Abstract
The search for biomarkers to diagnose psychiatric disorders such as schizophrenia has been underway for decades. Many molecular profiling studies in this field have focused on identifying individual marker signals that show significant differences in expression between patients and the normal population. However, signals for multiple analyte combinations that exhibit patterned behaviors have been less exploited. Here, we present a novel approach for identifying biomarkers of schizophrenia using expression of serum analytes from first onset, drug-naïve patients and normal controls. The strength of patterned signals was amplified by analyzing data in reproducing kernel spaces. This resulted in the identification of small sets of analytes referred to as targeted clusters that have discriminative power specifically for schizophrenia in both human and rat models. These clusters were associated with specific molecular signaling pathways and less strongly related to other neuropsychiatric disorders such as major depressive disorder and bipolar disorder. These results shed new light concerning how complex neuropsychiatric diseases behave at the pathway level and demonstrate the power of this approach in identification of disease-specific biomarkers and potential novel therapeutic strategies.
Schizophrenia is a debilitating neuropsychiatric disorder that affects more than 1% of the world population and costs hundreds of billions of United States dollars in healthcare provision and lost earnings (1). The diagnosis of this disease has not changed substantially over several decades and currently relies on subjective psychopathological ratings such as the Diagnostic and Statistical Manual of Mental Disorders (DSM)1-IV. Thus, diagnosis can be complicated by the presence of overlapping symptoms frequently occurring in other psychiatric illnesses such as bipolar disorder (BD) and major depressive disorder (MDD) and by the presence of confounding factors such as drug abuse and co-morbidities. This often results in diagnosis being delayed for several months to years. A delay in establishing an accurate diagnosis can have serious deleterious implications because a late or imprecise diagnosis can contribute to unsatisfactory outcomes to currently used drug therapies and to higher rates of relapse (2). Most importantly, more than half of schizophrenia subjects develop a progressive course of the disease associated with deficit symptoms (3).
In contrast, early therapeutic intervention holds promise in preventing or diminishing such effects (4–6). An empirical assay for early and accurate diagnosis of schizophrenia would deliver improved patient outcomes and reduce the costs of the disease for healthcare services and society (7–9). Such an assay could also provide a means of stratifying patients and monitoring drug responses and may also lead to the development of translational medicine tools that are critical for discovery of novel therapeutic strategies. Molecular profiling methods that afford the simultaneous measurement of multiple analytes in clinical and preclinical samples have considerable promise in this endeavor. These methods have been aimed predominantly at identifying individual molecules that show differences in expression between the disease and control conditions. However, such studies have often been fraught with small fold-changes in analyte levels, a common obstacle when investigating complex neuropsychiatric disorders (10–12). Thus, standard statistical techniques such as t tests will not be able to explore patterned behaviors involving proteins that have subtle expression changes but still contribute to the development of schizophrenia.
The main objective of this study was to determine whether unique patterns of biomarkers can be identified for subjects with first onset antipsychotic-naïve schizophrenia. Analyte expression lists were generated using the Multi-Analyte Profiling (MAP®) fluorescent bead-based technology for profiling serum samples from 77 male schizophrenia patients and 66 matched male controls. For comparison with other psychiatric disorders, we also analyzed the serum samples of 13 male BD and 17 male MDD patients. In parallel, serum samples from four relevant animal models were also profiled for comparison with the human disease state. Analysis of the respective expression lists was carried out using non-linear statistical analysis, which identifies small sets of analytes called targeted analyte clusters (TACs) that have the power to discriminate the patients from normal controls. We present here the performance of these clusters for diagnosis of schizophrenia. In addition, we show how this method can also contribute to increasing our understanding of the etiology of the disorder by determining its ability to classify various preclinical models of psychiatric disorders. The biological pathways associated with these clusters are discussed with their relevance to schizophrenia.
MATERIALS AND METHODS
Study Participants
Subjects were recruited with first onset schizophrenia (SZ; center 1: male, n = 42; female, n = 29; center 2: male, n = 35; female, n = 11), BD (male, n = 12; female, n = 19), and MDD (male, n = 10; female, n = 19) along with matching controls (SZ center 1: male, n = 31; SZ center 1: female, n = 28; SZ center 2: male, n = 35; SZ center 2: female, n = 11; BD male, n = 31; BD female, n = 28; MDD male, n = 44; MDD female, n = 44) (Table I). Only male subjects were chosen for the testing set to facilitate comparisons with animal model studies. Schizophrenia subjects were obtained from two clinical centers, namely the Department of Psychiatry and Psychotherapy, University of Cologne, Cologne, Germany (center 1) and the Department of Psychiatry, University Medical Faculty, Münster, Germany (center 2). The TAC was identified by the expression data of male subjects in center 1, whereas the rest of the data were used as test sets. In this way, we can evaluate the performance of the identified TAC in terms of identifying schizophrenia patients across gender and different centers. Also, the performance of the TAC in human models can be compared directly with animal models that include only male rats (see details under “Preclinical Models”). Schizophrenia was diagnosed using the Structured Clinical Interview for DSM-IV, and all subjects were identified as the paranoid subtype (classification 295.30). Type I and type II male euthymic BD patients (classifications 296.4 and 296.89, respectively) and male acute MDD subjects were diagnosed using DSM-IV criteria. Euthymic BD patients were selected as they can experience subjective cognitive deficit symptoms similar to those in schizophrenia (13, 14). These subjects had an average duration of illness of 9.9 ± 8.6 years, had an average of 10 episodes, and had received one or a combination of mood stabilizers and/or antidepressants. MDD patients were selected because of the conceptual overlap between depression and the negative symptoms of schizophrenia (15, 16). The MDD subjects had an average duration of illness of 14.0 ± 12.0 years.
Table I. Demographic details for clinical study.
| Center 1 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Schizophrenia |
Bipolar disorder |
|||||||
| Healthy controls |
Patients |
Healthy controls |
Patients |
|||||
| Male | Female | Male | Female | Male | Female | Male | Female | |
| Number | 31 | 28 | 42 | 29 | 31 | 28 | 12 | 19 |
| Age (years)a | 30.2 ± 7.8 | 29.9 ± 6.8 | 30.9 ± 10.1 | 33.3 ± 10.9 | 30.3 ± 8.8 | 29.9 ± 6.8 | 33.7 ± 12.7 | 33.7 ± 9.2 |
| BMI (kg/m2)a | 23.2 ± 3.6 | 22.1 ± 3.5 | 23.7 ± 4.9 | 23.0 ± 4.4 | 24.3 ± 3.4 | 22.1 ± 3.5 | 25.1 ± 2.9 | 24.3 ± 4.3 |
| Center 2 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Schizophrenia |
Major depressive disorderb |
|||||||
| Healthy controls |
Patients |
Healthy controls |
Patients |
|||||
| Male | Female | Male | Female | Male | Female | Male | Female | |
| Number | 35 | 11 | 35 | 11 | 44 | 44 | 10 | 19 |
| Age (years)a | 27.3 ± 9.3 | 23.6 ± 7.3 | 27.3 ± 9.4 | 23.6 ± 6.9 | 34.1 ± 14.7 | 39.7 ± 11.6 | 41.2 ± 12.4 | 42.8 ± 14.9 |
a Values are shown as mean ± S.D. Smoking, cannabis consumption, and date of sample collection are not shown. BMI data were absent for center 2.
b The patients were unmedicated and were not treated with electroconvulsive therapy.
