Abstract
Although various glycoforms appear to participate independently in multiple molecular interactions in cellular adhesion that contribute to embryogenesis and organogenesis, a full portrait of the glycome diversity and the effect of the structural variations of cellular glycoforms on individual cell stages in proliferation and differentiation remain unclear. Here we describe a novel concept for the characterization of dynamic glycoform alteration during cell differentiation by means of “glycoblotting-based cellular glycomics,” the only method allowing for rapid and quantitative glycan analysis. We demonstrated that processes of dynamic cellular differentiation of mouse embryonic carcinoma cells, P19CL6 and P19C6, and mouse embryonic stem cells into cardiomyocytes or neural cells can be monitored and characterized quantitatively by profiling entire N-glycan structures of total cell glycoproteins. Whole N-glycans enriched and identified by the glycoblotting method (67 glycans for P19CL6, 75 glycans for P19C6, and 72 glycans for embryonic stem cells) were profiled and bar-coded quantitatively with respect to the ratio of subgroups composed of characteristic glycoforms, namely glycotypes.
The global characterization of the genome, transcriptome, epigenome, and proteome of embryonic stem cells (ESCs)1 has contributed to basic understanding of ESC biology (1). Functional genomics and proteomics during cellular differentiation have also been intensively studied during recent years (2–4). However, it was suggested that there is no significant change of the protein expression profile in the differentiated cells in comparison with that of undifferentiated (progenitor) cells. It is generally recognized that work on ESCs has proceeded with a general lack of standards for adequate quantification of ESCs (5). It is likely that this has led to inconsistencies and difficulties in reproducing work from independent laboratories because this field has relied upon a variety of antibodies against a few cell surface antigens, which has been useful but not adequate. Therefore, it should be noted that there has been no reliable set of molecular markers that can easily establish the quality and differentiation status of a cell line.
Numerous important roles of mammalian glycans are now evident, and the variation in the cellular glycome is a molecular basis for modulating dynamic cellular mechanisms such as cell-cell adhesion, cell activation, and malignant alterations (6–9). Structural variations of the glycome in cell surface glycoproteins and/or glycosphingolipids appear to produce some useful biomarkers such as stage-specific embryonic antigens and ligands for endogenous lectins during cell proliferation and differentiation (10–13). Loss of some key N-glycans or expression of unusual N-glycans disrupts normal cell-cell adhesion in early mammalian embryos that is associated with fertilization (14–17). In addition, it seems likely that cellular responsiveness to growth or arrest is greatly dependent on total N-glycan number and the degree of branching of cell surface glycoproteins (12). Judging from the fact that N-glycans found in most proteins are an overwhelming majority of the glycome compared with other subgroups such as O-glycans classified as mucin glycoproteins or glycosphingolipids, our attention should be focused on the expression profiles of cellular N-glycans.
We hypothesized that drastic changes in the ratio of some key subtypes of the N-glycans with mutual core structure against total N-glycan expression level may be crucial to switch cell stages/types during cell differentiation in which generated N-glycan subtypes must exchange their major partner molecules in differentiated cell adhesion, namely cell surface carbohydrate-binding proteins or complementary glycans in sugar-sugar interaction (18). In other words, there might be a significant “threshold” or “critical point” in the expression level of the key N-glycan subtypes against cell surface area to initiate this dynamic cellular differentiation. For example, it is well documented that Lewis X trisaccharide antigen (SSEA-1), a prominent member of the Lewis blood group antigen family, is one of the most important subtypes that have key functional roles during developmental processes or cancer progression (10, 19). Glycosphingolipids SSEA-3 and SSEA-4 are also among the most commonly utilized markers to characterize ESCs, and they could also be classified as a sort of subtype composed of globoseries core oligosaccharide structures (13). Sialic acid-containing oligosaccharides are also supposed to be involved in some essential subtypes having various functions as potential markers in cell differentiation or malignant alteration (20). Therefore, it is not surprising that real time monitoring of entire N-glycan expression levels in the course of proliferation and differentiation, a full portrait of the cellular glycoforms, may allow for the identification of target cells and assessment of the quality of individual stem cells and differentiated cells. The advent of such a versatile and comprehensive protocol for quantitative cellular glycomics is now urgently needed because it should greatly contribute to the quality control of human ESCs and a variety of human induced pluripotent stem (iPS) cells in terms of the warranty of safety and reproducibility of required stem cell engineering (21–23).
Until now, technical limitations have restricted acquisition of the total glycan structures of mammalian cells and evaluation of the cell type-specific glycoforms (24). We report herein that the glycoblotting method (24), a PCR-like technology developed for rapid and large scale enrichment analysis of human serum glycans (25, 26), can be used for rapid and quantitative cellular glycomics to monitor dynamic glycoform alteration during differentiation of mouse embryonic carcinoma cells (P19CL6 and P19C6 cells) and mouse ESCs.
MATERIALS AND METHODS
Cell Culture and Differentiation
P19C6 and P19CL6 cells were subcloned from pluripotent mouse embryonal carcinoma, and both were obtained from RIKEN Cell Bank (Ibaragi, Japan) (27, 28). P19C6 and P19CL6 were maintained with Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 15% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel) and minimum Eagle's medium α supplemented with 10% FBS and penicillin-streptomycin (Invitrogen), respectively, at 37 °C in an atmosphere of 5% CO2. P19C6 cells were differentiated as described by Tang and co-workers (29). In brief, P19 cells were cultured by the hanging drop method and allowed to aggregate in bacterial grade Petri dishes at a cell density of 1 × 105 cells/ml in the presence of 1 μm retinoic acid (RA) (Sigma) in minimum Eagle's medium α supplemented with 10% FBS. After 4 days of aggregation, cells were collected in “aggregate,” an intermediate of differentiation, by treatment with 0.05% trypsin, EDTA (Invitrogen) and were replated onto a poly-l-lysine-coated tissue culture dish (6 cm inner diameter) at a density of 5 × 104 cells/cm2 in Dulbecco's modified Eagle's medium/F-12 medium (Invitrogen) containing N-2 supplement and fibronectin. The cells were then allowed to adhere and were cultured for 5 days. Cardiac muscle differentiation of P19CL6 cells was induced by DMSO under adherent conditions as described previously (27). Briefly, 3.7 × 105 cells were plated onto a tissue culture dish (6 cm inner diameter) (Corning) and cultured in a standard medium containing 1% DMSO for 16 days. As a control, undifferentiated cells were cultured in standard medium for 16 days. The media were changed every 2 days. All experiments were performed in triplicate and repeated twice independently (n = 6). Mouse ESCs were differentiated under conditions reported previously (30, 31).
Basic Protocol of Glycoblotting-based Quantitative Cellular N-Glycomics
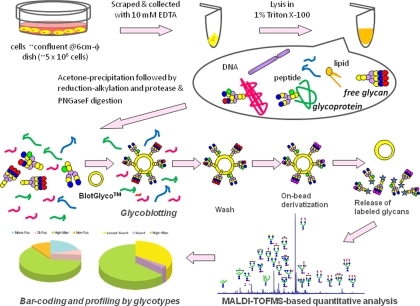
A preliminary trial for the cellular N-glycomics was reported previously in human prostate cancer cells (PC-3) and normal human prostate epithelial cells on the basis of the protocol designed for human serum N-glycomics using the BlotGlycoTM ABC bead, a prototype bead prepared by conjugating N-(2-aminobenzoyl)cysteine hydrazide and thiopropyl-Sepharose 6B (26). However, we had to re-examine and establish a comprehensive protocol feasible for mammalian cellular glycomics using BlotGlyco H, a commercially available synthetic polymer bead (Sumitomo Bakelite Co., Ltd., Tokyo, Japan) (see Fig. 1), because our recent studies on the functional glycomics using BlotGlyco H bead have demonstrated improved performance of this new platform in terms of quantification, reproducibility, and application (25).
Fig. 1.
Basic protocol of high throughput and quantitative cellular glycomics based on glycoblotting method using BlotGlyco H bead. φ, inner diameter; PNGaseF, peptide-N-glycosidase F.
