Abstract
Prostacyclin synthase (PGIS) is a membrane-bound class III cytochrome P450 that catalyzes an isomerization of prostaglandin H2, an endoperoxide, to prostacyclin. We report here the characterization of the PGIS intermediates in reactions with other peroxides, peracetic acid (PA), and iodosylbenzene. Rapid-scan stopped-flow experiments revealed an intermediate with an absorption spectrum similar to that of compound ES (Cpd ES), which is an oxo–ferryl (Fe(IV)=O) plus a protein-derived radical. Cpd ES, formed upon reaction with PA, has an X-band (9 GHz) EPR signal of g = 2.0047 and a half-saturation power, P1/2, of 0.73 mW. High-field (130 GHz) EPR reveals the presence of two species of tyrosyl radicals in Cpd ES with their g-tensor components (gx, gy, gz) of 2.00970, 2.00433, 2.00211 and 2.00700, 2.00433, 2.00211 at a 1:2 ratio, indicating that one is involved in hydrogen bonding and the other is not. The line width of the g = 2 signal becomes narrower, while its P1/2 value becomes smaller as the reaction proceeds, indicating migration of the unpaired electron to an alternative site. The rate of electron migration (~0.2 s−1) is similar to that of heme bleaching, suggesting the migration is associated with the enzymatic inactivation. Moreover, a g = 6 signal that is presumably a high-spin ferric species emerges after the appearance of the amino acid radical and subsequently decays at a rate comparable to that of enzymatic inactivation. This loss of the g = 6 species thus likely indicates another pathway leading to enzymatic inactivation. The inactivation, however, was prevented by the exogenous reductant guaiacol. The studies of PGIS with PA described herein provide a mechanistic model of a peroxidase reaction catalyzed by the class III cytochromes P450.
Cytochromes P450 (P450)1 play important roles in physiological processes, pharmaceutical metabolism, and catalysis of a variety of reactions, such as hydroxylations, epoxidations, N- and O-dealkylations, and isomerizations. P450 enzymes contain a low-spin ferric heme with a cysteinate residue as the proximal ligand (1). Binding of the substrate often induces the heme to convert to high spin with detachment of the distal ligand and an increase of the redox potential, thus facilitating the first electron transfer from a redox protein to the heme iron. The resulting ferrous heme binds an oxygen molecule to form an oxy–ferrous complex, which then accepts a second electron to form a peroxo–ferric intermediate. The subsequent steps, which occur rapidly, include protonation of the peroxo–ferric intermediate and heterolytic O–O bond cleavage with concomitant formation of water and an oxo–ferryl intermediate that is capable of catalyzing the oxidation of substrate. Cryogenic studies have detected the peroxo–ferric and hydroperoxo–ferric species in P450cam (2). Although a tentative oxo–ferryl species was also identified in a transient crystallographic study of P450cam (3), there has been an intensive pursuit to trap and identify the intermediate or its electronic equivalent in P450s. For decades, the intermediate containing oxo–ferryl heme of P450 was thought to be equivalent to compound I (Cpd I) of peroxidases (4), which contains an oxo–ferryl porphyrin π-cation radical (Fe(IV)=O + porphyrin+•) (5–7). However, some peroxidases produced a compound ES (Cpd ES) intermediate, which contains an oxo–ferryl (Fe(IV)=O) and an amino acid radical (8–10). Cpd ES species most likely result from intramolecular electron transfer between Cpd I and nearby amino acid residues, with the most common radical sites being tyrosine and tryptophan.
P450s can also directly interact with a peroxide or peracid to generate the oxo–-ferryl intermediate (4). Although the reaction readily leads to heme bleaching, the peroxide shunt provides an alternative pathway to oxo–ferryl species without the need for a reductase protein, reducing equivalents, and oxygen. Reactions of m-chloroperbenzoic acid with P450cam (11), P450BM3 (12) or CYP119 (13) produced Cpd I intermediates in which the Soret band was significantly diminished and its peak was shifted to ~370 nm, whereas the Q-band (collective name for the α- and β-bands) lost resolution of its vibronic transitions. These features closely resembled the Cpd I of chloroperoxidase, a well-characterized thiolate-ligated Cpd I (14). However, Cpd I in the reaction of P450cam with m-chloroperbenzoic acid was unstable and rapidly converted to Cpd ES with a Soret peak at ~407 nm (15). Cpd ES was also formed in the reaction of P450cam with peracetic acid (PA) and was further characterized by rapid freeze-quench EPR and Mössbauer spectroscopy (16–18). High-field EPR assigned the radical signal at g = 2 as a tyrosyl radical, and Mössbauer spectra confirmed the iron(IV) electronic configuration. In the reaction of P450BM3 with PA, a tyrosyl radical and a tryptophan radical were observed (19), suggesting that intramolecular electron transfer from aromatic amino acids to the porphyrin is a common feature in the reaction of P450s with PA. Efforts hitherto in identifying the chemical nature of the intermediates have been mainly restricted to the soluble microbial P450s. Studies on the membrane-bound, eukaryotic P450s suffer from kinetic heterogeneity, partly because the membranes are usually heterogeneous in the presence of detergent.
Prostacyclin synthase (also known as prostaglandin I2 synthase; PGIS) converts prostaglandin H2 to prostacyclin, a highly potent vasodilator and agonist for antiplatelet aggregation (20). PGIS is located in the membrane of endoplasmic reticulum and was assigned to the P450 superfamily as CYP8A1 (21, 22). Unlike other microsomal P450s that catalyze monooxygenation, PGIS catalyzes an isomerization of an endoperoxide and does not need molecular oxygen, reductase, or any other external electron donor and is thus classified as a class III P450. The catalytic mechanism of PGIS is therefore more akin to a peroxidase than to a monooxygenase (23). In spite of its catalytic difference from other P450s, PGIS retains the P450 features regarding active site hydrophobicity, UV/visible, magnetic circular dichroism, and EPR spectra, and the triangular prism-shaped tertiary structure (24, 25). We previously reported a high-yield prokaryotic system for expression of human PGIS (24). The recombinant PGIS is active, soluble, and monomeric (25), making it suitable for kinetic studies as a microsomal P450. In this study, kinetic measurements by UV/visible absorption and X- and D-band EPR spectroscopy were applied to characterize intermediates formed in the reaction of PGIS with PA or iodosylbenzene (PhIO).
MATERIALS AND METHODS
Materials
PA was purchased from Aldrich. Guaiacol was obtained from Sigma. PhIO was prepared as described previously (26). Recombinant human PGIS was expressed in Escherichia coli and purified as described previously (25). PGIS concentration was determined by using ε418 = 103 mM−1 cm−1 (24).
Steady-State Kinetic Analysis Using Guaiacol as Cosubstrate
UV/visible absorption spectra and kinetic traces of product formation were performed on a Shimadzu UV-2501PC spectrophotometer. Assays were carried out in 20 mM NaPi and 10% glycerol, pH 7.5, containing 1.78 mM guaiacol. To obtain steady-state parameters (KM and Vmax) of PGIS-catalyzed guaiacol oxidation, various concentrations of PA were added into 1 mL reaction buffer containing 1 μM enzyme. The initial rate of the product formation was calculated from ΔA470/min using Δε470 = 26.6 mM−1 cm−1 (27).
Kinetic Measurements by Stopped-Flow Spectroscopy
Single-wavelength kinetics and rapid-scan spectroscopy were performed on an Applied Photophysics (Leatherhead, U.K.) Bio-Sequential model DX-18MV stopped-flow spectrophotometer with diode array rapid-scan capability. Reactions of PGIS with PA were monitored by rapid scan using a 2 mm path optical cell, whereas the cell of 1 cm light path was used for reactions with PhIO. PGIS in 20 mM NaPi and 10% glycerol, pH 7.5, was loaded in one syringe, and PA or PhIO was in the other. Because PhIO is not stable in the presence of glycerol, glycerol was excluded when PhIO was used as an oxidant. The kinetic traces and spectral changes were recorded at 23 °C by both single-wavelength and rapid-scan modes. The absorption spectra of reaction intermediates were resolved by singular value decomposition using the ProK software package (Applied Photophysics Ltd.). In single-wavelength experiments, enzyme oxidation was monitored at 418 nm. Data were fit to the standard equation for two parallel reactions:
| (1) |
where t is the reaction time and k1 and k2 are the observed rate constants for the first and second phases, respectively. B1 and B2 are the amplitudes, and C is the final absorbance. Binding (kon) and dissociation (koff) rate constants were estimated from the slope and intercept, respectively, of the secondary plot of kobs versus reactant concentrations.
Reactivity of Cpd ES with Guaiacol
We performed sequential-mixing stopped-flow experiments at 23 °C to monitor the reactivity of Cpd ES with guaiacol. Briefly, PGIS (6.5 μM) was reacted with PA (150 μM) in the first mixing for a given time in an aging loop before further reaction with 3.46 mM guaiacol in the second mixing. Exponential decay of the initial rates of guaiacol oxidation determined by A470nm change is the rate of self-inactivation.
Preparation of EPR Samples
Samples of longer than 5 s reaction time were prepared by manual mixing of PGIS (a final concentration of 37.5 μM) with a 10-fold excess of PA or PhIO. The samples, after a given reaction time, were frozen immediately in a dry ice/acetone bath and then transferred to liquid nitrogen. Rapid-mixing/freeze-quench samples for X-band EPR analysis were prepared using a System 1000 chemical/freeze quench apparatus with a Model 1019 syringe ram, a Model 715 ram controller, and a 0.008 in. nozzle (Update Instrument, Inc., Madison, WI). PGIS (150 μM) in 20 mM NaPi and 10% glycerol, pH 7.5, was loaded in one syringe and a 10-fold excess of PA or PhIO in another. Glycerol was omitted in the PhIO reaction to prevent their direct reaction. Single-push mixing was used for reaction times of less than 263 ms, and a double-push program was used for longer reaction times. The shortest quenching time of the apparatus is about 5 ms at 2 cm/s ram velocity. The reaction of PGIS and PA (or PhIO) was initiated by mixing two reagents at room temperature and freeze-quenched by spraying the reaction mixtures through the nozzle into prechilled isopentane (130–135 K) (28). Due to the 1:1 mixing (2-fold dilution) and packing dilution factor (2-fold dilution), the final PGIS concentration was 37.5 μM. Rapid-mixing/freeze-quench samples were collected into 5 mm quartz EPR tubes and packed by a custom-built gas pressure device (29).
