Abstract
Regulation of the Class IA PI 3-kinase involves inhibition and stabilization of the catalytic subunit (p110) by the regulatory subunit (p85). Regulation is achieved by 2 major contacts: a stable interface involving the Adapter-Binding Domain (ABD) of p110 and the inter-SH2 (iSH2) domain of p85, and a regulatory interaction between the N-terminal SH2 (nSH2) domain of p85 and the helical domain of p110. In the present study, we have examined the relative orientation of the nSH2 and iSH2 of p85α using site-directed spin labeling and pulsed EPR. Surprisingly, both distance measurements and distance distributions suggest that the nSH2 domain is highly disordered relative to the iSH2 domain. Molecular modeling based on EPR distance restraints suggests that the nSH2 domain moves in a hinge-like manner, sampling a torus space around the proximal end of the iSH2 domain. These data have important implications for the mechanism by which p85/p110 dimers are regulated by phosphopeptides.
Keywords: PI3K, EPR, DEER, SDSL, Monte Carlo, Molecular dynamics, modeling
Phosphatidylinositol 3-kinases (PI3K) phosphorylate the 3’ position of the inositol ring of phosphatidylinositol, and are important in the regulation of cell motility, proliferation, apoptosis, and metabolism (1–4). The PI3K family comprises three classes of enzymes (I, II, and III) differing in structure and function. The Class IA subgroup of PI3K family, which generate PtdIns(3,4,5)P3 in vivo, consist of two subunits arranged in a heterodimer (1). The catalytic subunit, p110, has three isoforms: p110α and p110β, and p110δ. The α and β isoforms are found in abundance in mammalian cells, whereas p110δ is found mostly in hematopoietic cells. The regulatory domain (p85α, p85β, p55α, p50α and p55γ) forms a stable dimer with p110, stabilizing it against thermal denaturation and inhibiting its activity (1, 5). The regulatory subunit p85 consists of an N-terminal SH3 domain, a region homologous to the Breakpoint Cluster Region gene (BCR domain), and two Src homology 2 (SH2) domains linked by an antiparallel coiled coil, the inter-SH2 or iSH2 domain. The catalytic activity of the p85/p110 dimer is increased when the p85 SH2 domains bind to proteins or peptide sequences containing phosphorylated tyrosine peptides (YXXM motifs) such as those present in a variety of cell membrane receptors (1, 6–8). This regulation does not involve the dissociation of p85 from p110, as monomeric p110α (and presumably other isoforms) is unstable at 37°C and is rapidly degraded unless dimerized with p85 (5). Thus, the regulation of PI3K must occur via a conformational change in p85 that is transmitted to p110α. This conformational change is presumably communicated through the nSH2 domain upon phosphopeptide binding, either by changes in the intradomain structure, or by a repositioning of the entire domain, which subsequently relieves inhibition at the catalytic site of p110.
In order to obtain structural information regarding the regulatory subunit, we previously performed NMR experiments on p85ni, which consists of the iSH2 and nSH2 domains (9). p85ni is the minimal subunit that displays p110α binding and inhibition in the basal state and disinhibition upon phosphopeptide binding (8). Based on paramagnetic relaxation effects of spin probe-labeled cysteine residues in the iSH2 on backbone amide protons in the nSH2 domain, we predicted that the iSH2 residues 581–593 were in close proximity to one face of the nSH2 domain. More recently, crystal structures of wild type p110α and an His1047Arg p110α mutant complexed with p85ni have been solved (10–11). The nSH2 domain in the wt p110α/p85ni structure yielded weak electron density and is not resolved, but in the mutant p110α His1047Arg/p85ni structure the nSH2 domain is well ordered and forms a scaffold between the C2, helical and kinase domains of the p110α mutant. A comparison of this structure with one of an isolated p85α nSH2 domain bound to a phosphotyrosine peptide from PDGFR reveals that the loop of the p110α helical domain that contains the E545K hotspot mutation binds to nSH2 in the same location as the phosphopeptide (12). This data is consistent with our previous biochemical analysis of the nSH2 domain-helical domain interface (13), which predicted that the hotspot mutations in the helical domain interfere with nSH2 binding to p110α, releasing the inhibition in same manner that the phosphopeptide would.
This recent structural data make it clear that the nSH2 domain cannot bind to tyrosine phosphorylated proteins when present in the basal inhibited conformation, and must therefore exist in alternative conformations. Furthermore, the iSH2-nSH2 interdomain orientation determined by NMR in monomeric p85ni is not the same as in complex with p110α. These results, together with the lack of resolution of the nSH2 domain in the wt p110α-p85ni dimer crystal structure, suggest that a certain degree of flexibility may be required for the proper regulatory function of p85, and that the orientation of the nSH2 domain exhibits disorder in the basal state of p85. To test this hypothesis, we have employed site directed spin labeling (SDSL) in conjunction with a pulsed EPR technique named double electron-electron resonance (DEER, also known as pulsed electron double resonance, PELDOR) (14–16), which can measure distances between nitroxide pairs in the range of 15 to ~70 Å. We used a combination of Metropolis Monte Carlo minimization (MMCM) (17) and molecular dynamics (MD) simulations (18–19) to predict the spin labels’ conformation in the nSH2-iSH2 fragment of p85 (p85ni), and incorporated EPR distances as well as distance distributions as restraints into a rigid body motion modeling algorithm. As a result, we determined the possible orientations of the nSH2 domain of p85ni with respect to iSH2 domain.
MATERIALS AND METHODS
Protein Expression, Purification, and Spin Labeling
Double Cysteine mutants of p85ni (p85α residues 372–600) were generated on a cysteineless template. Protein was purified, expressed, and spin labeled with N-(1-oxy-2,2,5,5-tetramethyl-4-piperidinyl)maleimide spin label (MSL) as previously described (20). Labeling efficiency was calculated by comparing the number of spins in the sample measured by EPR to the number of protein molecules determined by UV absorption at 280 nm. All samples were labeled to >1.8 labels/protein. All p85ni cysteine mutants were assayed for both binding and inhibition of p110α; no significant differences were seen with any of the constructs relative to wild type, indicating that none of the mutated amino acids influences structure or function. Eight of the spin labeled double mutants used to generate the modeled structure were assayed for interactions with p110α. All bound p110α equivalently to wild type, and only one displayed a reduced capacity for inhibition (p85ni-C575-C330). Given these results, the structure determined here of the spin-labeled p85ni species reflects that of an essentially normal binding and inhibiting species.
