listed as hypothetical or putative uncharacterized. The total number of non-redundant proteins identified in the present study was 1756 (including both analytical and biological replicates, five individual LC/MS/MS runs) (supplemental Table 8). Eight hundred (45.5%) of these 1756 proteins were shared by all three experiments, and 419 (23.8%) were shared by two experiments; thus, more than two-thirds (1219 of 1756) of identified proteins were detected in at least two of the three experiments (Fig. 2A), showing a good reproducibility of the adopted proteomics platform in protein identification.
Fig. 2.
A, Venn diagram depicting the overlap of proteins identified in three independent iTRAQ experiments. The number in parentheses indicates the number of identified proteins for each sample (the numbers of samples 2 and 3 were proteins identified from duplicate analysis). B, the overall distribution of mean protein expression ratios of 1754 proteins identified during chondrogenic differentiation of C3H10T1/2 cells as measured by three independent iTRAQ experiments. Ratios were calculated as chondrogenic differentiation (117) versus undifferentiated control (114). The 95% confidence intervals are indicated by horizontal lines in the plot.
Identification of Differentially Expressed Proteins
Because the analytical and biological reproducibility of the present study was good, as stated above, the mean 117/114 ratios of three independent experiments were compared to identify altered expression proteins during chondrogenic differentiation of C3H10T1/2 cells. The mean, S.D., and R.S.D. of 117/114 ratios of proteins identified in three independent iTRAQ analyses were calculated (supplemental Table 8): the mean R.S.D. of the average ratios was 10.2%. In the present study, the threshold values for down- and up-regulated proteins were ≤0.3885 or ≥2.2392 (95% confidence interval) (Fig. 2B). Compared with undifferentiated cells, the mean 117/114 ratios of 17 proteins were up-regulated in the chondrogenically differentiated C3H10T1/2 cells, among which 13 proteins were identified in at least two of the three biological experiment (Table II), whereas the other four proteins only appeared in one of the three biological experiments (supplemental Table 9). Eighty-three proteins were found to be significantly down-regulated, among which 41 proteins were identified in at least two of the three biological experiments (Table III), and the other 42 proteins were identified in one of the three biological experiments (supplemental Table 10). The 117/114 ratio of most proteins listed in Tables II and III was in compliance with the thresholds, and the small number of outliers also manifested a change in the same direction that was close to the significance thresholds; these results further demonstrated the good analytical and biological reproducibility of the present study. Considering all 100 proteins listed in Tables II and III and supplemental Tables 9 and 10, the number of differentially expressed proteins merely accounted for 5.8% of the 1756 identified proteins. One potential drawback in using such highly stringent sets of criteria in protein identification and determination of differentially expressed proteins was the possible underestimation of proteins that existed and/or were differentially expressed during chondrogenic differentiation of C3H10T1/2 cells.
Table II. List of up-regulated proteins during chondrogenic differentiation of C3H10T 1/2 cells (chondrogenically differentiated C3H10T1/2 cells (117) versus undifferentiated control (114)).
| Accession no. | Gene symbol | Name | 117:114 sample 1 | 117:114 sample 2 | 117:114 sample 3 | Mean | S.D. | R.S.D. |
|---|---|---|---|---|---|---|---|---|
| IPI00762360.1 | Abcf1 | ATP-binding cassette, subfamily F (GCN20), member1 | 2.7533 | 3.2620 | 3.0077 | 0.3597 | 0.1196 | |
| IPI00396671.1 | ATP-binding cassette subfamily F member 1 | |||||||
| IPI00468203.3 | Anxa2 | Annexin A2 | 2.2679 | 2.7803 | 2.9859 | 2.6780 | 0.3698 | 0.1381 |
| IPI00317309.5 | Anxa5 | Annexin A5 | 2.1985 | 2.4189 | 2.5295 | 2.3823 | 0.1685 | 0.0707 |
| IPI00554894.2 | Anxa6 | Annexin A6 | 2.1822 | 2.9302 | 3.1376 | 2.7500 | 0.5025 | 0.1827 |
| IPI00310240.4 | Annexin A6 isoform b | 2.7181 | 2.9200 | 2.8191 | 0.1428 | 0.0506 | ||
| IPI00132539.3 | Btf3l4 | Transcription factor BTF3 homolog 4 | 1.9594 | 2.5395 | 2.9796 | 2.4928 | 0.5117 | 0.2053 |
| IPI00474367.2 | Col11a1 | A1(XI) collagen | 3.3549 | 1.9656 | 2.7260 | 2.6822 | 0.6957 | 0.2594 |
| IPI00468074.2 | Putative uncharacterized protein | |||||||
| IPI00828467.1 | Col2a1 | Isoform 4 of collagen α-1(II) chain | 3.1214 | 2.3228 | 2.9483 | 2.7975 | 0.4201 | 0.1685 |
| IPI00828653.1 | Isoform 1 of collagen α-1(II) chain | |||||||
| IPI00323035.3 | Fbln5 | Fibulin-5 | 2.2018 | 2.1689 | 3.2699 | 2.5469 | 0.6264 | 0.2459 |
| IPI00331707.1 | Hmgcs1; LOC100040592 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 2.8940 | 2.0397 | 2.1548 | 2.3628 | 0.4636 | 0.1962 |
| IPI00310056.3 | Lox | Protein-lysine 6-oxidase | 3.0691 | 4.1103 | 4.9188 | 4.0327 | 0.9273 | 0.2299 |
| IPI00876410.1 | Papss2 | 3′-Phosphoadenosine 5′-phosphosulfate synthase 2 | 3.9787 | 3.4589 | 3.7607 | 3.7328 | 0.2610 | 0.0699 |
| IPI00555059.2 | Prdx6 | Peroxiredoxin-6 | 2.3395 | 2.7116 | 2.5581 | 2.5364 | 0.1870 | 0.0737 |
| IPI00222557.5 | S100B | Protein S100-B | 2.9322 | 3.0373 | 2.9848 | 0.0743 | 0.0249 |
Table III. List of down-regulated proteins during chondrogenic differentiation of C3H10T 1/2 cells (chondrogenically differentiated C3H10T1/2 cells (117) versus undifferentiated control (114)) PEX, Phosphate regulating neutral endopeptidase; EH, Eps 15 homology.