For D-band sample collection, the nozzle used in X-band sample collection was replaced with a quartz capillary (0.6 mm inner diameter, 0.7 mm outer diameter, and 2 cm height). PGIS (240 μM) was placed in one syringe, and PA (2.4 mM) was placed in the other. Push-push mode was applied for sample collection. The first push allowed the reactants to incubate in the reaction tube for a given time, and the second push delivered the reaction sample to the capillary, which was placed immediately above a liquid nitrogen bath. As soon as the reaction sample entered the capillary, the capillary was immediately disconnected with a pair of prechilled forceps and dropped into a liquid nitrogen bath. Samples were reacted for 100 ms and 5 s, freeze-quenched for ~1 s, and stored in liquid nitrogen before analysis.
X-band EPR Spectroscopy
All X-band (9.6 or 9.3 GHz) EPR spectra were recorded at liquid helium (10 K) or liquid nitrogen (110 K) temperature on a Bruker EMX spectrometer. For the liquid helium system, temperature was maintained by a GFS600 transfer line and an ITC503 temperature controller. An Oxford ESP900 cryostat was used to accommodate the quartz sample tube. For liquid nitrogen transfer, a silver-coated double-jacketed glass transfer line and a BVT3000 temperature controller were used. Data were analyzed using WinEPR programs that were included with the EMX system. Spin quantification was performed by double integration of the g = 2 radical signal using the signal of 1 mM Cu2+ as a standard. EPR line width was defined by the width of magnetic field between the peak and trough of individual EPR signals (30). The data of microwave power dependent experiments were analyzed by nonlinear regression to the equation:
| (2) |
where P is the power, S is the peak to trough amplitude of the EPR signal, P1/2 is the power at half-saturation, b is assigned as 1 for powdered biological samples, and K is a floating parameter (31).
High-Frequency EPR Spectroscopy
D-band EPR spectra were performed on a spectrometer assembled at Albert Einstein College of Medicine, as previously described (32, 33). The magnetic field is generated using a 7 T Magnex superconducting magnet equipped with a 0.5 T sweep/active shielding coil. Field-swept spectra were obtained in the two-pulse echo-detected mode with the following parameters: temperature, 15 K; repetition rate, 100 Hz; 90 degree pulse, 40 ns; time τ between pulses, 130 ns. The magnetic field was calibrated to an accuracy of ~3 G using a sample of manganese doped into MgO. The temperature of the sample was maintained to an accuracy of approximately ±0.3 K using an Oxford Spectrostat continuous flow cryostat and ITC503 temperature controller.
X- and D-band EPR Spectral Simulations and Electron Pathway Analysis
The X-band and D-band spectra were simulated using software described previously (34, 35). The hyperfine interactions were treated to first order, and transition probabilities were taken as unity. The distribution in gx was modeled as a Gaussian: spectra with the appropriate g values were calculated, weighted according to position in the distribution, and summed to generate the final simulation. Electron transfer coupling efficiency was calculated using the PATHWAYS analysis in the HARLEM programs kindly provided by Drs. Kurniko and Beratan at Duke University (36).
RESULTS
Stopped-Flow Measurements of the Reaction of PGIS with PA
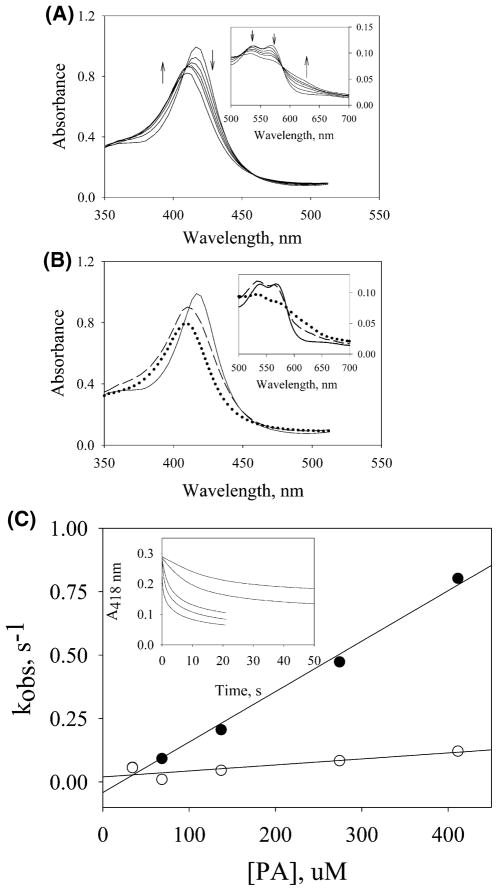
Stopped-flow analysis of PGIS reacting with PA was performed using both photodiode array and single-wavelength modes to follow the spectral changes in the Soret and Q-band regions. The combined approach aimed to study the kinetic steps associated with the substrate binding and intermediate formation. Kinetics of 110 μM ferric PGIS with a 10-fold excess PA was biphasic over a 1 s reaction time (Figure 1A). The first phase exhibited a Soret peak blue shifting from 418 to ~408 nm accompanied by a decrease in intensity. The α- and β-bands, however, showed slight decreases in absorbance (Figure 1A, inset). These features were different from Cpd I formation in which the Soret peak is blue shifted from 407 to 364 nm, whereas the original Q-band was diminished and a new charge transfer band at 690 nm was formed (37–39). With PGIS the spectral changes for the first 0.5 s revealed three isosbestic points at 410, 461, and 588 nm. Following the first phase, heme bleaching occurred, and the baseline was significantly shifted, making it difficult for further characterization. Singular value decomposition and global analyses (Figure 1B) were thus performed over the initial second of the reaction using an irreversible two-step model of A → B → C. The rate constants of the first and second phases were k1 = 4.3 s−1 and k2 = 1.0 s−1, respectively. The B → C step represents the bleaching of heme. However, characterization of species C and determination of k2 were not reliable because the simulation was limited to ~1 s due to the subsequent baseline shift. The spectrum of species B with the Soret peak at ~408 nm was very similar to that of Cpd ES in the reaction of P450cam with PA in which the Soret peak was ~406 nm (11, 15). The Q-band of species B exhibited the general feature of an oxo–ferryl intermediate and has a higher absorbance in the β-band than in the α-band, similar to those found for P450cam and P450BM3 (11, 12, 15) (Figure 1B, inset). However, the spectral features of Cpd I were not observed, suggesting that Cpd I, if formed, was rapidly converted to Cpd ES.
Figure 1.
Stopped-flow experiments of the reaction of PGIS (110 μM) with PA (1.1 mM) at 23 °C using a 2 mm light path. (A) Rapid-scan spectra were recorded at 0.0013, 0.129, 0.257, 0.385, 0.513, and 1.0 s after mixing. Arrows indicate the direction of spectral changes with increasing time. Inset: Eexpansion of data in the visible region. (B) Spectral intermediates were resolved by a two-step model A → B → C using ProK analysis. The spectrum for species A is shown with a solid line, B with a dashed line, and C with a dotted line in the Soret region and the visible region (inset). (C) Secondary plots of the observed rates determined at 418 nm versus PA concentration were analyzed for the first phase (filled circles) and the second phase (open circles). The inset shows the kinetic traces for reactions of PGIS with 35 (top), 70, 140, 280, and 420 (bottom) μM PA.
The kinetics of PGIS reacting with PA were investigated in more detail at 418 nm with the stopped-flow instrument in the single-wavelength mode. PGIS (~3 μM) was reacted with various concentrations of PA (35–420 μM) (Figure 1C, inset). All of the kinetic traces were best fit by a two-exponential function. The first phase was dependent on PA concentration (Figure 1C). The slope was used to estimate the second-order rate constant for PA association (kon) as 2200 M−1 s−1. The intercept, however, is a negative value, suggesting that the apparent irreversible first phase was not a simple binding step but included both the binding and the O–O bond cleavage steps. The second phase corresponded to heme bleaching and was fit with rate constants in the range of 0.1–0.2 s−1. The rates, however, are slightly higher at higher PA concentrations, thus making it somewhat concentration-dependent. This concentration dependence may be explained by the fact that heme bleaching (described below) proceeds via more than one pathway, one of which predominates at higher PA concentration.
Steady-state kinetic parameters for the PGIS-catalyzed PA reduction were also determined using a coupled assay that follows the absorbance change of guaiacol oxidation. Guaiacol is a well-known cosubstrate of peroxidases, catalyzing the reduction of Cpd I, Cpd ES, or Cpd II back to the ferric resting enzyme and generating a colored product with an absorption maximum at 470 nm (11, 27). A KM of 0.40 mM PA and a Vmax of 320 mol of consumed guaiacol (mol of PGIS)−1 min−1 were thus obtained (data not shown). The Vmax of 320 min−1, or 5.3 s−1, is close to the first-phase rate constant of 4.3 s−1 determined at high concentrations of PGIS and PA (Figure 1B). These results suggested that the rate-limiting step of PGIS in reaction with PA occurs during the first phase that includes both the binding and the O–O bond cleavage steps. Since no saturation behavior is observed for the plot of observed rate constant vs PA concentration (Figure 1C), we conclude that the binding step is rate-limiting in forming Cpd ES at all PA concentrations used in the kinetic measurements.
