Abstract
Knowledge of the subcellular localization of proteins is indispensable to understand their physiological roles. In the past decade, 18 studies have been performed to analyze the protein content of isolated organelles from Saccharomyces cerevisiae. Here, we integrate the data sets and compare them with other large scale studies on protein localization and abundance. We evaluate the completeness and reliability of the organelle proteomics studies. Reliability depends on the purity of the organelle preparations, which unavoidably contain (small) amounts of contaminants from different locations. Quantitative proteomics methods can be used to distinguish between true organellar constituents and contaminants. Completeness is compromised when loosely or dynamically associated proteins are lost during organelle preparation and also depends on the sensitivity of the analytical methods for protein detection. There is a clear trend in the data from the 18 organelle proteomics studies showing that proteins of low abundance frequently escape detection. Proteins with unknown function or cellular abundance are also infrequently detected, indicating that these proteins may not be expressed under the conditions used. We discuss that the yeast organelle proteomics studies provide powerful lead data for further detailed studies and that methodological advances in organelle preparation and in protein detection may help to improve the completeness and reliability of the data.
The yeast Saccharomyces cerevisiae is a widely used monocellular eukaryotic model organism. S. cerevisiae was the first eukaryotic organism of which the complete genome was sequenced (1). The genome contains 6,608 open reading frames of which 5,797 encode polypeptides. 4,666 proteins have annotated functions in the yeast genome database SGD.1
S. cerevisiae has intracellular compartments typical for eukaryotic cells (Table I). The subcellular localization of proteins in yeast has been investigated using genome-wide chromosomal GFP tagging approaches (2) in addition to immuno-EM and other low throughput methods. The genome-wide studies have provided an unprecedented wealth of information. Yet, for ∼17% of the 5,797 gene products (939 proteins), many of which have unknown biological function, the subcellular localization has not been assigned. In addition, tagging approaches can result in mislocalizations. Consequently, complementary proteome-wide studies, which avoid tagging, are required. Mass spectrometry-based proteomics, combined with subcellular fractionation, provides such alternative methodology. Yeast is uniquely suitable for this approach because of its relatively low complexity, infrequent occurrence of gene isoforms, and limited extent of posttranslational modifications. In the past decade, 18 studies of the protein content of isolated organelles by means of MS-based approaches (organelle proteomics) have been performed, covering all the subcellular compartments with the exceptions of the ER, the endosome, and the cytosol (Table II and Refs. 3–10). The protein composition of supramolecular assemblies from yeast, such as the nuclear pore complex, and the translation machineries (11–14) will not be discussed here, but the experimental design and the challenges are very similar to organelle proteomics.
Table I. Properties of yeast organelles.
Physical and biological properties of organelles are given. OM, outer membrane; IM, inner membrane; IMS, intermembrane space; GL, glycerol; OA, oleic acids; +/−Carb., with and without carbohydrates; SER, smooth ER; RER, rough ER; t-SNARE, target soluble N-ethylmaleimide-sensitive factor attachment protein receptor; TGN, trans-Golgi network.
| Organelle | Lipid-to-protein ratioa | Densityb | Marker enzyme | Antibody | Specific fluorescent dye | Range of protein abundancesc |
|---|---|---|---|---|---|---|
| mg/mg | g/cm3 | copies/cell | ||||
| Mitochondria | 0.92 (OM), 0.17 (IM) | 1.14–1.21 (60) | Aconitase, fumarase (matrix), cytochrome c peroxidase Ccp1 (IMS), cytochrome c oxidase (IM) | Anti-porin (OM) anti-cytochrome c oxidase subunit III (IM) | DAPId (pink-white staining), rhodamine, DASPMIe | 101–106 |
| Peroxisomes | 0.46 | 1.21–1.23 (GL + OA); 1.19–1.21 (GL) (61); 1.16–1.17 (Nycodenz; OA); 1.19–1.2 (OA) (62) | Catalase, isocitrate lyase | 102–104 | ||
| Lipid particle | 16.80 | <1.000 | Δ24-Sterol C-methyltransferase Erg6 | Nile red | 102–105 | |
| Nucleus | 0.11 | RNA polymerase | Anti-nucleoporin (e.g. Mab414) | DAPI | 101–105 | |
| Cell envelope | ||||||
| Cell wall including periplasm | >1.3 | Invertase | Aminoacridine (surface potential), concanavalin A (binds to mannans), calcofluor white (chitin in scars) | 101–106 | ||
| +Carb.: 1.24–1.26 (63, 64) | ||||||
| Plasma membrane | 0.65 | −Carb: 1.12–1.18 (65), 1.20–1.22 (5) | Vanadate-sensitive ATPase | Anti-PMA1 | FM4-64 and derivatives | 101–106 |
| Secretory system | ||||||
| ER (light microsomal fraction) | 0.13 | SER: 1.146; RER: 1.17–1.19 (66, 67) | NADPH-cytochrome c reductase NCP1,f dolichol-phosphate mannose synthase DPM1 (68) | Anti-Dpm1 (membrane) | 102–105 | |
| Golgi (heavy microsomal fraction) | 0.08 | 1.248 (sorbitol) (69) | Cis: ER-to-Golgi t-SNARE Sed5; α-1,6-mannosyltransferase Och1; medial/trans: α-1,3-mannosyltransferase Mnn1; α-1,2-mannosyltransferase Mnt1; trans/TGN: serine protease Kex2; serine carboxypeptidase Kex1; unclassified: β-glucan synthase; sorting receptor for HDEL-tagged proteins Erd2 | Anti-Vps10 (late Golgi membrane) | 101–105 | |
| Endosome | 1.17g | 1.12–1.16 (Nycodenz) (70) 1.030–1.075 (71) | Anti-Pep12 (membrane), α-mannosidase AMS1 (membrane) | Anti-alkaline phosphatase Pho8 (membrane) | 101–104 | |
| Vacuole | 0.58 | 1.011 (Ficoll) (7) | Proteinase A PEP4, proteinase B PRB1 (lumen) | Anti-carboxypeptidase Y (lumen), anti-ATPase subunits a, A, B (membrane) | Neutral red FM4-64 (vacuolar membrane) | 101–105 |
a The lipid-to-protein ratio describes the sum of phospholipids and sterols (33, 72). In the case of the heavy microsomal fraction, the phospholipid-to-protein ratio was taken from Kuchler et al. (73).
b The apparent density of organelles depends on the separation medium. Unless stated otherwise, the reported densities are for organelles in sucrose solution.
c From Ref. 35.
d DAPI, 4′,6-diamidino-2-phenylindole.
e Dimethyl-aminostyryl-methylpyridinium iodine.
f Although the enzyme is often used as an ER marker, it is also present at the mitochondrial outer membrane.
g Lipid-to-protein ratio refers of secretory vesicles.
Table II. Yeast organellar proteomics studies.
