Abstract
The enzyme mTOR (mammalian target of rapamycin) is a major target for therapeutic intervention to treat many human diseases, including cancer, but very little is known about the processes that control levels of mTOR protein. Here we show that mTOR is targeted for ubiquitination and consequent degradation by directly binding to the tumor suppressor protein FBXW7. Human breast cancer cell lines and primary tumors showed a reciprocal relationship between loss of FBXW7 and deletion or mutation of PTEN (phosphatase and tensin homologue), which also activates mTOR. Tumor cell lines harboring deletions or mutations in FBXW7 are particularly sensitive to Rapamycin treatment, suggesting that loss of FBXW7 may be a biomarker for human cancers susceptible to treatment with inhibitors of the mTOR pathway.
The FBXW7 gene (also known as hCDC4, FBW7 and hAGO) is a p53-dependent tumor suppressor gene that undergoes deletion and/or mutation in a variety of human tumors (1–3). Loss or mutation of FBXW7 has been associated with increased genetic instability or growth deregulation, due to its effects on ubiquitination and turnover of several oncoproteins (1–7), but the exact mechanism of tumor suppression remains unclear. We carried out a genome-wide search for Fbxw7 ubiquitination targets using the consensus CDC phosphodegron (CPD) sequence I/L-I/L/P-T-P-XXXX (where lysine and arginine are unfavorable in the X locations) (1) in a mouse protein database (http://www.ensembl.org/Mus_musculus). This search revealed a strong match within the HEAT domain of mTor (Fig. S1), which regulates cell growth, metabolism and proliferation. The mTor CPD region is evolutionarily conserved in different species ranging from humans to zebra fish (Fig. S2), suggesting its potential functional importance.
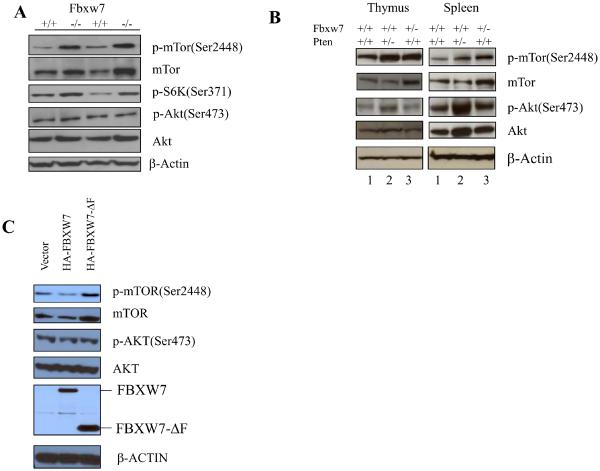
We tested the possibility that Fbxw7 may directly regulate levels of mTor by analyzing two independent preparations of mouse embryonic fibroblasts (MEFs) from Fbxw7−/− mice (Fig. 1A). Depletion of Fbxw7 increased the levels of both total mTor and phosphorylated mTor (p-mTor), as well as the downstream mTor target S6-kinase (PS6K). In contrast, there was no appreciable effect on upstream components of the mTor signaling pathway such as Akt and phosphorated Akt (p-Akt) (Fig. 1A).
Figure 1.
Depletion of FBXW7 increases mTOR and p-mTOR levels. (A) The levels of mTor and p-mTor, as well as the downstream mTor target S6-kinase (P-S6), are upregulated in Fbxw7−/− MEFs, whereas the levels of Akt and p-Akt (lower panels) were not appreciably affected. Two independent preparations were used. (B) Elevation of mTor in thymus (left panel) and spleen (right panel) from Fbxw7+/− mice. Lanes 1–3 contain extracts from wild-type, Pten+/−, and Fbxw7+/− mice respectively. (C) Overexpression of either full length FBXW7 (HA-FBXW7) or of a dominant-negative form (HA-FBXW7-ΔF) in 293T cells respectively decreases (lane 2) or increases (lane 3) the mTOR and p-mTOR levels..
Depletion of Fbxw7 in vivo also led to an increase in both mTor and p-mTor protein levels. Thymus and spleen from Fbxw7+/− mice (Fig. 1B) showed increased mTor and p-mTor in comparison with control wild type tissue (Fig. 1B). In contrast, levels of total Akt and p-Akt (at Ser437) did not change appreciably (Fig. 1B). However in Pten+/− mice (Fig. 1B), both thymus and spleen showed an increase in p-mTor without any obvious change in total mTor protein level. In agreement with these observations, p-Akt levels were elevated in thymus and spleen from Pten+/− mice and presumably were responsible for activation of mTor signaling (Fig. 1B).