Stopped-Flow Measurements of PGIS Reacting with PhIO
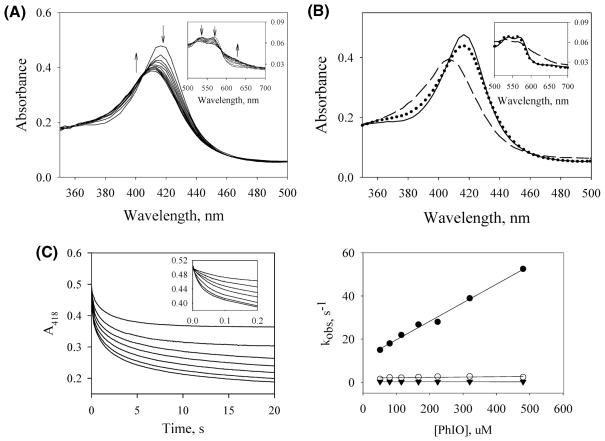
PhIO is a commonly used oxidant in peroxidase reactions because it is a highly efficient oxygen donor and thus is effective in the generation of Cpd I intermediates (40–43). The presence of only one oxygen atom in PhIO and the effectiveness of Ph–I− as a leaving group make PhIO a good reagent for producing Cpd I or Cpd ES. We reacted ferric PGIS with PhIO and monitored the reaction by stopped-flow rapid-scan spectroscopy. Figure 2A shows the spectral changes for the reaction of PGIS (5 μM) with a 25-fold excess of PhIO over 1 s. Longer incubation resulted in the heme bleaching (data not shown). Spectra in the first ~100 ms showed a slight decrease of the Soret band without any shift of its maximum, and the Q-band had very little change. Subsequent spectra showed a simultaneous blue shift and decrease of the Soret band accompanied by a diminishing Q-band that exhibited a higher absorbance of β compared to the α-band. Three isosbestic points at 407, 463, and 579 nm were also observed. The spectral intermediates were resolved by singular value decomposition and global analysis using the model of A′ → B′ → C′ over 0.5 s of reaction time (Figure 2B). The rate constants k1 and k2 were estimated to be 16.2 and 2.0 s−1, respectively. The spectrum of species C′ had a Soret peak at 406 nm and the Q-band features that are similar to those observed for the species B in the reaction of PGIS with PA (Figure 1B), suggesting the formation of the Cpd ES intermediate in the reaction with either PhIO or PA. Species B′ is thus likely a binding complex of PGIS and PhIO. To verify this, reactions with various concentrations of PhIO were subjected to single-wavelength kinetic measurements by following 418 nm absorbance changes over 20 s (Figure 2C, left panel). All data show triphasic kinetics. The first phase, completed in 100 ms, was dependent on PhIO concentration, whereas the subsequent two phases were not (Figure 2C, right panel). The kon and koff obtained from the slope and intercept of the first phase were 8.6 × 104 M−1 s−1 and 11.1 s−1, respectively. The dissociation constant thus estimated was 0.13 mM. Consistent with the rapid-scan results, the first phase is assigned to the formation of binding complex B′. The second and third phases with rate constants of 2.0 and 0.26 s−1 are assigned to the steps of I–O cleavage and heme bleaching, respectively.
Figure 2.
Stopped-flow experiments of the reaction of PGIS (10 μM) with PhIO (250 μM) at 23 °C using a 1 cm light path. (A) Rapid-scan spectra were recorded at 0.0013, 0.104, 0.206, 0.308, 0.411, 0.513, 0.616, 0.718, 0.821, 0.923, and 1.02 s. Arrows indicate the direction of spectral changes with increasing time. (B) Spectral intermediates A′ (solid line), B′ (dotted line), and C′ (dashed line) were resolved by a two-step model A′ → B′ → C′ over 0.5 s reaction using global analysis. (C) Left panel: Time courses of PGIS reacted with 51 (top), 80, 115, 166, 224, 320, and 480 (bottom) μM PhIO. The kinetic traces of the initial 0.2 s are shown in the inset. Right panel: Secondary plots of the observed rates determined at 418 nm versus PhIO concentration are shown for the first phase (filled circles), the second phase (open circles), and the third phase (filled triangles).
X-band EPR Studies of PGIS Reacting with PA
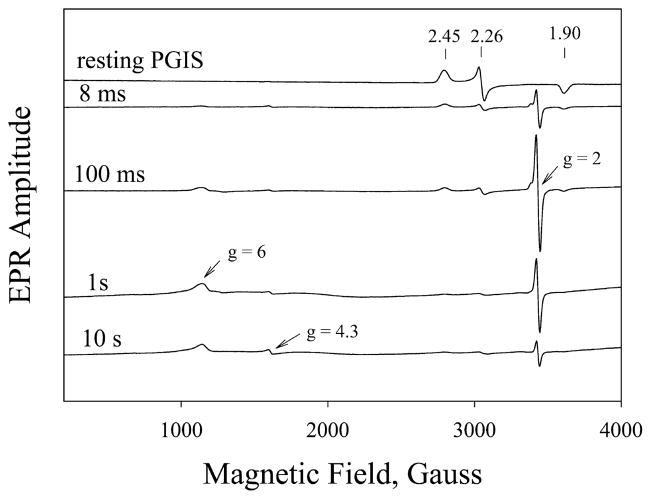
Although our stopped-flow results indicated that Cpd ES was an intermediate in the reaction of PGIS with PA or PhIO, UV/visible absorption spectroscopy alone is usually not sufficient for a clear assignment of the intermediate species. X-band EPR spectroscopy was thus employed to further characterize this transient species. Rapid freeze-quench samples were collected after incubation of PGIS (150 μM) with a 10-fold excess of PA over the reaction period of 8 ms to 5 s. Samples for the 5 to 120 s reaction were prepared by manual mixing. EPR spectra were recorded at both 10 and 110 K. No significant differences in the spectral features were observed at these two temperatures. Representative spectra taken at 8 ms, 100 ms, 1 s, and 10 s of reaction time are shown in Figure 3. An intermediate with a prominent signal at g = 2.0 appeared first, accompanied by a large decrease of the rhombic low-spin ferric heme signals at g = 2.45, 2.26, and 1.90, characteristics for the resting PGIS (44). Two small signals at g = 4.3 and g = 6 were also observed. The g = 4.3 is commonly thought to be a nonspecific iron signal and a marker for denatured heme, whereas g = 6 is likely the signal for one or more axial high-spin ferric heme forms resulting from modified coordination of the heme (45).
Figure 3.
X-band EPR spectra of the reaction of PGIS with PA. Resting PGIS (150 μM manually mixed with 4 volumes of NaPi buffer) or samples collected after PGIS (150 μM) was mixed with PA (1.5 mM) for 8 ms, 100 ms, 1 s, and 10 s. EPR conditions were microwave power, 4 mW; microwave frequency, 9.6 GHz; modulation amplitude, 10.87 G; and temperature, 10 K. Numbers above the vertical arrows are the g values of the rhombic low-spin heme center.
Identification of the g = 2 Species as a Cpd ES by X-band EPR
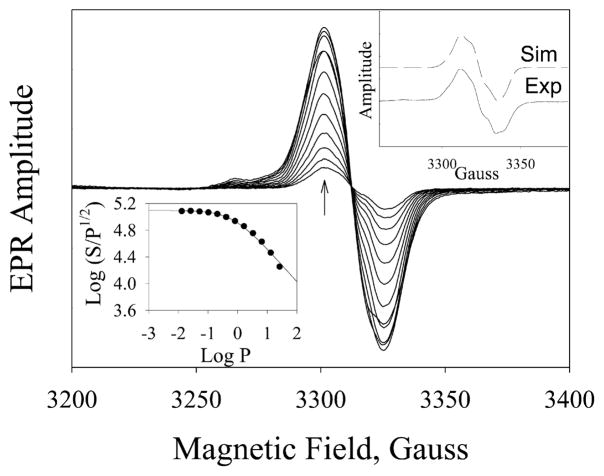
Cpd I and Cpd ES are both EPR active and have a signal at g = 2 (46, 47). However, their spin relaxation times are very different; Cpd I has a faster spin relaxation as the unpaired electron is in close proximity to the paramagnetic iron in the heme group, and thus a much higher P1/2, usually larger than 50 mW, is expected (Table 1) (48, 49). However, Cpd ES has a relatively slow spin relaxation with a P1/2 usually around 1 mW at liquid nitrogen temperature (17, 18). We thus performed power dependence experiments to distinguish the two intermediates. The rapid mixing/freeze-quenched sample collected at 100 ms reaction time gave, at 110 K, a 25 G wide symmetric spectrum centered at g = 2.0042 (Figure 4, upper right inset, solid line). Spectral changes of g = 2 radical at microwave power ranging from 12.9 μW to 25.8 mW are shown in Figure 4. The g = 2 species exhibited a homogeneous microwave power dependence and a very low P1/2 of 0.73 mW (Figure 4, lower left inset). This result strongly indicates that the intermediate is not Cpd I, but is Cpd ES, consistent with the result from UV/visible absorption spectroscopy.
Table 1.
EPR Half-Saturation Power, P1/2, at Different Temperatures and Reaction Times
| temp (K) | P1/2 (mW)a | reaction time (s) | P1/2(mW)b |
|---|---|---|---|
| 5 | 0.03 | 0.1 | 0.73 |
| 21 | 0.04 | 5 | 0.57 |
| 46 | 0.14 | 120 | 0.28 |
| 110 | 0.57 |
Sample was collected at 5 s reaction time.
Data were collected at 110 K.
Figure 4.
Progressive EPR power saturation for the radical intermediate trapped at 100 ms reaction of PGIS (150 μM) with PA (1.5 mM). EPR parameters were microwave power, 12.9 μW–25.8 mW; microwave frequency, 9.3 GHz; modulation amplitude, 2 G; and temperature, 110 K. Arrow indicates the direction of increasing microwave power. Lower left inset: microwave saturation curve of the g = 2 radical. The P1/2 value, fitted by eq 2 in which b was given as 1, is 0.73 mW. Upper right inset: Experimental (solid line) and simulated (dashed line; using the parameters in Table 2) X-band spectra of the freeze-quenched sample collected by reaction of 700 μM PGIS with 7 mM PA for 25 ms. Microwave power, 0.1 mW; microwave frequency, 9.3 GHz; modulation amplitude, 1 G; and temperature, 110 K.