Clinical tests were performed by psychiatrists under good clinical practice compliance, and the studies were approved by the appropriate ethics committees. Written informed consent was given by all participants, and clinical investigations were conducted according to the Declaration of Helsinki. Any patients whose clinical diagnosis required later revision were not used in the studies. Control subjects were matched to the respective patient populations based on social demographics. Those with a family history of mental disease or other medical conditions such as type II diabetes, hypertension, or cardiovascular or autoimmune diseases were not used. Along with matching for base-line characteristics of age, gender, BMI, smoking, cannabis consumption, and date of sample collection, patients and controls were matched for social status, lifestyle, and education level and recruited from the same geographic area surrounding the clinic. In addition, none of the subjects were taking any additional substances. Genetic stratification is not likely to be an important factor as the complexity of schizophrenia suggests that multiple genes contribute to the onset and manifestation of the disease.
Serum Samples
Blood was collected from all subjects between 8:00 and 12:00 (non-fasting) immediately after clinical diagnosis into S-Monovette 7.5-ml serum tubes (Sarstedt, Numbrecht, Germany). These were left at room temperature for 2 h to allow for blood coagulation and then centrifuged at 4000 × g for 5 min. The resulting supernatants were stored at −80 °C in Low Binding microcentrifuge tubes (Eppendorf, Hamburg, Germany).
Preclinical Models
Only male rats were used for these studies to avoid potential confounding factors due to hormonal fluctuations associated with females. Rats were maintained on a 12-h light/dark cycle (lights on from 06:00 to 18:00) under constant temperature (21 ± 1 °C) and humidity (50–58%). Food (Harlan Tekland 2014, Harlan UK Ltd., Bicester, UK) and water was available ad libitum.
For the phencyclidine (PCP) administration studies, rats were acclimated 7 days prior to experiments. PCP hydrochloride (Sigma-Aldrich) was dissolved in saline and administered subcutaneously into the flank. Control animals received saline by the same method. The hyperlocomotory effect of PCP administration was confirmed by measuring the number of infrared beam breaks performed by each rat using a Double IR Actimeter Harvard system (such as described in Ref. 17; Harvard Apparatus, Kent, UK). Two PCP models were investigated (below).
An acute PCP administration model was tested using 14-week-old Sprague-Dawley rats (Charles River). Eight rats were treated with PCP hydrochloride (5.0 mg/kg, intraperitoneal), eight were treated with vehicle, and these were killed via decapitation after 2 h.
We also analyzed a chronic PCP administration model using 12-week-old Sprague-Dawley rats. Eight rats were treated with PCP hydrochloride, and eight were treated with vehicle as above once a day for 15 days. Two hours after the last injection, rats were killed via decapitation.
A social isolation model was also analyzed using 16-week-old Lister Hooded rats (Charles River). Rats were fostered at birth, eight rats were housed singly, and eight were housed in groups of five (other littermates also included) on postnatal day 28. The efficacy of the model was confirmed as the isolated animals showed a significant reduction (p < 0.01) in prepulse inhibition of acoustic startle compared with their group-housed littermates as reported previously (18). Isolated and grouped animals were housed in the same holding room for 12 weeks (44). After this time, the rats were killed via decapitation.
We also tested a model using offspring rats from dams fed a low protein diet during pregnancy and lactation (19). Virgin Wistar rat dams were fed an isocalorific diet containing 8% protein throughout pregnancy and lactation. The offspring were delivered spontaneously, weaned onto a 20% protein diet on postnatal day 21, and killed at week 14 via decapitation. The behavioral effects on this model showing a reduced prepulse inhibition of acoustic startle have been reported previously (20). We demonstrated efficacy of the maternal protein restriction protocol by confirming significantly lower body and liver weights and an increased brain to body weight ratio in the low protein offspring rats compared with the control rats (data not shown). Eight low protein and eight control rats (dams fed standard 20% protein) were used.
For all four rat models, trunk blood was collected into BD Biosciences serum tubes, and serum was prepared as above for the clinical studies. All studies were conducted in compliance with the Home Office Guidance on the operation of the UK Animals (Scientific Procedures) Act 1986 and were approved by the GlaxoSmithKline Animal Procedures Review Panel.
Multianalyte Profiling
Analytes were measured in 25–50-μl serum samples using multiplexed immunoassays in a Clinical Laboratory Improved Amendments-certified laboratory at Rules-Based Medicine (Austin, TX) (21). Assays were calibrated using standards, and raw intensity measurements were converted to protein concentrations using proprietary software. Analyses were conducted under blinded conditions with respect to sample identities, and samples were analyzed randomly to avoid any sequential biases. The Human Metabolic MAP comprised adiponectin, ACTH, angiotensin-converting enzyme, angiotensinogen, C3 des-Arg, cortisol, follicle-stimulating hormone, galanin, glucagon, glucagon-like peptide-1 (GLP-1), growth hormone, insulin, IGF-1, leptin, luteinizing hormone (LH), pancreatic polypeptide, peptide YY, progesterone, prolactin, resistin, secretin, and testosterone. The Rat Metabolic MAP was comprised of adiponectin, ACTH, angiotensin-converting enzyme, angiotensinogen, C3a des-Arg, cortisol, galanin, glucagon, growth hormone, insulin, IGF-1, leptin, LH, peptide YY, plasminogen activator inhibitor 1, progesterone, prolactin, resistin, secretin, and testosterone. The multiplexed immunoassays are described in Ref. 21. Analytes were quantified by reference to eight-point calibration curves, and machine performance was verified using three quality control (QC) samples for each analyte. QC samples were distributed across the dynamic range of the assay at low, medium, and high levels and had coefficients of variance below 15%. Calibration standards and QC samples were in a complex serum-based matrix to match the sample background and were analyzed in duplicate. Assays were calibrated using standards, and raw intensity measurements were converted to protein concentrations using proprietary software. Analyses were conducted under blinded conditions with respect to sample identities, and samples were analyzed randomly to avoid any sequential biases. The Rules-Based Medicine metabolic assay panel was chosen as many of the constituent analytes have been associated previously with schizophrenia. The Human Metabolic MAP comprised adiponectin, ACTH, angiotensin-converting enzyme, angiotensinogen, C3 des-Arg, cortisol, follicle-stimulating hormone, galanin, glucagon, GLP-1, growth hormone, insulin, IGF-1, leptin, LH, pancreatic polypeptide, peptide YY, progesterone, prolactin, resistin, secretin, and testosterone. The Rat Metabolic MAP comprised adiponectin, ACTH, angiotensin-converting enzyme, angiotensinogen, C3a des-Arg, cortisol, galanin, glucagon, growth hormone, insulin, IGF-1, leptin, LH, peptide YY, plasminogen activator inhibitor 1, progesterone, prolactin, resistin, secretin, and testosterone. To carry out systematic comparisons between human and experimental models (rat), we chose to focus on the 17 analytes (Table II) that are common between the human and rat panels.