| Accession no. | Gene symbol | Name | 117:114 sample 1 | 117:114 sample 2 | 117:114 sample 3 | Mean | S.D. | R.S.D. |
|---|---|---|---|---|---|---|---|---|
| Energy metabolism | ||||||||
| IPI00468481.2 | Atp5b | ATP synthase subunit β, mitochondrial | 0.3878 | 0.3460 | 0.3401 | 0.3580 | 0.0260 | 0.0726 |
| IPI00128642.1 | Cbr2 | Carbonyl reductase (NADPH)2 | 0.3419 | 0.4184 | 0.3800 | 0.3801 | 0.0383 | 0.1006 |
| IPI00387379.1 | Decr1 | 2,4-Dienoyl-CoA reductase, mitochondrial | 0.3145 | 0.3053 | 0.3888 | 0.3362 | 0.0458 | 0.1362 |
| IPI00153660.4 | Dlat | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | 0.2524 | 0.3453 | 0.4393 | 0.3457 | 0.0935 | 0.2703 |
| IPI00223092.5 | Hadha | Trifunctional enzyme subunit α, mitochondrial; similar to hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A | 0.3405 | 0.3144 | 0.3277 | 0.3275 | 0.0131 | 0.0398 |
| IPI00474281.1; IPI00115607.3; Q99JY0 | Gm13910; Hadhb | Thiolase/enoyl-coenzyme A hydratase (trifunctional protein), β subunit; trifunctional enzyme subunit β, mitochondrial | 0.1833 | 0.2828 | 0.2917 | 0.2873 | 0.0063 | 0.0219 |
| IPI00876084.1 | Gpd2 | Glycerol-3-phosphate dehydrogenase, mitochondrial | 0.1997 | 0.2827 | 0.2416 | 0.2413 | 0.0415 | 0.172 |
| IPI00337893.2 | Pdha1 | Pyruvate dehydrogenase E1 component subunit α, somatic form, mitochondrial | 0.3482 | 0.3604 | 0.4302 | 0.3796 | 0.0442 | 0.1166 |
| IPI00132042.1 | Pdhb | Pyruvate dehydrogenase E1 component subunit β, mitochondrial | 0.3318 | 0.4388 | 0.3832 | 0.3846 | 0.0535 | 0.1391 |
| IPI00230351.1 | Sdha | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial | 0.3531 | 0.3691 | 0.3714 | 0.3645 | 0.0100 | 0.0273 |
| IPI00338536.1 | Sdhb | Succinate dehydrogenase (ubiquinone) iron-sulfur subunit, mitochondrial | 0.2565 | 0.2755 | 0.4815 | 0.3378 | 0.1248 | 0.3694 |
| Chromatin organization | ||||||||
| IPI00119959.1 | Banf1 | Barrier-to-autointegration factor | 0.3509 | 0.4390 | 0.3309 | 0.3736 | 0.0575 | 0.1539 |
| IPI00230264.5 | H2afx | Histone H2A.x | 0.1802 | 0.1214 | 0.1192 | 0.1403 | 0.0346 | 0.2467 |
| mRNA processing | ||||||||
| IPI00853914.1; B9EJ02 | LOC100045191; Hnrnpa2b1 | Similar to heterogeneous nuclear ribonucleoprotein A2/B1; Hnrnpa2b1 protein similar to heterogeneous nuclear ribonucleoprotein A3, isoform 3 similar to heterogeneous nuclear ribonucleoprotein A3, isoform 4 | 0.3587 | 0.3940 | 0.3923 | 0.3817 | 0.0199 | 0.0522 |
| IPI00750059.2; IPI00749630.1; A2AL12 | Gm9242; Hnrnpa3 | Heterogeneous nuclear ribonucleoprotein A3 | 0.3810 | 0.2901 | 0.3356 | 0.0643 | 0.1916 | |
| IPI00653643.3; Q8R081 | Hnrnpl | Putative uncharacterized protein; heterogeneous nuclear ribonucleoprotein L | 0.3995 | 0.2983 | 0.2882 | 0.3287 | 0.0616 | 0.1873 |
| IPI00919164.1; IPI00132443.3 | Hnrnpm | Heterogeneous nuclear ribonucleoprotein M; isoform 1 of heterogeneous nuclear ribonucleoprotein M | 0.3811 | 0.3242 | 0.4374 | 0.3809 | 0.0566 | 0.1486 |
| Transcriptional regulation | ||||||||
| IPI00339468.4 | Dhx9 | Isoform 2 of ATP-dependent RNA helicase A | 0.3223 | 0.3361 | 0.2444 | 0.3009 | 0.0494 | 0.1643 |
| IPI00453826.2 | Matr3 | Matrin-3 | 0.4054 | 0.2836 | 0.3339 | 0.3410 | 0.0612 | 0.1795 |
| IPI00466738.4; IPI00828976.1 | Tmpo | Isoform ϵ of lamina-associated polypeptide 2, isoforms β/δ/ϵ/γ; Tmpo thymopoietin isoform ϵ | 0.3702 | 0.2971 | 0.3337 | 0.0517 | 0.1549 | |
| IPI00312128.3 | Trim28 | Isoform 1 of transcription intermediary factor 1-β | 0.4460 | 0.3736 | 0.3278 | 0.3825 | 0.0596 | 0.1558 |
| Signal transduction | ||||||||
| IPI00123709.1; IPI00867770.1 | Akap12 | Isoform 1 of protein kinase A anchor protein 12; isoform 3 of protein kinase A anchor protein 12 | 0.3640 | 0.2585 | 0.2773 | 0.2999 | 0.0563 | 0.1876 |
| IPI00229534.5 | Marcks | Myristoylated alanine-rich protein kinase C substrate | 0.4394 | 0.2007 | 0.2339 | 0.2913 | 0.1293 | 0.4438 |
| Cytoskeleton | ||||||||
| IPI00223047.2 | Ckap4 | Cytoskeleton-associated protein 4 | 0.4214 | 0.3918 | 0.2995 | 0.3709 | 0.0636 | 0.1714 |
| IPI00553419.3; IPI00849835.1 | Dsp | Desmoplakin Dsp similar to desmoplakin | 0.2613 | 0.1939 | 0.2276 | 0.0477 | 0.2094 | |
| IPI00229475.1 | Jup | Junction plakoglobin | 0.2160 | 0.1397 | 0.1779 | 0.0540 | 0.3034 | |
| IPI00620256.3 | Lmna | Isoform A of lamin-A/C | 0.3967 | 0.2851 | 0.3022 | 0.3280 | 0.0601 | 0.1833 |
| Transport | ||||||||
| IPI00468924.5 | Slc25a24 | Calcium-binding mitochondrial carrier protein SCaMC-1 | 0.2265 | 0.2487 | 0.2376 | 0.0157 | 0.0661 | |