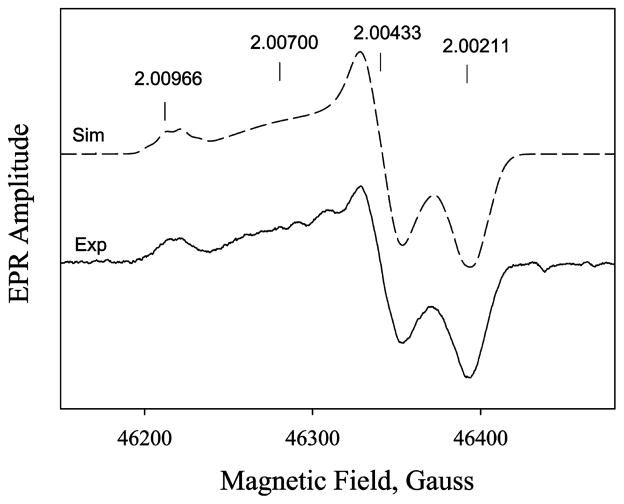
Identification of the g = 2 Species as a Tyrosyl Radical by D-band EPR
High-field EPR was performed to identify the amino acid species of the g = 2 radical. Samples were prepared by an equal mixing of 240 μM PGIS with a 10-fold excess of PA with 100 ms reaction tubing, but the freezing dead time is ~1 s. The D-band EPR (130 GHz) spectrum showed a clearly resolved g anisotropy (Table 2). Discrete gy and gz values of 2.00433 and 2.00200 are obtained. However, the gx value is not unique; the spectrum can be simulated using one discrete gx value at 2.00970 and a distributed gx centered at 2.00700. Thus, two spectral components coexisted in the 100 ms mixture (Figure 5, solid line). Both gy and gz values are very similar to gy = 2.0044 and gz = 2.00219 of Y96 in the P450cam and gy = 2.0043 and gz = 2.00219 of Y75 in the P450cam/Y96F mutant (18). The gx1 of 2.00970 indicates the tyrosyl radical is in a hydrophobic pocket and is not hydrogen bonded (50), and the gx2 of 2.0070 is characteristic of a strong hydrogen-bonded tyrosyl radical (50, 51). Notably, a significant Gaussian distribution with a width of 0.003 is found in gx2 = 2.00700. This phenomenon is similar to those observed for wild-type P450cam by 190 and 280 GHz EPR (18) in which the tyrosyl radical is affected either by a nearby water molecule or by the PA product remaining in the heme pocket, dispersing the gx component, whose principal axis orientation lies along the phenoxyl bond. Simulation of the spectrum, using the parameters shown in Table 2, further strengthened the assignment as a tyrosyl radical with two ortho protons and one β proton having major contributions to the hyperfine EPR features (52). The best-fit simulated spectrum, as shown in Figure 5, dashed line, is a weighted sum of the two components at a ratio of 1:2 for 2.00970 and 2.00700. The same parameter set also nicely fits the X-band spectrum (Figure 4, upper right inset, dashed line).
Table 2.
Parameters Used To Simulate both X- and D-band EPR Spectra of the Tyrosyl Radical in the Reaction of PGIS with PA
| component | valuea | percentagec/angled | |
|---|---|---|---|
| g values (species 1) | gx | 2.00700 (0.003)b | 67c |
| gy | 2.00433 | ||
| gz | 2.00200 | ||
| g values (species 2) | gx | 2.00970 | 33c |
| gy | 2.00433 | ||
| gz | 2.00200 | ||
| A3,5e | Ax | 9.6 | ±−25d |
| Ay | 2.8 | ||
| Az | 7.0 | ||
| A2,6e,f | Ax | 1.7 | ±−30d |
| Ay | 2.7 | ||
| Az | 0.4 | ||
| Aβ1e | Aiso | 11.0 | |
| Ae,f | Aiso | 0.60 |
Hyperfine coupling constants are absolute values, in gauss.
Full width at half-height of Gaussian distribution about gx value.
Percentage of species used in simulation.
Rotation about gz principal axis perpendicular to plane of ring, in degrees.
Used for both species 1 and 2.
These values not well determined in simulation and can be eliminated with a corresponding increase in broadening function without affecting spectral fit. Their inclusion indicates the experimental spectra are consistent with simulations containing these couplings which are typical of tyrosyl radicals (52).
Figure 5.
D-band (130 GHz) EPR spectra of 240 μM PGIS reacted with a 10-fold excess PA for 100 ms and freeze quenched in <1 s. Exp, the experimental spectrum (solid line); Sim, the simulated spectrum (dash line) using the parameter values shown in Table 2. The experimental spectrum is the first derivative of an average of five echo-detected spectra obtained at 15 K with ~22 min total acquisition time and parameters described in Materials and Methods.
D-band EPR spectroscopy performed on a 5 s reaction time sample produced a complicated spectrum from which radical assignment(s) could not be made (data not shown). In comparison with the 100 ms spectrum, however, the non-hydrogen-bonded tyrosyl radical disappeared while the spectral feature displaying the highest gvalue component shifted to ~2.0057. Thus, the contribution of a tyrosyl radical species to this 5 s sample spectrum seems unlikely, but other radical species, such as a tryptophan radical or other organic radical, cannot be ruled out.
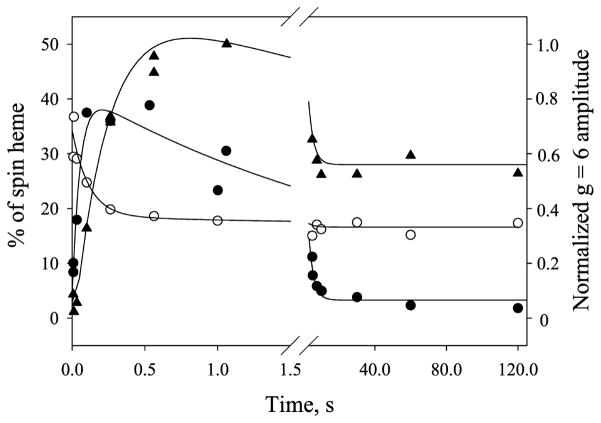
Kinetics of Radical and Low-Spin and High-Spin Heme Species
The amplitudes of the g = 2 and g = 6 signals, which initially increased and then decreased (Figure 3), prompted us to investigate the detailed kinetics of the two signals to correlate with the results from stopped-flow spectroscopy. The signals of low-spin heme decreased as the enzyme reaction started and eventually leveled off due to chemical conversion and heme bleaching. For direct comparison, the heme concentration of PGIS used in the stopped-flow rapid scan (110 μM) was similar to that in the EPR experiments (150 μM). The radical spin concentration, which was quantified against 1 mM copper standard, and the residual low-spin heme, which was normalized to the initial PGIS heme concentrations, showed bimodal kinetics (Figure 6, closed and open circle, respectively). Data were nicely fit to an irreversible three-species two-step reaction mechanism A → B → C. A numerical solution for the intermediate B was determined by the equation:
| (3) |
where k1 and k2 are the rate constants for the formation and decay of the intermediate, respectively. The g = 2 radical formed at a very early stage (~8 ms) of the reaction and increased to ~40% spin/heme at 100–500 ms (Figure 6, closed circle). The rate constants, k1 and k2, thus determined were 4.3 and 0.4 s−1, respectively. Consistent with the kinetics of g = 2 radical, changes of the low-spin heme signal (Figure 6, open circles) were also properly fit to the same kinetics. The rate constants, k1 and k2, thus obtained were of 6.5 and 0.3 s−1, respectively. The kinetics observed by EPR were comparable to those determined by the stopped-flow rapid-scan experiment in which the Cpd ES was formed at a rate of 4.3 s−1 and heme was bleached at 1.0 s−1, respectively, as determined by global analysis (Figure 1B). These data suggest that the 408 nm species observed by UV/visible absorption spectroscopy is kinetically equivalent to the g = 2 species measured by EPR spectroscopy. Together, our results indicate that the 408 nm intermediate is a Cpd ES.
Figure 6.
Kinetics of g = 2 radical (closed circle), residual low-spin heme (opened circle), and g = 6 high-spin heme (closed triangle) EPR signals in the reaction of PGIS (150 μM) with PA (1.5 mM). Data were analyzed as described in the Results.
The sum of the observed residual low-spin heme and g = 2 radical at any given time, however, is much less than the initial heme concentration. For example, at 100 ms, ~24% of the low-spin heme signal was retained and 38% of the radical signal of the heme was formed, indicating that ~40% of original heme signals were unaccountable. Notably, a signal at g = 6 also emerged first and then decayed, similar to the kinetics of the g = 2 signal. Because the g = 6 signal is not easily quantified due to its atypical line shape for a high-spin heme, only EPR amplitude was plotted against the reaction time (Figure 6, closed triangles). Kinetic analysis according to eq 3 yielded a formation rate constant of 3.8 s−1 and decay rate constant of 0.3 s−1. The g = 6 species that peaked at ~0.8–1 s (Figure 6, closed triangles) evolved later than the g = 2 species that peaked at ~0.1–0.5 s (Figure 6, closed circles), suggesting that they are kinetically distinct species. The g = 6 species is likely a high-spin ferric heme with heme axial ligand exchange. Therefore, at any given time, the g = 6 and EPR-silent oxo–ferryl species both contribute to the unaccounted heme concentration. It should be noted that the g = 6 species decayed at a rate close to that of the g = 2 species. Moreover, both g = 2 and g = 6 intermediates were similarly observed when PhIO reacted with PGIS (data not shown).
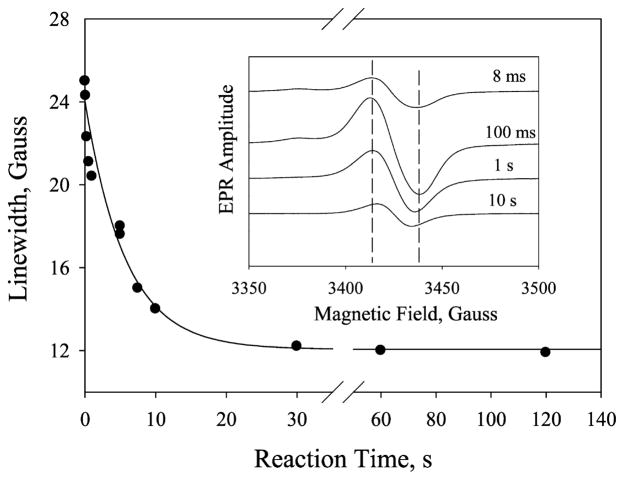
Temporal and Power-Dependent Changes of g = 2 EPR Line Width
The overall line width of the g = 2 signal changed from 26 to 14 G as the reaction time increased from 8 ms to 10 s (Figure 7, inset). Because the line width is an important indicator of radical species identity, this result suggests that the unpaired electron of Cpd ES migrated from one amino acid to another. As a result, the coupling strength between the radical and the nearby paramagnetic center(s) may be changed. This change is evident by the shift of the spin relaxation rate, which is characterized by its P1/2 value. We first examined by temperature-dependent microwave power saturation experiments whether coupling exists between the radical and the heme center. The temperature dependence of the P1/2 value of the g = 2 radical formed at 5 s reaction time is shown in Table 1, left column. The much higher P1/2 value as compared to a chemically generated model tyrosyl radical at various temperatures suggests that the radical is magnetically coupled to a paramagnetic center, as observed in P450cam (16), ribonucleotide reductase (RNR) (53), and prostaglandin H synthase (54). We then investigated whether the radical migrated to other amino acid residues. As shown in Table 1, right column, the P1/2 value decreased as the reaction time increased, indicating that distinct radical species were observed over the reaction period. Together, our results indicated that the radical migrates from a tyrosine residue to unidentified sites farther away from the heme.