Table II. Analytes showing statistically significant differences in expression between male schizophrenia and control subjects.
The expression changes were checked by t test where p < 0.05 is significant. Statistically significant values are shown in bold font. FC, -fold change.
| Analyte | Center 1 |
Center 2 |
||
|---|---|---|---|---|
| p value | FC | p value | FC | |
| Adiponectin | 0.41 | 1.08 | 0.27 | 1.10 |
| Angiotensin-converting enzyme | 0.33 | 0.91 | 0.95 | 1.00 |
| Angiotensinogen | 0.20 | 0.80 | 0.79 | 1.22 |
| Cortisol | <0.01 | 1.19 | <0.01 | 1.55 |
| Glucagon-like peptide-1a | 0.44 | 0.77 | <0.01 | 0.32 |
| Growth hormone | 0.01 | 1.76 | 0.83 | 0.46 |
| Insulin | 0.17 | 0.81 | 0.43 | 1.15 |
| Insulin like growth factor-1 | 0.15 | 0.79 | 0.07 | 0.71 |
| Leptin | 0.03 | 0.62 | 0.40 | 0.81 |
| Luteinizing hormone | 0.01 | 0.86 | 0.16 | 0.89 |
| Peptide YYa | 0.24 | 0.52 | 0.43 | 0.55 |
| Plasminogen activator inhibitor 1 | 0.98 | 1.00 | <0.01 | 1.04 |
| Progesterone | 0.03 | 0.87 | 0.03 | 1.27 |
| Prolactin | 0.96 | 1.09 | 0.16 | 0.79 |
| Resistin | 0.03 | 0.84 | 0.04 | 0.88 |
| Secretina | 0.45 | >10 | 0.94 | 1.04 |
| Testosterone | 0.95 | 0.99 | 0.01 | 1.22 |
a Proteins with many expression data equal to zero.
Factor Analysis and Feature Selection
Because of the potentially large number of analyte combinations involved in the analysis, we first sought to reduce the possibilities by looking for those that give maximal explanation power. For this purpose, we used factor analysis, which takes covariance and noise into account as a means of reducing the multidimensional data to fewer factors. Factor analysis (FA) is a statistical technique to reduce multidimensional data down to a few factors by considering their variances and noise. Formally, FA models the original data X as its mean behavior plus a linear combination (captured by matrix L) of a number of factors (matrix F) and noise, i.e. X = μ + LF + ε where Cov(F) = I, Cov(ε) = diag(ψ1, ψ2, … ), and F and ε are independent. Similar to principal component analysis (more details are given in the next section), FA seeks to decompose the data into factors that are linear combinations of the data attributes. However, FA takes into account the fact that noises incurred in different factors may have difference variances. Therefore, we used it to dissect the importance of proteins in terms of representing the overall variance of the diseased and control data.
Of the common 17 analytes measured in both the human and rat serum samples, FA indicated eight proteins exhibiting dominating explanatory power to the data. The next stage was to perform feature selection by considering all combinations of these eight proteins to identify a subset that gives the best classification results across the data sets.
Data Transformation
Principal component analysis (PCA) is a standard multivariate statistical technique for characterizing data variances. PCA recasts multidimensional data onto a new coordinate system such that the first eigenvector gives a projection of the original data displaying maximum variance, the second gives the second maximum, and so on. However, as standard PCA is linear in nature, these projections do not always yield meaningful results for classification purposes. For example, the data can form a quadratic line that is not adaptable to linear classifiers.
One possible solution is to introduce new dimensions, e.g. some non-linear combinations of the original features, to the data so that the data set may become linearly separable again. However, generating these new dimensions can add to the computational workload, and increasing dimensionality makes the calculation of eigenvalue decompositions computationally expensive. Kernel methods are one of the prominent approaches to tackle these problems. In theory, these methods analyze the data in an extended (potentially very high dimensional) feature space F that exhibits non-linear properties. F is sometimes referred to as augmented feature space in pattern recognition. For example, a straight line in F often becomes a curve or ellipsoid when projected down to two- or three-dimensional spaces. An interesting point is that there are a large number of families of F that are computationally tractable. This is made possible by exploiting the so-called “kernel functions.” In geometric terms, kernel functions in effect yield a new measure for the pairwise data point distances.
More formally, the kernel PCA technique essentially is a variant of linear PCA done in the kernel space. First, we introduce some notations concerning the kernel functions and spaces. Let φ:Rd → F be the feature function mapping X to F. The kernel method, rooted in functional analysis, computes a kernel function K(xi, xj) = 〈Φ(xi), Φ(xj)〉 that, as the equation implies, amounts to the inner product of xi, xj in an extended feature space F. This allows one to obtain the inner products without explicitly computing the high dimensional feature space provided that a kernel function exists. Although kernel functions may not exist for every possible feature space, popular kernel functions such as (〈xi, xj〉 + 1)d (polynomial kernel), exp(−|xi − xj|2/σ2) (Gaussian kernel), and tanh(xi·xj + b) (neural kernels) do provide strong classification power.
It has been shown that to obtain the principal components in a chosen F equipped with a suitable K(·) it suffices to compute eigenvalue decomposition over K(x) = [K(xi, xj)], specifically K(x)Y′ = nλY′ (22). Given a point x ϵ Rd and column vectors Y′ = {yi′}, the projection of x onto the ith principal component in the extended feature space yi can be computed through just the data space and the kernel function, namely 〈Φ(x), yi〉 = 〈yi′/√λi〉·(K(x1, xj), K(x2, xj), … , K(xn, xj)). All the PCA projections shown in this work were produced by a Gaussian kernel with a variance set to 4.
Classification of Samples Using Kernel Projections
We used Fisher's discrimination analysis (FDA) for classification because it gives Bayes optimal statistical classifiers for two classes. The input data to FDA are the first two projected dimensions due to kernel PCA. Samples were classified finally by considering discriminant analysis under Mahalanobis distance (23), which is a widely useful metric in statistical applications that assigns data to a specific group according to the normalized distance between the test sample and the center of mass of the data set.
We have considered using other methods such as kernel discriminant analysis, which would lead to the generation of prediction boundaries in four-dimensional space. However, this would result in a more complex signal that would be less intuitive in terms of an underlying biological signal than that produced by kernel PCA and FDA in a two-dimensional space. Furthermore, higher dimensional prediction boundaries may be more complicated for optimizing sensitivity or precision because this would increase the difficulty of assessing how much a change in the data will influence shifts in the decision boundaries. Recent studies have addressed this concern about maximal margin approaches as well as sample bias (24). For our purpose, reduction of the sampling space to as small as two dimensions is essential because the samples can be as small as n = 8 for diseased and control models (rat models). Although it is likely that some potential analyte clusters would be missed if the last N − 2 principal components are omitted, this should be a minor concern compared with the small sample size problem that affects the reliability of the prediction power across different data sets. In most cases, the first two principal components of PCA are accountable for most of the variance in the data; thus, the chance for missing analyte clusters in the remaining two principal components should not be high.