Release of total N-glycans was carried out directly using whole cell lysates as follows. After inducing differentiation, P19C6 and ESCs were cultured by using a poly-l-lysine-coated tissue culture dish (6 cm inner diameter) for the appropriate days indicated above, then were scraped in PBS containing 10 mm EDTA, and washed with PBS. Following suspension in PBS, cells were lysed by incubation with 1% Triton X-100 for 1 h on ice. The lysates were centrifuged at 15,000 rpm for 10 min at 4 °C, and the obtained supernatant was added to cold acetone (1:4) to precipitate proteinaceous materials. The precipitates were collected by centrifugation at 12,000 rpm for 15 min at 4 °C followed by serial washing with acetonitrile. The resulting precipitates were dissolved in 50 μl of 80 mm ammonium bicarbonate containing 0.2% of 1-propanesulfonic acid, 2-hydroxyl-3-myristamido and incubated at 60 °C for 10 min. The solubilized proteinaceous materials were reduced by 10 mm DTT at 60 °C for 30 min followed by alkylation with 20 mm iodoacetamide by incubation in the dark at room temperature for 30 min. The mixture was then treated with 400 units of trypsin (Sigma-Aldrich) at 37 °C overnight followed by heat inactivation of the enzyme at 90 °C for 10 min. After cooling to room temperature, N-glycans of glycopeptides were released from trypsin-digested samples by incubation with 2 units of peptide-N-glycosidase F (Roche Applied Science) at 37 °C overnight. Then the sample mixture was dried by SpeedVac and stored at −20 °C until use. Approximately 1–5 × 106 cells (∼200 μg of total protein) corresponding to cell confluence on the culture dish (6 cm inner diameter) were required for this procedure.
Glycoblotting of the sample mixtures containing whole cell N-glycans by means of BlotGlyco H bead was performed according to the procedure described previously (26). BlotGlyco H beads (500 μl) (10 mg/ml suspension; Sumitomo Bakelite Co., Ltd.) were aliquoted onto a well of a MultiScreen Solvinert filter plate (Millipore, Billerica, MA). Peptide-N-glycosidase F-digested samples were dissolved with 20 μl of water and applied to the well followed by the addition of 180 μl of 2% acetic acid in ACN. The plate was incubated at 80 °C for 45 min to capture total glycans in sample mixtures specifically onto beads via stable hydrazone bonds. The plate was washed by 200 μl of 2 m guanidine HCl in ammonium bicarbonate followed by washing with the same volume of water and 1% triethylamine in methanol (MeOH). Each washing step was performed twice, respectively. Unreacted hydrazide functional groups on beads were capped by incubation with 10% acetic anhydride in MeOH for 30 min at room temperature. Then the solution was removed by vacuum, and then the bead was serially washed by 2 × 200 μl of 10 mm HCl, MeOH, and dioxane, respectively. On-bead methyl esterification of carboxyl groups in sialic acids was carried out by incubation with 150 mm 3-methyl-1-p-tolyltriazene in dioxane at 60 °C to dryness. It usually took 90 min in a conventional oven. Then the bead was serially washed by 200 μl of dioxane, water, MeOH, and water. The glycans blotted on beads were subjected to the trans-iminization reaction with aoWR (aminooxy-functionalized peptide reagent) for 45 min at 80 °C. WR-tagged glycans were eluted by adding 100 μl of water and then purified by a Mass PREPTM hydrophilic interaction chromatography (HILIC) μElution Plate (Waters) according to the manufacturer's description.
The purified N-glycans were 10-fold concentrated by SpeedVac followed by direct dissolution with 2,5-dihydroxylbenzoic acid (10 mg/ml in 30% ACN) and were crystallized. Then the analytes were subjected to MALDI-TOF-MS analysis using an Ultraflex time-of-flight mass spectrometer III (Bruker Daltonics, Billerica, MA) in reflector, positive ion mode typically summing 1000 shots. The detected N-glycan peaks in MALDI-TOF-MS spectra were picked using the software FlexAnalysis version 3 (Bruker Daltonics) in independently performed experiments in P19C6, P19CL6, and ESCs, respectively. The intensity of the isotopic peaks of each glycan was normalized to 15 pmol of internal standard (A2 amide glycan) in each status. The changes of glycan expression levels were calculated as differentiated versus undifferentiated. Student's t test was used to calculate the statistical difference of cell status, namely differentiated versus undifferentiated. The glycan structures were speculated using GlycoMod Tool and GlycoSuite.
RT-PCR
RNA was extracted from 1.5–5 × 107 P19CL6 cells using an “R&D Quick” kit (Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) according to the manufacturer's instructions. RT-PCR was performed using the SuperScriptIII One-Step RT-PCR System with Platinum Taq DNA polymerase (Invitrogen) to reverse transcribe and amplify cDNAs coding the proteins described below. The primer sequences were as follows: β-actin, 3′-TGTGATGGTGGGAATGGGTCGG-5′ and 5′-TTTGATGTCACGCACGATTTCC-3′; α-cardiac myosin heavy chain, 3′-CTGCTGGAGAGGTTATTCCTCG-5′ and 5′-GGAAGAGTGAGCGGCGCATCAAGG-3′; and β-cardiac myosin heavy chain, 3′-TGCAAAGGCTCCAGGTCTGAGGGC-5′ and 5′-GCCAACACCAACCTGTCCAAGTTC-3′).
Immunohistochemistry
Sixteen days after incubation with or without 1% DMSO, P19CL6 cells were collected as described above and dissociated intensively by EDTA/PBS containing 0.05% trypsin. After washing, cells were dissolved in PBS and smeared onto silane-coated slide glass (SUPERFROST®, Matsunami Glass, Osaka, Japan). Cells were fixed in cold acetone and treated with MeOH containing 3% H2O2 for 10 min at room temperature to quench internal peroxidase activity. Nonspecific binding on cells was blocked by incubation with CAS-Block (Zymed Laboratories Inc.) for 10 min at room temperature. Cells were then incubated with MF20 (1:800) (The monoclonal antibody developed by Donald A. Fischman was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA.), which is a sarcomeric myosin-specific monoclonal antibody, overnight at 4 °C. They were then washed by PBS and incubated with HRP-labeled secondary antibody (Zymed Laboratories Inc.) for 10 min at room temperature. After washing, diaminobenzidine solution was added, and the reaction was stopped by washing with water. In the case of P19C6, anti-mouse neurofilament 160 was used as the primary antibody, and the other part of the procedure was performed similarly.
RESULTS
Concept
Our strategy of a glycoblotting-based rapid and quantitative glycomics designed for whole cellular N-glycans/free oligosaccharides using BlotGlyco (BlotGlyco H) beads is diagrammed in Fig. 1. The specific steps of this optimized protocol involve (i) enzymatic release of entire N-glycans from cellular glycoprotein fractions, including both cell surface and endogenous glycoproteins, (ii) glycoblotting (chemoselective enrichment) by BlotGlyco beads, (iii) on-bead derivatization and labeling with a reagent to enhance MS sensitivity by trans-iminization, and (iv) subsequent quantitative mass spectrometry-based glycomics and typing subgroups of characteristic glycoforms, namely glycotypes, in the presence of an internal standard. The key difference when compared with other published approaches for cellular glycomics is specific chemistry-based enrichment of entire N-glycans by a commercially available high density hydrazide bead (BlotGlyco) that allows for high throughput and quantitative glycan profiling. Through on-bead chemical protection of carboxyl groups, both human sialic acid Neu5Ac and non-human sialic acid Neu5Gc are stabilized to prevent the significant cleavage at the sensitive O-glycoside linkage of sialosides during the high energy ionization process in mass spectrometry. Given that human ESCs and iPS cells currently being developed are produced by using animal-derived materials such as serum and feeder layers or even fractionated glycoproteins, it is important to address potential contamination by introduction of non-human sialic acid Neu5Gc into human stem cell lines proposed for therapeutic applications in humans (13, 21). The reaction conditions and all procedures are carefully optimized by using not only mouse embryonic cells but also various human cancer cell lines to maximize efficacy of N-glycan enrichment and reproducibility of data acquisition. To facilitate quantitative N-glycan profiling analysis, the expression level of individual N-glycans was normalized and represented by using a standardized unit (pmol/200 μg of cellular proteins). We note that the present protocol is readily feasible for any type of mammalian cells, and the efficiency of glycoblotting is not dependent on cell type when the required cell numbers (for example, confluence on a 6-cm dish; ∼5 × 106 cells) can be prepared. Rapid and quantitative analysis of major N-glycans (60–80 major glycoforms) enriched by the glycoblotting method provided us with reliable and satisfactory information for investigating structural changes in entire N-glycans and comparing the ratio of significant glycoforms by bar coding with characteristic subtypes, namely stage-specific embryonic glycotypes, during dynamic cell differentiation and proliferation.
