Abstract
Gene silencing based on RNA interference is widely used in fundamental research and in practical applications. However, a commonly incomplete functional suppression represents a serious drawback of this technology. We describe a series of lentiviral vectors each containing a single or multiple shRNA expression cassette(s) driven by a RNA polymerase III specific promoter and localized within the 3′-LTR of the lentiviral DNA backbone. The vectors also contain an antibiotic-resistance gene that allows positive selection of recipient cells. The combined expression of three different shRNAs specific to a single mRNA was shown to improve dramatically the level of mRNA inhibition, while the use of three different RNA polymerase III specific promoters avoids the loss of shRNA expression cassettes through the homologous recombination. The vector system was used for successful simultaneous suppression of three related SESN1, SESN2 and SESN3 genes, which suggests its particular value for testing phenotypes of functionally redundant genes.
Keywords: multiplex shRNA expression, lentiviral vectors, RNAi mediated gene silencing
1. Introduction
RNA interference (RNAi) is a one of the natural ways of gene regulation that utilizes small interfering double stranded RNA (siRNA) for functional suppression of specific mRNAs. Introduction into cells of siRNA specific for particular mRNA has become a widespread tool in reverse genetics and for functional characterization of genes. The most straightforward approach is to introduce into cells or organisms siRNA oligonucleotides as it produces quick and robust suppression of a particular mRNA (Elbashir et al., 2001). However, the effect is transient and does not allow stable inhibition of the function. Expression of small hairpin RNAs (shRNAs), which are recognized by the RNAi machinery and processed into active siRNA, has become a preferable approach. It allows stable suppression of functions not only in cell culture, but also in animals (Brummelkamp et al., 2002; Tiscornia et al., 2003). Lentiviral vectors are currently the most appealing tool for efficient delivery and stable expression of genes in almost all cell types (Bos et al., 2009). This is why the development of convenient lentiviral vectors for expression of shRNAs is important for successful application of RNAi based technologies both in research, and in practical fields.
Despite many advantages of shRNA mediated mRNA suppression, the efficiency of inhibition varies substantially depending on particular mRNA sequence chosen for targeting. As result, partial phenotypes are produced impeding the definitive conclusions. The problem rises in particular when attempting to inhibit expression of vital functions, as there is potent selection against the cells with stronger level of suppression. In this case simultaneous introduction of several different shRNAs that target the same mRNA would help to achieve a more stable phenotype. The multiplex systems for the shRNA delivery would also benefit in cases when a function is maintained by few related genes, or when several functions need to be suppressed simultaneously. Recently, several lentiviral vectors were designed for delivery of multiple shRNAs to the cell using a single lentiviral backbone (Bos et al., 2009; Henry et al., 2006; Song et al., 2008; Stove et al., 2006; ter Brake et al., 2006; ter Brake et al., 2008; Xia et al., 2006). Here we describe the development of highly efficient lentiviral system for stable sustained expression of multiple shRNAs.
2. Materials and methods
2.1. Cells lines
Human colon carcinoma cells RKO and packaging cell line 293T were grown on Dulbecco MEM supplemented with 10% fetal bovine serum. Stable mtDNA-deficient (ρ0) RKO cells were obtained as described (King and Attardi, 1996) and maintained in the medium supplemented with 50 μg/ml uridine, and 100 μg/ml pyruvate.
2.2. Lentiviral vectors for expression of single shRNA expressing cassettes
For the introduction of shRNA expressing cassette driven by RNA polymerase III specific promoters we chose U3 region of distal LTR of HIV1 lacking the enhancer segment. Cloning sites for EcoRI, BamHI and ClaI restriction nucleases were introduced by PCR 102 bp downstream of the Acc65I site in pLV-H4 lentiviral vector (Razorenova et al., 2005). The BamHI and ClaI sites were used for the introduction of PCR-amplified fragments containing appropriate promoters, in reverse orientation. The following RNA polymerase III specific promoters were used – 215 bp human H1 RNA promoter, 256 bp human U6 RNA promoter and 241 bp human 7SL RNA promoter. A set of PCR-amplified DNA fragments each containing appropriate selective marker gene conferring resistance to antibiotics puromycin (603 bp “puro”), G418 (805 bp “neo”), phleomycin or zeocin (376 bp “bleo”) and hygromycin (1023 bp “hygro”) were then cloned under human histone H4 gene promoter using XbaI and SalI sites. The vectors pLSL(X), pLU6(X) and pLSL7(X) are designed for cloning of shRNA oligonucleotides at the EcoRI and BamHI sites. General scheme for the pLSL(X) vectors is shown on Figure 1a.