In Silico Pathway Analysis
To investigate further potential biomarkers and drug targets, we identified the interaction networks of the targeted analytes using the Ingenuity Pathways Knowledge Base (IPKB) (25). The IPKB uses computational algorithms to identify local networks that are particularly enriched in the data sets. Such local networks contain the most highly connected focus proteins that, in turn, have specific interactions with other proteins in computer-generated networks. The UniProt accession numbers and/or PubChem identifiers of the analytes were submitted to the IPKB for analysis. Significant networks show the input analytes and their associations with other markers in the IPKB database.
RESULTS
Biomarker Profiling
The first stage of this study was aimed at identifying a cluster of analytes capable of distinguishing schizophrenia serum samples from control subjects at a single clinical center. Center 1 was chosen for this as all recorded base-line characteristics (age, gender, BMI, smoking, cannabis consumption, and date of sample collection) were comparable between schizophrenia and control subjects. The levels of biomarkers in serum from male first onset antipsychotic-naïve schizophrenia (n = 42) and demographically matched control (n = 31) subjects (Table I) were measured using the Human Metabolic MAP platform. Expression data were obtained for 17 analytes (Table II), which could also be measured using the Rat Metabolic MAP (see below). Analysis of the data using standard statistical methods resulted in identification of six analytes that showed significant differences in expression (p < 0.05; a suitable two-tailed t test was chosen for checking each analyte according to the normality and homogeneity of the data) between the schizophrenia and control subjects. These were cortisol, growth hormone, leptin, luteinizing hormone, progesterone, and resistin. Three of these analytes (cortisol, progesterone, and resistin) also showed significant differences in clinical center 2 along with changes in glucagon-like peptide-1, plasminogen activator inhibitor 1, and testosterone.
Identification of TACs
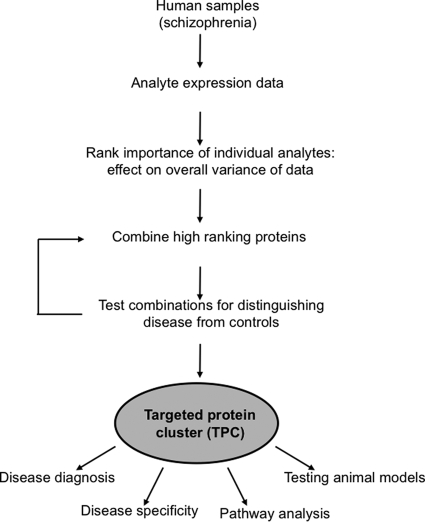
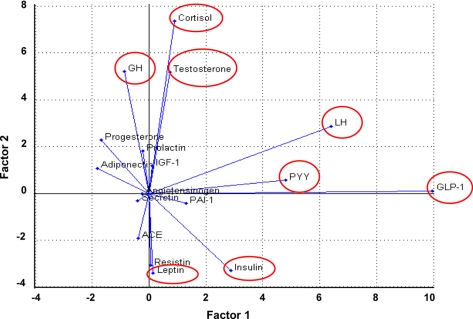
The schema used to identify TACs is shown in Fig. 1. As described above for identification of statistically significant analytes, this first required comparison of the expression levels of 17 serum molecules in schizophrenia patients and matched healthy controls. The next step involved dissecting the ability of each analyte to explain the data variance of the combined schizophrenia and control samples using factor analysis (see “Materials and Methods”). This identified a subgroup of eight analytes that had the strongest influence on data structure: GLP-1, LH, peptide YY, testosterone, cortisol, growth hormone (GH), leptin, and insulin (Fig. 2).
Fig. 1.
Scheme of identifying and validating TAC.
Fig. 2.
Factor analysis for identifying analytes that account for data variance. The expression levels of the 17 serum analytes in schizophrenia patients and controls were measured as described under “Materials and Methods.” The ability of each analyte to account for the data variance of the combined schizophrenia and control samples was then determined using factor analysis to reduce the multidimensional data to fewer factors that are linear combinations of data attributes. This led to classification of analytes according to their importance in representing the overall variance of the data. The most important analytes according to these criteria were GLP-1, LH, peptide YY (PYY), testosterone, cortisol, GH, leptin, and insulin (red enclosures). ACE, angiotensin-converting enzyme.
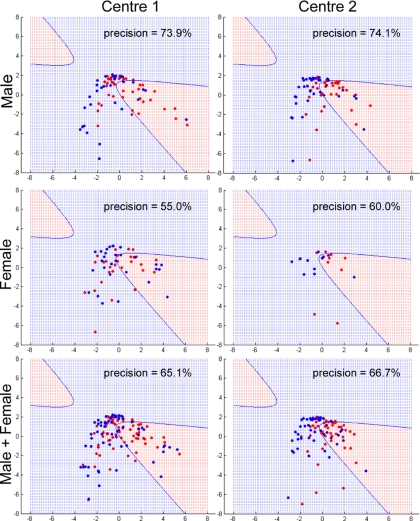
We then performed feature selection using these analyte readings in all combinations by brute force enumeration. This led to the identification of a specific cluster that included insulin, cortisol, leptin, and growth hormone, which gave the maximum discrimination power. Regarding the expression of these four proteins, six samples in clinic 1 gave low readings for growth hormone, and three samples gave low readings for insulin. Three samples in clinic 2 gave low readings for growth hormone, and three samples gave low readings for insulin. These samples were not included in the analysis. For all animal models, all of the analyte readings were within the linear range. Based on the discrimination analysis and taking into account variable correlations, this cluster yielded a precision of 73.9% using male samples in clinical center 1 (Fig. 3). The precision was calculated as the number of patients (red dots) in the prediction region (in red) divided by the total number of cases in the prediction region (or can be formulated as TP/(TP + FP) where TP and FP are the numbers of true positives and false positives, respectively). The sensitivity, calculated as TP/(TP + FN) where FN is the number of false negative cases, however, is as low as 40.5%. This shows that, although the specific TAC has a reasonably high accuracy in terms of identifying schizophrenia patients, it did miss some of the patients. The trade-off between precision and sensitivity is a problem encountered frequently in developing prediction algorithms. Future studies will be aimed at constructing statistical classifiers optimizing both precision and sensitivity for more clinical applications.
Fig. 3.
Gaussian kernel PCA projection of schizophrenia and control data using TAC. Data points (subjects) projected within the red region are considered schizophrenia, and those projected into the blue region are considered controls. Precision is the percentage of points in the red region that are real schizophrenia. Male, female, and combined male/female samples were tested from clinical centers 1 and 2. Red dots indicate schizophrenia patients, and blue dots are controls. Female subjects included 29 schizophrenia and 28 controls from clinical center 1 and 11 schizophrenia and 11 controls from clinical center 2. All of these were matched to male subjects for age and BMI.
The practical performance of the identified cluster was assessed by transforming the data of insulin, cortisol, leptin, and growth hormone from clinical center 2 into an independent kernel space and projecting these onto the first two principal components trained from center 1. The prediction power was then evaluated according to the prediction boundary trained by using the samples from center 1. The results showed a high precision of 74.1% (Fig. 3), and the sensitivity increased to 57.1%. This suggests that the TAC is able to maintain its predictive power across different data sets, whereas the coverage of the patients may not be consistent.