Monitoring Entire N-Glycan Expression during Mouse P19 Cell Differentiations
Herein we selected the mouse P19 subclone to evaluate efficiency and versatility of our glycoblotting-based strategy. P19C6 cells are mouse embryonic carcinoma cells and serve as a common model for studying neuronal differentiation after RA inducement (32). P19 cells treated with lower level RA or DMSO differentiate into muscle (33), and P19CL6 cells, a well established subclone derived from P19 cells, efficiently differentiate into beating cardiomyocytes by treatment with 1% DMSO (27, 34).
Differentiation of P19CL6 Cells to Cardiomyocytes
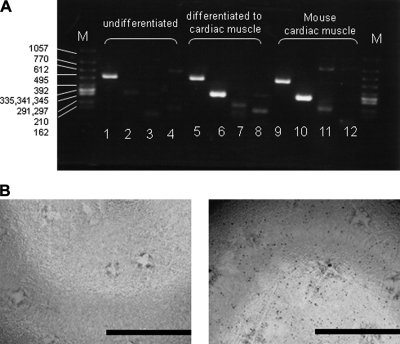
In the present study, we carefully characterized whole N-glycans of P19C6 cells with or without inducement into neural cells as well P19CL6 cells differentiated into cardiomyocytes. After 1% DMSO treatment, P19CL6 cells formed a monolayer at day 3 and multilayers on day 5 and started beating synchronously at day 16 of differentiation as reported (27), whereas control cells did not show any changes at the same period of culture. The differentiation into cardiac muscle was confirmed by conventional RT-PCR detecting mRNA coding cardiomyocyte-specific proteins and immunocytochemistry (Fig. 2, A and B). As shown in Fig. 2A, α- and β-cardiac myosin heavy chains, the markers of cardiomyocytes (35), were specifically detected in the DMSO-treated 16-day cultured P16CL6 cells and mouse cardiac muscles as a positive control. In addition, mouse monoclonal antibody MF20 reacted specifically with differentiated cardiomyocytes (Fig. 2B), indicating that differentiated cells readily express sarcomeric myosin.
Fig. 2.
Differentiation of P19CL6 cells to cardiomyocytes in presence of DMSO. A, RT-PCR analysis for the confirmation of cellular differentiation to cardiac muscle. M, DNA size marker (bp); lanes 1, 5, and 9, β-actin (503 bp), lanes 2, 6, and 10, α-cardiac myosin heavy chain (302 bp); lanes 3, 7, and 11, β-cardiac myosin heavy chain (205 bp); lanes 4, 8, and 12, embryonic skeletal muscle (151 bp). B, immunocytochemistry for the confirmation of P19CL6 cell differentiation to cardiac muscle. Primary antibody, MF20 (mouse monoclonal anti-sarcomere myosin); secondary antibody, HRP polymer-conjugated IgG; chromogenic substrate, diaminobenzidine. The bar represents 1.0 cm.
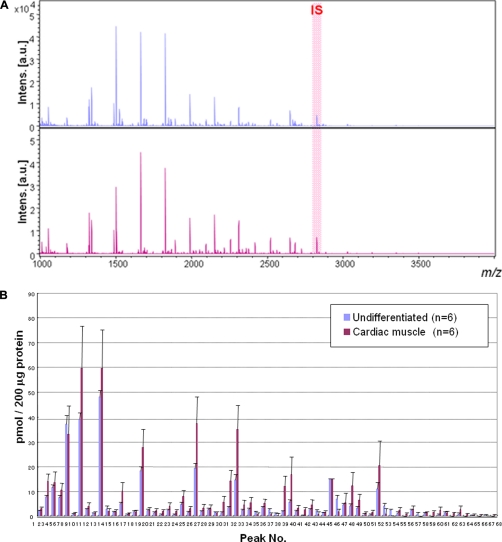
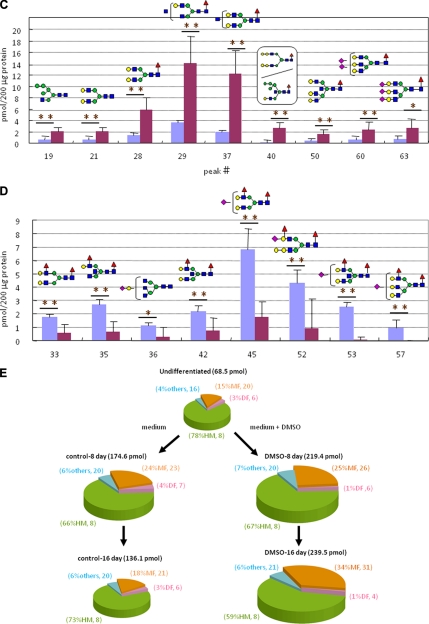
Whole N-glycans of P19CL6 cells (undifferentiated cells) and differentiated cells on day 18 (n = 6, six dishes each for both cells) were analyzed and identified for the first time by means of glycoblotting-based high throughput MALDI-TOF mass spectrometry (Fig. 3A). As summarized in Table I, 67 kinds of glycoforms were detected and quantified reproducibly in both cases. When the full portraits of N-glycan diversity of both cells were represented quantitatively (Fig. 3B), it seems likely that high mannose type N-glycans (peak numbers 1(M2), 3(M3), 5(M4), 7(M5), 10(M6), 16(M7), 24(M8), and 30(M9)) are major components throughout cardiomyocytic differentiation. However, it was also clearly suggested that expression levels of 26 N-glycans were significantly accompanied by cellular differentiation; 19 N-glycans, most of which are monofucosylated glycoforms, were increased concertedly, whereas seven difucosylated N-glycans decreased. Changes in two glycoforms (peak numbers 40 and 53) were especially significant (p < 0.001); their expression level was increased 16-fold (peak number 40) and decreased 30-fold (peak number 53) according to differentiation, respectively. Fig. 3, C and D, highlights dramatically changed N-glycan structures during cell differentiation; namely monofucosylated N-glycans were drastically increased, whereas difucosylated N-glycans decreased in comparison with the expression level of whole cellular N-glycans.
Fig. 3.
Large scale N-glycan analysis during P19CL6 cell differentiation. A, MALDI-TOF-MS of whole N-glycans of undifferentiated and differentiated cells. IS, internal standard. B, quantitative and total glycomics of undifferentiated and differentiated cells. C, magnification to visualize increased glycans (*, p < 0.01; **, p < 0.05). D, magnification to visualize decreased glycans (*, p < 0.01; **, p < 0.05). Error bars mean standard deviations. E, bar coding analysis. Intens., intensity; a.u., arbitrary units.
Table I. Glycoforms detected during P19CL6 cell differentiation.
Hex, hexose; dHex, deoxyhexose; HexNAc, N-acetylhexosamine.