A similar trend was seen in human cells from which the FBXW7 gene had been deleted by homologous recombination (4). Both HCT116 and DLD1 cells that lacked FBXW7 had slightly higher levels of mTOR and p-mTOR, as well as increased levels of p-S6K (Fig. S3). Again, no effect was seen on the upstream targets AKT and p-AKT (Fig. S3). Human 293T cells transfected with a dominant-negative form of FBXW7 (HA-FBXW7-ΔF) (8) also showed an increase in the level of both total mTOR and p-mTOR (Fig. 1C), whereas cells expressing the transfected normal HA-FBXW7 protein reproducibly had lower levels of both total mTOR and p-mTOR (Fig. 1C). In contrast, levels of AKT and p-AKT were not affected by overexpression of FBXW7 (Fig. 1C).
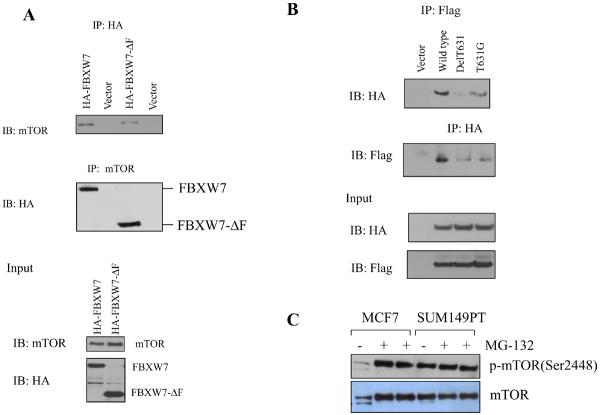
We next investigated possible interactions between mTOR and FBXW7 proteins. A vector encoding HA-tagged FBXW7 was transfected into 293T cells, followed by immunoprecipitation using antibodies directed against the HA tag or mTOR, and immuno blot analysis. Both showed an interaction between the mTOR and FBXW7 proteins (Fig. 2A). Immunoprecipitation also revealed an interaction between mTOR and FBXW7-ΔF (Fig. 2A), indicating that the WD40 domain of FBXW7 is the interaction site. Additional controls for the specificity of the interaction between FBXW7 and mTOR are shown in Figs. S4 and S5.
Figure 2.
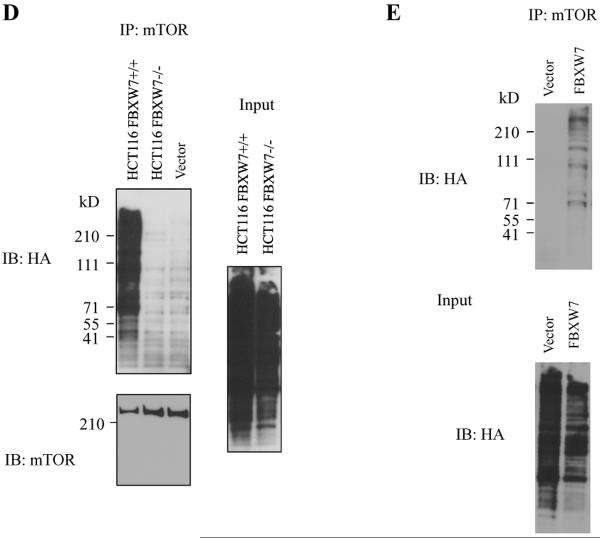
mTOR interacts with FBXW7. (A) Immunoprecipitation of HA tagged FBXW7 identifies mTOR as an interacting protein. HA-tagged FBXW7 or FBXW7-Δ were expressed in human 293T cells, followed by immunoprecipitation of the proteins with anti-HA antibodies and immunoblotting with antibodies against mTOR (top panel). The reciprocal experiment shows that the region of FBXW7 that interacts is the WD40 domain (middle panel). Lower panel shows the input levels of mTOR and FBXW7 proteins in the lysates. (B) FBXW7 binds the wild-type fragment of mTOR, but binding to the mTOR(delT631) and mTOR(T631G) mutants was dramatically reduced. (C) MCF7 breast cancer cells show increased mTOR levels upon treatment with MG-132, but this is not seen in SUM149PT cells which have no functional FBXW7 gene. (D) HCT116_WT and HCT116_FBXW7−/− cells were transfected with HA-ubiquitin. Immunoprecipitation of mTOR followed by Western blot analysis of the HA-ubiquitin showed that ubiquitination of mTOR was only seen in the HCT116_WT cells, and not in HCT116_FBXW7−/− cells. The vector lane shows HCT116_FBXW7−/− cells transfected with empty vector construct. (E) Ubiquitination of mTOR is restored by exogenous FBXW7 expression. HCT116_FBXW7−/− cells were transfected with an FBXW7-expressing construct and HA-tagged ubiquitin. Immunoprecipitation of mTOR showed increased ubiquitination compared to controls (fig. S7).