A summary of the 18 experimental proteomes discussed in this review is given. 2-DE, two-dimensional gel electrophoresis; BAC, N,N′-bisacrylylcystamine; CTAB, cetyltrimethylammonium bromide; GC, gas chromatography; HA, hemagglutinin; nLC, nanoflow LC; LDS, lithium dodecyl sulfate; MudPIT, multidimensional protein identification technology; PFP, peptide fingerprint; RP, reverse-phase; SAX, strong anion exchanger; SCX, strong cation exchanger; WB, Western blot; FFE, free flow electrophoresis; 1D, one-dimensional; 2D, two-dimensional; μLC, microcapillary LC; PMF, peptide mass fingerprint; EC, exchange chromatography; TMDs, transmembrane domains.
| Organelle | Straina and growth conditionsb | Spheroplasting and lysis | Purification separation enrichment | Stripping | Purity assessment | Protein and/or peptide separation | MS analysis | No. of identified proteins (%TMDs) | Detected abundance range | Validation | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| copies/cell | |||||||||||
| Mitochondria | CW04 (n), YPLc | Zymolyase; osmotic lysis | Differential centrifugation | No | EM | SDS-PAGE | nLC-ESI-MS/MS | 179 (12%) | 149–882,841 | GFP or HA tag, computation | 27 |
| Mitochondria | YPH499 (n), YPGd | Zymolyase; mechanical disruption | 1) Differential centrifugation, 2) sucrose density gradient (32/60% interface) | Salt, shaving with trypsin | WB | 1) 2-DE, 2) SCX-RP-nLC,e 3) SDS-PAGEe | 1) MALDI-MS/MS or nLC-ESI, 2) ESI-MS, 3) nLC-ESI-MS/MS | 750 (18%) | 99–1,255,722 | Protein import assay, presequence | 28 |
| Mitochondria | BJ1991 (n), YPD versus YPG | Zymolyase; mechanical disruption | Sucrose density gradient | EDTA | 2-DE | PFP | 251 (6%) | 149–1,018,216 | Transcriptome | 34 | |
| Mitochondria | W303 (n), S1001, YPL, YPD, SCL, and SCDf | 1) Nycodenz density gradient, 2) FFE, 3) differential centrifugation | Denaturation in 7 m urea, 2 m thiourea, 1% CHAPS | EM | RP-LC | ESI-MS/MS, LC-FTICR | 546 (17%) | 57–1,255,722 | Co-enrichment with known marker using WB | 26 | |
| Mitochondria | YPH499 (n), YPG | Zymolyase; mechanical disruption | 1) Differential centrifugation, 2) sucrose density gradient (32/60% interface)g | EDTA | WB | 1) 2-DE, 2) SAX- or SCX-RP-LC,e 3) SDS-PAGE | 1) MALDI-MS/MS or nLC-ESI, 2) ESI-MS, 3) nLC-ESI-MS/MS | 851 (18%) | 99–1,255,722 | 30 | |
| Mitochondria | YPH499 (n), YPG | Zymolyase; mechanical disruption | Intact mitochondria as in Ref. 28, Outer membranes: sucrose density gradient (interface 15/32%) | EDTA, trypsin | WB | 1) 2D BAC/SDS, 2) SCX-RP-LC,e 3) SDS-PAGE | 1) MALDI-MS/MS, 2) ESI-MS/MS, 3) nLC-ESI-MS/MS | 117 (20%) | 147–882,841 | Computational and biochemical analysis | 29 |
| Peroxisomes | BJ1991 (n), BJ1991Δpox1, YNOh | Lyticase | Sucrose density gradient (47–68%) | Salt, pH 11.3 | 1D SDS-PAGE | μLC- and nLC-ESI-MS/MS | 45 (18%) | 358–1,255,722 | No | 22 | |
| Peroxisomes | BY4743 (2n), SCIMi | Zymolyase; mechanical disruption | 1) Differential centrifugation, 2) Nycodenz density gradient (35–50% interface) | EDTA | μLC | GC-ESI-MS/MS | 44 (11%) | 238–111,935 | 23 | ||
| Peroxisomes | BY4743 (2n), SCIMi | Zymolyase; mechanical disruption | 1) Differential centrifugation, 2) Nycodenz density gradient (35–50% interface) | pH 11.3 | WB | ICAT-μLC | ESI-MS/MS | 72 (21%) | 238- 268,717 | GFP tagging, mutant phenotype | 21 |
| Lipid particles | X2180-1A (n), FY1679 (n, 2n), YPD | Zymolyase; osmotic lysis | Ficoll density gradient | Delipidation | Enzymatic assays, EM | 1D SDS-PAGE | nLC-ESI-MS/MS | 18 (28%) | 1,212–168,876 | Lipid and sterol composition in deletion mutants | 31 |
| Nucleus | BY 4741 (n), YPD | Zymolyase; mechanical disruption | DNase I, heparin, sucrose density gradient | SCX-RP-nLC | ESI-MS/MS | 2698 (11%) | 49–1,255,722 | 9 | |||
| Nucleus | YPD (Wickerham), BY 4743 (2n) | Zymolyase, lyticase, Glucanex; mechanical disruption | DNase I, heparin, sucrose density gradient | 1) Peptide IEF, 2) SCX-RP-nLC, 3) 1D SDS-PAGE, 4) phosphocellulose chromatography + SDS-PAGE | ESI-MS/MS | 1889 (5%) | 41–1,255,722 | 10 | |||
| Golgi | CJY119 (n), HIY1 (n), YPD | Lyticase; osmotic shock | 1) Differential centrifugation, 2) immunoprecipitation | Triton X-114 phase separation | WB | 1D SDS-PAGE | MALDI-MS/MS | 52 (48%) | 238–82,660 | Knock-out mutant analysis; HA tagging | 6 |
| Vacuolar lumen | SEY6210 (n), YPD | Zymolyase; mechanical disruption | Sucrose/sorbitol/Ficoll density gradient | Proteinase K | WB, marker enzymes assays, microscopy, flow cytometry | 2-DE | MALDI-MS/MS, nLC-IT-MS/MS, ESI-MS/MS | 118 (5%) | 172–1,018,216 | Internal | 8 |
| Vacuolar membrane | W303 (n), YPD | Zymolyase, lyticase, Glucanex; osmotic lysis | Ficoll density gradient | EDTA, pH | WB | SCX-RP-nLC | MALDI-MS/MS | 148 (57%) | 57–314,034 | LOPIT | 7 |
| Cell wall | FY833 (n), YPD | Direct mechanical disruption | Differential centrifugation, HF-pyridine extraction | Salt, EDTA, SDS, NaOH | No information | Anion-exchange chromatography | nLC-ESI-MS/MS | 19 (5%) | 600–42,000 | Phenotypic cell wall assay using knock-outs | 3 |
| Plasma membrane | FY1679 (n), YPD, 28 °C | Direct mechanical disruption | 1) Acid precipitation, 2) sucrose density gradient (34–56%) | 0.4% deoxycholate | SDS PAGE | CTAB/SDS PAGE | MALDI-MS/MS | 48 (19%) | 377–1,255,722 | 5 | |
| Plasma membrane | FY1679 (n), YPD | Direct mechanical disruption | 1) Acid precipitation, 2) Sucrose density gradient (43–53%) | EDTA, n-octyl glucoside | ATPase assays, SDS-PAGE | Anion EC/LDS-PAGE | PMF: MALDI-MS, ESI-MS/MS | 86 (21%) | 166–1,255,722 | 4 |
a (n) or (2n) after the strain name indicates haploid or diploid strain, respectively.
b Unless indicated otherwise, the cells were grown aerobically at 30 °C in YPD medium containing 1% yeast extract, 2% Bacto peptone, 2% glucose.
c YPL medium contained 1% yeast extract, 2% Bacto peptone, 0.5% lactic acid.
d YPG medium contained 1% yeast extract, 2% Bacto peptone, 3% glycerol, pH 5.0.
e Digestion was carried out using four proteases: trypsin, chymotrypsin, Glu-C, and subtilisin.
f SCL and SCD media are complete synthetic media supplemented with lactic acid or glucose, respectively.
g YNO contained 0.1% yeast extract, 0.5% ammonium sulfate, 1.7 g/liter yeast nitrogen base, 0.02% Tween 40, 0.3% glucose, 0.1% oleic acid.
h SCIM contained 0.5% yeast extract, 0.1% peptone, 0.79 g/liter complete synthetic medium mixture, 0.5% ammonium sulfate, 1.7 g/liter yeast nitrogen base, 0.1% Tween 40, 0.1% glucose, 0.15% oleic acid.