| IPI00115564.5 | Slc25a4 | ADP/ATP translocase 1 | 0.2294 | 0.2096 | 0.2195 | 0.0140 | 0.0638 | |
| IPI00402968.1 | Ehd2 | EH domain-containing protein 2 | 0.3709 | 0.3564 | 0.4246 | 0.3840 | 0.0359 | 0.0936 |
| Proteolysis | ||||||||
| IPI00111796.1 | Phex | Metalloendopeptidase homolog PEX | 0.2467 | 0.2207 | 0.2337 | 0.0184 | 0.0787 | |
| IPI00461861.5 | Mme | Neprilysin | 0.2038 | 0.3377 | 0.3487 | 0.2967 | 0.0807 | 0.2719 |
| IPI00460063.2 | Pcyox1 | Prenylcysteine oxidase | 0.3808 | 0.3620 | 0.3714 | 0.0133 | 0.0358 | |
| IPI00319509.5 | Anpep | Aminopeptidase N | 0.3419 | 0.2380 | 0.3767 | 0.3189 | 0.0722 | 0.2263 |
| IPI00221769.5 | Ak3 | GTP:AMP phosphotransferase, mitochondrial | 0.3492 | 0.4209 | 0.3773 | 0.3825 | 0.0361 | 0.0945 |
| Miscellaneous | ||||||||
| IPI00111218.1 | Aldh2 | Aldehyde dehydrogenase, mitochondrial | 0.3373 | 0.4029 | 0.4125 | 0.3842 | 0.0409 | 0.1065 |
| IPI00138190.1 | Cdh11 | Cadherin-11 | 0.3785 | 0.3966 | 0.3876 | 0.0128 | 0.033 | |
| IPI00115530.1 | Hexb | β-Hexosaminidase subunit β | 0.3898 | 0.3593 | 0.3746 | 0.0216 | 0.0576 | |
| IPI00608064.1 | Fech | Fech protein (fragment) | 0.2243 | 0.3986 | 0.2754 | 0.2994 | 0.0896 | 0.2992 |
| IPI00938517.1 | Sqrdl | Sulfide quinone reductase-like | 0.2079 | 0.4976 | 0.3528 | 0.2048 | 0.5807 | |
| Uncharacterized | ||||||||
| IPI00222228.5; Q148Q7 | 4732456N10Rik | Putative uncharacterized protein | 0.1442 | 0.1512 | 0.1477 | 0.0049 | 0.0335 |
Validation of Differentially Expressed Proteins by Western Blotting
The expression of three down-regulated proteins (HNRNPM, Histone H2A.x, and MARCKS), one up-regulated protein (LOX), and two constitutive proteins (ERK1/2 and β-tubulin) identified in the present proteomics study was validated by Western blotting in three independent experiments started from micromass culture and chondrogenesis induction. As shown in Fig. 3A, compared with the untreated control, ERK1/2 and β-tubulin exhibited no obvious difference; however, HNRNPM, Histone H2A.x, and MARCKS displayed a down-regulation, and LOX exhibited a significant up-regulation. To investigate the relationship between the expression ratios detected by Western blotting and iTRAQ experiments, the bands of Western blotting results were quantified by densitometric analysis. As shown in Fig. 3B, although the extent of change was different, the trend of the change in the ratios obtained from two distinct methods was the same. In short, Western blotting data confirmed the changing pattern observed in quantitative proteomics study.
Functional Category of Altered Proteins
To gain insight into the biological significance of the altered proteins during chondrogenic differentiation, these differentially expressed proteins were categorized according to their main biological functions collected from the UniProt protein knowledge database and PubMed. Up-regulated proteins comprised hallmarks of mature chondrocytes (collagen types II and XI), enzymes participating in chondrocyte ECM synthesis (3′-phosphoadenosine 5′-phosphosulfate synthase 2 (PAPSS2) and LOX), and a group of novel proteins that had not been associated with chondrogenesis, such as BTF3l4 and fibulin-5 (Table II and supplemental Table 9). Compared with the relatively simple biological functions of the up-regulated proteins, those of the 41 down-regulated proteins (Table III) were relatively complicated. According to their main biological functions, these proteins fell into 10 categories: energy metabolism (eleven), chromatin assembly (two), mRNA processing (four), transcriptional regulation (four), signaling transduction (two), cytoskeleton (four), transport (three), proteolysis (five), miscellaneous (five) and uncharacterized (one) (Table III). The other 42 down-regulated proteins in supplemental Table 9 were also similarly categorized (supplemental Table 10).
Biological Verifications of Up- and Down-regulated Proteins
Finally, we sought to functionally verify some intriguing proteins identified in the present quantitative proteomics study. Energy metabolism was the largest category of the down-regulated proteins (Table III); suppressed expression of these proteins implicated that aerobic energy metabolism was inhibited. This proposal was validated by lactate assay: the level of lactate in the medium of BMP-2-treated micromass cultures of C3H10T1/2 cells increased about 2.0-fold more than that in the untreated controls (11.24 ± 1.14 versus 5.30 ± 0.87 mm).