| Peak no. CL6 | m/z | Composition |
|---|---|---|
| 1 | 1178.50 | Hex2(HexNAc)2 |
| 2 | 1324.55 | Hex2(HexNAc)2dHex1 |
| 3 | 1340.55 | Hex3(HexNAc)2 |
| 4 | 1486.61 | Hex3(HexNAc)2dHex1 |
| 5 | 1502.60 | Hex4(HexNAc)2 |
| 6 | 1543.63 | Hex3(HexNAc)3 |
| 7 | 1664.65 | Hex5(HexNAc)2 |
| 8 | 1689.69 | Hex3(HexNAc)3dHex1 |
| 9 | 1746.71 | Hex3(HexNAc)4 |
| 10 | 1826.71 | Hex6(HexNAc)2 |
| 11 | 1848.74 | Hex3(HexNAc)4(NeuAc)1 |
| 12 | 1851.74 | Hex4(HexNAc)3dHex1 |
| 13 | 1892.76 | Hex3(HexNAc)4dHex1 |
| 14 | 1908.76 | Hex4(HexNAc)4 |
| 15 | 1949.79 | Hex3(HexNAc)5 |
| 16 | 1988.76 | Hex7(HexNAc)2 |
| 17 | 2010.79 | Hex4(HexNAc)3(NeuAc)1 |
| 18 | 2013.79 | Hex5(HexNAc)3dHex1 |
| 19 | 2029.79 | Hex6(HexNAc)3(NeuAc)1 |
| 20 | 2054.82 | Hex4(HexNAc)4dHex1 |
| 21 | 2070.81 | Hex5(HexNAc)4 |
| 22 | 2095.84 | Hex3(HexNAc)5dHex1 |
| 23 | 2111.84 | Hex4(HexNAc)5 |
| 24 | 2150.81 | Hex8(HexNAc)2 |
| 25 | 2156.85 | Hex4(HexNAc)3dHex1(NeuAc)1 |
| 26 | 2172.84 | Hex5(HexNAc)3(NeuAc)1 |
| 27 | 2213.87 | Hex4(HexNAc)4(NeuAc)1 |
| 28 | 2216.87 | Hex5(HexNAc)4dHex1 |
| 29 | 2257.90 | Hex4(HexNAc)5dHex1 |
| 30 | 2312.86 | Hex9(HexNAc)2 |
| 31 | 2334.90 | Hex6(HexNAc)3(NeuAc)1 |
| 32 | 2359.93 | Hex4(HexNAc)4dHex1(NeuAc)1 |
| 33 | 2362.93 | Hex5(HexNAc)4dHex2 |
| 34 | 2375.92 | Hex5(HexNAc)4(NeuAc)1 |
| 35 | 2403.95 | Hex4(HexNAc)5dHex2 |
| 36 | 2416.95 | Hex4(HexNAc)6(NeuAc)1 |
| 37 | 2419.95 | Hex5(HexNAc)5dHex1 |
| 38 | 2521.98 | Hex5(HexNAc)4dHex1(NeuAc)1 |
| 39 | 2537.98 | Hex6(HexNAc)4(NeuAc)1 |
| 40 | 2540.98 | Hex7(HexNAc)5dHex1 |
| 41 | 2563.01 | Hex4(HexNAc)5dHex1(NeuAc)1 |
| 42 | 2566.01 | Hex5(HexNAc)5dHex2 |
| 43 | 2579.00 | Hex5(HexNAc)5(NeuAc)1 |
| 44 | 2651.33 | Internal standard |
| 45 | 2668.04 | Hex5(HexNAc)4dHex2(NeuAc)1 |
| 46 | 2681.03 | Hex5(HexNAc)4(NeuAc)2 |
| 47 | 2684.03 | Hex6(HexNAc)4dHex1(NeuAc)1 |
| 48 | 2725.06 | Hex5(HexNAc)5dHex1(NeuAc)1 |
| 49 | 2741.06 | Hex6(HexNAc)6(NeuAc)1 |
| 50 | 2785.08 | Hex6(HexNAc)6dHex1 |
| 51 | 2827.09 | Hex5(HexNAc)4dHex1(NeuAc)2 |
| 52 | 2830.09 | Hex6(HexNAc)4dHex2(NeuAc)1 |
| 53 | 2871.12 | Hex5(HexNAc)5dHex2(NeuAc)1 |
| 54 | 2887.11 | Hex6(HexNAc)5dHex1(NeuAc)1 |
| 55 | 2928.14 | Hex5(HexNAc)6dHex1(NeuAc)1 |
| 56 | 3030.17 | Hex5(HexNAc)5dHex1(NeuAc)2 |
| 57 | 3033.17 | Hex6(HexNAc)5dHex2(NeuAc)1 |
| 58 | 3046.17 | Hex6(HexNAc)5(NeuAc)2 |
| 59 | 3090.19 | Hex6(HexNAc)6dHex1(NeuAc)1 |
| 60 | 3192.22 | Hex6(HexNAc)5dHex1(NeuAc)2 |
| 61 | 3351.28 | Hex6(HexNAc)5(NeuAc)3 |
| 62 | 3395.30 | Hex6(HexNAc)6dHex1(NeuAc)2 |
| 63 | 3497.34 | Hex6(HexNAc)5dHex1(NeuAc)3 |
| 64 | 3557.36 | Hex7(HexNAc)6dHex1(NeuAc)2 |
| 65 | 3700.42 | Hex6(HexNAc)6dHex1(NeuAc)3 |
| 66 | 3862.47 | Hex7(HexNAc)6dHex1(NeuAc)3 |
| 67 | 4021.52 | Hex7(HexNAc)6(NeuAc)4 |
| 68 | 4167.58 | Hex7(HexNAc)6dHex1(NeuAc)4 |
The significance of the dramatic changes in the expression level of two distinct glycotypes uncovered by quantitative glycomics of entire cellular N-glycans was revealed much more simply by bar coding individual glycotypes such as high mannose type (HM), monofucosylated type (MF), difucosylated type (DF), and others (O) (Fig. 3E). It is clear that the total N-glycan expression level was significantly increased from undifferentiated (68.5 pmol/200 μg of protein) to the intermediate cells at day 8 (219.4 pmol/200 μg of protein) and differentiated cells at day 16 (239.5 pmol/200 μg of protein). It was revealed that the ratio of glycotype MF was increased from 15 to 34%, and at least 11 glycoforms could be assigned as newly generated N-glycans involved in the glycotype MF after differentiation, whereas the others, glycotype HM, glycotype DF, and glycotype O, showed no significant change both in the ratio and the number of glycoforms. When P19CL6 cells were subjected to culture continuously until day 16 without DMSO induction, there was no notable change in entire N-glycan expression during this period. Surprisingly, these cells exhibited N-glycan profiles quite similar to each other in terms of not only the ratio of the above four glycotypes but also the numbers of glycoforms identified in the individual glycotypes.
Differentiation of P19C6 Cells and ESCs to Neural Cells
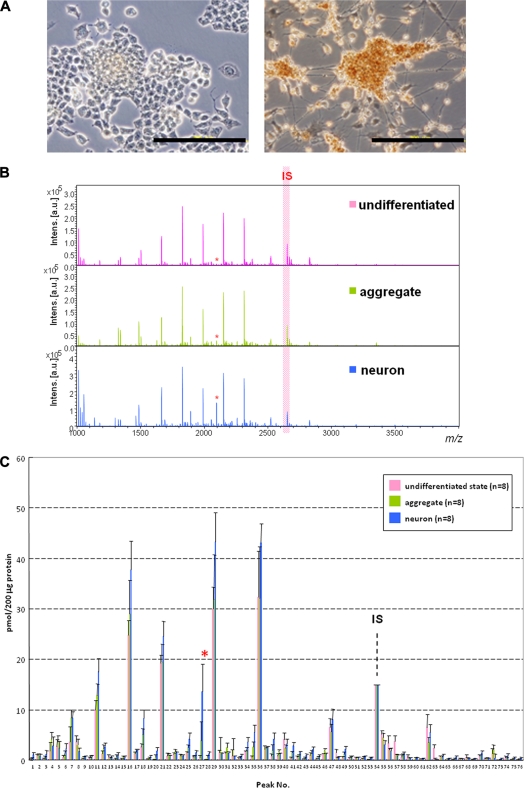
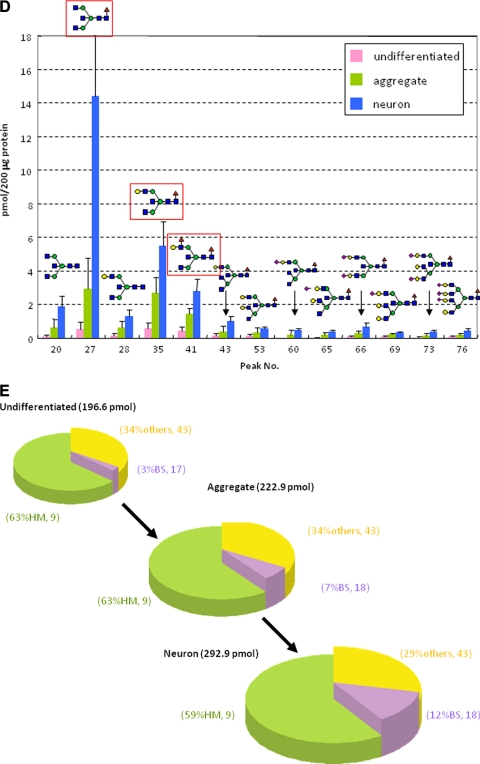
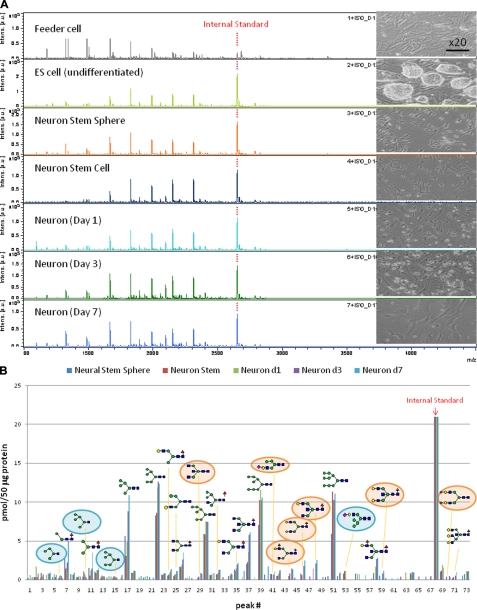
Versatility of the present concept and protocol was demonstrated by using P19C6 cells that differentiate into neural cells by RA inducement in which differentiation can be confirmed conventionally by immunocytochemistry using mouse monoclonal antibody (anti-neurofilament 160 monoclonal antibody) as shown in Fig. 4A. Fig. 4B shows MALDI-TOF mass spectra of whole N-glycans enriched by glycoblotting at three distinct stages observed during P19C6 cell differentiation to neural cells, namely undifferentiated, aggregate (day 4), and differentiated neural cells (day 9). A total of 75 N-glycans were identified; they are summarized in Table II and represented in a quantitative manner in Fig. 4C. In the case of P19C6 cell differentiation into neural cells, it was revealed that 12 glycoforms (peak numbers 20, 27, 28, 35, 41, 43, 53, 60, 65, 66, 69, and 73) were identified as bisect type (BS) N-glycans among 13 N-glycans that increased more than 5-fold after differentiation (Fig. 4D). The merit of bar coding analysis based on the characteristic subtypes is clear because profiling by focusing on glycotype BS clearly shows a drastic increase of this subtype (from 3 to 12%) compared with glycotype HM (from 63 to 59%) and glycotype O (from 34 to 29%) (Fig. 4E). On the contrary, l-fucose-focused bar coding analysis used in the case of the P19CL6/cardiomyocytes system did not show any meaningful N-glycans expression change during differentiation: glycotype HM, from 63 to 59%; glycotype DF, from 6 to 4%; glycotype MF, from 21 to 30%; and glycotype O, from 10 to 7%.