In this review, we will give an overview of results of the organelle proteome studies and try to answer the following questions. How complete is our current view of organelle proteomes? And how reliable are the proteomics data? Completeness and reliability of the organelle proteome data are dictated to a large extent by methodology. Although the different studies have used a plethora of methods (Table II), two steps are in common for all published organelle proteome analyses. First, pure organelles are isolated by biochemical fractionation methods. Second, the proteins in the purified organelles are identified. The reliability of organelle proteome analyses ultimately depends on the purity of the isolated organelles. In the ideal case, no proteins from different cellular locations (contaminants) should be present in the final preparation. Defining whether or not an identified protein is a contaminant is challenging as proteins may have multiple destinations in a cell, and the distribution over the different localizations may vary throughout the cell cycle and as a function of metabolic and environmental conditions. It is also important to realize that within a single culture cells of different age and cell cycle stages are present. Thus, organelle proteomics studies report “averaged” proteomes. The completeness of the organelle proteomes also depends on the quality of the organelle preparation because loosely attached proteins may be lost during the isolation. In addition, completeness depends on the sensitivity of the analytical methods that are used to identify the proteins. These methods include extraction of the proteins from the organelle preparation, generation and separation of peptides, and (tandem) mass spectrometry analysis to identify the proteins.
Because organelle proteomics is so intimately dependent on biochemical methods for subcellular fractionation, we will give a brief overview of the methodology to isolate subcellular compartments. We will not discuss the methods for protein extraction, peptide generation, and identification using tandem MS-based techniques. The analytical methods continue to evolve at a great pace because of ongoing technical advances. Consequently, the methodological details vary considerably between the different studies that were performed over a period of a decade. Recently, state of the art methods in proteomics were summarized in a comprehensive review by Speers and Wu (15). Mass spectrometry-based proteomics methods for organellar proteomics have been reviewed by Andersen and Mann (16) and Yates et al. (17). Methods dedicated to membrane proteomics have been reviewed by Wu et al. (18, 19), and recent instrumental developments in mass spectrometry for proteomics have been summarized in Han et al. (20).
ISOLATING ORGANELLES
All methods used to prepare different organelles have several steps in common: cells are cultivated and disrupted, and the cellular content including organelles is liberated. Then, the released organelles are separated into different populations according to their physical or chemical properties. In Table II, details for the organelle isolation in the 18 proteomics studies are given. Before integrating the results of the 18 organellar proteomics studies into a global view of protein localization in yeast, we will discuss a few experimental details of the isolation procedures that may affect the ensemble interpretation.
Considerations about Growth Conditions
The content and/or the abundance of organelle proteomes in yeast depends on the growth conditions. Care must be taken when integrating the results from different studies to obtain a global view of protein localization because different growth conditions have been used. It also must be noted that different strains of S. cerevisiae have been used in the various organelle proteomics studies (Table II).
For most organelle proteome analyses, yeast cultures were grown aerobically at 30 °C on a rich medium (YPD, 1% yeast extract, 2% Bacto peptone, 2% glucose) containing an excess of glucose as carbon source, and the cells were harvested in the mid-logarithmic phase. These are “standard” growth conditions for S. cerevisiae, but some organelles require specific growth conditions to be obtained in sufficient amounts (Table II and discussed below).
For the proteomics studies of peroxisomes, cells were grown in conditions that stimulate fatty acid β-oxidation (21–23), which leads to an increase in the number of peroxisomes from one to four per cell (under standard conditions) to as many as 14 per cell, occupying 8–10% of the cytoplasmic volume (24). Also, mitochondrial morphology and abundance depend on growth conditions (25). For the mitochondrial proteome analyses, yeast was grown on the non-fermentable carbon sources to activate respiration and induce many small mitochondria (26–30).
When grown on glucose-containing medium, the abundance of lipid particles increases toward the stationary phase. For the proteome analysis, the cells were grown on glucose and harvested in the late exponential phase (31).
Considerations about Organelle Isolation
When aiming at an inventory of proteins from organelles, a sensitive technique such as mass spectrometry will identify not only proteins that are truly located in the organelle of interest but also contaminating proteins. It is important to realize that, for physical and biological reasons, it is not possible to obtain 100% pure organelle preparations. One of the challenges in organelle proteomics is to obtain preparations of subcellular compartments as pure as possible. On the other hand, genuine organellar residents may have multiple locations and may be hard to discriminate from contaminants. Another challenge is to obtain complete/intact organelles. Because of generally lengthy isolation procedures, a fraction of the true resident proteins may be lost because they have a more dynamic or low affinity association with the organelle or because they are prone to proteolytic degradation.
In all of the 18 organelle proteomics studies, the crude cell lysates were subjected to differential centrifugation to obtain enriched crude organelles (Fig. 1). Differential centrifugation can involve multiple subsequent centrifugation steps that are performed at increasing g forces.
Fig. 1.
Centrifugation for subcellular fractionation. Differential centrifugation is usually the first step after cell lysis in subcellular fractionation protocols and is often followed by density gradient centrifugation. The fragments of the cell wall, unlysed cells, and the nuclei can be sedimented at low speed (3,000–5,000 × g). The pellet can be subjected to density gradient centrifugation for further purification of the nuclei. The postnuclear supernatant, containing all other organelles and the plasma membrane vesicles, is usually subjected to density centrifugation for further fractionation. The postnuclear supernatant can also be subjected to a second round of differential centrifugation at higher g force (20,000–30,000 × g). The mitochondria and peroxisomes are then further purified from the pellet using density gradient centrifugation, and the vacuoles, smooth ER (SER), and plasma membrane (PM) vesicles can be purified from the supernatant. Alternatively, the supernatant can be subjected to ultracentrifugation to sediment light organelles and organellar vesicles and separate these from the cytosol and lipid particles that remain in the supernatant. We emphasize that the figure is an oversimplification because of limitations to the designation of organelle names to crude differential fractions. RER, rough ER.
Organelles can be further separated from each other on the basis of their physical properties by the means of density gradient centrifugation or free flow electrophoresis or based on their antigenic properties by immunotechniques (6, 32). Density gradient centrifugation uses differences in buoyant density of the particles for separation (Table I) and was utilized in all yeast organelle proteomics studies performed thus far (Fig. 1 and Table II) except for the proteomes of the Golgi (where immunoprecipitation was used) and the cell wall (where differential centrifugation only was used). In the case of peroxisomes, density gradient centrifugation was used in combination with immunologically based pullouts (21).
Suborganelle Preparations
Membrane-enclosed organelles such as mitochondria, peroxisomes, vacuoles, ER, Golgi, and nuclei can be fractionated further into subcompartments. The internal (luminal) content can be separated from the membrane upon osmotic and/or mechanical lysis of the organelles and subsequent differential centrifugation. Because membrane proteins are usually of a lower abundance than luminal enzymes, enrichment of the membrane fraction of an organelle is important for a comprehensive identification of these proteins. In only two cases (vacuoles and mitochondria) have suborganellar proteomes been studied separately (7, 8, 29, 33).
ORGANELLE PROTEOMES: ORGANELLE PROFILING
The proteomes of eight subcellular compartments (further referred to as experimental organelle proteomes) have been analyzed by mass spectrometry-based techniques in 18 separate studies (Table II). The eight experimental organelle proteomes combine results of the corresponding individual studies dedicated to the same organelle. These are proteomes of the cell wall (3), the plasma membrane (4, 5), the Golgi apparatus (6), the vacuoles (7, 8), the nucleus (9, 10), the peroxisomes (21–23), the mitochondria (26–30, 34), and the lipid particles (31) (Table II and supplemental Table 1).