We cloned a fragment of mTOR (from 1 to 898 AAs) that contained the putative CPD, and generated two mutants, one carrying a deletion of T631 [mTOR(delT631)], and the other a point mutation converting T631 to G [mTOR(T631G)]. The wild type and mutant mTOR fragments were co-expressed with HA-FBXW7 in 293T cells, followed by immunoprecipitation with antibodies against one protein and immuno blot analysis of the second. Although the wild type fragment of mTOR immunoprecipitated efficiently with FBXW7, binding to the mTOR (delT631) and mTOR(T631G) mutants was dramatically reduced (Fig. 2B). Residual binding could be due to the presence of an additional, but weaker, consensus binding site in mTOR (located at position AA 314–319). Thus FBXW7 binds mTOR predominantly through the major conserved CPD site.
To determine whether the regulation of mTOR by FBXW7 is through the proteasome-dependent degradation pathway, MCF7 and SUM149PT cells (9, and Table S1) were treated with the proteasome inhibitor MG-132. Proteasome inhibition caused a dramatic increase in the mTOR levels in MCF7 cells, which retain FBXW7, but not in SUM147PT cells, which have a homozygous mutation (Fig. 2C), suggesting that degradation of mTOR is FBXW7-dependent. To examine whether the ubiquitination status of mTOR was FBXW7-dependent, 293T and SUM149PT were transfected with a vector containing CMV promoter-driven HA-ubiquitin. Immunoprecipitation of mTOR followed by immuno blot analysis of the HA-ubiquitin showed that mTOR ubiquitination was found only in cells retaining a functional FBXW7 gene (Fig. S6). The same experiment in HCT116_WT and HCT116_FBXW7−/− cells (Fig. 2D) again showed efficient ubiquitination of mTOR only in HCT116_WT cells. Finally, we co-transfected SUM149PT cells with constructs encoding both FBXW7 and HA-ubiquitin, and found that ubiquitination of mTOR was restored by exogenous FBXW7 expression (Fig. 2E). Thus ubiquitination of mTOR is largely, if not exclusively, mediated by binding to FBXW7.
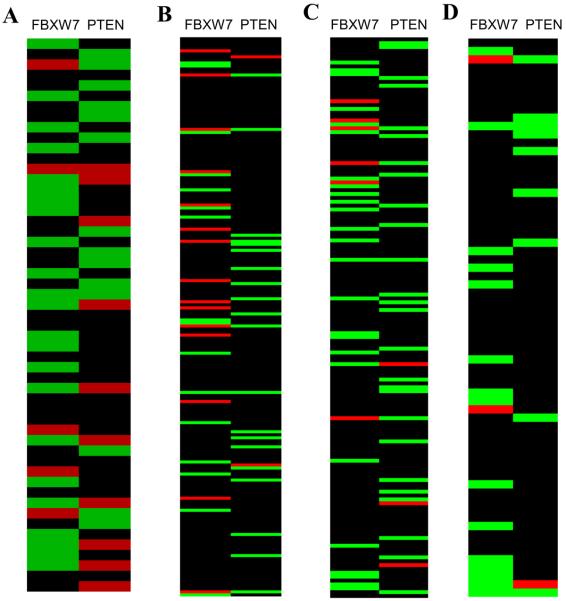
Since FBXW7 and PTEN both affect signaling through mTOR, we examined the genetic status of both genes in a panel of 53 breast cancer cell lines (10). Quantitative TaqMan assays of the number of copies of FBXW7 and PTEN genes in each of the cell lines were in good concordance with data found by BAC CGH Microarray (see Table S1). Most of the breast cancer cell lines that exhibited loss of a single copy of FBXW7 (23 out of 53, shown in green in the FBXW7 column, Fig. 3A) did not show corresponding loss of PTEN. In contrast, of the 14 lines that showed loss of a single copy of PTEN (green bars in the PTEN column, Fig. 3A), only one had also lost a copy of FBXW7, suggesting that FBXW7 and PTEN show some functional redundancy in tumor development. Similar results were obtained by examination of the copy number status of genomic regions containing FBXW7 and PTEN genes in three independent human primary breast cancer sets for which BAC CGH microarray data were available (11–13). From a total of 450 tumor and cell line DNA samples shown in Fig.3A–D, only 4 had lost a copies of the regions containing both genes, a result that is unlikely to be a consequence of random genetic alterations (p= 4.9 × 10−7).