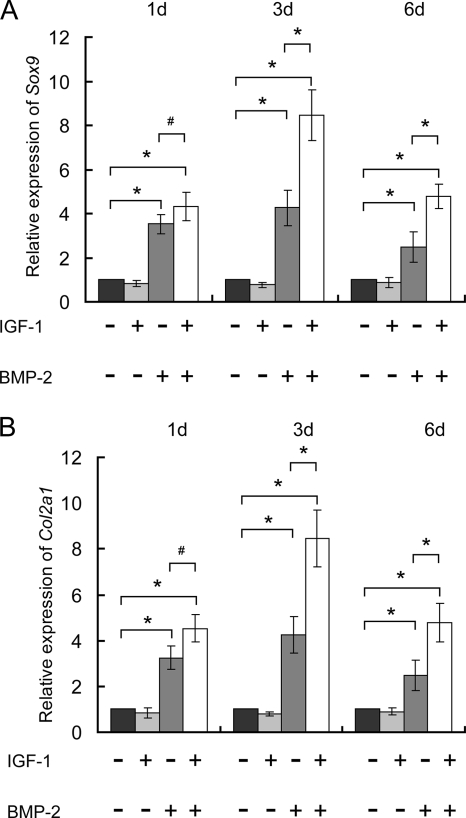
Insulin-like growth factor-1 (IGF-1), a key regulator involved in cartilage and bone development (25), was found to be remarkably up-regulated (117/114 ratio = 3.3916) (supplemental Table 8) in this in vitro chondrogenesis model for the first time. To investigate the roles of IGF-1 in chondrogenesis of C3H10T1/2 cells, 10 ng/ml IGF-1 (R&D Systems) and 100 ng/ml BMP-2 were added separately or simultaneously in the beginning of chondrogenic induction. Real time PCR experiments showed that, unlike the strong stimulation effects of 100 ng/ml BMP-2, 10 ng/ml IGF-1 alone had no obvious effects on transcription of Col2a1 and Sox9 at all three examined time points. However, the combination of 10 ng/ml IGF-1 and 100 ng/ml BMP-2 was more potent than 100 ng/ml BMP-2 alone: they could further enhance the expression of these two genes at days 3 and 6 (Fig. 4).
Fig. 4.
Synergistic effects of IGF-1 on chondrogenic differentiation of C3H10T1/2 cells induced by BMP-2. In the beginning of chondrogenic induction, 10 ng/ml IGF-1 and 100 ng/ml BMP-2 were simultaneously or separately added to the micromass culture of C3H10T1/2 cells, and transcription of Sox9 and Col2a1 was determined by real time PCR at days (d) 1, 3, and 6. Unlike 100 ng/ml BMP-2, 10 ng/ml exogenous IGF-1 had no obvious effects on Sox9 (A) and Col2a1 (B) expression at days 1, 3, and 6, but it enhanced the stimulatory effects of BMP-2 on Sox9 (A) and Col2a1 (B) at days 3 and 6. *, p < 0.05; #, p > 0.05. Bars showed the S.D.
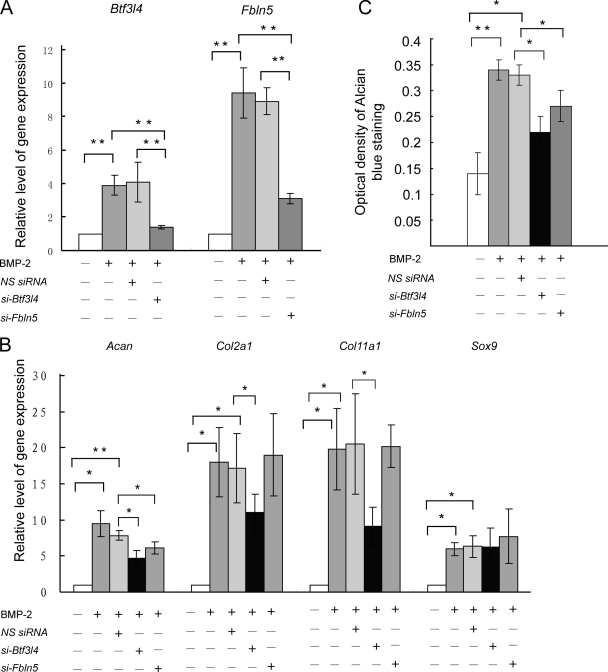
In the present study, BTF3l4 and fibulin-5 were first reported to be associated with chondrogenic differentiation. Transient transfection of siRNAs against Btf3l4 and Fbln5 successfully restricted the up-regulation of endogenous mRNA levels induced by BMP-2 (Fig. 5A). Consequently, BMP-2 induction of chondrogenesis, as evidenced by quantifying Alcian blue-positive material at an OD of 650 nm, was also inhibited by siRNAs (Fig. 5C). The expression of chondrocyte-specific genes was further examined. As shown in Fig. 5B, BMP-2-induced up-regulation of Col2a1, aggrecan, and Col11a1, three differentiation markers of mature chondrocytes, was suppressed 40, 37, and 66%, respectively, in response to Btf3l4 siRNA transfection; however, with Fbln5 siRNA transfection, only the up-regulation of aggrecan was significantly inhibited. Notably, neither Btf3l4 nor Fbln5 siRNA affected the expression level of Sox9, a master transcription factor in chondrogenesis.
Fig. 5.
Up-regulation of Btf3l4 and Fbln5 genes was indispensable for chondrogenesis of C3H10T1/2 cells induced by BMP-2. Cells were transfected with 20 nm Btf3l4 siRNA or Fbln5 siRNA, respectively. Six hours after transfection, cells were subcultured in micromass cultures and treated with or without BMP-2 (100 ng/ml) for 6 days. A and B, measurement of gene expression by real time PCR. *, p < 0.05; **, p < 0.01. C, cells were extracted with 4 m guanidine HCl, and OD of the extracted dye was measured at 650 nm for GAG analysis. *, p < 0.05; **, p < 0.01. NS siRNA, nonspecific siRNA; si-Btf3l4, siRNAs against Btf3l4; si-Fbln5, siRNAs against Fbln5. Bars showed the S.D.
DISCUSSION
Multipotent MSCs are an attractive cell source for cell-based cartilage repair and also an ideal in vitro model for the study of skeletal development (26). Since Pittenger et al. (1) reported the multilineage potential of human MSCs in 1999, knowledge about isolation, purification, and expansion of MSCs derived from different tissues and species has expanded greatly, and a panel of growth factors, adhesion molecules, and components of ECM playing important roles in chondrocyte differentiation and self-renewal of MSCs have been identified and investigated (3). However, the underlying intracellular molecular mechanisms remain an important subject of current investigation. To unravel this complex biological process, a large scale profiling should be the preferred choice. Previous large scale studies on chondrogenesis were mainly restricted to the transcriptome level (12, 27). Because of the poor concordance between mRNA and protein expression, transcriptome changes may account for only 50% of proteome changes (28). A comprehensive proteomics analysis is a necessity for a better understanding of the chondrogenic differentiation as well as self-renewal of MSCs.