Fig. 4.
Differentiation of P19C6 cells to neural cells. A, immunocytochemistry for the confirmation of neural differentiation. Primary antibody, mouse monoclonal anti-neurofilament 160; secondary antibody, HRP polymer-conjugated IgG; chromogenic substrate, diaminobenzidine. The bar represents 200 μm. B, MALDI-TOF-MS of whole N-glycans of undifferentiated and differentiated cells. IS, internal standard. * represents peak number 27. C, quantitative and total glycomics of undifferentiated and differentiated cells. IS, internal standard. * represents peak number 27. D, magnification to visualize increased glycans. E, bar coding analysis.
Table II. Glycoforms detected during P19C6 cell differentiation.
Hex, hexose; dHex, deoxyhexose; HexNAc, N-acetylhexosamine.
| Peak no. | m/z | Composition |
|---|---|---|
| 1 | 1137.47 | Hex3(HexNAc)1 |
| 2 | 1178.50 | Hex2(HexNAc)2 |
| 3 | 1299.52 | Hex4(HexNAc)1 |
| 4 | 1324.55 | Hex2(HexNAc)2dHex1 |
| 5 | 1340.55 | Hex3(HexNAc)2 |
| 6 | 1461.57 | Hex5(HexNAc)1 |
| 7 | 1486.61 | Hex3(HexNAc)2dHex1 |
| 8 | 1502.60 | Hex4(HexNAc)2 |
| 9 | 1543.63 | Hex3(HexNAc)3 |
| 10 | 1623.63 | Hex6(HexNAc)1 |
| 11 | 1664.65 | Hex5(HexNAc)2 |
| 12 | 1689.69 | Hex3(HexNAc)3dHex1 |
| 13 | 1705.68 | Hex4(HexNAc)3 |
| 14 | 1746.71 | Hex3(HexNAc)4 |
| 15 | 1785.68 | Hex7(HexNAc)1 |
| 16 | 1826.71 | Hex6(HexNAc)2 |
| 17 | 1851.74 | Hex4(HexNAc)3dHex1 |
| 18 | 1892.76 | Hex3(HexNAc)4dHex1 |
| 19 | 1908.76 | Hex4(HexNAc)4 |
| 20 | 1949.79 | Hex3(HexNAc)5 |
| 21 | 1988.76 | Hex7(HexNAc)2 |
| 22 | 2010.79 | Hex4(HexNAc)3(NeuAc)1 |
| 23 | 2013.79 | Hex5(HexNAc)3dHex1 |
| 24 | 2029.79 | Hex6(HexNAc)3(NeuAc)1 |
| 25 | 2054.82 | Hex4(HexNAc)4dHex1 |
| 26 | 2070.81 | Hex5(HexNAc)4 |
| 27 | 2095.84 | Hex3(HexNAc)5dHex1 |
| 28 | 2111.84 | Hex4(HexNAc)5 |
| 29 | 2150.81 | Hex8(HexNAc)2 |
| 30 | 2172.84 | Hex5(HexNAc)3(NeuAc)1 |
| 31 | 2175.84 | Hex6(HexNAc)3dHex1 |
| 32 | 2200.88 | Hex4(HexNAc)4dHex2 |
| 33 | 2213.87 | Hex4(HexNAc)4(NeuAc)1 |
| 34 | 2216.87 | Hex5(HexNAc)4dHex1 |
| 35 | 2257.90 | Hex4(HexNAc)5dHex1 |
| 36 | 2312.86 | Hex9(HexNAc)2 |
| 37 | 2334.90 | Hex6(HexNAc)3(NeuAc)1 |
| 38 | 2359.93 | Hex4(HexNAc)4dHex1(NeuAc)1 |
| 39 | 2362.93 | Hex5(HexNAc)4dHex2 |
| 40 | 2375.92 | Hex5(HexNAc)4(NeuAc)1 |
| 41 | 2403.95 | Hex4(HexNAc)5dHex2 |
| 42 | 2419.95 | Hex5(HexNAc)5dHex1 |
| 43 | 2460.98 | Hex4(HexNAc)6dHex1 |
| 44 | 2474.92 | Hex10(HexNAc)2 |
| 45 | 2480.95 | Hex6(HexNAc)3dHex1(NeuAc)1 |
| 46 | 2508.99 | Hex5(HexNAc)4dHex3 |
| 47 | 2521.98 | Hex5(HexNAc)4dHex1(NeuAc)1 |
| 48 | 2537.98 | Hex6(HexNAc)4(NeuAc)1 |
| 49 | 2563.01 | Hex4(HexNAc)5dHex1(NeuAc)1 |
| 50 | 2566.01 | Hex5(HexNAc)5dHex2 |
| 51 | 2579.00 | Hex5(HexNAc)5(NeuAc)1 |
| 52 | 2582.00 | Hex6(HexNAc)5dHex1 |
| 53 | 2623.03 | Hex5(HexNAc)6dHex1 |
| 54 | 2651.33 | Internal standard |
| 55 | 2668.04 | Hex5(HexNAc)4dHex2(NeuAc)1 |
| 56 | 2681.03 | Hex5(HexNAc)4(NeuAc)2 |
| 57 | 2684.03 | Hex6(HexNAc)4dHex1(NeuAc)1 |
| 58 | 2725.06 | Hex5(HexNAc)5dHex1(NeuAc)1 |
| 59 | 2741.06 | Hex6(HexNAc)6(NeuAc)1 |
| 60 | 2766.09 | Hex4(HexNAc)6dHex1(NeuAc)1 |
| 61 | 2785.08 | Hex6(HexNAc)6dHex1 |
| 62 | 2827.09 | Hex5(HexNAc)4dHex1(NeuAc)2 |
| 63 | 2830.09 | Hex6(HexNAc)4dHex2(NeuAc)1 |
| 64 | 2887.11 | Hex6(HexNAc)5dHex1(NeuAc)1 |
| 65 | 2928.14 | Hex5(HexNAc)6dHex1(NeuAc)1 |
| 66 | 3030.17 | Hex5(HexNAc)5dHex1(NeuAc)2 |
| 67 | 3033.17 | Hex6(HexNAc)5dHex2(NeuAc)1 |
| 68 | 3046.17 | Hex6(HexNAc)5(NeuAc)2 |
| 69 | 3090.19 | Hex6(HexNAc)6dHex1(NeuAc)1 |
| 70 | 3192.22 | Hex6(HexNAc)5dHex1(NeuAc)2 |
| 71 | 3338.28 | Hex6(HexNAc)5dHex2(NeuAc)2 |
| 72 | 3351.28 | Hex6(HexNAc)5(NeuAc)3 |
| 73 | 3395.30 | Hex6(HexNAc)6dHex1(NeuAc)2 |
| 74 | 3497.34 | Hex6(HexNAc)5dHex1(NeuAc)3 |
| 75 | 3557.36 | Hex7(HexNAc)6dHex1(NeuAc)2 |
| 76 | 3862.47 | Hex7(HexNAc)6dHex1(NeuAc)3 |
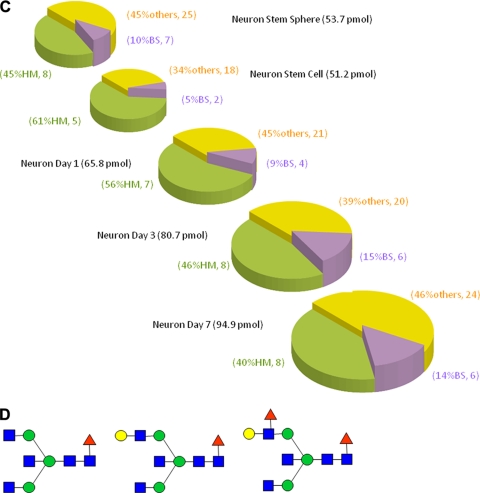