To see whether the predictive power of this TAC generalizes across gender, we used the same kernel space for prediction of samples from female subjects. This resulted in lower but still acceptable precision values for female samples from centers 1 and 2 with values of 55 and 60%, respectively (Fig. 3), and with sensitivities of 37.9 and 54.6%, respectively. The same analysis was repeated for combined male and female samples from centers 1 and 2, and this yielded good results with precisions of 65 and 67%, respectively (Fig. 3), and with sensitivities 39.4 and 52.2%, respectively. These findings suggest that the precision of this particular cluster can generalize beyond gender and samples acquired from different clinical centers, whereas further development of other complementary methods with low false positive rates would be required to maintain a higher sensitivity across different databases without sacrificing the precisions.
We also determined the robustness of the TAC by using center 2 male samples as a training set to define the kernel PCA projection and prediction boundary that were subsequently used to predict the samples from different centers and of different gender. The results (supplemental Table S1) show that the combination of cortisol, leptin, GH, and insulin was still identified as a useful TAC with high precision. Although other analyte clusters showed better precision on the training set, they had lower prediction results across the two clinical centers and gender (see an example of cluster angiotensin-converting enzyme, cortisol, IGF-1, and PAI-1 in supplemental Table S1). Thus, the TAC presented here is likely to be a stable signal for analyzing and diagnosing the disease.
Comparison of TAC Method with Standard Statistical Approach
The cluster comprised of insulin, cortisol, leptin, and growth hormone gave good precision and predictive power for diagnosing schizophrenia subjects compared with controls. This performance was not explained by the individual proteins as only one of these analytes (cortisol) showed statistically significant differences in both centers (Table II). Therefore, the strong statistical signal of the TAC is due to the collective features of the four analytes in reproducing Gaussian kernel space. Moreover, the precision and prediction results for the six proteins that were identified as statistically significant in clinic center 1 (see Table II; cortisol, growth hormone, leptin, luteinizing hormone, progesterone, and resistin) were lower than those achieved by the cluster (Table III). This was despite the fact that a new prediction boundary was produced and optimized for these analytes.
Table III. Comparison of TAC and standard statistical method with respect to precision and predictive power for diagnosis of schizophrenia.
Values for the TAC were extracted from Fig. 3 for male schizophrenia subjects in clinical centers 1 and 2. Individually significant analytes were identified using a two-tailed non-parametric t test and then tested as for the TAC for male schizophrenia subjects in clinical centers 1 and 2.
| Method | Precision | Prediction |
|---|---|---|
| % | % | |
| TAC | 73.9 | 74.1 |
| 6 significant analytes | 66.7 | 62.5 |
Specificity of TAC for Schizophrenia
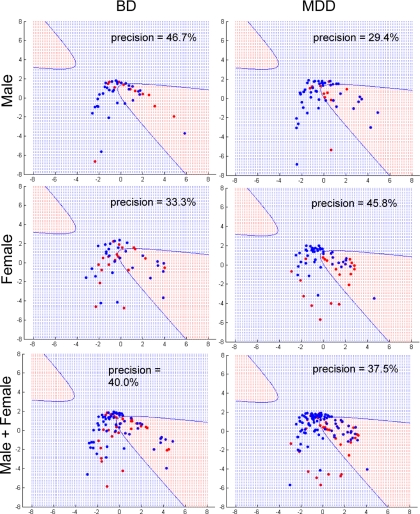
Schizophrenia appears to have etiology, pathophysiology, and symptomology similar to those aspects of BD and MDD (10–13). To determine whether the prediction power of the cluster is specific for schizophrenia, we generated Gaussian kernel PCA projections for BD and MDD patients compared with controls using the same settings. The precision for predicting the BD patients was consistently lower than those of the schizophrenia cases regardless of gender with values for male, female, and both sexes of 47, 33, and 40%, respectively (Fig. 4). Similarly, there was no clear separation between the MDD and control groups with values for male, female, and both sexes of 29, 46, and 48%, respectively (Fig. 4). This demonstrated that the insulin, cortisol, leptin, and growth hormone cluster gives good posterior conditions for detecting schizophrenia.
Fig. 4.
Gaussian kernel PCA projection of BD, MDD, and control data using TAC. Data points (subjects) projected within the red region are considered disease (BD or MDD), and those projected into the blue region are considered controls. Precision is the percentage of points in the red region that are actual disease. Male, female, and combined male/female samples were tested. Red dots indicate schizophrenia patients, and blue dots are controls.
Specificity of TAC for Preclinical Models of Schizophrenia
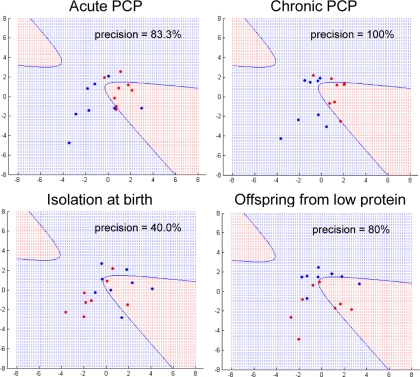
Animal models are essential for studying human diseases and for the discovery and development of novel pharmaceuticals. Therefore, we tested the insulin, cortisol, leptin, and growth hormone cluster to determine whether the discriminatory power can be applied to animal models. For this, the expression levels of the same 17 serum proteins were determined in four different rat models using the Rodent MAP technology. We then generated Gaussian kernel projections for the data using the same settings as for the human studies. This showed that the acute and chronic PCP rat models, which are used routinely in studies of schizophrenia (26), gave high predictive results with precisions of greater than 80% (Fig. 5). However, the isolation rat model, which is considered to be relevant as a model of depression (27), gave a lower precision value of only 40% (Fig. 5). Interestingly, we found that another rat model based on the maternal effects of a low protein diet (19) gave prediction values that were similar to the standard schizophrenia models (Fig. 5). This is consistent with reports of epidemiological studies showing a higher incidence of schizophrenia, antisocial behaviors, and other disorders in the offspring of mothers who suffered from nutrient deprivation in times of famine (28, 29). The results require confirmation on whether other interventions produce a similar TAC signature.
Fig. 5.
Gaussian kernel PCA projection of preclinical animal models using TAC. Data points (animals) projected within the red region are considered model animals, and those projected into the blue region are considered controls. Precision is the percentage of points in the red region that are actual model animals. Male acute PCP, chronic PCP, isolation, and low protein rats were tested (see “Materials and Methods”). Red dots indicate model animals, and blue dots are controls.