To avoid the complicated problems caused by heterogeneity of primary cultured MSCs (29) in our preliminary proteomics study, we selected C3H/10T1/2 mouse embryo cell lines, which have been demonstrated to possess multiple differentiation potency (9), as a model for MSCs. According to previous studies (1), chondrogenic differentiation of C3H10T1/2 cells was validated by the expression of GAG and collagen type II (Fig. 1, A and B), two markers for mature chondrocytes. Flow cytometry analyses further revealed that more than 85% of cells were collagen type II-positive at the time when the cells were collected for comparative proteomics analysis (Fig. 1C). The appearance of collagen type II-negative cells should be ascribed to the culture condition and cannot be completely avoided in this model. Considering that the number of these negative cells is small and hence their influences on the final results are limited, their potential effects have not been taken into account in the present study.
Another factor that could influence proteomics result is cell cycle. Previous studies have revealed the proteome dynamics during the cell cycle of mammalian cells (30). To assess the effects of BMP-2 treatment on cell cycle distribution of C3H10T1/2 cells in micromass culture, propidium iodide staining and fluorescence-activated cell sorting analysis were performed. As shown in supplemental Fig. 3, although the cell proliferation is significantly suppressed under high cell density conditions, there is no significant difference in cell cycle stage between BMP-2-treated and untreated C3H10T1/2 cells.
Comparative Proteomics Analysis of Chondrogenic Differentiation of MSCs
Our study presents the first and most comprehensive proteomic profile of C3H10T1/2 cells, containing 1756 proteins with extremely high confidence. However, a number of proteins previously identified in MSCs by two-dimensional gel electrophoresis (16) and antibody array (13) are not included, highlighting the complementarity of different proteomics strategies. Both MSCs and embryonic stem cells possess the self-renewal and differentiation potential, but their potentials are quite different. A global comparison of the cellular proteome between the mesenchymal stem cell line and ES cell line would provide some valuable clues for understanding these differences. Excluding 316 protein entries that only appeared in the mouse IPI 3.62 database (adopted in the present study), at least 80% of the proteins identified in the C3H10T1/2 cells are also detected in the ES cells (containing 5111 proteins, by searching against the mouse IPI 3.24 database) (31), showing the higher similarity between these two cell lines. However, 288 proteins (20%) are only detected in C3H10T1/2 cells, including markers for MSCs, VCAM-1 and CD109 (3, 32); a series of collagens, COL1A2, COL2A1, COL3A1, COL5A1, COL5A2, COL6A1, COL6A2, COL11A1, and COL15A1, which have been demonstrated to be expressed by MSCs (17); special isoforms of housekeeping proteins, such as ACTA1 (actin, α skeletal muscle) and ACTA2 (actin, aortic smooth muscle), and some others (supplemental Table 11), demonstrating the unique features of C3H10T1/2 cells. In addition, these proteins also reflect the more mature state of C3H10T1/2 cells, which are derived from embryo cells of C3H mice (8), and may account for the different potential in self-renewal and differentiation of C3H10T1/2 cells.
More importantly, a list of 100 significantly altered proteins was identified. According to their expression pattern, these proteins can be simply categorized into two groups: up- and down-regulated (Tables II and III and supplemental Tables 8 and 9). In turn, an obvious feature of the changing pattern appears: in comparison with the number of up-regulated proteins, many more proteins are down-regulated in this process (83 versus 17). This unbalanced pattern has also been revealed by a previous transcriptomics comparison of ES cells with progenitor and mature cells (33) and could be proteomics evidence for the “stem state” concept (34, 35). Stem state cells express promiscuous genes and possess a number of differentiation options; whereas the expression of a major part of them is reduced during the differentiation state, and “options” are greatly reduced, at the same time, expression of a small collection of lineage-specific genes is increased to higher levels. Accordingly, these increased proteins are most likely involved in chondrogenic commitment, whereas those that are decreased should be potential candidates for self-renewal of MSCs or commitment of MSCs to other lineages. Functional categories partially support this proposal: a number of up-regulated proteins are cartilage-specific ECM components and enzymes involved in synthesis of such components (Table II), whereas some of the down-regulated proteins are known to be related to self-renewal of MSCs, such as epidermal growth factor receptor and platelet-derived growth factor receptor (7) (supplemental Table 10). However, the majority of these altered proteins have not been reported in chondrogenesis and/or self-renewal of MSCs. Functional categorization shows that they are involved in a variety of biological functions, such as chromatin organization, transcription, mRNA processing, signaling transduction, cytoskeleton, ECM and ECM synthesis, and so on (Tables II and III and supplemental Tables 9 and 10), exemplifying the multiple layers of coordinated molecular control in chondrogenic differentiation of MSCs.
Proteomics View of the Characterizations of Chondrogenesis
The predominant feature of chondrogenic differentiation of mesenchymal is the deposition of intense cartilage ECM (36). Therefore, it is expected that collagen types II and XI (Table II), main components and hallmarks of mature chondrocytes (4), are up-regulated. In addition, we also identified two major enzymes involved in posttranslational modification of chondrocyte ECM components, PAPSS2 and LOX (Table II). Sulfation of proteoglycans is an essential posttranslational modification in chondrocytes. In mammals, PAPSS2 is the dominant isoform in developing cartilage and plays a major role in cartilage proteoglycan sulfation (37, 38); PAPSS2 has also been identified in a previous proteomics study based on two-dimensional gel electrophoresis (39). LOX is an enzyme responsible for cross-linking of collagen by catalyzing the formation of lysine-derived aldehyde (40). In short, concomitantly enhanced expression of LOX and PAPSS2 will accommodate the robust ECM synthesis during chondrogenesis.
Because cartilage is avascular and chondrocytes reside in a hypoxic milieu, anaerobic glycolysis is dominant in generating ATP to support chondrocyte matrix synthesis and viability in vivo (36). This unique metabolic feature is confirmed in this in vitro chondrogenesis model by our proteomic study as the most enriched category of down-regulated proteins is energy metabolism (Table III). In addition, remarkably increased lactate content in the differentiated C3H10T1/2 cells further validated this fundamental metabolic switch. Metabolism flexibility of MSCs has also been demonstrated by a recent report (41): in the absence of oxygen, MSCs can survive using anaerobic ATP production and maintain their ability to differentiate into chondrocytes.