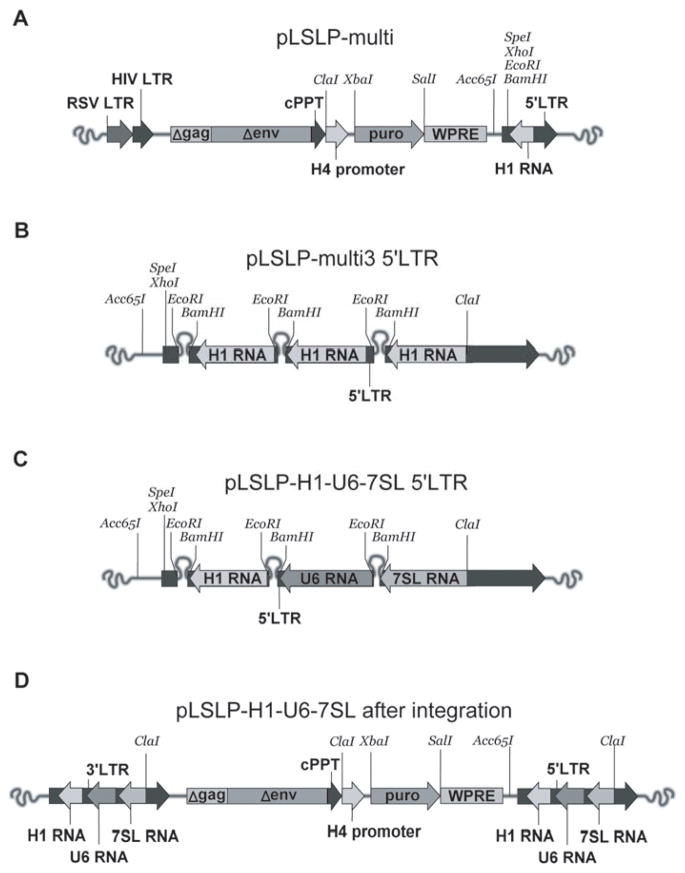
Figure 1.
Development of lentiviral vectors for multiplex expression of shRNAs. A. General scheme of the lentiviral cassette of the pLSL(X)multi series lentiviral vector pLSLP. After the introduction into a packaging cell line the transcript for the lentiviral cassette is initiated on promoter from the RSV virus LTR, and terminated by polyadenylation in the region corresponding to the right LTR. The transcript is being packaged into viral-like particle after binding to HIV1 and VSV proteins co-expressed in the packaging cells from the packaging plasmids. The transcripts includes the sequences of H1RNA promoter, which is placed in opposite direction. B. Scheme of the left LTR of the pLSLPmulti vector with three consecutively introduced cassettes expressing shRNAs under control of the H1 RNA promoter; C. Scheme of the left LTR of the pLSLPmulti/H1/U6/7SL vector carrying three different types of RNA polymerase III specific promoters; D. Scheme of integrated proviral DNA carrying sequences of the lentiviral vector pLSLPmulti/H1/U6/7SL. Note that the LTR carrying three promoter cassettes is duplicated during the integration process.
The following design was used for 60 nt synthetic self-complementary oliogonucletides carrying small interfering hairpin RNAs (shRNAs):
5′-gatccg---19 nt sense---cttcctgtca---19 nt antisense---tttttg
5′-aattcaaaaa---19 nt sense---tgacaggaag---19 nt antisense---cg
For designing optimal shRNA structures we used the “si-shRNASelector” program (Biopolymer design LLC, UT). The program considers optimal thresholds for the free energy defining terminal duplex asymmetry and optimal ranges for free energies of fully-paired shRNA duplexes, which is based on extensive statistical analysis of databases derived from successful and non successful silencing experiments (Matveeva et al., 2010).