Figure 3.
Genetic interaction between FBXW7 and PTEN in human breast cancers. The green bar indicates loss, the red bar indicates gain, and the black bar indicates no changes. (A) The data from 53 human breast cancer cell lines, ordered in the vertical axis from 1–53 (10). The copy number of FBXW7 and PTEN was determined by quantitative PCR TaqMan. Deletion of FBXW7 rarely occurred in tumors that also show deletion of PTEN (p=0.014). (B), (C) and (D) are three independent sets of human primary breast cancers, analyzed by the same BAC CGH microarray platform. (B) 185 human primary breast cancers (11). (C) 145 human primary breast cancers (12). (D) 67 human primary breast cancers (13). The copy number was determined based on the published CGH data (FBXW7 was based on BAC RP11-73G16, PTEN on BAC RP11-380G5). Loss is defined as log2(ratio) <−0.25 and gain as log2(ratio) >0.25.
We also considered the possibility that other somatic changes such as point mutations or gene silencing events could impact the results. The FBXW7 gene continued to be expressed in all 25 breast cancer cell lines examined (Fig. S8), indicating that no gene silencing had occurred, although very low levels were found in 5 cell lines (lanes 10, 13, 14, 16, 20; fig. S8). All of these lines had lost one copy of the FBXW7 gene except one (SUM149PT, lane 16), in which a point mutation was detected (Table S1). The PTEN gene was found to be silent in two cell lines (Fig. S8, lane 11 and 12), and both had lost one copy of the PTEN gene. Three mutations in PTEN were found (Fig. S8, and Table S1). Thus gene silencing (for example by promoter methylation) or point mutations in FBXW7 and PTEN are relatively rare mechanisms of inactivation of these genes, in comparison to single copy deletions. These data are further compatible with the identification of both genes as haplo-insufficient tumor suppressors (3, 14, 15).
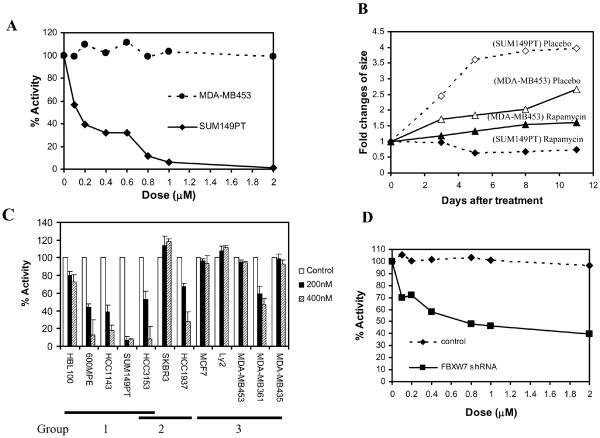
Because deletion or mutation of FBXW7 in human breast cancer cells leads to increased levels of mTOR, we tested the possibility that cells harboring these deletions may show increased sensitivity to the mTOR inhibitor rapamycin. We treated two breast cancer cell lines, SUM149PT cells (homozygous FBXW7 mutations) and MDA-MB453 cells (wild type FBXW7) with rapamycin and counted numbers of viable cells. SUM149PT cells proved to be very sensitive to this treatment (IC50 < 200 nM), whereas MDA-MB453 cells were relatively resistant (IC50 > 2 μM) (Fig. 4A). In nude mouse xenografts, groups of 5 mice were injected with both cell lines, one on each flank, and were treated by intraperitoneal injection with rapamycin over an 11-day time period. The SUM149PT cells showed a relative decrease in size followed by stable tumor growth, whereas the MDA-MB453 cells were relatively unaffected by treatment (Fig. 4B).