Taken together, besides the high confidence of protein identification and quantification validated by the aforementioned stringent criteria, FDR analyses, and high correlation coefficients, identification of these well documented proteins provides another solid quality control for the accuracy of detection of the true altered proteins during chondrogenesis. Meanwhile, these data also show in a proteomics view that the in vitro system used here can recapitulate the main features of in vivo chondrogenesis and demonstrate that iTRAQ labeling coupled with on-line 2D LC/MS/MS is a reliable proteomics technique for identification of differentially expressed proteins.
Synergetic Effects of IGF-1 on BMP-2-induced Chondrogenic Differentiation of C3H10T1/2 Cells
Growth factors are one of the three primary requirements for tissue engineering (the other two are seed cells and scaffolds for delivering and retaining seed cells) (42). To optimize the therapeutic use of MSCs in cartilage regeneration, we need to understand the mechanisms by which growth factors induce and maintain the chondrogenic differentiation of MSCs. In the present study, IGF-1 was detected, and its expression was significantly up-regulated by BMP-2 in C3H10T1/2 cells (supplemental Table 8); therefore, IGF-1 could be a downstream target modulated by BMP-2. Regulated paracrine or autocrine action of IGF-1 has also been discovered in human MSCs recently (43). Because IGF-1 is a critical growth factor for cartilage and bone growth (25), it is reasonable to postulate that IGF-1 and BMP-2 could have additive effects on chondrogenesis of C3H10T1/2 cells. This proposal was confirmed by our real time PCR experiments. As expected, 10 ng/ml IGF-1 alone had no obvious effects on Col2a1 and Sox9 transcription; this intentionally adopted dosage was just one-tenth of that routinely used in chondrogenesis studies (44), but it could significantly enhance the expression of these two genes in the presence of 100 ng/ml BMP-2, and the effects were more potent than BMP-2 alone (Fig. 4). Thus, IGF-1 did have a synergistic effect on BMP-2-induced chondrogenesis. Moreover, as IGF-1 had no obvious effects on day 1 cells, its synergistic effects seemed to depend on the differentiation state of C3H10T1/2 cells.
Up-regulated Fibulin-5 and Chondrogenesis
ECM is not only the molecular basis for chondrocytes in executing their biological functions but also critical for maintaining and/or regulating their phenotype. Fibulins, a newly recognized family of ECM proteins (45), are distributed in a restricted manner during embryonic development. All five members of this family are detected in perichondrium of the developing bone in E15 mouse embryo (46). In our study, three members (fibulin-1, -2, and -5) of this family were identified (supplemental Table 8), among which only fibulin-5 was up-regulated, suggesting different functions of the members of this family in chondrogenic differentiation. It has been reported that mutations (47, 48) or altered expression (49) of fibulin-5 impairs elastic fiber assembly (50, 51); therefore, elevated fibulin-5 could be a benefit for ECM synthesis and organization and in turn promote maturation of chondrocytes. In our work, a loss-of-function RNA interference analysis validated that fibulin-5 was necessary for BMP-2-elicited chondrogenesis as demonstrated by the inhibition of Alcian blue staining and aggrecan expression (Fig. 5).
Transcriptional Factor BTF3l4 Is Indispensable for Chondrogenesis
Previous experimental studies have clarified that transcription factor SOX9 plays pivotal roles in the onset of chondrogenesis (52). However, the commitment of MSCs to chondrocytes is such a complex biological process that SOX9 alone cannot fulfill this process (4, 22), which should depend on the integration of multiple transcription factors rather than merely SOX9. In the present study, an up-regulated general transcription factor, BTF3l4, was identified (Table II). To our knowledge, this is the first report regarding the biological function of BTF3l4. BTF3l4 is a homolog of the basic transcription factor BTF3, which is involved in the initiation of transcription by RNA polymerase from proximal promoter elements such as the TATA box and CAAT box sequences (53, 54). The insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice (55), pointing to an important role of this gene during normal embryonic development. Considering that the TATA box and CAAT box are generally contained in gene promoters and BTF3l4 was induced by BMP-2 treatment as demonstrated by our proteomics analysis, BTF3l4 could contribute to chondrogenic differentiation to some extent. The function of BTF3l4 in chondrogenesis was preliminarily validated by an siRNA knockdown experiment: suppression of endogenous Btf3l4 expression by siRNA transfection resulted in inhibition of expression of two chondrogenic marker genes, Col2a1 and Col11a1, and synthesis of ECM (Fig. 5).
Conclusions
In summary, the present study provided the first and most comprehensive proteomic profile of the chondrogenically differentiated and undifferentiated mesenchymal stem cell line C3H10T1/2 whereby 100 differentially expressed proteins were identified. Functional categorization of these altered proteins partially confirmed the stem state concept and exemplified the multiple layers of coordinated protein expression regulation during chondrogenic differentiation of MSCs. However, except for a number of well documented proteins, the majority of these differentially expressed proteins have not been reported and could be potential candidates involved in chondrogenesis and self-renewal of MSCs. In the present study, the biological functions of a few of them were preliminarily characterized in the context of chondrogenesis, but the definite biological roles of these altered proteins deserve in-depth investigation as it will provide valuable clues for unraveling the molecular mechanisms underlying chondrogenic differentiation as well as self-renewal of MSCs. Finally, the results of the present study also verified that iTRAQ labeling coupled with on-line 2D LC/MS/MS is a robust, reliable, and automated proteomics technology that is suitable for high throughput quantitative proteomics studies.
Supplementary Material
Acknowledgments
We thank Qing-ping Liu, a member of the Institutes of Biomedical Sciences, Fudan University, for technical assistance in establishing the on-line 2D LC/MS/MS proteomics platform and Yang Wan and Su-mei Shi, members of the Institute of Biology Engineering, Jinan University, for assistance in cell culture. We are highly grateful to Dr. Debra M. Ferraro, a member of the Department of Microbiology, University of Iowa, for critical review of the manuscript.