Two questions arise in the light of these studies: how complete are these organelle proteomes, and how reliable are the results of these studies? In other words, what level of sensitivity has been achieved, and how selective were these studies? Sensitivity is determined by the analytical methods used as well as by the isolation procedure in which proteins may be lost. Selectivity is determined by the quality of the organelle preparation: how many contaminating proteins from different cellular locations were present in the purified organelles? To be able to answer these questions accurately, we would need to know the subcellular localization of all protein in yeast. But, of course, we do not have this information because the knowledge of the subcellular localizations was the purpose of organelle proteomics. Fortunately, for yeast, we can use a good approximation because of the wealth of information organized in SGD, a manually curated database that integrates information on all genes and gene products of S. cerevisiae.
Defining the Reference Proteomes
For the purpose of this review and based on the annotation of subcellular locations from the yeast genome database SGD, we define that proteins can be located in 11 different subcellular locations: cell wall, plasma membrane, cytosol, nucleus, mitochondrion, peroxisomes, lipid particle, endosome, ER, Golgi, and vacuole (supplemental Table 2). We define the content of each reference proteome as “all proteins that according to the annotations in the SGD are present in the subcellular location.” The annotations for localization in the SGD are derived from a variety of genome-wide high throughput studies (2, 35), low throughput studies on individual proteins (e.g. immuno-EM), and global computational predictions. Localization data obtained in the 18 mass spectrometry-based organelle proteomics studies had not been included in the SGD at the time of our analysis, apart from the mitochondrial proteomics data, the implications of which will be discussed below.
It is possible that a protein has SGD annotations for localization in more than one organelle. Multiple annotated locations of a protein could be biologically relevant as some proteins reside in more than one subcellular location. But dual or multiple localizations may also be artifacts of the methods used, e.g. a tagged protein may be targeted to the wrong location. So, we emphasize that the reference proteomes are not complete and contain mistakes. Nonetheless, by and large they give us a good approximation of the numbers of proteins to expect. Because we have to take into account dual/multiple localizations, proteins with more than one annotated localization in the SGD were counted in each separate reference organelle proteome.
The reference organelle proteomes defined in this way have very different sizes (Fig. 2, large pie diagram, bottom right). The two smallest annotated proteomes harbor only 35 (lipid particles) and 58 proteins (peroxisomes). In contrast, the two largest reference proteomes contain 1,820 and 1,870 proteins (cytosol and nucleus, respectively) of which 576 proteins have annotations for both organelles. The mitochondrial proteome with 1,056 proteins is the third largest, containing 594 proteins with unique mitochondrial annotation. Other organelle proteomes are of intermediate size, containing between 100 and 300 annotated proteins.
Fig. 2.
Abundance, annotated localization, and experimental coverage of yeast proteins. The yeast proteins were split into six groups according to their cellular copy numbers. The top six bars represent the abundance groups and show the percentage of proteins identified by experimental proteomics studies in each group. The pie diagrams to the right of the bars show the annotated localizations of the proteins (identified plus unidentified) in each abundance group. The total number of unique proteins per group is indicated as the “SUM,” and the number of identified proteins is indicated in parentheses. Because proteins with multiple annotations were counted more than once, the sum of the proteins from all the segments of each pie is not the same as the SUM of unique proteins. The bottom bar shows the percentage of identified proteins for the entire yeast proteome, and the large pie at the bottom right shows the annotated localizations of all 5,797 yeast proteins. The localization of the 3,516 yeast proteins identified in at least one proteome study and the localization of the 2,281 unidentified proteins are shown in the bottom pies (left and middle, respectively). Each color indicates a subcellular location: orange, cytosol; pink, plasma membrane (PM); beige, cell wall; red, mitochondria; green, vacuole; light violet, endosome; violet, Golgi apparatus; purple, endoplasmic reticulum; gray, nucleus; blue, peroxisome; magenta, lipid particles; yellow, no localization assigned; black, others (e.g. extracellular).
For many of the proteins in the reference proteomes, not only their subcellular localizations but also their copy numbers in the cell have been determined. The copy numbers are largely based on a genome-wide study using TAP fusions in conjunction with quantitative Western blot analysis, which yielded copy number estimates for 3,837 of the 5,797 yeast proteins (35). Again, just like the annotation of subcellular localizations, the annotated copy numbers for some of the proteins may be incorrect (for instance, when the tag interferes with targeting, it is also likely to affect the copy number). However, for the purpose of a global analysis of the organelle proteomes, the data provide us with extremely useful information. We distributed all quantified proteins over five abundance bins (copy numbers: <500, 500–2,000, 2,000–5,000, 5,000–20,000, and 20,000–2,000,000). The bin widths were chosen to have approximately equal numbers of proteins per bin. The small pie charts in Fig. 2 show the distribution of cellular localizations of the proteins in each abundance bin. The distribution varies among the bins: the reference peroxisomal and nuclear proteomes are slightly biased toward lower copy numbers (only 75 nuclear proteins and no peroxisomal proteins are present in the cell at a copy number >20,000). The bias for low abundance proteins in the nuclear proteome may be caused by the presence of many low copy number transcription factors and regulatory proteins. The low abundance of peroxisomal proteins is caused in part by the growth conditions used in the studies to determine abundance: peroxisomal proliferation was not induced (with oleic acid). The reference proteomes for the cytosol, the cell wall, and lipid particles have relatively many high copy number proteins, i.e. highly abundant glycolytic and other metabolic enzymes and free ribosomes of the cytosol.
The protein abundances range from 101 to 106 molecules per cell in four of the reference proteomes (cell wall, plasma membrane, mitochondria, and cytosol), from 101 to 105 in three organelles (vacuole, nucleus, and Golgi), from 101 to 104 molecules per cell in lipid particles and ER, and from 102 to 105 for endosomal proteins. The reference peroxisomal proteome has the narrowest range, covering only 2 orders of magnitude from 102 to 104 molecules per cell (Table I).
Overall Experimental Coverage
The 18 proteomics studies of yeast subcellular compartments have collectively led to a coverage of 61% of the proteins from the entire predicted yeast proteome (3–10, 21–23, 26–31, 34). In other words, 3,516 different proteins were found in at least one of 18 individual proteome analyses. It is noteworthy that ∼80% (4,517 proteins) of the yeast proteome has been detected under the standard growth conditions by chromosomal TAP and GFP fusions (35). The lower fraction of identified proteins by the ensemble of proteomics studies may have several causes. 1) Some subcellular compartments have not been analyzed in dedicated proteomics studies (cytosol, ER, and endosome). 2) Loosely attached proteins may be lost in the organellar isolation procedures. 3) Some proteins escape identification in the proteomics studies because of their sequence properties. For example, small and hydrophobic proteins may not yield suitable peptides for identification by mass spectrometry when the common proteolytic enzyme trypsin is used. Trypsin cleaves C-terminally of arginine and lysine residues, and small/hydrophobic proteins may lack these residues at suitable positions.
As indicated in the Fig. 2, the fraction of identified proteins in the collective proteome studies increases with protein abundances. Whereas only 54% of yeast proteins with a copy number <500 are covered by the proteome analyses, nearly 95% of the proteins with copy number >20,000 have been identified. These numbers indicate that abundance is the dominant determinant for protein identification.