In Silico Pathway Analysis
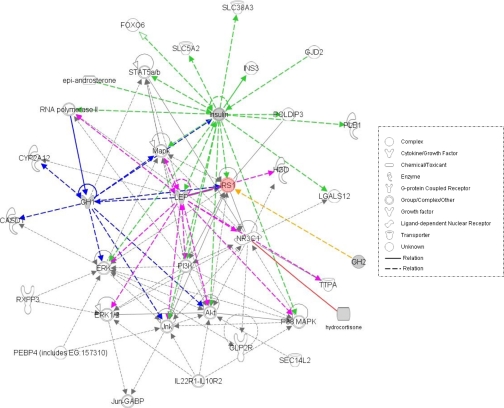
Pathway functional analysis was carried out on the four components of the TAC using the IPKB tool as described under “Materials and Methods.” This software allows data to be analyzed in a systematic way using published molecular interaction data to determine the most significant biological functions and pathways (25). One major network was identified after uploading the analyte identifiers. This contained the four analytes and displayed their associations with other molecules input by the IPKB software (Fig. 6). This showed that all four analytes were associated with insulin receptor substrate 1 (IRS1), whereas three of the analytes (insulin, leptin, and growth hormone) were also associated with ERK and protein kinase B (AKT) signaling. On the pathway level, the insulin signaling pathway seems to be the best match with a p value of 7.06 × 10−3 calculated by the IPKB software, considering the proteins in the identified network (see supplemental Fig. S1). This is consistent with our recent study showing that at least some schizophrenia subjects show signs of alterations in insulin signaling (30).
Fig. 6.
In silico pathway mapping of TAC analytes. LEP, leptin; HGD, homogentisate 1,2-dioxygenase; TTPA, alpha-tocopherol transfer protein; GABP, GA-binding protein; the UniProt accession numbers (insulin, leptin, and growth hormone) and PubChem identifiers (cortisol) of the analytes were submitted to the IPKB for analysis as described under “Materials and Methods.” The interaction network suggests that IRS1 is highly associated with the TAC proteins. MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase; Jnk, c-Jun N-terminal kinase.
DISCUSSION
Traditional biomarker studies have focused on identifying molecules that show significantly different expression levels between the test cases and controls. Although this approach is intuitively straightforward, it may miss those molecules that form consistent group patterns. To this end, the approach of using multianalyte fingerprints to distinguish patients from controls has increased in usage. A promising example is the recent application of a profile consisting of 18 biomarkers for predicting patients with Alzheimer disease (31). The multiplex profile-based approach has been applied broadly to other complex diseases such as breast cancer and autoimmune disorders, but most have focused on gene expression data rather than protein or metabolite levels (32–34).
A major obstacle of applying the profiles on disease classification and prediction is the poor generalizability of the resulting profiles across different data sets. Usually the validated molecular profiles consist of tens to hundreds of features, and therefore these can collapse when a different data set is introduced for testing. One reason for this could be due to the difficulty of collecting enough data for comparative studies. Given a random data set, the expression data of all the molecules in the profile must be available; otherwise, the predictive value may not be reliable. The TAC approach described in this study is less likely to suffer from this problem because these clusters contain only small numbers of molecules.
Another important issue is the low precision in terms of analyzing disease etiology. By definition, large profiles incorporate more analytes, which clearly make these studies expensive to carry out. Furthermore, a TAC provides reliable precision that originates from the fact that redundancy is minimized because of reduced noise that can confuse the classifier. We confirmed this by showing that the predictive results are lower with a cluster constructed from all 17 analytes that were measured in the study (supplemental Fig. S2). More analytes also make statistical modeling more difficult as the required data set size can grow exponentially with the size of the profile. This phenomenon is known as “the curse of dimensionality” (35). In contrast, application of small clusters reduces the complexity of analyzing disease pathways and therefore minimizes the chances of following up false positives. In this way, networks arising from cluster analysis are likely to serve as a better reference for designing follow-up experiments.
That the TAC approach can still perform well while including analytes that show inconsistent behavior across the different centers is due to the nature of Gaussian kernel space used in our kernel PCA model. In Gaussian kernel space, the variances between every pair of individuals (no matter whether they are patients or controls) are considered through Gaussian kernel function: K(xi, xj) = exp(−((‖xi − xj‖2)/σ2)) where xi and xj are expression data of a specific analyte in individuals i and j and σ is the regularization parameter for the kernel space. This should not be linked directly to the overall fold-change and thus t test. Fold-change is the ratio of average values between the patients and controls and thus is too crude to describe the detailed variances between individual samples. t test, which compares the distribution and variance on each sample group, is not sensitive enough to capture patterned behavior considering individual samples. The kernel function is better in terms of identifying analytes that have collective behaviors across different clinical centers due to the ‖xi − xj‖ term, which essentially puts vectors (protein readings) of the same direction closer. For example, given two analytes A and B, if they have a certain patterned behavior (say A is up-regulated when B is down-regulated), then they may tend to be identified by the cluster approach no matter whether they are both significantly different between the patients and controls or not.
Another important benefit of the cluster approach was the accuracy across gender and species. Although large panel profiles often suffer from numerical instabilities and require significant data to be statistically reliable, the cluster approach avoids this by extracting the maximum information out of a small panel. This highlights the point that inclusion of additional analytes into a cluster leads to the necessity of increasing the size of the training sets exponentially to obtain unbiased and reliable results. It also indicates that classifiers based on sparse points with many input features make generalization more difficult.
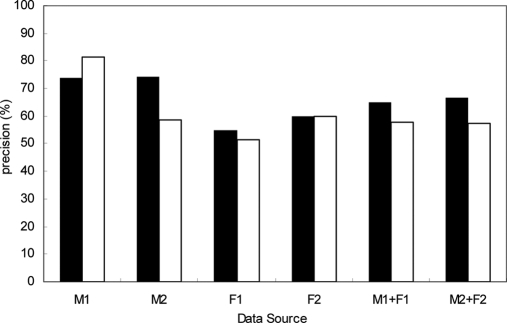
The TAC that we present here was constructed through analyzing the data in reproducing Gaussian kernel space. This approach is known for its robustness in pattern recognition and its ability to produce better classification of data that cannot be achieved by linear algorithms. Linear PCA is widely used for analyses of high dimensional data as it provides a low dimensional approximation and thus makes classification simple. The adopted kernel PCA in this study combines the advantages of both methods by using linear PCA in the extended reproducing Gaussian kernel space. As a reference, we also carried out a projection of schizophrenia and control data using the cluster with linear PCA. In the case of the linear PCA, the prediction results were in general worse than those of the Gaussian kernel PCA (Fig. 7 and supplemental Fig. S3). Also, there is less consistency across clinical centers 1 and 2, especially for the male samples. This indicates that it is harder to evaluate the prediction results on a test set based on the performance of a training set in the linear model. Therefore, although both linear and kernel PCAs are useful for reducing high dimensional data to a low dimensional approximation, kernel PCA is more powerful for recognizing non-linear patterns and may provide more consistent results across different data sources.
Fig. 7.
Comparison between precisions of linear and kernel PCAs. The black bars are the precision of TAC based on kernel PCA; the white bars are the precision of TAC based on linear PCA. M1 (F1) and M2 (F2) stand for male (female) samples in hospital centers 1 and 2, respectively.
The finding of a discriminatory signal in analytes relating to insulin, cortisol, leptin, and growth hormone signaling lends support to our previous findings of insulin resistance (30) and perturbations in metabolism and glucose handling in schizophrenia (36, 37). All four molecules are known to interact with each other and are involved in regulation of metabolic signaling pathways (38). The biological characteristics of this cluster also conform to the fact that homeostatic regulation of metabolism requires interaction of multiple hormonal pathways. In this study, the collective signal was manifested in a non-linear fashion and would therefore be missed by standard statistical methods.