Annealing of the two oligonucleotides would give sticky ends compatible with the EcoRI and BamHI sites in the vector. The microRNA-based motif was used for loop region as it supposedly facilitates the transportation of siRNA to the cytoplasm (Kawasaki and Taira, 2003). For cloning of shRNA oligonucleotides 1 μg (40 pmoles) of each the two oligos were annealed in 50 μl of 0.1 M NaCl, 10 mM Tris HCl pH 7.6 by bringing to 95°C and then to room temperature. The mixture was diluted 1000x in water and 1 μl (40 pg) was taken for ligation with 1 ng of EcoRI and BamHI-cut vector in 50 μl reaction containing 1 μl of T4 DNA ligase (Roche) for 30 minutes at room temperature.
The following 19 bp stretches of mRNA sequences were used for designing shRNAs: Human POLRMT gene mRNA (NM_005035): M1- 5′-gcgggaccaatgggagaaa; M2 – 5′-gggcaaagatactggagaa; M3 – 5′-gcaaataggaggtggaatt; M4 – 5′-ggagctggtatatgtgtta; M5 – 5′-ccaacacacgtaagcagaa. Human SESN1 gene mRNA (NM_014454): SESN1-1 - 5′-ggcattagaattcctcgat; SESN1-2 - 5′-cgcacagatgcatgcttta. Human SESN2 gene mRNA (NM_031459): SESN2-1 – 5′-gaccatggctactcgctga; SESN2-2 – 5′-ctgactactttaccagctt. Human SESN3 gene (NM_144665): SESN3-1 – 5′-gacgaggagaagagcattt; SESN3-2 – 5′-ccagagagagatccagaaa.
2.3. Lentiviral vectors for expression of multiple shRNA expressing cassettes
To allow insertion of additional shRNA-expressing cassettes pLSLPmulti vector was constructed by PCR-assisted introduction of SpeI and XhoI sites upstream of the EcoRI and BamHI shRNA cloning sites in the pLSLP vector. For the introduction of second shRNA expression cassettes, the appropriate shRNA oligonucleotides were first cloned and tested in a single-shRNA-expression vector. Then the whole shRNA-expression cassette containing H1 RNA promoter and shRNA was PCR-amplified with the following primers: containing SalI cloning site and proximal segment of the H1 RNA promoter – 5′-gagagtcgacgaacgctgacgtcatcaac; containing SpeI, XhoI sites and complementary to the region in the LTR located just upstream of the EcoRI site and shRNA cassette in the pLSLP vector- agagactagtagactcgagcccaacgaagacaagaattc. The PCR product was digested with SpeI and SalI and the 314 bp fragment was ligated into the pLSLPmulti vector linearized with SpeI and XhoI restriction nucleases. The ligation of SalI and XhoI sites resulted in inactivation of the original XhoI site in the vector, and creation of new cloning sites for the third expression cassette (Figure 1b). For cloning of the human U6 RNA and 7SL RNA promoter cassettes a similar scheme was used. However, the appropriate single-shRNA expressing constructs were amplified with corresponding promoter-specific primers containing SalI site were used, together with the common second primer containing the SpeI and SalI sites. The following promoter-specific primers were used: for U6 promoter – 5′-gagagtcgacgatgggcaggaagagggc; for 7SL RNA – 5′-gagagtcgacatccagtatttagcatgcc. The composition of pLSLmulti construct with three different promoter-specific shRNA expression cassettes is shown on Figure 1c.
Lentivirus vector packaging and lentiviral transduction into target cells
To obtain lentiviral vector virions, lentiviral constructs were transduced into 293T cells with the use of Lipofectamine-Plus (Invitrogen). The transfected cells were incubated in DMEM supplemented with 2% fetal bovine serum. Viral stocks were harvested 24 hours after transfection, with 8 h intervals during 72 hours, filtered through 0.22 μm syringe filters, mixed with 0.3 volumes of 40% PEG-8000/PBS solution and incubated for 8 hours on ice. The formed precipitate was collected by centrifugation at 5000 g for 15 minutes, and the pellets were dissolved in PBS (1/10 of the original volume). The resulting viral stocks supplemented with 1 μg/ml polybrene were used for infection of target cells with multiplicity of infection (MOI) of 1 colony forming unit per cell or less. Two days after the infection the cells were subjected to selection with appropriate antibiotic - 1 μg/ml of puromycin or 200 μg/ml of hygromycin.