Figure 7.
Plot of line width versus reaction time. Data were fitted using one-exponential decay. Inset: Narrow scan EPR spectra of g = 2 radical of the reaction between PGIS (150 μM) and PA (1.5 mM) collected at 8 ms, 100 ms, 1 s, and 10 s. EPR parameters were microwave power, 1 mW; microwave frequency, 9.3 GHz; modulation amplitude, 2 G; and temperature, 110 K.
The finding of radical migration prompted us to investigate the time-dependent line width change. The rate of the line width narrowing was calculated to be 0.2 s−1 (Figure 7), comparable to that of heme bleaching (0.1–0.2 s−1), implying that radical migration is associated with heme bleaching.
Self-Inactivation of PGIS upon Reaction with PA
To examine whether narrowing of the EPR line width and heme bleaching are correlated with self-inactivation of PGIS upon reaction with PA, we carried out the stopped-flow sequential mixing experiments to measure the remaining activity of ~3.2 μM PGIS after incubation for a given time with a 23-fold excess of PA, as described under Materials and Methods. The residual activity of PGIS follows a one-exponential kinetics with a rate constant of 0.13 s−1. This self-inactivation rate constant is comparable to those observed for the narrowing of the EPR line width (~0.2 s−1) and for the heme bleaching (0.1–0.2 s−1), suggesting that amino acid radical migration occurred in parallel with heme bleaching and self-inactivation. Indeed, when PGIS was mixed with guaiacol prior to reaction with PA, more oxidized guaiacol products were generated than those without premixing with guaiacol (data not shown). These results suggest that guaiacol reacts with Cpd ES before its initial amino acid radical migrates to other residue(s) and efficiently recycles the heme, thus preventing inactivation.
DISCUSSION
In this study we characterized reaction intermediates and, on the basis of stopped-flow UV/vis spectroscopy and rapid freeze-quench EPR kinetic experiments, proposed plausible mechanisms for the reactions of PGIS with PA and PhIO. Moreover, we identified the amino acid radical species of the Cpd ES as a tyrosyl radical. Reacting with either oxidant, PGIS yielded an intermediate whose absorption spectrum is very different from those observed with lipid hydroperoxides (24, 44). Lipid hydroperoxides do not easily undergo heterolysis because alkoxide products are poorer leaving groups. Therefore, homolysis of the peroxide O–O bond is more likely to occur. As a result, Cpd II (or Fe(IV)=O) is formed, and this intermediate exhibited a red-shifted Soret peak at ~422 nm. The intermediate of the reaction of PGIS and PA, however, showed an absorption spectrum featuring Cpd ES with the Soret peak at 408 nm, similar to the Cpd ES formed in the reaction of P450cam with peroxy acid (15). Given that the electronic configurations of the porphyrin in Cpd II and Cpd ES are identical, it is puzzling that their Soret peaks are considerably different. These findings were also observed for P450cam in reactions with PA and cumene hydroperoxide (55) but were rationalized as being due to different states of protonation of the Fe(IV)=O. The product of PA, acetate, is a good leaving group and may promote conditions in the active site that favors protonation of the ferryl intermediate (Fe(IV)–OH) for Cpd ES. In contrast, Cpd II formed from reactions with lipid hydroperoxides appears to be an unprotonated ferryl complex. Although no acylperoxo complex or Cpd I intermediate was observed in our studies, it cannot be ruled out that the very short lived porphyrin cation radical of Cpd I forms and accepts an electron from a nearby amino acid residue in PGIS to generate the metastable Cpd ES intermediate.
While reactions of PGIS with PA and PhIO share a common intermediate, their kinetic behaviors differ. The PhIO binding is much faster, making the binding step (16 s−1) and the subsequent chemical step (2 s−1) well separated. In contrast, the binding step in the PA reaction is rather slow. The difference in the binding rate can be partly attributed to the relative hydrophobicities of these two compounds. PhIO is more hydrophobic than PA and therefore exhibits a higher affinity for the hydrophobic active site of PGIS (25). A difference in the binding rate was also seen in the reactions of P450cam with PA and m-chloroperbenzoic acid, as the more hydrophobic m-chloroperbenzoic acid bound faster than PA to the hydrophobic active site of P450cam (11).
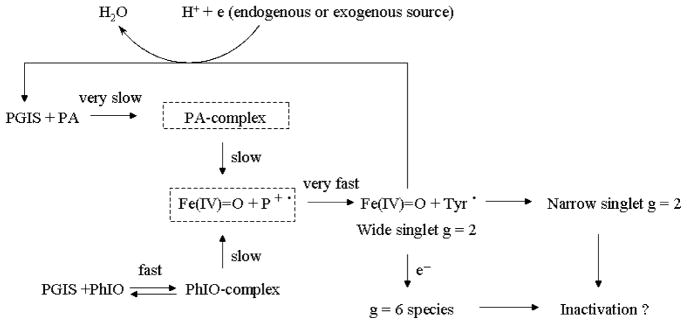
Figure 8 summarizes our proposed mechanism for PGIS reacting with PA or PhIO. Upon formation of the enzyme–substrate complex, a heterolytic cleavage of the O–O or I–O bond takes place to form Cpd ES, presumably via Cpd I. Cpd ES can be reduced by an exogenous substrate or endogenous reductant to the resting enzyme with concomitant production of water and formation of the g = 6 species. The exogenous cosubstrate guaiacol reduces Cpd ES more efficiently than endogenous reductant. Peracid itself can be a cosubstrate similar to that observed in the catalytic reactions of both horseradish peroxidase and P450cam (11, 56). In addition to the recycling pathway, Cpd ES has been shown to have two possible fates: (1) the tyrosyl radical (26 G line width) may migrate to another residue (with a 12 G line width), resulting in heme modification and enzymatic inactivation, and (2) the ferryl intermediate may be reduced to a high-spin ferric form (g = 6 species) that also leads to heme structural changes and enzymatic inactivation. Because the supply of endogenous reductant is limited and PA is not an efficient cosubstrate, the heme is eventually modified during multiple turnovers.
Figure 8.
The peroxidase reaction mechanism for PGIS with PA and PhIO. The boxes in broken line represent the putative intermediates not observed in this study.
The g values obtained from high-field EPR data clearly indicate that the g = 2 radicals formed at the early stage of the reaction are indeed tyrosyl radicals with spectroscopic parameters comparable to those tyrosyl radicals formed in other enzymes (18, 50, 51, 57–60). These parameters differ from those for the tryptophan radical found in the E. coli RNR Y122F mutant (gx = 2.0035, gy = 2.0024, and gz = 2.0020) and mouse RNR Y177W mutant (gx = 2.0033, gy = 2.0024, and gz = 2.0021) (50). The gx distribution width of 0.003 is also consistent with the assignment of a tyrosyl radical rather than a tryptophan radical. This is because the value of gx in tyrosyl radicals is dependent on the environment of the phenolic oxygen, with a heterogeneous environment leading to a distribution in gx values. The gx value of the tyrosyl radical is more sensitive to the environment of phenolic oxygen than is the gx value of the tryptophan radical to the environment of the indole ring. Thus, the distribution in gx values of tyrosyl radicals is significantly larger than that of tryptophan radicals, which is on the order of ~0.0015 when it is observed (50). Furthermore, the gx values of tyrosyl radicals from other enzymes range from 2.0091 for a non-hydrogen-bonding environment in RNR (50, 61, 62) to 2.0066 for a strong hydrogen-bonding environment in turnip peroxidase, γ-irradiated crystals, and prostaglandin H synthase (51, 63, 64). P450cam also has tyrosyl radicals with two gx values of 2.0078 and 2.0064 that are correlated to weakly and very strongly hydrogen-bonded environments, respectively (18).
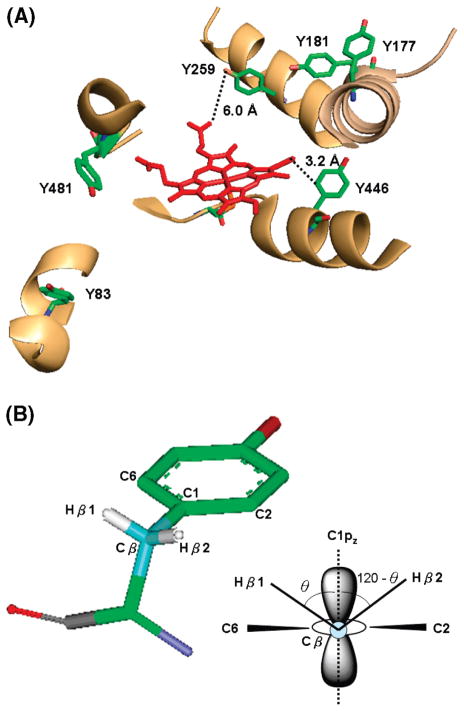
Tyrosine residues near the PGIS heme are the obvious candidates for the tyrosyl radical of Cpd ES. A distance within ~4 Å is shown to be a good range for electron hopping (65). In this regard, Tyr446, located at the edge of the heme with a distance of 3.2 Å from the vinyl group of the C-pyrrole ring (PDB code 2IAG) (25), is a likely candidate. Notably, intramolecular electron transfer within a moderate distance (<10 Å) can also proceed via neighboring residues. For example, Tyr96 of P450cam, which is 7.4 Å from a porphyrin meso-carbon atom, can donate an electron through Thr101 to the propionate of the heme and from there to the porphyrin ring (18). If Tyr96 is replaced by a phenylalanine, the radical is then located at Tyr75, ~7.6 Å from the heme edge. Electron transfer is proposed to proceed via Gln322 to the nearest methyl group of the heme. The efficiency of such intramolecular electron transfer depends greatly on the chemical nature and position of neighboring residues. Thus, we tested every tyrosine residue in the PGIS crystal structure by the PATHWAYS analysis using the HARLEM programs (36). The calculated edge-to-edge distances and electron transfer coupling efficiencies are shown in Table 3. Six tyrosine residues with a coupling efficiency of greater than 10−4 are within 10 Å from the heme edge (Figure 9A). Three (Tyr259, Tyr446, and Tyr481) are non-hydrogen-bonded phenoxyl radicals, and three (Tyr83, Tyr177, and Tyr181) are hydrogen-bonded phenoxyl radicals. These residues will be useful targets for our future mutagenesis and functional studies.