EPR Spectroscopy
Continuous wave EPR spectra for spin counting were recorded at room temperature (298 K) using a Varian E112 series X-band spectrometer (Varian Inc, CA) equipped with a TE102 rectangular cavity at 9 GHz. Samples were sealed in 50 µL glass capillaries. For DEER experiments, spin labeled p85ni was resuspended in PBS buffer containing 40% glycerol for glass formation. 100 µL samples, varying in concentration from 20 to 100 µM, were transferred to 4 mm O.D. 3.2 mm I.D. quartz capillaries, flash frozen by dipping into liquid nitrogen and stored in liquid nitrogen. Inter-spin distances were measured by 4 pulse DEER experiment (16) on a home made spectrometer (21) with split ring resonator at 20 K. In the DEER experiment, the observer frequency was set to the resonator dip, and the static field was fixed at the low field resonance of the spin label spectrum. The pump pulse was set to 65 MHz lower than the observer frequency in order to be on resonance with the central (maximal) peak in the nitroxide EPR spectrum. The delay time between the first two pulses was set to 200 ns in order to minimize proton modulation. The second delay time was varied in each experiment to ensure at least 1/4 of recorded data was inter-molecular background. Typically, a long scan was acquired to ensure no low frequency modulation (corresponding to a long interspin distance) was present. If no such long period oscillation was observed shorter scans were averaged in order to maximize signal to noise. Pulse lengths were 20 ns for π/2 pulse and 40 ns for π pulse. The pump pulse was adjusted to 60 ns which maximized the DEER-induced echo modulation for the distances observed here: under no experimental conditions (pulse widths, acquisition time, temperature) were distances below 2.5 nm or above 6 nm observed. The acquisition parameters used here are slightly different than optimal parameters reported for other spectrometers (22–24). For example other groups using a Bruker spectrometer report an optimal temperature of 65 K and pump pulse length of 32 ns. For the spectrometer used here, we have determined that S/N is optimized by the balance of absolute echo intensity and repetition rate achieved with the pulse parameters and temperature given above.
We determined the lower boundaries for observable mean distance and distance distribution, and best parameters for the DEER experiment in our instrument by running a series of control experiments on two rigid biradicals (16, 25) and a short helical peptide containing two TOAC residues (26). The excitation bandwidth of the DEER pulses was sufficient to observe distances as short as 2 nm. Moreover, we observed distance distributions as sharp as 0.01 nm (supplemental material). Thus, any distance distributions observed in the p85ni samples derive from the structure of the protein under study and not from artifacts introduced by the spectrometer. In order to gauge experimentally the inter-spin distance distribution expected to originate from the conformational heterogeneity of the spin label side chains themselves, we performed DEER experiments on samples in which both labels were attached to the nSH2 domain. This labeling scheme eliminates any contributions from the inter-domain disorder from the measurement, and because the nSH2 domain has been shown to be well structured in solution, the majority of any distance distribution observed originates from conformational heterogeneity of the spin label side chains. The DEER pattern obtained for a p85ni (337,410 MSL) intra-nSH2 domain mutant shows a clear oscillation and corresponds to a distance distribution of 1.37 nm. Although this distance distribution is more than that predicted based on MD of the side chains alone (vide infra), it is less than all but one of the 16 distributions measured for the inter-domain distances, and is significantly less than the largest distribution measured (3.16 nm). We also recorded dipolar evolution trace of single labeled protein, p85ni(546MTSSL), to exclude the possibility of oligomerization or other specific inter molecular interactions contributing significantly to the DEER pattern (see supplemental data).
DEER data were analyzed using DEFit, a MATLAB software which assumes a sum of Gaussian distributed distances contributing to the observed echo modulation (24). In DEFit, a 3D homogenous inter-molecular background is fit to the baseline and subtracted to achieve pure intra-molecular echo modulation. Data fitting is performed via the Monte Carlo/SIMPLEX algorithm to find the distance distribution which best describes the experimental data (26). Selection of the number of Gaussian populations used to describe the experimental data is based on the statistical F-test. This method was tested for simulated dipolar evolutions and experimental data corresponding to known distance distributions. The main advantage of using DEFit to analyze dipolar data is that the model dependent analyses allow error values to be assigned to the mean distance and distance distribution values. The software is available at http://kadirilkersen.googlepages.com and at http://fajerpc.magnet.fsu.edu.