Real-time-PCR detection of template
RNA from cell cultures was extracted with TRIsol Reagent (Invitrogen). Quantitative measurement of specific RNA regions was performed after reverse transcription of total RNA samples with iScript cDNA synthesis kit (Bio-Rad) and the resulting cDNA was amplified using primers and a probe specific to the α-tubulin gene. Triplicate reactions for each RNA sample (100 ng/reaction) were amplified along with no-template controls on the iCycler iQ real-time system using the iQ SYBR Green Supermix (Bio-Rad). Thermal cycling conditions were as follows: 2 min at 50°C, 30 min at 60°C, 5 min at 95°C, followed by 50 cycles of 15 sec at 95°C and 1 min at 60°C. Setting baseline and threshold values was performed manually and the data were analyzed using the iCycler iQ optical system software. Primers for RT-PCR were: for POLRMT gene transcript encoding mitochondrial RNA polymerase, 5′-ggactccaaggtcaagcaaataggag and 5′-aggtcgaaggcccctggcttg, producing a 405 bp PCR fragment; for SESN1 gene transcripts: 5′-cttctggaggcagttcaagc and 5′-tgaatggcagcctgtcttcac < producing a 341 bp PCR fragment; for SESN2 gene transcripts: 5′-caagctcggaattaatgtgcc and 5′-ctcacaccattaagcatggag, producing a 323 bp PCR fragment; for SESN3 gene transcripts: 5′-gttcactgtatgtttggaatcagg and 5′-gggtgatacttcaggtcaaatg, producing 265 bp PCR fragment.
Detection of the integrity of recombinant vector DNA inserts
Isolation of genomic DNA from cultured cells was performed with Qiagen DNeasy Blood & Tissue Kit. Recombination events leading to elimination of shRNA-expressing cassettes were detected by PCR amplification of DNA with primers: 5′-ctcccaacgaagacaactag and 5′-gtacaagcaaaaagcagaatcg.
RESULTS AND DISCUSSION
Introduction of multiple lentiviral constructs expressing shRNAs corresponding to different regions of mRNA improves the suppression efficiency
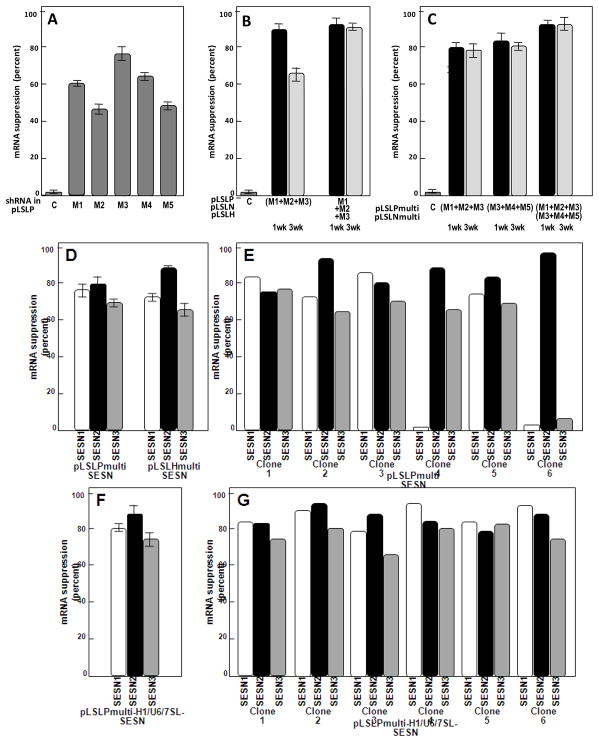
Introduction of a single shRNA rarely produces the level of inhibition greater than 80%. To test the efficiency of five individual shRNAs specific for mitochondrial RNA polymerase gene (POLRMT) transcript we used lentiviral vector pLSLP carrying a single shRNA expressing cassette driven by the H1 RNA promoter. We introduced the constructs into mitochondrial DNA-depleted ρ0 cells (Marusich et al., 1997) derived from the RKO human colon carcinoma cells to avoid toxic effects related to mitochondrial dysfunction after inhibition of mitochondrial RNA polymerase (Kravchenko et al., 2005). The efficiency of inhibition tested by RT-PCR ranged from 45 to 75%, compared to the control cell line carrying empty vector (Figure 2a).