The specificity of the TAC for schizophrenia compared with BD and MDD was of particular interest not only from a diagnostic point of view but also from a mechanistic one. Previous studies have identified systemic metabolic dysfunction in first onset antipsychotic-naïve schizophrenia subjects, including increased prevalence of metabolic syndrome (30, 39, 40), although similar disturbances have been reported widely for other mental illnesses including BD and MDD (41). The current results suggest that these pathways may differ between schizophrenia and the other neuropsychiatric disorders.
In addition, the TAC was able to distinguish the pharmaceutical industry standard PCP rat models of schizophrenia from the social isolation model, which has traditionally been used as a model of depression and anxiety. This suggests that this approach may be useful in identifying additional models of schizophrenia. It was of interest in this case that the cluster rated the low protein rat model with high precision. The similarity between a known metabolic model and a model of schizophrenia is intriguing.
By definition, a TAC is a non-redundant molecular profile associated with a specific disease due to its ability to distinguish disease from controls. Therefore, an interaction network containing the components of these clusters could provide insights into the core pathological mechanisms. We used the IPKB network analysis tool to visualize a molecular interaction network incorporating the four analytes. The highly associated protein IRS1 together with other proteins such as AKT, ERK, and phosphatidylinositol 3-kinase shows that the TAC may be associated with insulin signaling pathways that play an important role in regulating processes such as glucose and lipid homeostasis, apoptosis, protein synthesis, cell proliferation, and differentiation (42, 43). Thus, investigation of other molecules that impinge upon these functions may be worth further investigation as potential biomarkers and could lead to the identification of potential novel therapeutic strategies for treatment of schizophrenia.
In summary, we have shown that the TAC approach affords a promising new direction in characterization and potential diagnosis of complex psychiatric disorders such as schizophrenia. Our results show that the conventional biomarker approach, which emphasizes individual proteins that have significant differences in expression between disease and controls, has low generalizability across multiple investigations. On the other hand, the TAC identified here was generalizable in multiple studies across gender, species, and disease boundaries. Therefore, this approach merits further investigation as a tool for gaining additional insights into disease mechanisms and for identifying potential disease biomarkers or therapeutic intervention strategies.
Supplementary Material
* This work was supported by the Stanley Medical Research Institute.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S3 and Table S1.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S3 and Table S1.
1 The abbreviations used are:
- DSM
- Diagnostic and Statistical Manual of Mental Disorders
- AKT
- protein kinase B
- BD
- bipolar disorder
- ERK
- extracellular signal-regulated kinase
- FDA
- Fisher's discrimination analysis
- GH
- growth hormone
- GLP-1
- glucagon-like peptide-1
- LH
- luteinizing hormone
- MDD
- major depressive disorder
- PCA
- principal component analysis
- PCP
- phencyclidine
- TAC
- targeted analyte cluster
- MAP
- Multi-Analyte Profiling
- SZ
- schizophrenia
- BMI
- body mass index
- ACTH
- adrenocorticotropic hormone
- QC
- quality control
- FA
- factor analysis
- IPKB
- Ingenuity Pathways Knowledge Base
- IRS1
- insulin receptor substrate 1.
REFERENCES
- 1.Thaker G. K., Carpenter W. T., Jr. (2001) Advances in schizophrenia. Nat. Med 7, 667–671 [DOI] [PubMed] [Google Scholar]
- 2.Csernansky J. G., Schuchart E. K. (2002) Relapse and rehospitalisation rates in patients with schizophrenia: effects of second generation antipsychotics. CNS Drugs 16, 473–484 [DOI] [PubMed] [Google Scholar]
- 3.Möller H. J. (2004) Course and long-term treatment of schizophrenic psychoses. Pharmacopsychiatry 37, 126–135 [DOI] [PubMed] [Google Scholar]
- 4.van Haren N. E., Hulshoff Pol H. E., Schnack H. G., Cahn W., Brans R., Carati I., Rais M., Kahn R. S. (2008) Progressive brain volume loss in schizophrenia over the course of the illness: evidence of maturational abnormalities in early adulthood. Biol. Psychiatry 63, 106–113 [DOI] [PubMed] [Google Scholar]
- 5.van Haren N. E., Hulshoff Pol H. E., Schnack H. G., Cahn W., Mandl R. C., Collins D. L., Evans A. C., Kahn R. S. (2007) Focal gray matter changes in schizophrenia across the course of the illness: a 5-year follow-up study. Neuropsychopharmacology 32, 2057–2066 [DOI] [PubMed] [Google Scholar]
- 6.Keefe R. S., Sweeney J. A., Gu H., Hamer R. M., Perkins D. O., McEvoy J. P., Lieberman J. A. (2007) Effects of olanzapine, quetiapine, and risperidone on neurocognitive function in early psychosis: a randomized, double-blind 52-week comparison. Am. J. Psychiatry 164, 1061–1071 [DOI] [PubMed] [Google Scholar]
- 7.Norman R. M., Malla A. K. (2001) Duration of untreated psychosis: a critical examination of the concept and its importance. Psychol. Med 31, 381–400 [DOI] [PubMed] [Google Scholar]
- 8.McGorry P. D., Warner R. (2002) Consensus on early intervention in schizophrenia. Schizophr. Bull 28, 543–544 [DOI] [PubMed] [Google Scholar]
- 9.Harrigan S. M., McGorry P. D., Krstev H. (2003) Does treatment delay in first-episode psychosis really matter? Psychol. Med 33, 97–110 [DOI] [PubMed] [Google Scholar]
- 10.Grünblatt E. (2004) The benefits of microarrays as tools for studying neuropsychiatric disorders. Drugs Today 40, 147–156 [PubMed] [Google Scholar]
- 11.Pennington K., Dicker P., Dunn M. J., Cotter D. R. (2008) Proteomic analysis reveals protein changes within layer 2 of the insular cortex in schizophrenia. Proteomics 8, 5097–5107 [DOI] [PubMed] [Google Scholar]
- 12.English J. A., Dicker P., Föcking M., Dunn M. J., Cotter D. R. (2009) 2-D DIGE analysis implicates cytoskeletal abnormalities in psychiatric disease. Proteomics 9, 3368–3382 [DOI] [PubMed] [Google Scholar]
- 13.Ferrier I. N., Stanton B. R., Kelly T. P., Scott J. (1999) Neuropsychological function in euthymic patients with bipolar disorder. Br. J. Psychiatry 175, 246–251 [DOI] [PubMed] [Google Scholar]
- 14.Gitlin M. J., Swendsen J., Heller T. L., Hammen C. (1995) Relapse and impairment in bipolar disorder. Am. J. Psychiatry 152, 1635–1640 [DOI] [PubMed] [Google Scholar]