* This work was supported by National Natural Science Foundation of China Grants 30600587 and 30900563 and Grant 2010CB833603 from the Major State Basic Research Development Program of China (973 Program).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–11 and Figs. 1–3.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–11 and Figs. 1–3.
1 The abbreviations used are:
- MSC
- mesenchymal stem cell
- 2D
- two-dimensional
- BMP-2
- bone morphogenetic protein 2
- ECM
- extracellular matrix
- FA
- formic acid
- FDR
- false discovery rate
- iTRAQ
- isobaric tag for relative and absolute quantitation
- SCX
- strong cation exchange
- ES
- embryonic stem
- LOX
- protein-lysine 6-oxidase
- MARCKS
- myristoylated alanine-rich protein kinase C substrate
- HNRNP
- heterogeneous nuclear ribonucleoprotein
- nano-LC
- nanoscale LC
- IPI
- International Protein Index
- siRNA
- short interfering RNA
- GAG
- glycosaminoglycan
- IGF-1
- Insulin-like growth factor-1
- PAPSS2
- 3′-phosphoadenosine 5′-phosphosulfate synthase 2
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- R.S.D.
- relative S.D.
REFERENCES
- 1.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 2.DeLise A. M., Fischer L., Tuan R. S. (2000) Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage 8, 309–334 [DOI] [PubMed] [Google Scholar]
- 3.Kolf C. M., Cho E., Tuan R. S. (2007) Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res. Ther 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldring M. B., Tsuchimochi K., Ijiri K. (2006) The control of chondrogenesis. J. Cell. Biochem 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 5.Steinert A. F., Ghivizzani S. C., Rethwilm A., Tuan R. S., Evans C. H., Nöth U. (2007) Major biological obstacles for persistent cell-based regeneration of articular cartilage. Arthritis Res. Ther 9, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baksh D., Song L., Tuan R. S. (2004) Adult mesenchymal stem cells: characterization, differentiation, and application in cell and gene therapy. J. Cell. Mol. Med 8, 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satomura K., Derubeis A. R., Fedarko N. S., Ibaraki-O'Connor K., Kuznetsov S. A., Rowe D. W., Young M. F., Gehron Robey P. (1998) Receptor tyrosine kinase expression in human bone marrow stromal cells. J. Cell. Physiol 177, 426–438 [DOI] [PubMed] [Google Scholar]
- 8.Reznikoff C. A., Brankow D. W., Heidelberger C. (1973) Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res 33, 3231–3238 [PubMed] [Google Scholar]
- 9.Shea C. M., Edgar C. M., Einhorn T. A., Gerstenfeld L. C. (2003) BMP treatment of C3H10T1/2 mesenchymal stem cells induces both chondrogenesis and osteogenesis. J. Cell. Biochem 90, 1112–1127 [DOI] [PubMed] [Google Scholar]
- 10.Denker A. E., Haas A. R., Nicoll S. B., Tuan R. S. (1999) Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation 64, 67–76 [DOI] [PubMed] [Google Scholar]
- 11.Zhao L., Li G., Chan K. M., Wang Y., Tang P. F. (2009) Comparison of multipotent differentiation potentials of murine primary bone marrow stromal cells and mesenchymal stem cell line C3H10T1/2. Calcif. Tissue Int 84, 56–64 [DOI] [PubMed] [Google Scholar]
- 12.Izzo M. W., Pucci B., Tuan R. S., Hall D. J. (2002) Gene expression profiling following BMP-2 induction of mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage 10, 23–33 [DOI] [PubMed] [Google Scholar]
- 13.Sze S. K., de Kleijn D. P., Lai R. C., Khia Way Tan E., Zhao H., Yeo K. S., Low T. Y., Lian Q., Lee C. N., Mitchell W., El Oakley R. M., Lim S. K. (2007) Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Mol. Cell. Proteomics 6, 1680–1689 [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Park J. S., Chu J. S., Krakowski A., Luo K., Chen D. J., Li S. (2004) Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor beta1 stimulation. J. Biol. Chem 279, 43725–43734 [DOI] [PubMed] [Google Scholar]
- 15.Foster L. J., Zeemann P. A., Li C., Mann M., Jensen O. N., Kassem M. (2005) Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation. Stem Cells 23, 1367–1377 [DOI] [PubMed] [Google Scholar]
- 16.Celebi B., Elçin Y. M. (2009) Proteome analysis of rat bone marrow mesenchymal stem cell subcultures. J. Proteome Res 8, 2164–2172 [DOI] [PubMed] [Google Scholar]
- 17.Park H. W., Shin J. S., Kim C. W. (2007) Proteome of mesenchymal stem cells. Proteomics 7, 2881–2894 [DOI] [PubMed] [Google Scholar]
- 18.Wiese S., Reidegeld K. A., Meyer H. E., Warscheid B. (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 [DOI] [PubMed] [Google Scholar]
- 19.Ahrens P. B., Solursh M., Reiter R. S. (1977) Stage-related capacity for limb chondrogenesis in cell culture. Dev. Biol 60, 69–82 [DOI] [PubMed] [Google Scholar]
- 20.Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 21.Reidegeld K. A., Eisenacher M., Kohl M., Chamrad D., Körting G., Blüggel M., Meyer H. E., Stephan C. (2008) An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications. Proteomics 8, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 22.Pan Q., Yu Y., Chen Q., Li C., Wu H., Wan Y., Ma J., Sun F. (2008) Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J. Cell. Physiol 217, 228–241 [DOI] [PubMed] [Google Scholar]
- 23.Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 24.Bradshaw R. A., Burlingame A. L., Carr S., Aebersold R. (2006) Reporting protein identification data: the next generation of guidelines. Mol. Cell. Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 25.Baker J., Liu J. P., Robertson E. J., Efstratiadis A. (1993) Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75, 73–82 [PubMed] [Google Scholar]
- 26.Magne D., Vinatier C., Julien M., Weiss P., Guicheux J. (2005) Mesenchymal stem cell therapy to rebuild cartilage. Trends Mol. Med 11, 519–526 [DOI] [PubMed] [Google Scholar]