Computational Modeling
We used two structural templates in computational studies, the crystal structures of iSH2 domain (pdb code 2V1Y, (27)) and nSH2 domain (pdb code 2IUG, (12)). In silico mutations of desired sites to MSL in individual domains of p85ni were carried out using Modeller program (28) and Charmm (29) topologies. Once the side chain was mutated to spin label, its minimum energy conformation was predicted using Metropolis Monte Carlo simulations in Charmm (17). In this method, all atoms of the protein except for the spin label are held in place while the conformational space of the spin label side chain was searched by rotating dihedral angles by a random angle followed by energy minimization using implicit solution. Four of the lowest energy conformations are selected for Molecular dynamics simulations. In MD, the rigid cage is removed and all atoms within 15 Å of the spin label atoms are allowed to move (18). The protein was slowly heated (12 ps simulation with 2 fs time steps) to 300 K and equilibrated for 200 ps with 1 fs time steps. The production run was performed for 10 ns with 2 fs time steps. The trajectory was analyzed using a home written Matlab program to generate a volume surface within which the nitroxide can be found in the simulation. This program extracts the N and O coordinates of the nitroxide from the MD trajectory and assumes their midpoint to be location of the unpaired electron. Then the space occupied by these coordinates is divided into 25 equally spaced intervals along each axis, resulting in 253 rectangular bins in 3D. Nitroxide coordinates falling into each bin were counted, and together with the coordinates of the bins, stored in a 4×3 volume data matrix that was subjected to smoothing. The final volume data was visualized by VMD (30) as isosurfaces Figure 1. Upon comparing the volume surfaces of MD simulations corresponding to four initial rotamer seeds, we used only the sites whose trajectories converged to the same volume from different initial structures at 300 or 500 K MD simulations as restraints in domain-domain conformation modeling. To find the conformation of the nSH2 domain that satisfies the experimental distances, we randomly rotated and translated the entire domain’s coordinates with respect to the iSH2 domain. Among millions of structures generated this way, those that satisfy the experimental distances within the experimental distance distributions were pooled. These distributions were redefined as square wells with widths equal to the full width at half maximum of Gaussian curves of experimental data. More than 2000 nSH2 structures were generated this way and connected to iSH2 via 12-residue linker for which experimental data is not available. The linker was constructed with Modeller’s loopmodel routine (31) where the iSH2 and nSH2 domains’ coordinates were fixed. We performed a final conjugate gradient energy minimization to all p85ni models and sorted them with respect to total energy. We calculated probabilities of each structure assuming a Gaussian distribution to energies and selected those that are in the lowest 5% of the energy distribution. This method eliminates the structures that have an extremely stretched linker or those with van der Waals clashes. In our experience, this modeling scheme was in overall superior to constructing the linker first and running simulated annealing and/or molecular dynamics simulations with the distance restraints on p85ni (data not shown). In the latter case, converging to local minima was extremely likely. In order to estimate via modeling the impact that spin label side chain conformational heterogeneity may have on the measured distance distribution, we performed MD on the spin labels assuming a fixed iSH2-nSH2 interdomain structure. The internitroxide distance distribution was found to be 0.3 – 0.7 nm depending on the specific spin label pair. We then attempted to remove this contribution from the distance distribution used to generate the iSH2-nSH2 conformational structures via subtraction from the measured distributions. This method did not produce a conformational space spanned by the calculated structures that was significantly smaller than that produced without removing the side chain distribution estimate. This seems to be due to two reasons: 1) the measured distributions (1.3 – 3.2 nm) are significantly larger than those predicted from side chain conformational distributions alone; and 2) the mean distances measured are incompatible with a single relative orientation of the nSH2 - iSH2 domains, requiring the inclusion of a distribution in order to achieve convergence.
Figure 1.
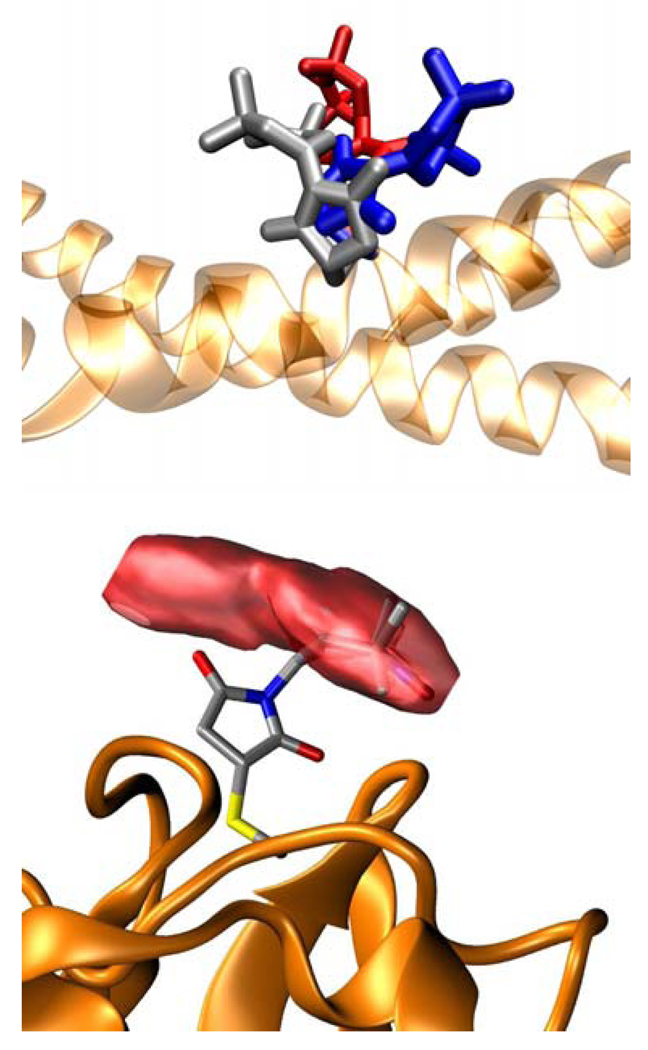
Modeling spin label conformations. Top: Three distinct lowest energy conformations of p85ni_575MSL. Bottom: p85ni_422MSL and the volume occupied by its nitroxide during MD trajectory.