Table 3.
Edge-to-Edge Distance from a Specific Tyrosine Residue to the PGIS Heme and Best Coupling Efficiency Estimated from Electron Pathway Analysis
| Tyr residue | edge distance (Å) | best coupling value |
|---|---|---|
| Tyr42 | 20.0 | 3.30 × 10−8 |
| Tyr73 | 17.5 | 4.38 × 10−7 |
| Tyr83 | 6.5 | 3.62 × 10−4 |
| Tyr99 | 7.3 | 2.66 × 10−6 |
| Tyr115 | 5.6 | 1.56 × 10−5 |
| Tyr141 | 12.3 | 1.83 × 10−5 |
| Tyr169 | 10.1 | 4.14 × 10−5 |
| Tyr177 | 8.1 | 2.92 × 10−4 |
| Tyr181 | 8.1 | 8.12 × 10−4 |
| Tyr259 | 6.5 | 3.62 × 10−4 |
| Tyr398 | 14.5 | 4.81 × 10−6 |
| Tyr406 | 16.1 | 2.32 × 10−7 |
| Tyr421 | 21.7 | 6.36 × 10−8 |
| Tyr430 | 13.2 | 1.08 × 10−4 |
| Tyr446 | 3.2 | 2.89 × 10−2 |
| Tyr481 | 9.6 | 4.58 × 10−4 |
| Tyr496 | 21.4 | 9.51 × 10−9 |
Figure 9.
(A) The position of tyrosyl radical candidates nearby the PGIS heme (PDB code 2IAG). Distances of the two most likely candidates, Tyr259 and Tyr446, from the heme are indicated. (B) Molecular structure of the tyrosyl radical with atomic numbering (left) and end-on view along the C1–Cβ bond showing the dihedral angle, θ, and the orientation of the β protons.
For a detailed assignment of different EPR features, we applied the simulated parameters obtained from the X- and D-band EPR to the McConnell relations (66–68):
where Aiso is the isotropic hydrogen hyperfine coupling constant arising from the Cβ hydrogen (Figure 9B), ρC1 is the spin density at C1 of the phenoxyl ring, θ is the dihedral angle between the axis of the pz orbital of C1 and the Cβ–Hβ bond, and B0 and B1 are empirical constants (B0 = 1–2 G and B1 = 50–52.4 G (18, 69)). From the simulated isotropic hydrogen hyperfine coupling constants of 11.0 and 0.6 G (Table 2), the dihedral angle θ of 32.1–37.2° and spin density of 0.28–0.31 are calculated. The uncertainty of the small value for Aβ2,iso was further assessed. We varied the value of Aβ2,iso from 0.4 to ~2.0, without causing visible line shape changes of the simulated EPR, but kept other parameters constant (Aβ1, iso = 11, B0 = 1, and B1 = 50) to calculate the dihedral angle and found its value varied in a small range of 30.7–37.4°, indicating the small contribution of Aβ2,iso to the overall line shape. The calculated spin density is comparable to the theoretical calculation of spin density on the C1 of ~0.32 (70, 71) and the experimentally measured spin density for mouse RNR (0.33), photosystem II YD (0.14–0.34), and the tyrosine model (0.20–0.44) (72). Together, these results lead to the prediction of Tyr259 and Tyr446 as the most likely residues since these two residues, revealed from the crystal structure, have the dihedral angles of 31° and 34.9° with a reasonable distance to the heme edge (Table 3). The prediction thus further focuses our potential targets for site-directed mutagenesis in order to locate the origin of the observed tyrosyl radical species.
An important finding in this study is that the line width of the g = 2 signal narrowed from 26 to 12 G as the reaction proceeded (Figure 7). Similar line width narrowing was observed in the studies of Tyr and Trp radicals in the Mtb KatG (33), the polarized P700+ radical in photosystem I (73), and Tyr radicals in prostaglandin H synthase (74, 75) and was attributed to a process of electron migration. This phenomenon has not been reported for P450s. By comparing the P1/2 value with that in the P450cam (18), we anticipated that the amino acid radical formed at 100 ms was within 10 Å from the heme. As the g = 2 signal narrowed, the P1/2 value becomes smaller, indicating that the electron is being distributed to new locations. Moreover, the rate of electron migration agrees well with that of the heme bleaching, possibly suggesting that electron migration may lead to enzyme inactivation. The cause/effect relationship of PGIS inactivation is difficult to pin down in this study as heme bleaching, activity loss, and radical decay all showed similar kinetics. For example, the g = 6 signal is likely to arise from heme destruction, which itself is rather complicated. Therefore, directly comparing it with appearance and disappearance of the amino acid radical may simply be due to a combination of catalytic events and of heme destruction proceeding simultaneously. In sharp contrast to a recent study on prostaglandin H synthase, loss of peroxidase activity correlates well with the disappearance of the oxyferryl heme and is at least an order of magnitude faster than the tyrosyl radical decay (76).
In conclusion, parallel experiments with stopped-flow and rapid freeze-quench EPR demonstrated for the first time that a Cpd ES intermediate is formed in the reaction of PGIS with PA or PhIO and that Cpd ES is Fe(IV)=O plus a tyrosyl radical. Results of line width narrowing and time-dependent changes of P1/2 implied that the unpaired electron migrated to other residues on a second time scale. Our results indicate that multiple radical species are present in PGIS catalysis. Electron migration in Cpd ES upon reduction of oxo–ferryl may result in enzymatic inactivation. Although the biological role of PGIS in peroxidase reactions is unclear, PGIS acting as a peroxidase to generate a Cpd ES intermediate that leads to self-inactivation may have physiological implications in the inflammatory tissues.
Footnotes
This work is supported by Grants HL60625 (L.-H.W.), GM44911 (A.-L.T.), and GM075920 (G.J.G.) from the National Institutes of Health.
Abbreviations: P450, cytochrome P450; PGIS, prostacyclin synthase; Cpd I, compound I; Cpd ES, compound ES; Cpd II, compound II; PA, peracetic acid; PhIO, iodosylbenzene; EPR, electron paramagnetic resonance; P450cam, cytochrome P450 from Pseudomonas putida, catalyzing hydroxylation of camphor; P450 BM3, cytochrome P450 from Bacillus megaterium, catalyzing oxidation of a variety of fatty acids; RNR, ribonucleotide reductase.