- 15.Chemerinski E., Bowie C., Anderson H., Harvey P. D. (2008) Depression in schizophrenia: methodological artifact or distinct feature of the illness? J. Neuropsychiatry Clin. Neurosci 20, 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischhacker W. (2000) Negative symptoms in patients with schizophrenia with special reference to the primary versus secondary distinction. Encephale 26, 12–14 [PubMed] [Google Scholar]
- 17.Guo Y., Zhang H., Chen X., Cai W., Cheng J., Yang Y., Jin G., Zhen X. (2009) Evaluation of the antipsychotic effect of bi-acetylated l-stepholidine (l-SPD-A), a novel dopamine and serotonin receptor dual ligand. Schizophr. Res 115, 41–49 [DOI] [PubMed] [Google Scholar]
- 18.Schubert M. I., Porkess M. V., Dashdorj N., Fone K. C., Auer D. P. (2009) Effects of social isolation rearing on the limbic brain: a combined behavioral and magnetic resonance imaging volumetry study in rats. Neuroscience 159, 21–30 [DOI] [PubMed] [Google Scholar]
- 19.Ozanne S. E. (2009) The long term effects of early postnatal diet on adult health. Adv. Exp. Med. Biol 639, 135–144 [DOI] [PubMed] [Google Scholar]
- 20.Palmer A. A., Printz D. J., Butler P. D., Dulawa S. C., Printz M. P. (2004) Prenatal protein deprivation in rats induces changes in prepulse inhibition and NMDA receptor binding. Brain Res 996, 193–201 [DOI] [PubMed] [Google Scholar]
- 21.Bertenshaw G. P., Yip P., Seshaiah P., Zhao J., Chen T. H., Wiggins W. S., Mapes J. P., Mansfield B. C. (2008) Multianalyte profiling of serum antigens and autoimmune and infectious disease molecules to identify biomarkers dysregulated in epithelial ovarian cancer. Cancer Epidemiol. Biomarkers Prev 17, 2872–2881 [DOI] [PubMed] [Google Scholar]
- 22.Schölkopf B., Smola A. J., Müller K. (1998) Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput 10, 1299–1319 [Google Scholar]
- 23.Mahalanobis P. C. (1936) On the generalised distance in statistics. Proc. Natl. Inst. Sci. India 2, 49–55 [Google Scholar]
- 24.Elkan C., Noto K. (2008) Learning classifiers from only positive and unlabeled data, in Proceeding of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Las Vegas, August 24–27, 2008, pp. 213–220, Association for Computing Machinery Special Interest Group on Knowledge Discovery and Data Mining, New York [Google Scholar]
- 25.Salim K., Guest P. C., Skynner H. A., Bilsland J. G., Bonnert T. P., McAllister G., Munoz-Sanjuan I. (2007) Identification of proteomic changes during differentiation of adult mouse subventricular zone progenitor cells. Stem Cells Dev 16, 143–165 [DOI] [PubMed] [Google Scholar]
- 26.Pratt J. A., Winchester C., Egerton A., Cochran S. M., Morris B. J. (2008) Modelling prefrontal cortex deficits in schizophrenia: implications for treatment. Br. J. Pharmacol 153, S465–S470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fone K. C., Porkess M. V. (2008) Behavioural and neurochemical effects of post-weaning social isolation in rodents-relevance to developmental neuropsychiatric disorders. Neurosci. Biobehav. Rev 32, 1087–1102 [DOI] [PubMed] [Google Scholar]
- 28.Susser E. S., Lin S. P. (1992) Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944–1945. Arch. Gen. Psychiatry 49, 983–988 [DOI] [PubMed] [Google Scholar]
- 29.Smil V. (1999) China's great famine: 40 years later. BMJ 319, 1619–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guest P. C., Wang L., Harris L. W., Burling K., Levin Y., Ernst A., Wayland M. T., Umrania Y., Herberth M., Koethe D., van Beveren J. M., Rothermundt M., McAllister G., Leweke F. M., Steiner J., Bahn S. (2010) Increased levels of circulating insulin-related peptides in first-onset, antipsychotic naïve schizophrenia patients. Mol. Psychiatry 15, 118–119 [DOI] [PubMed] [Google Scholar]
- 31.Ray S., Britschgi M., Herbert C., Takeda-Uchimura Y., Boxer A., Blennow K., Friedman L. F., Galasko D. R., Jutel M., Karydas A., Kaye J. A., Leszek J., Miller B. L., Minthon L., Quinn J. F., Rabinovici G. D., Robinson W. H., Sabbagh M. N., So Y. T., Sparks D. L., Tabaton M., Tinklenberg J., Yesavage J. A., Tibshirani R., Wyss-Coray T. (2007) Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat. Med 13, 1359–1362 [DOI] [PubMed] [Google Scholar]
- 32.Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A. L., Brown P. O., Botstein D. (2000) Molecular portraits of human breast tumours. Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 33.Pusztai L. (2008) Current status of prognostic profiling in breast cancer. Oncologist 13, 350–360 [DOI] [PubMed] [Google Scholar]
- 34.Dowsett M., Dunbier A. K. (2008) Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin Cancer Res 14, 8019–8026 [DOI] [PubMed] [Google Scholar]
- 35.Bellman R. E. (1961) Adaptive Control Processes, Princeton University Press, Princeton, NJ [Google Scholar]
- 36.Khaitovich P., Lockstone H. E., Wayland M. T., Tsang T. M., Jayatilaka S. D., Guo A. J., Zhou J., Somel M., Harris L. W., Holmes E., Pääbo S., Bahn S. (2008) Metabolic changes in schizophrenia and human brain evolution. Genome Biol 9, R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prabakaran S., Swatton J. E., Ryan M. M., Huffaker S. J., Huang J. T., Griffin J. L., Wayland M., Freeman T., Dudbridge F., Lilley K. S., Karp N. A., Hester S., Tkachev D., Mimmack M. L., Yolken R. H., Webster M. J., Torrey E. F., Bahn S. (2004) Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 9, 684–697, 643 [DOI] [PubMed] [Google Scholar]
- 38.McMurray R. G., Hackney A. C. (2005) Interactions of metabolic hormones, adipose tissue and exercise. Sports Med 35, 393–412 [DOI] [PubMed] [Google Scholar]
- 39.Ryan M. C., Sharifi N., Condren R., Thakore J. H. (2004) Evidence of basal pituitary-adrenal overactivity in first episode, drug naïve patients with schizophrenia. Psychoneuroendocrinology 29, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 40.Spelman L. M., Walsh P. I., Sharifi N., Collins P., Thakore J. H. (2007) Impaired glucose tolerance in first-episode drug-naïve patients with schizophrenia. Diabet. Med 24, 481–485 [DOI] [PubMed] [Google Scholar]
- 41.Toalson P., Ahmed S., Hardy T., Kabinoff G. (2004) The metabolic syndrome in patients with severe mental illnesses. Prim. Care Companion J. Clin. Psychiatry 6, 152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogata H., Goto S., Fujibuchi W., Kanehisa M. (1998) Computation with the KEGG pathway database. Biosystems 47, 119–128 [DOI] [PubMed] [Google Scholar]
- 43.Kanehisa M. (2002) The KEGG database. Novartis Found. Symp 247, 91–101; discussion 101–103, 119–128, 244–252 [PubMed] [Google Scholar]
- 44.Cilia J., Cluderay J. E., Robbins M. J., Reavill C., Southam E., Kew J. N., Jones D. N. (2005) Reversal of isolation-rearing-induced PPI deficits by an alpha7 nicotinic receptor agonist. Psychopharmacology 182, 214–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.