RESULTS
Domain-domain distances in p85ni
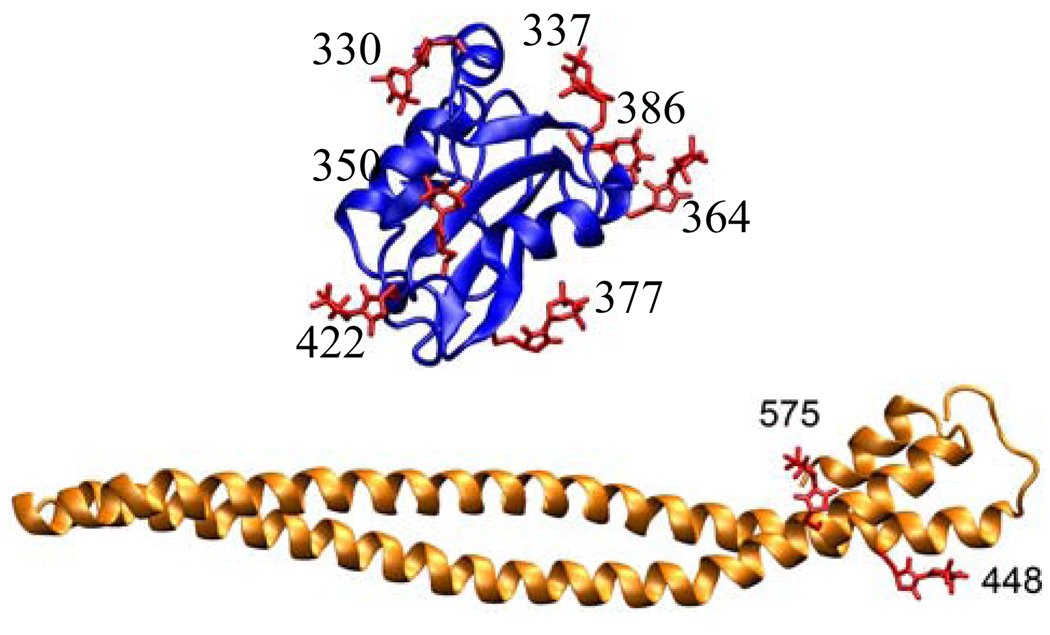
In order to determine the relative orientation of nSH2 domain with respect to the iSH2 domain in the regulatory subunit construct p85ni, we measured inter domain distances by site directed spin labeling and 4-Pulse DEER. Spin labels were attached to p85ni as pairs on strategically selected sites so that they would be within the sensitivity range of EPR distance measurements, which is between 15 and 70 Å for DEER experiments and 2–25 Å for exchange or dipolar broadened cw EPR spectra. Double spin labeled p85ni samples consisted of a combination of three sites on iSH2 domain (448, 575, and 584) and 7 sites on nSH2 (330, 337, 350, 364, 377, 386, 422), as listed in Table 1. We obtained DEER traces of a series of double and single labeled controls to ensure that the distributions reported here did not result from instrumental artifacts, inter-molecular interactions or spin label side chain conformational heterogeneity (see Materials and Methods). The dipolar evolution of all double mutants had characteristics of a mid range distance (~3–4 nm) as observed from the initial decay of the echo modulation, and broad distance distribution as observed from the dampened oscillations in the time domain data (Figure 2, left column). Quantitative analyses were performed with the DEFit software (24). In p85ni samples, all echo modulation was fit best with a distance distribution curve of a single Gaussian. The mean distances (r) and width (Δ) of Gaussian curves which result in the best fit to data are tabulated in Table 1 and plotted in Figure 2 center column. Errors in r and Δ are determined via random sampling of parameter space around the best fit. The goodness of fit value (χ2) corresponding to each randomly generated parameter is used to generate a surface plot against r and Δ parameters (Figure 2, right column) in a confidence interval of 68%. The size of the χ2 surface along r or Δ axes is indicative of the error in the corresponding parameter. A large surface as in 448-330 corresponds to a large error, whereas a small surface as in 575-330 corresponds to a smaller error. The error values in Table 1 are the standard deviation of all sampled parameters.
Table 1.
Distances from DEER experiments.
| Site 1 | Site 2 | r (nm) | Δ (nm) |
|---|---|---|---|
| 448 | 330 | 3.41±0.54 | 1.26±2.10 |
| 448 | 337 | 3.47±0.21 | 1.57±0.70 |
| 448 | 350 | 3.32±0.13 | 1.86±0.39 |
| 448 | 364 | 3.42±0.60 | 3.10±1.90 |
| 448 | 377 | 3.89±0.23 | 2.81±0.65 |
| 448 | 386 | 3.48±0.42 | 3.16±1.33 |
| 575 | 330 | 3.84±0.17 | 1.65±0.52 |
| 575 | 337 | 3.92±0.31 | 1.40±1.04 |
| 575 | 350 | 4.02±0.20 | 1.47±0.63 |
| 575 | 364 | 3.73±0.19 | 2.02±0.56 |
| 575 | 377 | 3.85±0.18 | 1.58±0.60 |
| 575 | 386 | 3.78±0.19 | 1.68±0.58 |
| 575 | 422 | 4.31±0.25 | 2.86±0.78 |
| 584 | 337 | 3.67±0.21 | 1.79±0.65 |
| 584 | 350 | 3.29±0.29 | 2.03±0.88 |
| 584 | 351 | 3.28±0.13 | 2.05±0.39 |
Figure 2.
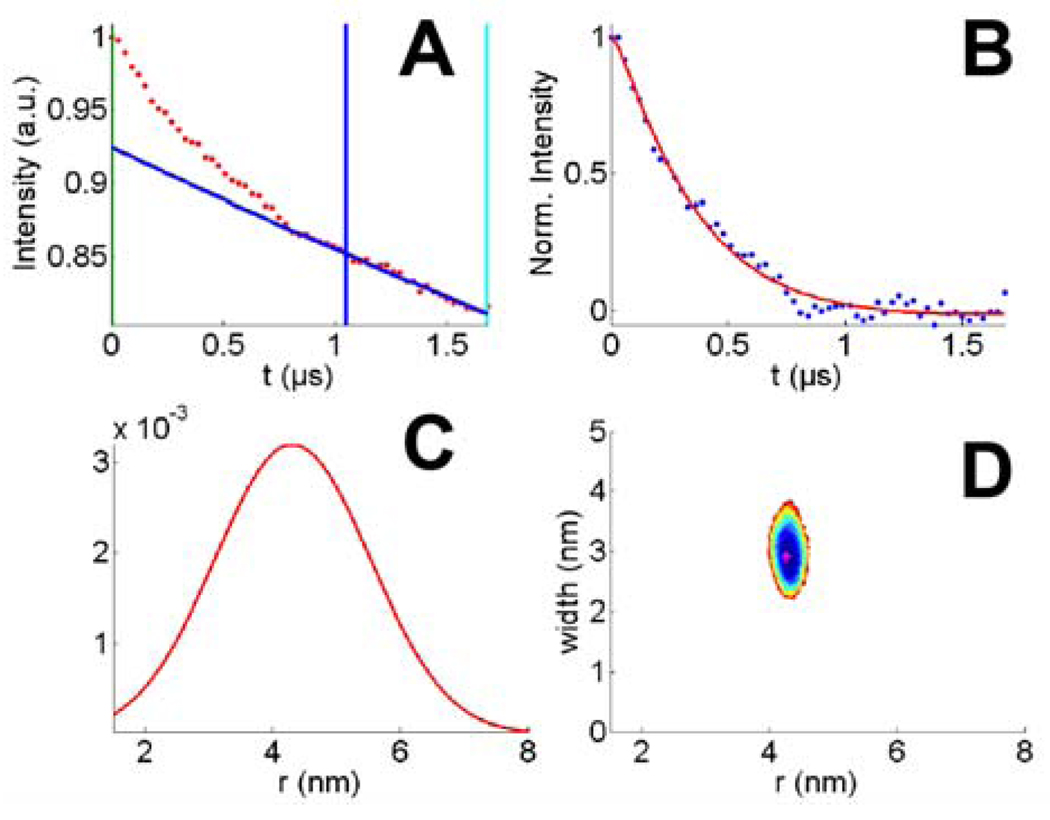
DEER data and analysis of p85ni(575/422 MSL). A: Original data showing echo modulation (red points), background fit region (vertical blue and cyan lines), and the fitted 3D homogenous background (blue line); B: Pure modulation after background subtraction (blue points) and the fit (red line); C: The distance distribution which describes the echo modulation; D: Error surface of the fit.