References
- 1.Poulos TL, Howard AJ. Crystal structures of metyrapone- and phenylimidazole-inhibited complexes of cytochrome P-450cam. Biochemistry. 1987;26:8165–8174. doi: 10.1021/bi00399a022. [DOI] [PubMed] [Google Scholar]
- 2.Davydov R, Perera R, Jin S, Yang TC, Bryson TA, Sono M, Dawson JH, Hoffman BM. Substrate modulation of the properties and reactivity of the oxy-ferrous and hydroperoxo-ferric intermediates of cytochrome P450cam as shown by cryoreduction-EPR/ENDOR spectroscopy. J Am Chem Soc. 2005;127:1403–1413. doi: 10.1021/ja045351i. [DOI] [PubMed] [Google Scholar]
- 3.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome P450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 4.Oritz de Montellano PR. Oxygen Activation and Reactivity. In: Oritz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Plenum; NY: 1995. pp. 245–303. [Google Scholar]
- 5.Hiner AN, Raven EL, Thorneley RN, Garcia-Canovas F, Rodriguez-Lopez JN. Mechanisms of compound I formation in heme peroxidases. J Inorg Biochem. 2002;91:27–34. doi: 10.1016/s0162-0134(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 6.Jung C, Schunemann V, Lendzian F. Freeze-quenched iron-oxo intermediates in cytochromes P450. Biochem Biophys Res Commun. 2005;338:355–364. doi: 10.1016/j.bbrc.2005.08.166. [DOI] [PubMed] [Google Scholar]
- 7.Makris TM, von Koenig K, Schlichting I, Sligar SG. The status of high-valent metal oxo complexes in the P450 cytochromes. J Inorg Biochem. 2006;100:507–518. doi: 10.1016/j.jinorgbio.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Sivaraja M, Goodin DB, Smith M, Hoffman BM. Identification by ENDOR of Trp191 as the free-radical site in cytochrome c peroxidase compound ES. Science. 1989;245:738–740. doi: 10.1126/science.2549632. [DOI] [PubMed] [Google Scholar]
- 9.Barrows TP, Poulos TL. Role of electrostatics and salt bridges in stabilizing the compound I radical in ascorbate peroxidase. Biochemistry. 2005;44:14062–14068. doi: 10.1021/bi0507128. [DOI] [PubMed] [Google Scholar]
- 10.Tsai AL, Wu G, Palmer G, Bambai B, Koehn JA, Marshall PJ, Kulmacz RJ. Rapid kinetics of tyrosyl radical formation and heme redox state changes in prostaglandin H synthase-1 and -2. J Biol Chem. 1999;274:21695–21700. doi: 10.1074/jbc.274.31.21695. [DOI] [PubMed] [Google Scholar]
- 11.Spolitak T, Dawson JH, Ballou DP. Reaction of ferric cytochrome P450cam with peracids: Kinetic characterization of intermediates on the reaction pathway. J Biol Chem. 2005;280:20300–20309. doi: 10.1074/jbc.M501761200. [DOI] [PubMed] [Google Scholar]
- 12.Raner GM, Thompson JI, Haddy A, Tangham V, Bynum N, Ramachandra Reddy G, Ballou DP, Dawson JH. Spectroscopic investigations of intermediates in the reaction of cytochrome P450(BM3)-F87G with surrogate oxygen atom donors. J Inorg Biochem. 2006;100:2045–2053. doi: 10.1016/j.jinorgbio.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Kellner DG, Hung SC, Weiss KE, Sligar SG. Kinetic characterization of compound I formation in the thermostable cytochrome P450 CYP119. J Biol Chem. 2002;277:9641–9644. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 14.Palcic MM, Rutter R, Araiso T, Hager LP, Dunford HB. Spectrum of chloroperoxidase compound I. Biochem Biophys Res Commun. 1980;94:1123–1127. doi: 10.1016/0006-291x(80)90535-5. [DOI] [PubMed] [Google Scholar]
- 15.Spolitak T, Dawson JH, Ballou DP. Rapid kinetics investigations of peracid oxidation of ferric cytochrome P450cam: Nature and possible function of compound ES. J Inorg Biochem. 2006;100:2034–2044. doi: 10.1016/j.jinorgbio.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Schunemann V, Jung C, Trautwein AX, Mandon D, Weiss R. Intermediates in the reaction of substrate-free cytochrome P450cam with peroxy acetic acid. FEBS Lett. 2000;479:149–154. doi: 10.1016/s0014-5793(00)01886-x. [DOI] [PubMed] [Google Scholar]
- 17.Schunemann V, Jung C, Terner J, Trautwein AX, Weiss R. Spectroscopic studies of peroxyacetic acid reaction intermediates of cytochrome P450cam and chloroperoxidase. J Inorg Biochem. 2002;91:586–596. doi: 10.1016/s0162-0134(02)00476-2. [DOI] [PubMed] [Google Scholar]
- 18.Schunemann V, Lendzian F, Jung C, Contzen J, Barra AL, Sligar SG, Trautwein AX. Tyrosine radical formation in the reaction of wild type and mutant cytochrome P450cam with peroxy acids: A multifrequency EPR study of intermediates on the millisecond time scale. J Biol Chem. 2004;279:10919–10930. doi: 10.1074/jbc.M307884200. [DOI] [PubMed] [Google Scholar]
- 19.Jung C, Schunemann V, Lendzian F, Trautwein AX, Contzen J, Galander M, Bottger LH, Richter M, Barra AL. Spectroscopic characterization of the iron-oxo intermediate in cytochrome P450. Biol Chem. 2005;386:1043–1053. doi: 10.1515/BC.2005.120. [DOI] [PubMed] [Google Scholar]
- 20.Moncada S, Gryglewski R, Bunting S, Vane JR. An enzyme isolated from arteries transforms prostaglandin endoperoxides to an unstable substance that inhibits platelet aggregation. Nature. 1976;263:663–665. doi: 10.1038/263663a0. [DOI] [PubMed] [Google Scholar]
- 21.Hara S, Miyata A, Yokoyama C, Inoue H, Brugger R, Lottspeich F, Ullrich V, Tanabe T. Isolation and molecular cloning of prostacyclin synthase from bovine endothelial cells. J Biol Chem. 1994;269:19897–19903. [PubMed] [Google Scholar]
- 22.Pereira B, Wu KK, Wang LH. Molecular cloning and characterization of bovine prostacyclin synthase. Biochem Biophys Res Commun. 1994;203:59–66. doi: 10.1006/bbrc.1994.2148. [DOI] [PubMed] [Google Scholar]
- 23.Hecker M, Ullrich V. On the mechanism of prostacyclin and thromboxane A2 biosynthesis. J Biol Chem. 1989;264:141–150. [PubMed] [Google Scholar]
- 24.Yeh HC, Hsu PY, Wang JS, Tsai AL, Wang LH. Characterization of heme environment and mechanism of peroxide bond cleavage in human prostacyclin synthase. Biochim Biophys Acta. 2005;1738:121–132. doi: 10.1016/j.bbalip.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang CW, Yeh HC, Wang LH, Chan NL. Crystal structure of the human prostacyclin synthase. J Mol Biol. 2006;364:266–274. doi: 10.1016/j.jmb.2006.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake RC, II, Coon MJ. On the mechanism of action of cytochrome P-450. Spectral intermediates in the reaction with iodosobenzene and its derivatives. J Biol Chem. 1989;264:3694–3701. [PubMed] [Google Scholar]
- 27.Baldwin DA, Marques HM, Pratt JM. Hemes and hemoproteins. 5: Kinetics of the peroxidatic activity of microperoxidase-8: Model for the peroxidase enzymes. J Inorg Biochem. 1987;30:203–217. doi: 10.1016/0162-0134(87)80064-8. [DOI] [PubMed] [Google Scholar]
- 28.Du M, Yeh HC, Berka V, Wang LH, Tsai AL. Redox properties of human endothelial nitric-oxide synthase oxygenase and reductase domains purified from yeast expression system. J Biol Chem. 2003;278:6002–6011. doi: 10.1074/jbc.M209606200. [DOI] [PubMed] [Google Scholar]
- 29.Tsai AL, Berka V, Kulmacz RJ, Wu G, Palmer G. An improved sample packing device for rapid freeze-trap electron paramagnetic resonance spectroscopy kinetic measurements. Anal Biochem. 1998;264:165–171. doi: 10.1006/abio.1998.2774. [DOI] [PubMed] [Google Scholar]
- 30.Barry BA, El-Deeb MK, Sandusky PO, Babcock GT. Tyrosine radicals in photosystem II and related model compounds. Characterization by isotopic labeling and EPR spectroscopy. J Biol Chem. 1990;265:20139–20143. [PubMed] [Google Scholar]
- 31.Hales BJ. Intrinsic and extrinsic parameters as probes of metal clusters. Methods Enzymol. 1993;227:384–395. [Google Scholar]
- 32.Dorlet P, Seibold SA, Babcock GT, Gerfen GJ, Smith WL, Tsai AL, Un S. High-field EPR study of tyrosyl radicals in prostaglandin H2 synthase-1. Biochemistry. 2002;41:6107–6114. doi: 10.1021/bi015871f. [DOI] [PubMed] [Google Scholar]
- 33.Ranguelova K, Girotto S, Gerfen GJ, Yu S, Suarez J, Metlitsky L, Magliozzo RS. Radical sites in Mycobacterium tuberculosis KatG identified using electron paramagnetic resonance spectroscopy, the three-dimensional crystal structure, and electron transfer couplings. J Biol Chem. 2007;282:6255–6264. doi: 10.1074/jbc.M607309200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerfen GJ, Bellew BF, Griffin RG, Singel DJ, Ekberg CA, Whittaker JW. High-frequency electron paramagnetic resonance spectroscopy of the apogalactose oxidase radical. J Phys Chem. 1996;100:16739–16748. [Google Scholar]
- 35.Van der Donk WA, Stubbe J, Gerfen GJ, Bellew BF, Griffin RG. EPR investigations of the inactivation of the E. coli ribonucleotide reductase with 2′-azido-2′-deoxyuridine 5′-diphosphate: Evidence for the involvement of the thiyl radical of C225-R1. J Am Chem Soc. 1995;117:8908–8916. [Google Scholar]
- 36.Onuchic JN, Beratan DN, Winkler JR, Gray HB. Pathway analysis of protein electron-transfer reactions. Annu Rev Biophys Biomol Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]
- 37.Egawa T, Proshlyakov DA, Miki H, Makino R, Ogura T, Kitagawa T, Ishimura Y. Effects of a thiolate axial ligand on the pi→pi* electronic states of oxoferryl porphyrins: A study of the optical and resonance Raman spectra of compounds I and II of chloroperoxidase. J Biol Inorg Chem. 2001;6:46–54. doi: 10.1007/s007750000181. [DOI] [PubMed] [Google Scholar]
- 38.Hewson WD, Hager LP. Oxidation of horseradish peroxidase compound II to compound I. J Biol Chem. 1979;254:3182–3186. [PubMed] [Google Scholar]
- 39.Yeh HC, Wang JS, Su YO, Lin WY. Stopped-flow kinetic study of the H2O2 oxidation of substrates catalyzed by microperoxidase-8. J Biol Inorg Chem. 2001;6:770–777. doi: 10.1007/s007750100253. [DOI] [PubMed] [Google Scholar]