Figure 2.
Levels of inhibition of transcripts after introduction of single and multiple shRNA expressing cassettes. A. Inhibition of the POLRMT transcript in ρ0 RKO cell lines carrying different shRNAs (M1–M5) and a control pLSLP vector. B. Levels of the inhibition after introduction of a mixture of pLSLP constructs carrying shRNA M1, M2 and M3, and three different constructs pLSLP, pLSLH and pLSLN carrying the shRNA and selected in the medium containing G418, hygromycin and puromycin. The levels were measured 1 and 3 weeks after shRNA introduction. C. Levels of transcript inhibition in the cells after introduction of pLSLPmulti expressing three different shRNA (M1 + M2 + M3) and (M3 + M4 + M5), and in the cells expressing two constructs – pLSLPmultiM1M2M3 and pLSLNmultiM3M4M5 and selected in a medium containing puromycin and G418. The levels were measured 1 and 3 weeks after shRNA introduction. The suppression levels were calculated from the results of real-time PCR analysis. D. In RKO cells after introduction of pLSLPmultiSESN and pLSLHmultiSESN that carry different shRNA sequences. E. Inhibition of transcripts in individual clones of RKO cells carrying the pLSLPmultiSESN construct. F. Inhibition of transcripts on mass culture of RKO cells with introduced pLSLPmulti-H1/U6/7SL-SESN construct. G. Inhibition of transcripts in individual clones of RKO cells carrying the pLSLPmulti-H1/U6/7SL-SESN construct.
To improve the efficiency of mRNA inhibition, a mixture of three high-titer stocks of lentiviral shRNA constructs in pLSLP vector (shRNAs 1, 2 and 3) was introduced into the cells. Although one week after the infection and puromycin selection the efficiency of mRNA inhibition reached 90%, the effect was not stable, and after further two weeks the inhibition efficiency dropped to 65% (Figure 2b). Apparently, the deficiency for the mitochondrial RNA polymerase following shRNA introduction resulted in selection against the cells with maximal level of PORMT mRNA suppression.
To achieve stable and efficient inhibition of mRNA the expression cassettes for individual shRNAs were introduced into cells by sequential infection and selection with lentiviral constructs carrying three different selection markers. In a preliminary study we have established that scaling of shRNA level by sequential introduction of three constructs carrying the same type of shRNA did not improve the efficiency of mRNA inhibition (not shown). We therefore designed five different shRNA to same mRNA type (transcript of the POLRMT gene) and introduced consequently two combinations three different shRNA constructs into cell lines. The cell lines carrying shRNAs 1,2 and 3, and 3, 4 and 5 demonstrated approximately 90% inhibition of POLRMT mRNA and the effect remained stable for at least three weeks (Figure 2b). However, the approach is time-consuming due to multiple rounds of selection with different antibiotics.