Mean distances of all 16 double spin labeled samples were within ~1 nm of each other (in Table 1 minimum r is 3.28 nm, maximum r is 4.31 nm), despite the fact that the spin labels on nSH2 were distributed about the domain’s periphery and the domain’s diameter is ~2 nm. Structural studies of the iSH2 and the nSH2 domains in solution indicated that these individual domains are well structured (32–33). Thus both the mean distances and the distance distributions measured here suggest a disorder, which is likely due to inter domain flexibility.
The distance distributions in each double mutant are extremely large with 1.26 to 3.16 nm width. The distribution may be a combination of spin label side chain and protein backbone conformational heterogeneity. The simulated distance distribution of two MSL side chains on a Glycine α-helix is 1.2 nm (data not shown), which corresponds well to the experimentally determined intra-domain distance distribution of 1.37 nm measured for two spin labels attached to the nSH2 domain (Fig. S2F). Hence the greater inter-domain distance distributions must be due to multiple protein conformations. Structural studies on independent nSH2 and iSH2 domains revealed stable structures for the individual backbone folds (34) hence the conformational distribution is likely due to heterogeneity in the relative orientations of nSH2 and iSH2 domains.
Predicting spin label conformation
The distances observed in DEER experiments are between the unpaired electrons of the spin label. The spin label side chain’s conformation directly affects the mean distance and the distance distribution observed via EPR. Thus one needs to predict the spin labels’ conformation in order to convert experimental distances into structural parameters. We modeled the spin label on all mutation sites via in silico mutagenesis and Metropolis Monte Carlo minimization. The predicted lowest energy rotamers of MSL are shown in Figure 3. These conformers were seeded into Molecular Dynamics simulations in which the atoms within 15 Å of the spin label were allowed to sample the local environment. In the cases of 364MSL, 377MSL, and 422MSL on the nSH2 domain, the MD trajectory largely diverted from the initial MMCM conformation due to backbone fluctuations. Because of uncertainty in side chain conformation, these residues were not used in modeling domain-domain orientation. Residue 584 of iSH2 domain was observed to be buried in the crystal structure by Williams et al (27), and not observed at all in the more recent p110α/p85ni crystal structure of Amzel et al (10). In the Williams structure, there is insufficient space at the coiled coil interface for the MSL side chain. As a result, the minimum energy conformation generated via MMCM had impossibly long bond lengths. MD simulation of 584MSL resulted in changes in the secondary structure and the breaking of coiled-coil. Hence, 584MSL is also excluded from domain-domain modeling.
Figure 3.
Energy minimized MSL rotamers attached to mutation sites on the nSH2 domain (top) and the iSH2 domain (bottom).
MD trajectories were also analyzed to show the possible distribution of distances due to spin label side chain motion alone. Within the final pairs of spin labels, the largest predicted distance distribution ranges from 3 to 7 Å, supporting that the large experimental distance distributions are due to protein conformational heterogeneity.