The coverage of the proteomes of the individual organelles, expressed as the percentage of identified proteins of each reference proteome, varies considerably (Fig. 3). The reference proteomes with the highest coverage are the mitochondrion (88%) and peroxisomes (84%). The proteomes of the nucleus, Golgi, ER, and lipid particles are covered to 70–75%. The vacuolar and cytosolic proteomes with 60 and 65% coverage, respectively, are close to the average of 61%. The proteomes of the cell wall (46% coverage), the endosome (50% coverage), and the plasma membrane (51% coverage) are covered below average. The low coverage of the cell wall proteome is caused by the experimental design, which was aimed at identification of covalently bound (mainly glycosylphosphatidylinositol-anchored) proteins. Soluble cell wall proteins were discarded by applying harsh extraction methods (3). The proteins without known localization were covered to only 16%. The low coverage may indicate that these proteins are not usually expressed under the conditions tested. Just like for the entire yeast proteome, the coverage of proteins in the individual organelles tends to be better for more abundant proteins (Fig. 3).
Fig. 3.
Coverage of organellar proteomes and sensitivity of proteomics studies. Analogous to the presentation in Fig. 2, the coverage of each organellar proteome is depicted as the total coverage of the reference proteome (the last bar in each small diagram) or for each abundance group separately (first six bars from left to right represent copy numbers: <500, 500–2,000, 2,000–5,000, 5,000–20,000, 20,000–2,000,000, and no abundance data available). The coverage is presented as the fraction of proteins in each reference proteome that was identified by any of the 18 experimental proteome studies. The fraction of each reference proteome that is covered by the dedicated organelle proteome study is indicated as the striped area of the total organelle coverage and represents the apparent sensitivity of the experimental proteomics experiments.
2,281 proteins (∼⅓ of the predicted proteome) still remain to be identified by mass spectrometry. The largest group of unidentified proteins (785) consists of proteins without known localization of which the majority does not have assigned biological and/or molecular function. The abundance of unidentified proteins is biased toward low copy numbers: more than ⅓ (700 proteins) of the unidentified proteins have copy numbers of less than 2,000 per cell, and for more than a half (1,233 proteins), the copy number could not be determined. It seems that all high throughput methods (both mass spectrometry-based proteomics and tagging techniques such as GFP and TAP fusions) have so far failed to record these proteins. The reason could be that the copy numbers are below the detection limit (∼40 copies per cell) or that these proteins are not expressed under the conditions used. Among the unidentified proteins, there are many regulatory proteins that are involved in signal transduction, such as GTPases, and transcription and translation factors, which might be expressed only under specific conditions.
Sensitivity of Experimental Proteomes
The coverage of the reference proteomes described above is the sum of all the proteins identified in the different experimental studies. It does not imply that all of the identified proteins were found in experimental studies aimed at the organelle of interest; some proteins may have been found only as contaminants in experimental studies aimed at studying different organelles.
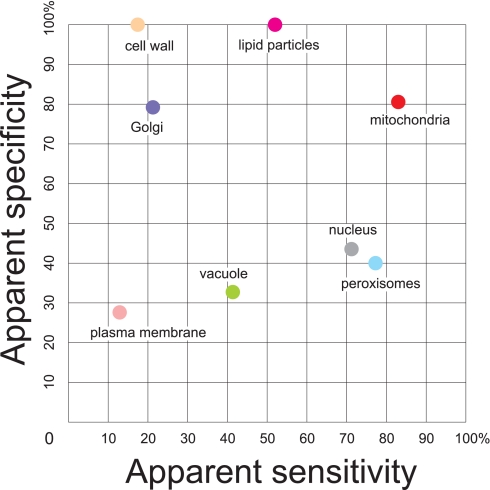
If the reference proteomes did not contain mistakes and the sensitivity of the analytical techniques was infinite, each experimental proteome study would find all proteins of the corresponding reference proteome. In reality, the overlap is never complete because the reference proteomes are not perfect, and the sensitivity of the analytical techniques is limited. For the eight cellular locations that have been targeted experimentally by proteomics studies, the overlap (apparent sensitivity) is indicated in Fig. 3 as striped areas of each organelle reference proteome. Clearly, there is a great variety in the apparent sensitivities of the different proteomics studies. The dedicated investigations of the plasma membrane, the cell wall, and Golgi found less than ¼ of their respective reference proteomes (32 of 270 proteins for plasma membrane, 19 of 110 proteins for the cell wall, and 41 of 199 proteins for Golgi). The reference proteomes of lipid particles and vacuoles are moderately covered by the dedicated proteome studies with about 40–50% coverage. The majority (over 70%) of the reference mitochondrial (881 of 1,056 proteins), peroxisomal (45 of 58 proteins), and nuclear proteins (1,303 of 1,870 proteins) were found in the dedicated experimental studies, indicating a high sensitivity.
Specificity of Experimental Proteomes
From Fig. 3, it is clear that for all organelles fewer proteins have been identified by dedicated studies (striped parts of the bars) than by the ensemble of all 18 organelle proteome studies (total bars). Strikingly, in some organelles, the majority of covered proteins was found in different studies than the one(s) targeted at the specific organelle (i.e. endosome, cytosol, and ER). The extra proteins found in the ensemble could be contaminants from proteome studies dedicated at other organelles or could indicate incomplete or wrong database annotations of the reference proteomes. Here we discuss some striking examples, which highlight the difficulty of distinguishing between the two possibilities.
One way to obtain an indication about specificity is to calculate the fraction of proteins identified in each of the eight experimental proteomes that was indeed annotated in the SGD as localized in the organelle of interest. In Fig. 4, this apparent specificity is indicated. The apparent specificity ranges from 100% (all proteins identified in the lipid particle and cell wall proteomes have corresponding annotations) to less than 30% (more than 70% of the identified proteins in the plasma membrane proteome have annotations for different cellular locations). The high apparent specificities of the lipid particle and cell wall proteomes are a direct consequence of the biochemical fractionation methods, which resulted in very clean preparations because of the characteristic physical properties of these subcellular isolates. But a low apparent specificity does not necessarily indicate low quality of the biochemical fractionation. For instance, when highly sensitive analytical methods are used, the apparent specificity is likely to become lower, because low abundance contaminants are more efficiently detected (e.g. in the case of the nuclear proteome). Also, SGD annotations play a crucial role in the apparent specificity. For example, the outcome of the mitochondrial proteome analyses has been included in annotations for localization in the SGD, which obviously will increase the apparent specificity. In contrast, data from another proteome analysis have not (yet) been included in the SGD, which lowers the apparent specificity for those proteomics studies that identify a large number of new organelle proteins (for example, the vacuolar proteome; see below).
Fig. 4.
Apparent sensitivity and specificity of experimental organellar proteomics studies. The apparent sensitivity was calculated as the percentage of proteins from the reference proteome that was identified by the dedicated experimental organellar proteome study. The apparent specificity was calculated as the fraction of proteins identified in the dedicated experimental proteomics studies that was annotated as localized in the same subcellular location.
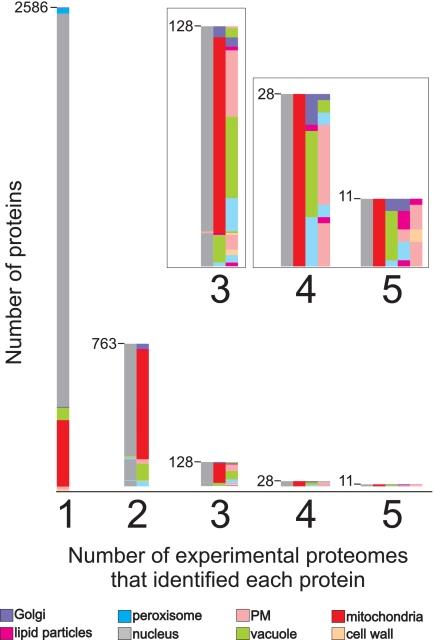
A rough indication about the overall specificity can also be drawn from the number of uniquely identified proteins in each of the eight experimental proteome analyses (Fig. 5). Nearly 74% of all identified proteins were found in one proteomics study only, and 22% were found in two experimental organelle proteomes. The remaining 4% of the identified proteins were found in three to five proteomes with a clear trend (∼⅔) toward a high abundance (copy number >20,000) for those proteins. Some of these proteins are likely nonspecific contaminations, e.g. P-type ATPase Pma1 of the plasma membrane, ribosomal proteins, and many glycolytic enzymes. But among the proteins present in multiple proteomes are also proteins that are known to have multiple localizations, e.g. proteins involved in signal transduction like GTPases Rho1, YPT1, and CDC42 or the protein kinase TOR2. No proteins, not even very high abundance proteins, were found in more than five organelle proteomes.