Our interest was next directed toward alteration of N-glycan expression of mouse ESC differentiation to neural cells. We considered that neural cells differentiated from mouse ESCs should also exhibit a structural alteration in the stage-specific glycotypes, an increase of glycotype BS, similar to those observed in P19C6 cell differentiation. Fig. 5, A and B, and Table III show the results of MALDI-TOF-MS of all typical cellular stages during mouse ESC differentiation into neural cells. As expected, bar coding analysis by three glycotypes used in the P19C6 cells demonstrated the significant increase of glycotype BS (10% at neural stem sphere or 5% at neural stem cell to 14% at neural cell day 7) in comparison with glycotype HM (45% at neural stem sphere or 61% at neural stem cell to 40%) and glycotype O (45% at neural stem sphere or 34% at neural stem cell to 46%) (Fig. 5C). During the differentiation from neural stem sphere into neural stem cells, it seems likely that an increase of glycotype HM (from 45 to 61%) and a decrease of glycotype BS (from 10 to 5%) occurred concurrently, although the reason is not clear. Surprisingly, it was revealed that neural cells differentiated from ESCs also exhibited up-regulated high level expression of the same three glycoforms (Fig. 5D; peak numbers 37, 48, and 58 in Fig. 5B) as those observed in P19C6 differentiation (peak numbers 27, 35, and 41 in Fig. 4D). We could not detect any Neu5Gc residue in whole N-glycans identified in the present study, and the expression levels of any N-glycans containing Neu5Ac residue(s) did not accompany P19 series cell differentiation. This means that bar coding analysis focusing on the expression level of Neu5Ac did not work for identifying stage-specific embryonic glycotypes in these cell lines. Our preliminary result (n = 3) indicates clearly the importance of the stage-specific embryonic glycotype BS as a new class of biomarkers for identifying and monitoring processes of mouse ESC differentiation into neural cells, although the effect of feeder cells and other various factors of individual culture conditions used on the ratio of these glycotypes must be examined carefully.
Fig. 5.
Differentiation of mouse ESCs to neural cells. A, MALDI-TOF-MS spectra during cell differentiation. B, quantitative glycan profiling during ESC differentiation. C, bar coding analysis. D, novel glycan biomarkers for identifying and monitoring the processes of mouse neural cell differentiation. Intens., intensity; a.u., arbitrary units; d, day.
Table III. Glycoforms detected during ESC differentiation.
Hex, hexose; dHex, deoxyhexose; HexNAc, N-acetylhexosamine.
| Peak no. ESC | m/z | Composition |
|---|---|---|
| 1 | 934.39 | Hex3 |
| 2 | 1096.44 | Hex4 |
| 3 | 1137.47 | Hex3(HexNAc)1 |
| 4 | 1178.50 | Hex2(HexNAc)2 |
| 5 | 1258.49 | Hex5 |
| 6 | 1299.52 | Hex4(HexNAc)1 |
| 7 | 1324.55 | Hex2(HexNAc)2dHex1 |
| 8 | 1340.55 | Hex3(HexNAc)2 |
| 9 | 1420.55 | Hex6 |
| 10 | 1461.57 | Hex5(HexNAc)1 |
| 11 | 1486.61 | Hex3(HexNAc)2dHex1 |
| 12 | 1502.60 | Hex4(HexNAc)2 |
| 13 | 1543.63 | Hex3(HexNAc)3 |
| 14 | 1582.60 | Hex7 |
| 15 | 1623.63 | Hex6(HexNAc)1 |
| 16 | 1645.66 | Hex2(HexNAc)2dHex1 |
| 17 | 1664.65 | Hex5(HexNAc)2 |
| 18 | 1689.69 | Hex3(HexNAc)3dHex1 |
| 19 | 1705.68 | Hex4(HexNAc)3 |
| 20 | 1746.71 | Hex3(HexNAc)4 |
| 21 | 1785.68 | Hex7(HexNAc)1 |
| 22 | 1826.71 | Hex6(HexNAc)2 |
| 23 | 1848.74 | Hex3(HexNAc)3(NeuAc)1 |
| 24 | 1851.74 | Hex4(HexNAc)3dHex1 |
| 25 | 1867.73 | Hex5(HexNAc)3 |
| 26 | 1892.76 | Hex3(HexNAc)4dHex1 |
| 27 | 1908.76 | Hex4(HexNAc)4 |
| 28 | 1947.73 | Hex8(HexNAc)1 |
| 29 | 1949.79 | Hex3(HexNAc)5 |
| 30 | 1988.76 | Hex7(HexNAc)2 |
| 31 | 2010.79 | Hex4(HexNAc)3(NeuAc)1 |
| 32 | 2013.79 | Hex5(HexNAc)3dHex1 |
| 33 | 2029.79 | Hex6(HexNAc)3 |
| 35 | 2054.82 | Hex4(HexNAc)4dHex1 |
| 36 | 2070.81 | Hex5(HexNAc)4 |
| 37 | 2095.84 | Hex3(HexNAc)5dHex1 |
| 38 | 2111.84 | Hex4(HexNAc)5 |
| 39 | 2150.81 | Hex8(HexNAc)2 |
| 40 | 2156.85 | Hex4(HexNAc)3dHex1(NeuAc)1 |
| 41 | 2172.84 | Hex5(HexNAc)3(NeuAc)1 |
| 42 | 2175.84 | Hex6(HexNAc)3dHex1 |
| 43 | 2191.84 | Hex7(HexNAc)3 |
| 44 | 2200.88 | Hex4(HexNAc)4dHex2 |
| 45 | 2213.87 | Hex4(HexNAc)4(NeuAc)1 |
| 46 | 2216.87 | Hex5(HexNAc)4dHex1 |
| 47 | 2232.92 | Hex6(HexNAc)4 |
| 48 | 2257.90 | Hex4(HexNAc)5dHex1 |
| 49 | 2273.90 | Hex5(HexNAc)5 |
| 50 | 2298.92 | Hex3(HexNAc)6dHex1 |
| 51 | 2312.86 | Hex9(HexNAc)2 |
| 52 | 2318.90 | Hex5(HexNAc)3dHex1(NeuAc)1 |
| 53 | 2334.90 | Hex6(HexNAc)3(NeuAc)1 |
| 54 | 2359.93 | Hex4(HexNAc)4dHex1(NeuAc)1 |
| 55 | 2375.92 | Hex5(HexNAc)4(NeuAc)1 |
| 56 | 2378.92 | Hex6(HexNAc)4dHex1 |
| 57 | 2394.92 | Hex7(HexNAc)4 |
| 58 | 2403.95 | Hex4(HexNAc)5dHex2 |
| 59 | 2419.95 | Hex5(HexNAc)5dHex1 |
| 60 | 2460.98 | Hex4(HexNAc)6dHex1 |
| 61 | 2474.92 | Hex10(HexNAc)2 |
| 62 | 2480.95 | Hex6(HexNAc)3dHex1(NeuAc)1 |
| 63 | 2508.99 | Hex5(HexNAc)4dHex3 |
| 64 | 2521.98 | Hex5(HexNAc)4dHex1(NeuAc)1 |
| 65 | 2537.98 | Hex6(HexNAc)4(NeuAc)1 |
| 66 | 2540.98 | Hex7(HexNAc)5dHex1 |
| 67 | 2566.01 | Hex5(HexNAc)5dHex2 |
| 68 | 2651.33 | Internal standard |
| 69 | 2668.04 | Hex5(HexNAc)4dHex2(NeuAc)1 |
| 70 | 2681.03 | Hex5(HexNAc)4(NeuAc)2 |
| 71 | 2684.03 | Hex6(HexNAc)4dHex1(NeuAc)1 |
| 72 | 2712.79 | Hex5(HexNAc)4(NeuGc)2 |
| 73 | 2827.09 | Hex5(HexNAc)4dHex1(NeuAc)2 |
DISCUSSION
For most mammalian cell types, it is not known which proteins are expressed at each cellular stage and how these protein expression patterns change quantitatively upon differentiation and proliferation. Flow cytometry and immunohistochemistry have been generally used for the identification of cell surface proteins such as cell differentiation markers. However, it is not currently possible to profile a global view of the cell surface protein landscape due to the limitation of feasible antibodies with validated specificity and affinity strength and the difficulty in the development of multiplexed assays for identifying sets of cell surface proteins in a single experiment.