Modeling of domain-domain orientation
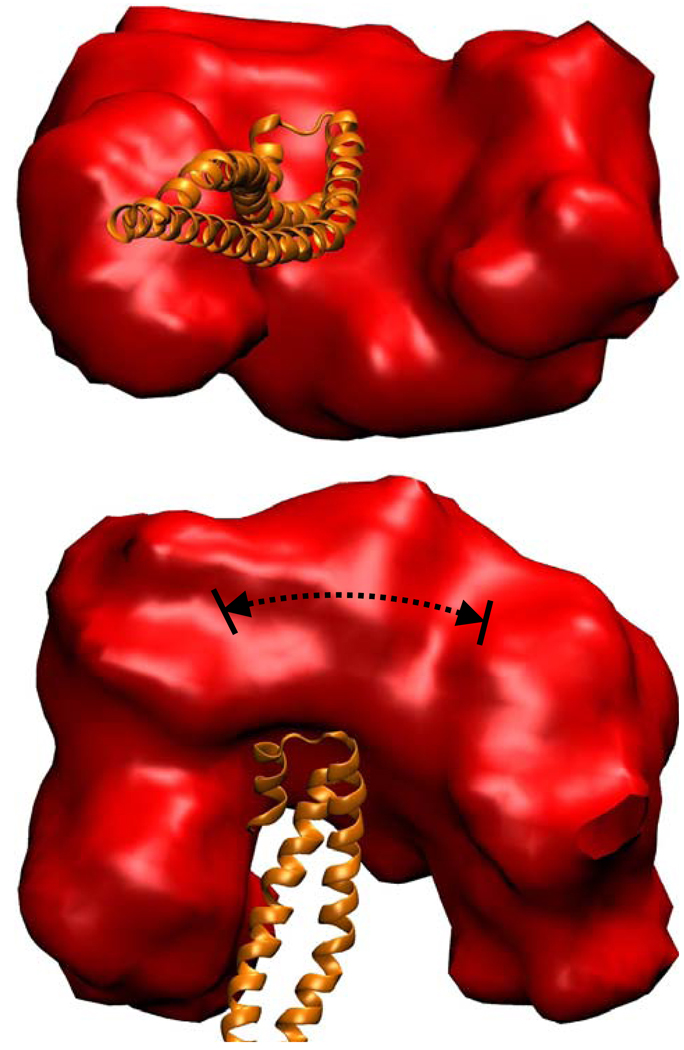
We reconstructed nSH2 and iSH2 domains with all the spin labels for which there was confidence in the predicted conformation (575, 448, 386, 350, 337, and 330). We then performed rigid body modeling and pooled structures that satisfied the experimental distances and experimental distance distributions. The spatial volume that can be occupied by these conformers is shown in Figure 4. The conformers do not converge to a single position, but rather to a ring-like volume around the iSH2 domain. This is mostly due to the wide distributions in distances, but also due to the computational artifacts of incomplete triangulation. In order to completely determine the location of a particular residue on nSH2 domain, three linearly independent distances between this residue and three residues on the iSH2 domain are required (35–36). In the case of p85ni, the modeling involved only two residues on the iSH2 domain; hence the axis connecting the two spin labels’ nitroxides on the iSH2 domain serves as an artificial symmetry axis. To determine the exact effect of symmetry in our results, we picked an arbitrary starting p85ni structure and subjected it to the side-chain modeling algorithm described above. From the initial pdb, we measured the resulting inter nitroxide distances, and assumed those distances were experimental. We then repeated our inter-domain modeling scheme and pooled structures that satisfied the pseudo distances within a distribution of 4 Ǻ. We were able to reproduce the starting model structure, but also others with a radial distribution of ~30 Ǻ (indicated by arrows on Figure 4) around the artificial symmetry axis. This radial distribution indicates the extent to which incomplete triangulation contributes to the total conformational heterogeneity. However, this conformational heterogeneity due to improper triangulation does not explain the large volume in Figure 4, which has an approximately 100 Å distribution. Hence the wide range of conformational possibilities for the nSH2 domain with respect to iSH2 domain is primarily due to a true distribution in experimentally determined distances.
Figure 4.
The space volume that may be occupied by the nSH2 domain (red) around the iSH2 domain (orange) based on EPR distances. The dashed line shows contribution from improper triangulation as explained in the narrative.
DISCUSSION
Double electron-electron resonance (DEER)(16) as been used to investigate the structures and/or conformational changes in a number of biological systems including myosin actin-binding cleft (37), maltose ABC-importer (38), Rod Arrestin (39), Na+/proline transporter PutP (40), Anthracis Repressor (24), Light-harvesting Chlorophyll a/b Protein (41), T4-lysozyme (39), and vonWillebrand Factor A (vWF-A) (42). The DEER technique is unique in the sense that it provides a positive means to measure disorder, and hence is sensitive to the conformational heterogeneity of the protein. For instance, in the Anthracis Repressor protein from Bacillus anthracis, a disordered state in the protein’s DNA binding region was observed by DEER, which was removed following the activation of the protein by metal binding, suggesting a tweezers-like conformational change (24).
We have now used site-specific spin labeling and pulsed EPR (DEER) to measure distances and distance distributions between pairs of labeled cysteine residues in the nSH2 and iSH2 domains of p85ni in solution. The iSH2 residues K575, K448, and nSH2 residues D337, T350, M364, N377, R386, and V422 were selected for spin labeling because they are surface exposed (hence accessible for spin label attachment) and because they are most probably within the experimental distance range of DEER (2–7 nm). Surprisingly, the mean distance between each pair (3 distinct iSH2 residues and 6 or 7 distinct nSH2 residues, total 13 distances) varied by no more than 1 nm, despite the fact that the nSH2 domain has an approximate diameter of 2 nm. The widths of the distance distributions range from 1.2 nm to 3.6 nm, despite that the distribution of distances between any two MSL side chains used in this study are estimated to be 0.3–0.7 nm on backbone restrained p85ni, and 1.2 nm on a glycine alpha helix based on molecular dynamics simulations (data not shown), well below the observed experimental distributions. Since both the iSH2 and nSH2 domains are inherently rigid, these results suggest a large disorder in the relative orientation of the nSH2 domain with respect to the iSH2 domain.
To quantify the degree of disorder in p85ni, we modeled the domain-domain orientation, restrained only by internal domain structures and EPR distances. The distances observed in DEER experiments are between the unpaired electrons of the spin label. The spin label side chain conformation directly affects the mean distance and the distance distribution observed via EPR. Thus we first predicted the conformation of the spin labels in order to understand the experimental distances in structural terms via Metropolis Monte Carlo Minimization and Molecular Dynamics simulations. Fixing the orientation of the spin label at its most probable conformation, we the modeled the domain-domain orientation. We used a Monte Carlo search of possible orientations that satisfy the EPR distance distributions, followed by linker construction and Simulated Annealing. This method proved more efficient than restrained Simulated Annealing and restrained MD. In our experience, restrained simulations are more suitable for narrow distance distributions where the local minima are not abundant on the energy landscape. The Monte Carlo search, which excludes the linker between domains, removes the possible restraining from linker conformation while searching for agreement with EPR distances. Improbable linker conformations can be removed by reconstructing the linker residues on a library of possibilities, which is primarily due to the experimental distance distributions. The data strongly suggest that the nSH2 domain does not assume a preferred orientation relative to the iSH2 domain in p85ni. A parallel study (43) supports this model by measuring the apparent rotational correlation time of the nSH2 domain as 13 ns in the p85ni construct based on NMR relaxation methods. This is faster than the calculated rotational correlation time of 45–52 ns for a completely rigid p85ni (several possible geometries), suggesting that in solution the nSH2 domain is highly mobile compared to the iSH2 domain.