Figure 4.
Loss of FBXW7 increases sensitivity to an mTOR inhibitor (rapamycin). (A) The breast cancer SUM149PT cells which have a homozygous mutation in FBXW7 were killed at a rapamycin concentration of 100nM, whereas MDA-MB453 cells with wild type FBXW7 were resistant. (B) Treatment of nude mouse xenografts with rapamycin. The SUM149PT cells showed a relative decrease in size followed by stable tumor growth, whereas the MDA-MB453 cells were unaffected by treatment. (C) Sensitivity to mTOR rapamycin in a range of breast cancer cell lines. Tumor cells with deletion or mutation of FBXW7 are in Group 1, those with deletion or mutation of PTEN are Group 2, and cells with wild type copies of both genes are in Group 3. (D) Downregulation of FBXW7 using specific shRNA in rapamycin resistant MDA-MB453 cells increases the sensitivity to this treatment.
An additional set of 10 breast cancer cell lines were treated with rapamycin at concentrations of 200nM and 400nM. Cells with deletion or mutation of FBXW7 (HBL100, 600MPE, SUM149PT, HCC3153 and HCC1143) or PTEN (HCC1937 and HCC3153) showed significant sensitivity to killing by rapamycin, although the magnitude of the effect varied (16) (Fig. 4C). To establish a direct link between loss of FBXW7 and rapamycin sensitivity, we downregulated expression levels of FBXW7 using shRNA (17) in the rapamycin-resistant MDA-MB453 cells, resulting in an increase in sensitivity to this drug (IC50< 0.8μM; Fig. 4D).
Our findings implicate FBXW7 in an evolutionarily conserved pathway that controls regulation of mTOR protein levels. Because FBXW7 is a haploinsufficient tumor suppressor that undergoes heterozygous loss in a substantial proportion of human tumours, the data suggest new approaches to reduce mTOR levels in cancers by the use of drugs that may re-activate the remaining copy of FBXW7 in a similar way that nutlins have been shown to activate wild type copies of p53 in human tumors (18). Loss of FBXW7 may also be a useful biomarker for sensitivity of human tumors to inhibitors of the mTOR pathway.
Supplementary Material
Acknowledgments
We are grateful to B. Vogelstein for providing us with the HCT116 WT, HCT116 FBXW7−/−, DLD1 wild type and DLD1 FBXW7−/− cell lines, KI Nakayama for providing Fbxw7 knockout mice and vectors (HA-FBXW7 and HA-FBXW7Δ) and O. Tetsu for vector encoding HA-ubiquitin. These studies were supported by NCI grant U01 CA84244 and the DOE (DE-FG02-03ER63630) to AB, the UCSF Research-Evaluation Allocation Committee (REAC) to JHM. AB acknowledges support from the Barbara Bass Bakar Chair of Cancer Genetics.
References and Notes
- 1.Welcker M, Clurman BE. Nat. Rev. Cancer. 2008;8:83. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 2.Akhoondi S, et al. Cancer Res. 2007;67:9006. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 3.Mao JH, et al. Nature. 2004;432:775. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 4.Rajagopalan H, et al. Nature. 2004;428:77. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 5.Yada M, et al. EMBO J. 2004;23:2116. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welcker M, et al. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9085. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemp Z, et al. Cancer Res. 2005;65:11361. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, et al. Mol. Cell Biol. 2001;21:7403. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Nature. 2001;413:316. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 10.Neve RM, et al. Cancer Cell. 2006;10:515. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Climent J, et al. Cancer Res. 2007;67:818. doi: 10.1158/0008-5472.CAN-06-3307. [DOI] [PubMed] [Google Scholar]
- 12.Chin K, et al. Cancer Cell. 2006;10:529. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Fridlyand J, et al. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao JH, Wu D, Perez-Losada J, Nagase H, DelRosario R, Balmain A. Oncogene. 2003;22:8379. doi: 10.1038/sj.onc.1207083. [DOI] [PubMed] [Google Scholar]
- 15.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Nat. Genet. 1998;19:348. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 16.Steelman LS, et al. Oncogene. 2008;27:4086. doi: 10.1038/onc.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. Curr. Biol. 2004;14:1852. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 18.Buolamwini JK, Addo J, Kamath S, Patil S, Mason D, Ores M. Curr. Cancer Drug Targets. 2005;5:57. doi: 10.2174/1568009053332672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.