Large scale proteomics analysis by two-dimensional gel electrophoresis MALDI-TOF-MS suggested that only 17 proteins (0.7% of total detected ∼2200 proteins) with different expression patterns may be involved in the DMSO-induced cardiac differentiation of P19CL6 cells (36). They also reported that real time PCR data showed discrepancies from that of proteomics in at least three kinds of proteins that reflected the importance of posttranslational modifications in expressed proteins. On the other hand, it was also reported that only 0.8% of total detected proteins (28 proteins of ∼3500 proteins) were increased or decreased during the 8-day differentiation of P19 cells to neural cells (34, 37). Consequently, it was concluded that changes in the expression level of detected proteins are not helpful for identifying or monitoring the processes of cellular differentiation of both P19 and P19C6 cells. Compared with the results of proteome-based analysis, the high potential of the glycome-based approach is clear because our results revealed for the first time that 28% of glycoforms (19 N-glycans of 67 total N-glycans; peak numbers 13, 20, 21, 22, 25, 28, 29, 32, 34, 37, 38, 39, 40, 41, 47, 48, 54, 60, and 63) were increased, and 10% (7 of 67) were decreased during P19CL6 cell differentiation to cardiomyocytes. Furthermore, the expression level of 31 N-glycans involved in the glycotype MF was up-regulated to 34% (78.2 pmol/200 μg of protein) against total detected N-glycan expression (239.5 pmol/200 μg of protein), whereas undifferentiated cells expressed only 15% glycotype MF (20 glycoforms). Interestingly, differentiated cardiomyocytes lost most Lewis X trisaccharide (SSEA-1) and sialyl-Lewis X tetrasaccharide moieties in major N-glycans as shown in Fig. 3C, whereas monofucosylation occurred specifically in the above 31 glycoforms at the GlcNAc residue involved in core chitobiose moiety after differentiation, suggesting that loss of cell adhesion through the interaction with selectins may be key to the differentiation of P19CL6 cells toward cardiomyocytes. It was also demonstrated that both P19C6 cells and ESCs showed quite similar drastic changes in the profiles of entire N-glycan expression during cell differentiation into neural cells in which the expression level of glycotype BS was up-regulated to 12 and 14% from 3 and 5%, respectively. Among N-glycans involved in the glycotype BS, three common bisect type glycoforms detected in neural cells were found to become new potential markers to identify and monitor the process of mouse neural cell differentiation. Given that the glycoforms corresponding to these three bisect type N- glycans have been known to exist in the mouse brain system (38, 39), this drastic up-regulation of the N-glycans involved in glycotype BS seems to be crucial for the differentiation to neural cells. Actually, GlcNAc-transferase III (Mgat3 gene), a glycosyltransferase responsible for the synthesis of bisecting GlcNAc linkage, appears to be normally expressed at high levels in mammalian brain and kidney tissues (40, 41). However, the relationship between threshold in the expression level of glycotype BS and mechanism in P19C6 cell differentiation remains unclear.
In the present study, we demonstrated the versatility of glycoblotting-based quantitative glycomics in the investigation of dynamic glycoform alteration during mammalian cell proliferation and differentiation. A full portrait of N-glycan expression at each cell stage allowed identification of the characteristic glycotypes showing drastic and concerted expression changes during cell differentiation, termed stage-specific embryonic glycotypes. The present results indicate the existence of a threshold in expression level of the characteristic glycotypes required for initiating individual cell differentiations, although functional roles, mechanism, and designated partner molecules remain unknown. A microarray displaying major glycotypes expressed at individual cell stages should be a useful tool to elicit candidate partner molecules as well as the adhesion mechanism of the differentiated cells. However, we consider that most glycoproteins at the specific cell stage might share some biosynthetic pathways at the Golgi, resulting in the dynamic and large scale expression changes of glycotypes during cell differentiation. We should set a goal to compare and accumulate a database of whole N-glycan expression levels of feasible human ESC and iPS cell lines established by different laboratories and to make these resources readily available to the scientific community as soon as possible.
* This work was supported in part by a grant for “Innovative program for future drug discovery and medical care” from the Japan Science and Technology Agency and the Ministry of Education, Culture, Science, Sports, and Technology of Japan.
1 The abbreviations used are:
- ESC
- embryonic stem cell
- iPS
- induced pluripotent stem
- FBS
- fetal bovine serum
- RA
- retinoic acid
- HRP
- horseradish peroxidase
- NeuGc
- N-glycolylneuraminic acid
- HM
- high mannose type
- MF
- monofucosylated type
- DF
- difucosylated type
- O
- others
- BS
- bisect type.