Fig. 5.
Proteins identified in multiple experimental proteomes. Each of the 3,516 identified proteins was found in one to five experimental proteomes. The bars indicate the total number of proteins found in one, two, three, four, or five proteomes. The inset shows an enlargement of the bars for the proteins identified in three, four, or five proteomes. The colors indicate in which experimental proteomics studies the proteins were identified: pink, plasma membranes; beige, cell walls; red, mitochondria; green, vacuoles; violet, Golgi apparatus; gray, nuclei; blue, peroxisomes; magenta, lipid particles.
The coverage of the reference proteomes for ER, endosome, and cytosol are above 50%, although no dedicated proteomics studies were performed for these organelles. This could be indicative of large amounts of contaminants in the organellar preparations. However, there are also alternative explanations for the detection of these proteins. The largest experimental organelle proteomes, the nuclear proteome, contributed the most to their coverage. Almost all proteins (236 of 253 identified proteins with ER annotations, 42 of 52 endosomal proteins, and 1,079 of 1,174 cytosolic proteins) were identified in the nuclear proteomics studies. There is a straightforward biological explanation for the appearance of ER proteins in the nuclear proteome as the nuclear envelope is continuous with the ER. The perinuclear ER has general ER function and covers ∼20% of the ER surface (36, 37).
We took a more detailed look into the coverage of the reference ER proteome and found that of the 253 identified proteins 160 ER-annotated proteins were uniquely detected in the nuclear proteomes. As indicated above, finding these ER-annotated proteins in the experimental nuclear proteomes is biologically relevant. Of the remaining proteins, 62 were detected in two experimental proteomes, nine proteins were detected in three proteomes, and five proteins were detected in four proteomes, mainly from the experimental nuclear, mitochondrial, vacuolar, and peroxisomal proteomes. The annotated functions of the ER proteins found in multiple experimental proteomes are biased toward sterol and phospholipid biosynthesis and protein trafficking across the secretory system. The proteins found in multiple organelle proteomes are relatively low abundance proteins and, in the light of the mentioned functions, might be of biological relevance; i.e. these proteins have multiple localizations in the cell.
The proteins with endosomal annotation make up 14% of the experimental Golgi proteome and 5% of the vacuolar proteome, respectively. In the other experimental proteomes, endosomal proteins account for a very minor fraction (<1%, even though the absolute number in the nuclear proteome is large). The endosome is an integral part of the secretory machinery and shares many components of the vesicle fusion and protein trafficking complexes with vacuoles and the ER-Golgi network. It is not surprising, therefore, that many of the proteins that are annotated as “endosomal” in the yeast database and that are found in the Golgi and vacuolar proteomes have functions in ER-to-Golgi transport. The identified endosomal proteins in the Golgi and vacuolar proteomes are therefore unlikely to be contaminants.
The experimental proteome of the plasma membrane has the lowest apparent sensitivity and specificity of all eight proteomes (Fig. 4). In two dedicated proteomics studies, 121 proteins could be identified of which 73 proteins are annotated as cytosolic in the SGD. Those 73 proteins are mostly high abundance contaminations such as ribosomal proteins and glycolytic enzymes.
How to Deal with Contaminants?
The above examples show the need to deal explicitly with contaminants because no biochemical separation technique will yield 100% pure organelles. In all organellar proteomics studies, the obtained list of identified proteins was evaluated very carefully by comparison with existing database and literature information. Proteins that had not previously been assigned to the studied organelle were either regarded as contaminants or considered as novel proteins. An example is the peroxisomal proteome (23) where the authors could identify ∼240 proteins. After careful analysis, only 46 of these proteins were assigned as peroxisomal based on prior knowledge about the peroxisomal content. However, when aiming at identification of new proteins not previously annotated to be associated with an organelle, a database- and literature-based evaluation may not be sufficient.
One way to deal with contaminants would be to obtain purer organelle preparations. The mitochondrial proteome, which is of high apparent sensitivity and selectivity, indicates that very good purity can be achieved. However, for physical and biological reasons (38–41), it is unlikely that organelles can be isolated to an absolute purity and homogeneity; rather, the final preparations are enriched to a variable extent depending on the fractionation methods. In many cases, marker proteins for the organelle of interest were used to assess the purity (either by Western blotting or by activity assays; Table II). Marker enzymes for different organelles are indicated in Table I. Marker enzymes are useful for qualitative assessment of organelle enrichment, but the assays are not sufficiently sensitive to judge the purity of a preparation for MS purposes. Even when organelle preparations appear completely pure based upon immunological detection of the markers, they may still be contaminated, and these contaminations will be revealed by mass spectrometry because of the higher sensitivity of the method. The issue of contaminating proteins is particularly problematic because proteins in cells and organelles are present in abundances that can vary by 6 orders of magnitude. Consequently, even if a very small fraction of a contaminating organelle is present in the final preparation, highly abundant proteins originating from the contaminating organelle may be as easily detectable by MS as the low abundance proteins from the organelle of interest.
Instead of trying to avoid contaminants by using (apparently) very pure preparations, the contaminations can also be accounted for. One way of doing so is to use additional quality assessments, for instance GFP-tagged fusions. However, as mentioned above, GFP-tagged proteins are not always targeted to their native destinations, which results in mislocalizations.
A robust approach to account for contaminants is to use relative quantification-based techniques (for reviews see, Refs. 16, 17, and 42). The original idea of this approach (for a review, see Ref. 43) is that proteins that are physically associated with an organelle become enriched upon purification of this organelle, for instance by density gradient centrifugation. The contaminants, in contrast, although still present and detectable by mass spectrometry, will become depleted upon purification. The specific enrichment or depletion can be quantified by comparing protein abundances in crude and pure preparations of the organelle of interest (7, 21, 44) or by comparing abundances in different fractions of a density gradient after centrifugation (Fig. 6). When the protein abundances in all of the fractions are compared, proteins with multiple localizations can be recognized because they will be enriched in more than one part of the gradient. Enrichment in multiple fractions of a density gradient may also occur when the preparation of an organelle is heterogeneous. Such heterogeneity may be the result of the presence of slightly different organelle composition within a single cell or because the final preparation contains organelles derived from cells of different age and cell cycle stage.
Fig. 6.
Principle of LOPIT and label-free quantification. The LOPIT technology is based on the notion that biochemical purification of an organelle never results in an absolutely homogeneous preparation. Instead, proteins that are specifically associated with the organelle of interest (green dots) become enriched upon purification (top layer; a), and contaminants (red dots), although still present in the enriched fraction a, become depleted. The LOPIT technology uses labeling of peptides with stable isotopes such as ICAT or iTRAQ (right-hand side of the figure). For this, peptides from the enriched (a) and depleted (b and c) samples are labeled with reagents of different masses, mixed together, and processed by liquid chromatography and mass spectrometry. The isotope labeling allows relative quantitation of the protein abundance in the enriched and depleted fractions simultaneously (a, b, and c). For the label-free quantitative approach (left-hand side of the figure), peptides from the enriched (a) and depleted (b and c) fractions are processed by liquid chromatography and mass spectrometry separately without labeling or mixing. In this case, the protein abundance in each fraction is quantified based on the number of observed peptides divided by the number of observable peptides (protein abundance index), the peptide peak intensity (protein correlation profiling), or the spectral count (number of identified spectra for each protein).