Based on NMR relaxation studies, we previously proposed that the C-terminal end of the iSH2 domain makes a contact with the nSH2 domain, positioning it so as to inhibit p110α (9, 33). However, the data presented here are not consistent with this model. The pulsed EPR measurements show that the orientation of the nSH2 domain is in fact highly disordered, distributed through an arc around a hinge-like fashion at the end of the iSH2 domain. This hinge-like motion may in fact be important for the regulation of p110α. We have previously shown that basic residues near the phosphopeptide binding site of the nSH2 domain make an inhibitory contact with an acidic patch in the helical domain of p110α, and proposed that this contact is disrupted when tyrosine phosphoproteins bind to the SH2 domain (13). This model was supported by the crystal structure of the p85ni/p110α-H1047R dimer, which shows that the basal conformation of the nSH2 domain was not compatible with phosphopeptide binding (11). The hinge-like motion of the nSH2 domain provides a mechanism by which the nSH2 domain can maintain a specific inhibitory interaction with p110, while still “sampling” the external environment for potential SH2 domain ligands. Indeed, we find that rigid fixation of the nSH2 domain relative of the iSH2 domain, through the introduction of an engineered disulfide bond, allows inhibition of p110 but not a release of inhibition upon SH2 domain occupancy (Wu, H. and Backer, J.M., unpublished observations).
The hinge-like motion of the nSH2 domain also provides a possible explanation for the NMR relaxation data reported previously(44). The NMR model was determined using paramagnetic relaxation effects on backbone amide protons in the nSH2 domain of p85ni caused by nitroxide spin labels in the iSH2 domain. Interestingly, the attenuation of NMR crosspeaks by the iSH2 domain spin labels was restricted to a single face of the nSH2 domain, and resulted in almost complete loss of signal intensity from some nSH2 residues. These data were interpreted as reflecting a close contact between the iSH2 and one face of the SH2 domain. However, given the hinge-like movement of the SH2 domain, a single face of the nSH2 domain samples this orientation with a time constant (13 ns) that is significantly shorter than the length of time that the proton spins are transverse during the 2D NMR pulse sequence (~11 ms). Thus, the NMR relaxation data are almost certainly biased towards these shorter distances, and reflect a subset of structures out of the larger distribution seen in the DEER experiments.
The present study cannot rule out the possibility that a distributed orientation of nSH2 is due to the lack of cSH2, BCR, SH3 domains in the protein construct. However, we consider this scenario unlikely due to the fact that p85ni retains regulatory function of full-length p85. Moreover, preliminary studies in our lab suggest that the nSH2-iSH2 orientation in full-length p85α is similar to that seen in p85ni (data not shown).
The DEER data indicate that in solution, the iSH2-nSH2 interdomain structure is not fixed in a single structure but rather displays a distribution of orientations. The long linker region connecting the two domains presumably facilitates this distribution. Indeed, in the mutant p110α-p85ni crystal structure this linker region is almost fully extended to facilitate both the contact of the iSH2 and ABD domains as well as the positioning of the nSH2 domain in its central “scaffolding” position in contact with the kinase, helical and C2 domains (11). Moreover, the authors speculate that phosphopeptide-binding induced release of inhibition involves the shift in nSH2 orientation relative to the helical domain. Together, these data suggest that iSH2-nSH2 interdomain flexibility is important for both proper binding and regulation of p110α.
DEER provides a means to measure not only long range inter spin distances but also their distributions, making it one of the few techniques that can quantify protein disorder. Interpretation of DEER distances in structural terms is complicated due to the inherent flexibility of the spin label. When convolved with backbone disorder, non-trivial computational modeling is essential to elucidate structural information. Indeed, structural modeling using DEER distances has very recently gained popularity (35, 41, 45). To our knowledge, modeling backbone induced large distance distributions has currently not been accomplished. In this paper, we have demonstrated that modeling of tertiary/quaternary protein structure from sparse EPR distance restraints is feasible even in presence of disorder. We determined that the nSH2 domain of p85α does not adopt a rigid conformation but moves in an arc around a hinge like fashion around the distal end of the iSH2 domain. These data have important implications for the mechanism by which p85α regulates the activity of PI3K.
Supplementary Material
ABBREVIATIONS
- PI3K
Phosphoinositide 3-kinase
- EPR
Electron Paramagnetic Resonance
- DEER
Double electron-electron resonance
Footnotes
This work was supported by NIH grants GM075920 (G.J.G.) and GM55692 (J.M.B.), Albert Einstein College of Medicine Cancer Center grant#: 5P30CA013330-36 (J.M.B.), and Albert Einstein College of Medicine Diabetes Research Training Center grant#: DT20541 (J.M.B.)