A single vector can be used for the delivery of multiple shRNA expression cassettes
To simplify the task we introduced into single vector a tandem of three different shRNAs driven by three identical H1 RNA promoters. Previously, as similar approach was tested by Henry et al.(Henry et al., 2006), although the expression cassettes were placed to the region between two the LTRs of lentiviral construct. In the pLSLPmulti vector we placed the tandem of shRNA expressing cassettes to the U3 region o 3′-terminal LTR. As the capacity of lentiviral LTR is known to be not less than 800 bp (Bordignon et al., 1995; Junker et al., 1995) we were able to insert three copies of the H1 RNA promoter cassettes. After the reverse transcription of vector RNAs the DNA insets of such vector constructs should contain two copies of 3′-LTR surrounding the central region of the lentiviral backbone (Figure 1d). This means that the total number of shRNA expressing cassettes is doubled, which does not take place in the vector designs that contain the centrally-localized expression cassettes used by others (Tiscornia et al., 2003; Tiscornia et al., 2006). Apparently, the duplication of shRNA cassettes would give a stronger expression and would lower the probability of potential epigenetic silencing (Lotti et al., 2002). The efficiency of POLRMT mRNA inhibition in the ρ0 RKO cells carrying either pLSLPmulti-shRNA 1,2,3, or pLSLHmulti3,4,5 was 80 and 85%, respectively. A 90% inhibition was observed in the cell line carrying the two constructs simultaneously, and the level of inhibition was stable for at least three weeks (Figure 2c).
Next we decided to test the pLSLPmulti vector system in simultaneous inhibition of three different genes from the sestrin family. The genes specify functionally similar products (Budanov et al., 2004), and in such a case the vector system would be particularly useful, being able to suppress a function supported by redundant genes. By testing in a single shRNA-expressing vector pLSLP we selected two sets of shRNAs for each of the Sestrin genes that inhibit the corresponding mRNAs with efficiencies exceeding 70%. Using the pLSLPmulti vector we have assembled two constructs each carrying three shRNA expressing cassettes for the SESN1, SESN2 and SESN3 genes (pLSLPmulti-SESN and pLSLHmulti-SESN). After introduction into RKO cells and selection for 7 days the mass cultures demonstrated strong inhibition (65–85%) of each of the three sestrin genes (Figure 2d).
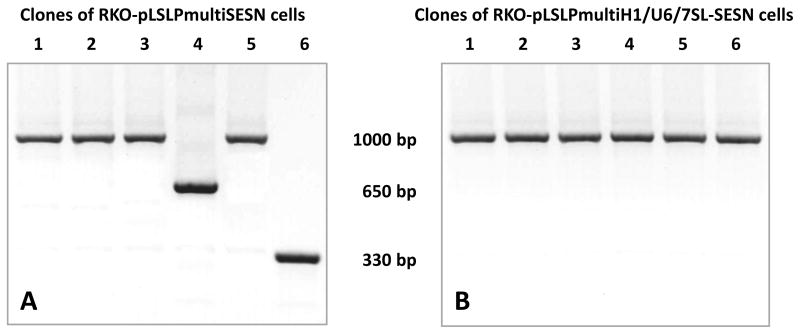
Previously it was reported that the introduction into a single vector of multiple shRNA-expression cassettes carrying identical promoter sequences results in frequent recombination events that remove the repeated sequence fragments. To test whether such events might take place in RKO cells with the introduced pLSLPmultiSESN construct we obtained several individual clones derived from the mass culture. Although four of the six clones displayed strong inhibition of all three sestrin genes, in one clone there was no inhibition of SESN1, and in one – no inhibition of SESN1 and SESN3 genes (Figure 2e). PCR amplification of the LTR region containing the shRNA expressing cassettes have revealed a shortened band corresponding to approximately two H1 RNA promoter cassettes in clone 3, and an even shorter band in clone 5 (Figure 3a). As deletions occur in both copies of integrated LTRs most likely the homologous recombination events take place before packaging of vector RNAs.
Picture 3.
PCR amplification of the LTR region carrying shRNA expressing cassettes in individual clones of RKO cells carrying A. pLSLPmultiSESN and B. pLSLPmulti-H1/U6/7SL-SESN constructs.