For quantification, isotope tags such as iTRAQ or ICAT can be used as developed by Lilley and co-workers (44–46) to assess the localization of organelle proteins by isotope tagging (LOPIT). Alternatively, label-free quantification can be performed by protein correlation profiling, which is based on the intensities of the peptide peaks in different MS analyses (47–49) using the proteins abundance index, which represents the number of peptides identified divided by the number of theoretically observable peptides (50, 51), or spectral counting where the number of mass spectra identified for a protein is used as a measure of the abundance of a protein (52). Use of the spectral count-based quantification for absolute and relative protein abundance determination in complex mixtures is described in Vogel and Marcotte (53). These different methods to account for contaminants are globally outlined in Fig. 6.
Label-based quantification in the yeast proteomics studies was successfully utilized for the vacuolar membrane and peroxisomal proteomes (7, 21). The most valuable result of these studies is that many proteins with annotated localizations other then vacuolar or peroxisomal were found to be specifically co-enriched with vacuoles or peroxisomes, which strongly indicates that they are part of the true proteomes of these organelles. Thus, the low apparent specificities of the peroxisomal and vacuolar proteome studies (Fig. 4) appear not to be caused by contaminants but rather by incomplete database annotations for localization. Proteomics studies in which quantification was used to assess subcellular localization allow reassignment of the subcellular localizations of ambiguously annotated proteins.
CONCLUSIONS
Mass spectrometry-based proteomics is a powerful technique to study the subcellular localization of native, non-tagged proteins. The currently available yeast organellar proteomes have yielded a tremendous amount of valuable information. The proteomics studies complement high throughput localization studies based on GFP tagging and imaging. Both techniques have their advantages: proteomics studies do not require tagging, which may cause targeting failure, and GFP tagging studies do not require subcellular fractionation. Subcellular fractionation of the cell lysate is an essential part of organellar proteomics. Robust methods to isolate subcellular proteomes are available and produce highly purified preparations. But the isolation procedures are lengthy and may lead to loss of proteins that are loosely attached or susceptible to degradation. Therefore, more rapid isolation procedures, e.g. based on affinity pullouts, as used for Golgi isolation (6) are desirable (54).
No organellar preparation will be completely free of contaminants from different subcellular locations. Quantitative proteomics techniques that assay enrichment profiles allow for the discrimination between contaminants and genuine organellar residents and facilitate the assignment of cellular localizations to the identified proteins (7, 21, 44, 47, 55, 56). With the ongoing developments in quantification and identification methods, it will become feasible to perform a global analysis of protein localization in yeasts from a single culture. One could envision that such an experiment would use a multidimensional fractionation scheme and extensive analysis of co-fractionation profiles that follow from quantitative mass spectrometry using e.g. the multiple reaction monitoring method (57, 58). We expect that combining these data with those from co-purification strategies where protein complexes are isolated using tagged proteins will provide a more complete and accurate description of protein localization. Ultimately, dynamics will also have to be included in the localization experiments because proteins may be associated only temporarily, and dependent on the environmental conditions, with an organelle proteome (59).
Supplementary Material
* This work was supported by the Netherlands Organisation for Scientific Research NWO (VIDI fellowships (to D. J. S. and L. M. V.) and TopSubsidy (to B. P.)), the Netherlands Proteomics Centre, and the Zernike Institute for Advanced Materials.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1 and 2.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1 and 2.
1 The abbreviations used are:
- SGD
- Saccharomyces genome database
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LOPIT
- localization of organelle proteins by isotope tagging
- TAP
- tandem affinity purification
- EM
- electron microscopy
- YPD
- yeast peptone dextrose.
REFERENCES
- 1.Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. (1996) Life with 6000 genes. Science 274, 546–567 [DOI] [PubMed] [Google Scholar]
- 2.Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Cheung K. H., Miller P., Gerstein M., Roeder G. S., Snyder M. (2002) Subcellular localization of the yeast proteome. Genes Dev 16, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin Q. Y., de Groot P. W., Dekker H. L., de Jong L., Klis F. M., de Koster C. G. (2005) Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem 280, 20894–20901 [DOI] [PubMed] [Google Scholar]
- 4.Delom F., Szponarski W., Sommerer N., Boyer J. C., Bruneau J. M., Rossignol M., Gibrat R. (2006) The plasma membrane proteome of Saccharomyces cerevisiae and its response to the antifungal calcofluor. Proteomics 6, 3029–3039 [DOI] [PubMed] [Google Scholar]
- 5.Navarre C., Degand H., Bennett K. L., Crawford J. S., Mørtz E., Boutry M. (2002) Subproteomics: identification of plasma membrane proteins from the yeast Saccharomyces cerevisiae. Proteomics 2, 1706–1714 [DOI] [PubMed] [Google Scholar]
- 6.Inadome H., Noda Y., Adachi H., Yoda K. (2005) Immunoisolaton of the yeast Golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments. Mol. Cell. Biol 25, 7696–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiederhold E., Gandhi T., Permentier H. P., Breitling R., Poolman B., Slotboom D. J. (2009) The yeast vacuolar membrane proteome. Mol. Cell. Proteomics 8, 380–392 [DOI] [PubMed] [Google Scholar]
- 8.Sarry J. E., Chen S., Collum R. P., Liang S., Peng M., Lang A., Naumann B., Dzierszinski F., Yuan C. X., Hippler M., Rea P. A. (2007) Analysis of the vacuolar luminal proteome of Saccharomyces cerevisiae. FEBS J 274, 4287–4305 [DOI] [PubMed] [Google Scholar]
- 9.Mosley A. L., Florens L., Wen Z., Washburn M. P. (2009) A label free quantitative proteomic analysis of the Saccharomyces cerevisiae nucleus. J. Proteomics 72, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauci S., Veenhoff L. M., Heck A. J., Krijgsveld J. (2009) Orthogonal separation techniques for the characterization of the yeast nuclear proteome. J. Proteome Res 8, 3451–3463 [DOI] [PubMed] [Google Scholar]
- 11.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 12.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 14.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 15.Speers A. E., Wu C. C. (2007) Proteomics of integral membrane proteins—theory and application. Chem. Rev 107, 3687–3714 [DOI] [PubMed] [Google Scholar]
- 16.Andersen J. S., Mann M. (2006) Organellar proteomics: turning inventories into insights. EMBO Rep 7, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 18.Wu C. C., MacCoss M. J., Howell K. E., Yates J. R., 3rd (2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol 21, 532–538 [DOI] [PubMed] [Google Scholar]
- 19.Wu C. C., Yates J. R., 3rd (2003) The application of mass spectrometry to membrane proteomics. Nat. Biotechnol 21, 262–267 [DOI] [PubMed] [Google Scholar]
- 20.Han X., Aslanian A., Yates J. R., 3rd (2008) Mass spectrometry for proteomics. Curr. Opin. Chem. Biol 12, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marelli M., Smith J. J., Jung S., Yi E., Nesvizhskii A. I., Christmas R. H., Saleem R. A., Tam Y. Y., Fagarasanu A., Goodlett D. R., Aebersold R., Rachubinski R. A., Aitchison J. D. (2004) Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol 167, 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer H., Nau K., Sickmann A., Erdmann R., Meyer H. E. (2001) Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22, 2955–2968 [DOI] [PubMed] [Google Scholar]