- 40.Anzenbacher P, Niwa T, Tolbert LM, Sirimanne SR, Guengerich FP. Oxidation of 9-alkylanthracenes by cytochrome P450 2B1, horseradish peroxidase, and iron tetraphenylporphine/iodosylbenzene systems: Anaerobic and aerobic mechanisms. Biochemistry. 1996;35:2512–2520. doi: 10.1021/bi952330f. [DOI] [PubMed] [Google Scholar]
- 41.Guengerich FP. Oxidation of halogenated compounds by cytochrome P-450, peroxidases, and model metalloporphyrins. J Biol Chem. 1989;264:17198–17205. [PubMed] [Google Scholar]
- 42.Gustafsson JA, Rondahl L, Bergman J. Iodosyl-benzene derivatives as oxygen donors in cytochrome P-450 catalyzed steroid hydroxylations. Biochemistry. 1979;18:865–870. doi: 10.1021/bi00572a020. [DOI] [PubMed] [Google Scholar]
- 43.Guengerich FP, Yun CH, Macdonald TL. Evidence for a 1-electron oxidation mechanism in N-dealkylation of N,N-dialkylanilines by cytochrome P450 2B1. Kinetic hydrogen isotope effects, linear free energy relationships, comparisons with horseradish peroxidase, and studies with oxygen surrogates. J Biol Chem. 1996;271:27321–27329. doi: 10.1074/jbc.271.44.27321. [DOI] [PubMed] [Google Scholar]
- 44.Yeh HC, Tsai AL, Wang LH. Reaction mechanisms of 15-hydroperoxyeicosatetraenoic acid catalyzed by human prostacyclin and thromboxane synthases. Arch Biochem Biophys. 2007;461:159–168. doi: 10.1016/j.abb.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigman JA, Pond AE, Dawson JH, Lu Y. Engineering cytochrome c peroxidase into cytochrome P450: A proximal effect on heme-thiolate ligation. Biochemistry. 1999;38:11122–11129. doi: 10.1021/bi990815o. [DOI] [PubMed] [Google Scholar]
- 46.Davydov R, Kofman V, Fujii H, Yoshida T, Ikeda-Saito M, Hoffman BM. Catalytic mechanism of heme oxygenase through EPR and ENDOR of cryoreduced oxy-heme oxygenase and its Asp 140 mutants. J Am Chem Soc. 2002;124:1798–1808. doi: 10.1021/ja0122391. [DOI] [PubMed] [Google Scholar]
- 47.Erman JE, Vitello LB, Mauro JM, Kraut J. Detection of an oxyferryl porphyrin pi-cation-radical intermediate in the reaction between hydrogen peroxide and a mutant yeast cytochrome c peroxidase. Evidence for tryptophan-191 involvement in the radical site of compound I. Biochemistry. 1989;28:7992–7995. doi: 10.1021/bi00446a004. [DOI] [PubMed] [Google Scholar]
- 48.Khindaria A, Aust SD. EPR detection and characterization of lignin peroxidase porphyrin pi-cation radical. Biochemistry. 1996;35:13107–13111. doi: 10.1021/bi961297+. [DOI] [PubMed] [Google Scholar]
- 49.Fujii H, Yoshimura T, Kamada H. ESR studies of A1u and A2u oxoiron(IV) porphyrin pi-cation radical complexes. Spin coupling between ferryl iron and A1u/A2u orbitals. Inorg Chem. 1996;35:2373–2377. doi: 10.1021/ic9513752. [DOI] [PubMed] [Google Scholar]
- 50.Bleifuss G, Kolberg M, Potsch S, Hofbauer W, Bittl R, Lubitz W, Graslund A, Lassmann G, Lendzian F. Tryptophan and tyrosine radicals in ribonucleotide reductase: A comparative high-field EPR study at 94 GHz. Biochemistry. 2001;40:15362–15368. doi: 10.1021/bi010707d. [DOI] [PubMed] [Google Scholar]
- 51.Ivancich A, Dorlet P, Goodin DB, Un S. Multifrequency high-field EPR study of the tryptophanyl and tyrosyl radical intermediates in wild-type and the W191G mutant of cytochrome c peroxidase. J Am Chem Soc. 2001;123:5050–5058. doi: 10.1021/ja0036514. [DOI] [PubMed] [Google Scholar]
- 52.Cappuccio JA, Ayala I, Elliott GI, Szundi I, Lewis J, Konopelski JP, Barry BA, Einarsdottir O. Modeling the active site of cytochrome oxidase: Synthesis and characterization of a cross-linked histidine-phenol. J Am Chem Soc. 2002;124:1750–1760. doi: 10.1021/ja011852h. [DOI] [PubMed] [Google Scholar]
- 53.Liu A, Potsch S, Davydov A, Barra AL, Rubin H, Graslund A. The tyrosyl free radical of recombinant ribonucleotide reductase from Mycobacterium tuberculosis is located in a rigid hydrophobic pocket. Biochemistry. 1998;37:16369–16377. doi: 10.1021/bi981471p. [DOI] [PubMed] [Google Scholar]
- 54.Karthein R, Dietz R, Nastainczyk W, Ruf HH. Higher oxidation states of prostaglandin H synthase. EPR study of a transient tyrosyl radical in the enzyme during the peroxidase reaction. Eur J Biochem. 1988;171:313–320. doi: 10.1111/j.1432-1033.1988.tb13792.x. [DOI] [PubMed] [Google Scholar]
- 55.Spolitak T, Dawson JH, Ballou DP. Replacement of tyrosine residues by phenylalanine in cytochrome P450cam alters the formation of Cpd II-like species in reactions with artificial oxidants. J Biol Inorg Chem. 2008;13:599–611. doi: 10.1007/s00775-008-0348-9. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Lopez JN, Hernandez-Ruiz J, Garcia-Canovas F, Thorneley RN, Acosta M, Arnao MB. The inactivation and catalytic pathways of horseradish peroxidase with m-chloroperoxybenzoic acid. A spectrophotometric and transient kinetic study. J Biol Chem. 1997;272:5469–5476. doi: 10.1074/jbc.272.9.5469. [DOI] [PubMed] [Google Scholar]
- 57.Ivancich A, Mazza G, Desbois A. Comparative electron paramagnetic resonance study of radical intermediates in turnip peroxidase isozymes. Biochemistry. 2001;40:6860–6866. doi: 10.1021/bi002826j. [DOI] [PubMed] [Google Scholar]
- 58.Dorlet P, Rutherford AW, Un S. Orientation of the tyrosyl D, pheophytin anion, and semiquinone QA*− radicals in photosystem II determined by high-field electron paramagnetic resonance. Biochemistry. 2000;39:7826–7834. doi: 10.1021/bi000175l. [DOI] [PubMed] [Google Scholar]
- 59.Hofbauer W, Zouni A, Bittl R, Kern J, Orth P, Lendzian F, Fromme P, Witt HT, Lubitz W. Photosystem II single crystals studied by EPR spectroscopy at 94 GHz: The tyrosine radical YD*. Proc Natl Acad Sci USA. 2001;98:6623–6628. doi: 10.1073/pnas.101127598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivancich A, Jouve HM, Gaillard J. EPR evidence for a tyrosyl radical intermediate in bovine liver catalase. J Am Chem Soc. 1996;118:12852–12853. [Google Scholar]
- 61.Gerfen GJ, Bellew BF, Un S, Bollinger JM, Stubbe J, Griffin RG, Singel DJ. High-frequency (139.5 GHz) EPR spectroscopy of the tyrosyl radical in Escherichia coli ribonucleotide reductase. J Am Chem Soc. 1993;115:6420–6421. [Google Scholar]
- 62.van Dam PJ, Willems JP, Schmidt PP, Potsch S, Barra AL, Hagen WR, Hoffman BM, Andersson KK, Graslund A. High-frequency EPR and pulsed Q-band ENDOR studies on the origin of the hydrogen bond in tyrosyl radicals of ribonucleotide reductase R2 proteins from mouse and herpes simplex virus type 1. J Am Chem Soc. 1998;120:5080–5085. [Google Scholar]
- 63.Fasanella EL, Gordy W. Electron spin resonance of an irradiated single crystal of L-tyrosine-HCl. Proc Natl Acad Sci USA. 1969;62:299–304. doi: 10.1073/pnas.62.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson JC, Wu G, Tsai AL, Gerfen GJ. Determination of the structural environment of the tyrosyl radical in prostaglandin H2 synthase-1: A high frequency ENDOR/EPR study. J Am Chem Soc. 2005;127:1618–1619. doi: 10.1021/ja043853q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamm RN, Turner JE, Stabin MG. Monte Carlo simulation of diffusion and reaction in water radiolysis—A study of reactant “jump through” and jump distances. Radiat Environ Biophys. 1998;36:229–234. doi: 10.1007/s004110050076. [DOI] [PubMed] [Google Scholar]
- 66.McLachlan AD. Hyperconjugation in the electron resonance spectra of free radicals. Mol Phys. 1958;1:233–240. [Google Scholar]
- 67.Lykos PG. On the electron spin resonance spectra of hydrocarbon radicals. J Chem Phys. 1960;32:625–626. [Google Scholar]
- 68.Heller H, McConnell HM. Radiation damage in organic crystals. II Electron spin resonance of (CO2H)CH2-CH(CO2H) in beta-succinic acid. J Chem Phys. 1960;32:1535–1539. [Google Scholar]
- 69.Stone EW, Maki AH. Hindered internal rotation and ESR spectroscopy. J Chem Phys. 1962;37:1326–1333. [Google Scholar]
- 70.O’Malley PJ, MacFarlane AJ, Rigby SEJ, Nugent JHA. The geometry and spin density distribution of the tyrosyl radical: A molecular orbital study. Biochim Biophys Acta. 1995;1232:175–179. [Google Scholar]
- 71.Qin Y, Wheeler RA. Similarities and differences between phenoxyl and tyrosine phenoxyl radical structures, vibrational frequencies, and spin densities. J Am Chem Soc. 1995;117:6083–6092. [Google Scholar]
- 72.Hoganson CW, Babcock GT. Protein-tyrosyl radical interactions in photosystem II studied by electron spin resonance and electron nuclear double resonance spectroscopy: Comparison with ribonucleotide reductase and in vitro tyrosine. Biochemistry. 1992;31:11874–11880. doi: 10.1021/bi00162a028. [DOI] [PubMed] [Google Scholar]
- 73.Dismukes GC, McGuire A, Blankenship R, Sauer K. Electron spin polarization in photosynthesis and the mechanism of electron transfer in photosystem I. Experimental observations. Biophys J. 1978;21:239–256. doi: 10.1016/S0006-3495(78)85522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogge CE, Ho B, Liu W, Kulmacz RJ, Tsai AL. Role of Tyr348 in Tyr385 radical dynamics and cyclooxygenase inhibitor interactions in prostaglandin H synthase-2. Biochemistry. 2006;45:523–532. doi: 10.1021/bi051235w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogge CE, Liu W, Wu G, Wang LH, Kulmacz RJ, Tsai AL. Identification of Tyr504 as an alternative tyrosyl radical site in human prostaglandin H synthase-2. Biochemistry. 2004;43:1560–1568. doi: 10.1021/bi035717o. [DOI] [PubMed] [Google Scholar]
- 76.Wu G, Rogge CE, Wang JS, Kulmacz RJ, Palmer G, Tsai AL. Oxyferryl heme and not tyrosyl radical is the likely culprit in prostaglandin H synthase-1 peroxidase inactivation. Biochemistry. 2007;46:534–542. doi: 10.1021/bi061859h. [DOI] [PMC free article] [PubMed] [Google Scholar]