A single vector carrying three different RNA polymerase III specific promoters is suitable for stable inhibition of multiple shRNA targets
As the elimination of some promoter cassettes could originate from homologous recombination between two lentiviral RNAs carrying repeats of identical promoter sequences we decided to use three different RNA-polymerase III specific promoters. Promoters of U6 RNA and 7SL RNA have been previously shown to be as efficient for the expression of shRNAs, as the H1RNA promoter (Brummelkamp et al., 2002; Koper-Emde et al., 2004; Miyagishi and Taira, 2002; ter Brake et al., 2008; Yu et al., 2002). We created single-cassette vectors pLU6P and pL7SLP in which the H1 RNA gene promoter of pLSLP was replaced with 256 bp fragment corresponding to the human U6 RNA gene promoter, or with 241 bp fragment corresponding to the human 7SL RNA gene promoter, respectively. Similar to the pLSLP, the vectors demonstrated good efficiency in inhibiting the SESN2 and SESN3 mRNAs when used for the delivery of appropriate shRNAs into RKO cells (not shown). The cassettes U6-shRNA-SESN2-1 and 7SL-shRNA-SESN3-1 were PCR amplified and inserted into pLSLPmulti-shRNA-SESN1-1 and the resulting construct pLSLPmulti-H1/U6/7SL-SESN was tested for suppression of each of the three sestrin genes. There was more than 70% inhibition of the sestrin genes both in the mass culture and in six individual clones (Figure 2f,g). PCR amplification of the LTR region containing the shRNA expressing cassettes have revealed uniform band corresponding to intact triple cassettes in all six clones tested (Figure 3b).
The improved lentiviral based vector system can be used for stable and efficient inhibition of individual mRNAs, including the mRNAs specifying vital functions, as well as for simultaneous suppression of several genes in the same or different pathways, and for inhibition of a function specified by redundant genes.
Acknowledgments
The study was supported by NIH grants R01 CA104903 and AG025276 (P.M.C.), from HHMI grant 55005603 (P.M.C.), from the Russian Foundation for Basic Research (to P.M.C., V.S.P., E.I.F. and J.E.K.), from the Russian Academy of Sciences Program on Molecular and Cellular Biology grant (to P.M.C., V.S.P. and J.E.K.).
Footnotes
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bordignon C, et al. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA- immunodeficient patients. Science. 1995;270:470–475. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- Bos TJ, et al. In Search of the Most Suitable Lentiviral shRNA System. Curr Gene Ther. 2009;9:192–211. doi: 10.2174/156652309788488578. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, et al. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Budanov AV, et al. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Henry SD, et al. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Junker U, et al. Genetic instability of a MoMLV-based antisense double-copy retroviral vector designed for HIV-1 gene therapy. Gene Ther. 1995;2:639–646. [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- Koper-Emde D, et al. RNA interference by small hairpin RNAs synthesised under control of the human 7S K RNA promoter. Biol Chem. 2004;385:791–794. doi: 10.1515/BC.2004.103. [DOI] [PubMed] [Google Scholar]
- Kravchenko JE, et al. Transcription of mammalian messenger RNAs by a nuclear RNA polymerase of mitochondrial origin. Nature. 2005;436:735–739. doi: 10.1038/nature03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, et al. Transcriptional targeting of lentiviral vectors by long terminal repeat enhancer replacement. J Virol. 2002;76:3996–4007. doi: 10.1128/JVI.76.8.3996-4007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusich MF, et al. Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived Rho0 human fibroblasts: a comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochim Biophys Acta. 1997;1362:145–159. doi: 10.1016/s0925-4439(97)00061-6. [DOI] [PubMed] [Google Scholar]
- Matveeva OV, et al. Optimization of duplex stability and terminal assymmetry for shRNA design. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0010180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Razorenova OV, et al. Virus-based reporter systems for monitoring transcriptional activity of hypoxia-inducible factor 1. Gene. 2005;350:89–98. doi: 10.1016/j.gene.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, et al. Multiple shRNA expressing vector enhances efficiency of gene silencing. BMB Rep. 2008;41:358–362. doi: 10.5483/bmbrep.2008.41.5.358. [DOI] [PubMed] [Google Scholar]
- Stove V, et al. Multiple gene knock-out by a single lentiviral vector expressing an array of short hairpin RNAs. Electronic Journal of Biotechnology. 2006;9:572–579. [Google Scholar]
- ter Brake O, et al. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- ter Brake O, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, et al. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci U S A. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, et al. Design and cloning of lentiviral vectors expressing small interfering RNAs. Nat Protoc. 2006;1:234–240. doi: 10.1038/nprot.2006.36. [DOI] [PubMed] [Google Scholar]
- Xia XG, et al. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- Yu JY, et al. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]