- 23.Yi E. C., Marelli M., Lee H., Purvine S. O., Aebersold R., Aitchison J. D., Goodlett D. R. (2002) Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis 23, 3205–3216 [DOI] [PubMed] [Google Scholar]
- 24.Veenhuis M., Mateblowski M., Kunau W. H., Harder W. (1987) Proliferation of microbodies in Saccharomyces cerevisiae. Yeast 3, 77–84 [DOI] [PubMed] [Google Scholar]
- 25.Visser W., van Spronsen E. A., Nanninga N., Pronk J. T., Gijs Kuenen J., van Dijken J. P. (1995) Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Leeuwenhoek 67, 243–253 [DOI] [PubMed] [Google Scholar]
- 26.Prokisch H., Scharfe C., Camp D. G., 2nd, Xiao W., David L., Andreoli C., Monroe M. E., Moore R. J., Gritsenko M. A., Kozany C., Hixson K. K., Mottaz H. M., Zischka H., Ueffing M., Herman Z. S., Davis R. W., Meitinger T., Oefner P. J., Smith R. D., Steinmetz L. M. (2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pflieger D., Le Caer J. P., Lemaire C., Bernard B. A., Dujardin G., Rossier J. (2002) Systematic identification of mitochondrial proteins by LC-MS/MS. Anal. Chem 74, 2400–2406 [DOI] [PubMed] [Google Scholar]
- 28.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schönfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., Meisinger C. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A 100, 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahedi R. P., Sickmann A., Boehm A. M., Winkler C., Zufall N., Schönfisch B., Guiard B., Pfanner N., Meisinger C. (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinders J., Zahedi R. P., Pfanner N., Meisinger C., Sickmann A. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res 5, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 31.Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol 181, 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lüers G. H., Hartig R., Mohr H., Hausmann M., Fahimi H. D., Cremer C., Völkl A. (1998) Immuno-isolation of highly purified peroxisomes using magnetic beads and continuous immunomagnetic sorting. Electrophoresis 19, 1205–1210 [DOI] [PubMed] [Google Scholar]
- 33.Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohlmeier S., Kastaniotis A. J., Hiltunen J. K., Bergmann U. (2004) The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J. Biol. Chem 279, 3956–3979 [DOI] [PubMed] [Google Scholar]
- 35.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 36.Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., Botstein D. (1991) Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 7, 891–911 [DOI] [PubMed] [Google Scholar]
- 37.Strambio-de-Castillia C., Blobel G., Rout M. P. (1995) Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol 131, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumann U., Subramani S. (2008) Special delivery from mitochondria to peroxisomes. Trends Cell Biol 18, 253–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. (2006) An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perktold A., Zechmann B., Daum G., Zellnig G. (2007) Organelle association visualized by three-dimensional ultrastructural imaging of the yeast cell. FEMS Yeast Res 7, 629–638 [DOI] [PubMed] [Google Scholar]
- 41.Kvam E., Goldfarb D. S. (2006) Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site. Biochem. Soc. Trans 34, 340–342 [DOI] [PubMed] [Google Scholar]
- 42.Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 43.De Duve C., Beaufay H. (1981) A short history of tissue fractionation. J. Cell Biol 91, 293s–299s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., Hanton S. L., Griffin J. L., Bessant C., Brandizzi F., Hawes C., Watson R. B., Dupree P., Lilley K. S. (2006) Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. U.S.A 103, 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunkley T. P., Dupree P., Watson R. B., Lilley K. S. (2004) The use of isotope-coded affinity tags (ICAT) to study organelle proteomes in Arabidopsis thaliana. Biochem. Soc. Trans 32, 520–523 [DOI] [PubMed] [Google Scholar]
- 46.Sadowski P. G., Dunkley T. P., Shadforth I. P., Dupree P., Bessant C., Griffin J. L., Lilley K. S. (2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat. Protoc 1, 1778–1789 [DOI] [PubMed] [Google Scholar]
- 47.Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 48.Wiese S., Reidegeld K. A., Meyer H. E., Warscheid B. (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 [DOI] [PubMed] [Google Scholar]
- 49.Yan W., Hwang D., Aebersold R. (2008) Quantitative proteomic analysis to profile dynamic changes in the spatial distribution of cellular proteins. Methods Mol. Biol 432, 389–401 [DOI] [PubMed] [Google Scholar]
- 50.Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 52.Pang J. X., Ginanni N., Dongre A. R., Hefta S. A., Opitek G. J. (2002) Biomarker discovery in urine by proteomics. J. Proteome Res 1, 161–169 [DOI] [PubMed] [Google Scholar]
- 53.Vogel C., Marcotte E. M. (2008) Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat. Protoc 3, 1444–1451 [DOI] [PubMed] [Google Scholar]
- 54.Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 55.Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 56.Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fortin T., Salvador A., Charrier J. P., Lenz C., Bettsworth F., Lacoux X., Choquet-Kastylevsky G., Lemoine J. (2009) Multiple reaction monitoring cubed for protein quantification at the low nanogram/milliliter level in nondepleted human serum. Anal. Chem 81, 9343–9352 [DOI] [PubMed] [Google Scholar]
- 58.Kuhn E., Wu J., Karl J., Liao H., Zolg W., Guild B. (2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 59.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 60.Nurminen T., Taskinen L., Suomalainen H. (1976) Distribution of membranes, especially of plasma-membrane fragments, during zonal centrifugations of homogenates from glucose-repressed Saccharomyces cerevisiae. Biochem. J 154, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCammon M. T., Veenhuis M., Trapp S. B., Goodman J. M. (1990) Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J. Bacteriol 172, 5816–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thieringer R., Shio H., Han Y. S., Cohen G., Lazarow P. B. (1991) Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol. Cell. Biol 11, 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welten-Verstegen G. W., Boer P., Steyn-Parvé E. P. (1980) Lipid-mediated glycosylation of endogenous proteins in isolated plasma membrane of Saccharomyces cerevisiae. J. Bacteriol 141, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuhrmann G. F., Wehrli E., Boehm C. (1974) Preparation and identification of yeast plasma membrane vesicles. Biochim. Biophys. Acta 363, 295–310 [DOI] [PubMed] [Google Scholar]
- 65.Hossack J. A., Wheeler G. E., Rose A. H. (1973) in Yeast, Mould and Plant Protoplasts: Proceedings. Held at Salamanca, Spain, in Oct. 1972 (Villanueva J. R., Garcia-Acha I., Cascon S., Uruburu F. eds) Academic Press, London [Google Scholar]
- 66.Swida U., Kreutzfeldt C., Ramezani-Rad M., Käufer N. (1982) Isolation and characterisation of rough and smooth endoplasmic reticulum from Saccharomyces cerevisiae. FEMS Microbiol. Lett 15, 313–318 [Google Scholar]
- 67.Sanderson C. M., Meyer D. I. (1991) Purification and functional characterization of membranes derived from the rough endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem 266, 13423–13430 [PubMed] [Google Scholar]
- 68.Braell W. A. (1988) Two sensitive, convenient, and widely applicable assays for marker enzyme activities specific to endoplasmic reticulum. Anal. Biochem 170, 328–334 [DOI] [PubMed] [Google Scholar]
- 69.Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. (1991) Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singer-Krüger B., Frank R., Crausaz F., Riezman H. (1993) Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J. Biol. Chem 268, 14376–14386 [PubMed] [Google Scholar]
- 71.Wiemken A., Dürr M. (1974) Characterization of amino acid pools in the vacuolar compartment of Saccharomyces cerevisiae. Arch. Microbiol 101, 45–57 [DOI] [PubMed] [Google Scholar]
- 72.Zinser E., Paltauf F., Daum G. (1993) Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol 175, 2853–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuchler K., Daum G., Paltauf F. (1986) Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol 165, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.