- 27.Shinomura T., Ito K., Kimura J. H., Höök M. (2006) Screening for genes preferentially expressed in the early phase of chondrogenesis. Biochem. Biophys. Res. Commun 341, 167–174 [DOI] [PubMed] [Google Scholar]
- 28.Unwin R. D., Smith D. L., Blinco D., Wilson C. L., Miller C. J., Evans C. A., Jaworska E., Baldwin S. A., Barnes K., Pierce A., Spooncer E., Whetton A. D. (2006) Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood 107, 4687–4694 [DOI] [PubMed] [Google Scholar]
- 29.Phinney D. G. (2007) Biochemical heterogeneity of mesenchymal stem cell populations: clues to their therapeutic efficacy. Cell Cycle 6, 2884–2889 [DOI] [PubMed] [Google Scholar]
- 30.Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Alaluf I., Swerdlin N., Perzov N., Danon T., Liron Y., Raveh T., Carpenter A. E., Lahav G., Alon U. (2006) Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat. Methods 3, 525–531 [DOI] [PubMed] [Google Scholar]
- 31.Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–68318045802 [Google Scholar]
- 32.Vogel W., Grünebach F., Messam C. A., Kanz L., Brugger W., Bühring H. J. (2003) Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica 88, 126–133 [PubMed] [Google Scholar]
- 33.Golan-Mashiach M., Dazard J. E., Gerecht-Nir S., Amariglio N., Fisher T., Jacob-Hirsch J., Bielorai B., Osenberg S., Barad O., Getz G., Toren A., Rechavi G., Itskovitz-Eldor J., Domany E., Givol D. (2005) Design principle of gene expression used by human stem cells: implication for pluripotency. FASEB J 19, 147–149 [DOI] [PubMed] [Google Scholar]
- 34.Zipori D. (2004) The nature of stem cells: state rather than entity. Nat. Rev. Genet 5, 873–878 [DOI] [PubMed] [Google Scholar]
- 35.Zipori D. (2006) The stem state: mesenchymal plasticity as a paradigm. Curr. Stem Cell Res. Ther 1, 95–102 [DOI] [PubMed] [Google Scholar]
- 36.Archer C. W., Francis-West P. (2003) The chondrocyte. Int. J. Biochem. Cell Biol 35, 401–404 [DOI] [PubMed] [Google Scholar]
- 37.Stelzer C., Brimmer A., Hermanns P., Zabel B., Dietz U. H. (2007) Expression profile of Papss2 (3′-phosphoadenosine 5′-phosphosulfate synthase 2) during cartilage formation and skeletal development in the mouse embryo. Dev. Dyn 236, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 38.Fuda H., Shimizu C., Lee Y. C., Akita H., Strott C. A. (2002) Characterization and expression of human bifunctional 3′-phosphoadenosine 5′-phosphosulphate synthase isoforms. Biochem. J 365, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S. J., Jeon H. B., Lee J. H., Yoo J. S., Chun J. S., Yoo Y. J. (2004) Identification of proteins differentially expressed during chondrogenesis of mesenchymal cells. FEBS Lett 563, 35–40 [DOI] [PubMed] [Google Scholar]
- 40.Trackman P. C. (2005) Diverse biological functions of extracellular collagen processing enzymes. J. Cell. Biochem 96, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mylotte L. A., Duffy A. M., Murphy M., O'Brien T., Samali A., Barry F., Szegezdi E. (2008) Metabolic flexibility permits mesenchymal stem cell survival in an ischemic environment. Stem Cells 26, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 42.Langer R., Vacanti J. P. (1993) Tissue engineering. Science 260, 920–926 [DOI] [PubMed] [Google Scholar]
- 43.Crisostomo P. R., Wang Y., Markel T. A., Wang M., Lahm T., Meldrum D. R. (2008) Human mesenchymal stem cells stimulated by TNF-alpha, LPS, or hypoxia produce growth factors by an NF kappa B- but not JNK-dependent mechanism. Am. J. Physiol. Cell Physiol 294, C675–C682 [DOI] [PubMed] [Google Scholar]
- 44.Longobardi L., O'Rear L., Aakula S., Johnstone B., Shimer K., Chytil A., Horton W. A., Moses H. L., Spagnoli A. (2006) Effect of IGF-I in the chondrogenesis of bone marrow mesenchymal stem cells in the presence or absence of TGF-beta signaling. J. Bone Miner. Res 21, 626–636 [DOI] [PubMed] [Google Scholar]
- 45.Timpl R., Sasaki T., Kostka G., Chu M. L. (2003) Fibulins: a versatile family of extracellular matrix proteins. Nat. Rev. Mol. Cell Biol 4, 479–489 [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi N., Kostka G., Garbe J. H., Keene D. R., Bächinger H. P., Hanisch F. G., Markova D., Tsuda T., Timpl R., Chu M. L., Sasaki T. (2007) A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J. Biol. Chem 282, 11805–11816 [DOI] [PubMed] [Google Scholar]
- 47.Hucthagowder V., Sausgruber N., Kim K. H., Angle B., Marmorstein L. Y., Urban Z. (2006) Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am. J. Hum. Genet 78, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stone E. M., Braun T. A., Russell S. R., Kuehn M. H., Lotery A. J., Moore P. A., Eastman C. G., Casavant T. L., Sheffield V. C. (2004) Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl. J. Med 351, 346–353 [DOI] [PubMed] [Google Scholar]
- 49.Wlazlinski A., Engers R., Hoffmann M. J., Hader C., Jung V., Müller M., Schulz W. A. (2007) Downregulation of several fibulin genes in prostate cancer. Prostate 67, 1770–1780 [DOI] [PubMed] [Google Scholar]
- 50.Zheng Q., Davis E. C., Richardson J. A., Starcher B. C., Li T., Gerard R. D., Yanagisawa H. (2007) Molecular analysis of fibulin-5 function during de novo synthesis of elastic fibers. Mol. Cell. Biol 27, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. (2002) Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 415, 168–171 [DOI] [PubMed] [Google Scholar]
- 52.Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell. Biol 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng X. M., Moncollin V., Egly J. M., Chambon P. (1987) A general transcription factor forms a stable complex with RNA polymerase B (II). Cell 50, 361–368 [DOI] [PubMed] [Google Scholar]
- 54.Kanno M., Chalut C., Egly J. M. (1992) Genomic structure of the putative BTF3 transcription factor. Gene 117, 219–228 [DOI] [PubMed] [Google Scholar]
- 55.Deng J. M., Behringer R. R. (1995) An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res 4, 264–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.