REFERENCES
- 1.Stanton L. W., Bakre M. M. (2007) Genomic and proteomic characterization of embryonic stem cells. Curr. Opin. Chem. Biol 11, 399–404 [DOI] [PubMed] [Google Scholar]
- 2.Kuramitsu Y., Nakamura K. (2006) Proteomic analysis of cancer tissues: shedding light on carcinogenesis and possible biomarkers. Proteomics 6, 5650–5661 [DOI] [PubMed] [Google Scholar]
- 3.Gonnet F., Bouazza B., Millot G. A., Ziaei S., Garcia L., Butler-Browne G. S., Mouly V., Tortajada J., Danos O., Svinartchouk F. (2008) Proteome analysis of differentiating human myoblasts by dialysis-assisted two-dimensional gel electrophoresis (DAGE). Proteomics 8, 264–278 [DOI] [PubMed] [Google Scholar]
- 4.Watkins J., Basu S., Bogenhagen D. F. (2008) A quantitative proteomic analysis of mitochondrial participation in p19 cell neuronal differentiation. J. Proteome Res 7, 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loring J. F., Rao M. S. (2006) Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells 24, 145–150 [DOI] [PubMed] [Google Scholar]
- 6.Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haltiwanger R. S., Lowe J. B. (2004) Role of glycosylation in development. Annu. Rev. Biochem 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 8.Guérardel Y., Chang L. Y., Maes E., Huang C. J., Khoo K. H. (2006) Glycomic survey mapping of zebrafish identifies unique sialylation pattern. Glycobiology 16, 244–257 [DOI] [PubMed] [Google Scholar]
- 9.Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 10.Solter D., Knowles B. B. (1978) Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc. Natl. Acad. Sci. U.S.A 75, 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannagi R., Cochran N. A., Ishigami F., Hakomori S., Andrews P. W., Knowles B. B., Solter D. (1983) Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J 2, 2355–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 13.Lanctot P. M., Gage F. H., Varki A. P. (2007) The glycans of stem cells. Curr. Opin. Chem. Biol 11, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surani M. A. (1979) Glycoprotein synthesis and inhibition of glycosylation by tunicamycin in preimplantation mouse embryos: compaction and trophoblast adhesion. Cell 18, 217–227 [DOI] [PubMed] [Google Scholar]
- 15.Akama T. O., Nakagawa H., Sugihara K., Narisawa S., Ohyama C., Nishimura S., O'Brien D. A., Moremen K. W., Millan J. L., Fukuda M. N. (2002) Germ cell survival through carbohydrate-mediated interaction with Sertoli cells. Science 295, 124–127 [DOI] [PubMed] [Google Scholar]
- 16.Shur B. D., Rodeheffer C., Ensslin M. A. (2004) Mammalian fertilization. Curr. Biol 14, R691–R692 [DOI] [PubMed] [Google Scholar]
- 17.Hato M., Nakagawa H., Kurogochi M., Akama T. O., Marth J. D., Fukuda M. N., Nishimura S. I. (2006) Unusual N-glycan structures in alpha-mannosidase II/IIx double null embryos identified by a systematic glycomics approach based on two-dimensional LC mapping and matrix-dependent selective fragmentation method in MALDI-TOF/TOF mass spectrometry. Mol. Cell. Proteomics 5, 2146–2157 [DOI] [PubMed] [Google Scholar]
- 18.Eggens I., Fenderson B., Toyokuni T., Dean B., Stroud M., Hakomori S. (1989) Specific interaction between Lex and Lex determinants. J. Biol. Chem 264, 9476–9484 [PubMed] [Google Scholar]
- 19.Muramatsu T., Muramatsu H. (2004) Carbohydrate antigens expressed on stem cells and early embryonic cells. Glycoconj. J 21, 41–45 [DOI] [PubMed] [Google Scholar]
- 20.Varki A. (2007) Glycan-based interactions involving vertebrate sialic acid-recognizing proteins. Nature 446, 1023–1029 [DOI] [PubMed] [Google Scholar]
- 21.Martin M. J., Muotri A., Gage F., Varki A. P. (2005) Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat. Med 11, 228–232 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 23.Sakurada K., McDonald F. M., Shimada F. (2008) Regenerative medicine and stem cell based drug discovery. Angew. Chem. Int. Ed. Engl 47, 5718–5738 [DOI] [PubMed] [Google Scholar]
- 24.Pilobello K. T., Mahal L. K. (2007) Deciphering the glycocode: the complexity and analytical challenge of glycomics. Curr. Opin. Chem. Biol 11, 300–305 [DOI] [PubMed] [Google Scholar]
- 25.Nishimura S., Niikura K., Kurogochi M., Matsushita T., Fumoto M., Hinou H., Kamitani R., Nakagawa H., Deguchi K., Miura N., Monde K., Kondo H. (2004) High-throughput protein glycomics: combined use of chemoselective glycoblotting and MALDI-TOF/TOF mass spectrometry. Angew. Chem. Int. Ed. Engl 44, 91–96 [DOI] [PubMed] [Google Scholar]
- 26.Furukawa J., Shinohara Y., Kuramoto H., Miura Y., Shimaoka H., Kurogochi M., Nakano M., Nishimura S. I. (2008) Comprehensive approach to structural and functional glycomics based on chemoselective glycoblotting and sequential tag conversion. Anal. Chem 80, 1094–1101 [DOI] [PubMed] [Google Scholar]
- 27.Edwards M. K., Harris J. F., McBurney M. W. (1983) Induced muscle differentiation in an embryonal carcinoma cell line. Mol. Cell. Biol 3, 2280–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley P. (2002) Biological consequences of overexpressing or eliminating N-acetylglucosaminyltransferase-TIII in mouse. Biochim. Biophys. Acta 1573, 363–368 [DOI] [PubMed] [Google Scholar]
- 29.Gao X., Bian W., Yang J., Tang K., Kitani H., Atsumi T., Jing N. A. (2001) role of N-cadherin in neuronal differentiation of embryonic carcinoma P19 cells. Biochem. Biophys. Res. Commun 284, 1098–1103 [DOI] [PubMed] [Google Scholar]
- 30.Tang K., Yang J., Gao X., Wang C., Liu L., Kitani H., Atsumi T., Jing N. (2002) Wnt-1 promotes neuronal differentiation and inhibits gliogenesis in P19 cells. Biochem. Biophys. Res. Commun 293, 167–173 [DOI] [PubMed] [Google Scholar]
- 31.Yagi T., Tokunaga T., Furuta Y., Nada S., Yoshida M., Tsukada T., Saga Y., Takeda N., Ikawa Y., Aizawa S. (1993) A novel ES cell line, TT2, with high germ line-differentiating potency. Anal. Biochem 214, 70–76 [DOI] [PubMed] [Google Scholar]
- 32.Miura Y., Hato M., Shinohara Y., Kuramoto H., Furukawa J., Kurogochi M., Shimaoka H., Tada M., Nakanishi K., Ozaki M., Todo S., Nishimura S. I. (2008) BlotGlycoABCTM, an integrated glycoblotting technique for rapid and large scale clinical glycomics. Mol. Cell. Proteomics 7, 370–377 [DOI] [PubMed] [Google Scholar]
- 33.McBurney M. W., Jones-Villeneuve E. M., Edwards M. K., Anderson P. J. (1982) Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature 299, 165–167 [DOI] [PubMed] [Google Scholar]
- 34.Habara-Ohkubo A. (1996) Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct. Funct 21, 101–110 [DOI] [PubMed] [Google Scholar]
- 35.Inberg A., Bogoch Y., Bledi Y., Linial M. (2007) Cellular processes underlying maturation of P19 neurons: changes in protein folding regimen and cytoskeleton organization. Proteomics 7, 910–920 [DOI] [PubMed] [Google Scholar]
- 36.Baharvand H., Piryaei A., Rohani R., Taei A., Heidari M. H., Hosseini A. (2006) Ultrastructural comparison of developing mouse embryonic stem cell- and in vivo-derived cardiomyocytes. Cell Biol. Int 30, 800–807 [DOI] [PubMed] [Google Scholar]
- 37.Wen J., Xia Q., Lu C., Yin L., Hu J., Gong Y., Yin B., Monzen K., Yuan J., Qiang B., Zhang X., Peng X. (2007) Proteomic analysis of cardiomyocytes differentiation in mouse embryonic carcinoma P19CL6 cells. J. Cell. Biochem 102, 149–160 [DOI] [PubMed] [Google Scholar]
- 38.An J., Yuan Q., Wang C., Liu L., Tang K., Tian H. Y., Jing N. H., Zhao F. K. (2005) Differential display of proteins involved in the neural differentiation of mouse embryonic carcinoma P19 cells by comparative proteomic analysis. Proteomics 5, 1656–1668 [DOI] [PubMed] [Google Scholar]
- 39.Zamze S., Harvey D. J., Pesheva P., Mattu T. S., Schachner M., Dwek R. A., Wing D. R. (1999) Glycosylation of a CNS-specific extracellular matrix glycoprotein, tenascin-R, is dominated by O-linked sialylated glycans and “brain-type” neutral N-glycans. Glycobiology 9, 823–831 [DOI] [PubMed] [Google Scholar]
- 40.Shimizu H., Ochiai K., Ikenaka K., Mikoshiba K., Hase S. (1993) Structures of N-linked sugar chains expressed mainly in mouse brain. J. Biochem 114, 334–338 [DOI] [PubMed] [Google Scholar]
- 41.Priatel J. J., Sarkar M., Schachter H., Marth J. D. (1997) Isolation, characterization and inactivation of the mouse Mgat3 gene: the bisecting N-acetylglucosamine in asparagines-linked oligosaccharides appears dispensable for viability and reproduction. Glycobiology 7, 45–56 [DOI] [PubMed] [Google Scholar]