SUPPORTING INFORMATION AVAILABLE
Raw DEER data, fits, distance distributions, and error surfaces of all double spin labeled p85ni mutants (Figure S1), DEER control experiments (Figure S2), and EPR lineshapes (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annual Review of Biochemistry. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Experimental Cell Research. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 4.Wymann MP, Pirola L. Structure and function of phosphoinositide 3-kinases. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 1998;1436:127–150. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3'-kinase: Stabilization and inhibition of the p110-alpha catalytic subunit by the p85 regulatory subunit. Mol.Cell.Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rordorf-Nikolic T, Van Horn DJ, Chen D, White MF, Backer JM. Regulation of phosphatidylinositol 3'-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J.Biol.Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 7.Backer JM, Myers MG, Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, Hu P, Margolis B, Skolnik EY, Schlessinger J, White MF. Phosphatidylinositol 3'-Kinase Is Activated by Association with Irs-1 During Insulin Stimulation. Embo Journal. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu JH, Wjasow C, Backer JM. Regulation of the p85/p110 alpha Phosphatidylinositol 3 '-kinase - Distinct roles for the N-terminal and C-terminal SH2 domains. J Biol Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 9.Shekar SC, Wu H, Fu Z, Yip SC, Cahill SM, Girvin ME, Backer JM. Mechanism of constitutive PI 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. Journal of Biological Chemistry. 2005 doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 10.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110 alpha/p85 alpha complex elucidates the effects of oncogenic PI3K alpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 11.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu JX, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. A frequent kinase domain mutation that changes the interaction between PI3K alpha and the membrane. P Natl Acad Sci USA. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolte RT, Eck MJ, Schlessinger J, Shoelson SE, Harrison SC. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nature Structural Biology. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 13.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 14.Milov AD, Ponomarev AB, Tsvetkov YD. Electron Electron Double-Resonance in Electron-Spin Echo - Model Biradical Systems and the Sensitized Photolysis of Decalin. Chem Phys Lett. 1984;110:67–72. [Google Scholar]
- 15.Larsen RG, Singel DJ. Double electron-electron resonance spin-echo modulation: Spectroscopic measurement of electron spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993;98:5134–5146. [Google Scholar]
- 16.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. Journal of Magnetic Resonance. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 17.Sale K, Sar C, Sharp KA, Hideg K, Fajer PG. Structural determination of spin label immobilization and orientation: A Monte Carlo minimization approach. Journal of Magnetic Resonance. 2002;156:104–112. doi: 10.1006/jmre.2002.2529. [DOI] [PubMed] [Google Scholar]
- 18.Sale K, Song LK, Liu YS, Perozo E, Fajer P. Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J Am Chem Soc. 2005;127:9334–9335. doi: 10.1021/ja051652w. [DOI] [PubMed] [Google Scholar]
- 19.Fajer M, Fajer PG, Sale KL. Molecular Modeling of Spin Labels. In: Hemminga M, Berliner L, editors. ESR Spectroscopy in Membrane Biophysics. New York: Springer Verlag; 2007. pp. 254–260. [Google Scholar]
- 20.Fu Z, Aronoff-Spencer E, Backer JM, Gerfen GJ. The structure of the inter-SH2 domain of class IA phosphoinositide 3-kinase determined by site-directed spin labeling EPR and homology modeling. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3275–3280. doi: 10.1073/pnas.0535975100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avdievich NI, Gerfen GJ. Multifrequency probe for pulsed EPR and ENDOR spectroscopy. Journal of Magnetic Resonance. 2001;153:178–185. doi: 10.1006/jmre.2001.2450. [DOI] [PubMed] [Google Scholar]
- 22.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. Sensitivity enhancement in pulse EPR distance measurements. Journal of Magnetic Resonance. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 23.Fajer PG, Brown L, Song L. Practical Pulsed Dipolar EPR (DEER) In: Hemminga MA, Berliner LJ, editors. Biological Magnetic Resonance 27: ESR Spectroscopy in Membrane Biophysics. Springer Verlag; 2007. pp. 95–128. [Google Scholar]
- 24.Sen KI, Logan TM, Fajer PG. Protein dynamics and monomer-monomer interactions in AntR activation by electron paramagnetic resonance and double electron-electron resonance. Biochemistry. 2007;46:11639–11649. doi: 10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- 25.Godt A, Franzen C, Veit S, Enkelmann V, Pannier M, Jeschke G. EPR probes with well-defined, long distances between two or three unpaired electrons. Journal of Organic Chemistry. 2000;65:7575–7582. doi: 10.1021/jo005556j. [DOI] [PubMed] [Google Scholar]
- 26.Monaco V, Formaggio F, Crisma M, Toniolo C, Hanson P, Millhauser GL. Orientation and immersion depth of a helical lipopeptaibol in membranes using TOAC as an ESR probe. Biopolymers. 1999;50:239–253. doi: 10.1002/(SICI)1097-0282(199909)50:3<239::AID-BIP2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 28.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 29.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14 doi: 10.1016/0263-7855(96)00018-5. 33-&. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meike H, Grant WB, George P, Jonathan B, Jeffrey L, Mike W, Iain DC. Phosphopeptide binding to the N-terminal SH2 domain of the p85alpha subunit of PI 3prime-kinase: A heteronuclear NMR study. Protein Science. 1994;3:1020–1030. doi: 10.1002/pro.5560030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Z, Aronoff-Spencer E, Wu HY, Gerfen GJ, Backer JM. The iSH2 domain of PI 3-kinase is a rigid tether for p110 and not a conformational switch. Arch Biochem Biophys. 2004;432:244–251. doi: 10.1016/j.abb.2004.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolte RT, Eck MJ, Schlessinger J, Shoelson SE, Harrison SC. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat Struct Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 35.Borbat PP, McHaourab HS, Freed JH. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J Am Chem Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 36.Borbat PP, Freed JH. Measuring distances by pulsed dipolar ESR spectroscopy: Spin-labeled histidine kinases. Two-Component Signaling Systems, Pt B. 2007;423 doi: 10.1016/S0076-6879(07)23003-4. 52-+ [DOI] [PubMed] [Google Scholar]
- 37.Fajer PG, Gyimesi M, Malnasi-Csizmadia A, Bagshaw CR, Sen KI, Song LK. Myosin cleft closure by double electron-electron resonance and dipolar EPR. Journal of Physics-Condensed Matter. 2007;19:10. [Google Scholar]
- 38.Grote M, Bordignon E, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E. A comparative electron paramagnetic resonance study of the nucleotide-binding domains' catalytic cycle in the assembled maltose ATP-binding cassette importer. Biophys J. 2008;95:2924–2938. doi: 10.1529/biophysj.108.132456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanson SM, Dawson ES, Francis DJ, Van Eps N, Klug CS, Hubbell WL, Meiler J, Gurevich VV. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeschke G, Wegener C, Nietschke M, Jung H, Steinhoff HJ. Interresidual distance determination by four-pulse double electron-electron resonance in an integral membrane protein: the Na+/proline transporter PutP of Escherichia coli. Biophys J. 2004;86:2551–2557. doi: 10.1016/S0006-3495(04)74310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeschke G, Bender A, Schweikardt T, Panek G, Decker H, Paulsen H. Localization of the N-terminal domain in light-harvesting chlorophyll a/b protein by EPR measurements. J Biol Chem. 2005;280:18623–18630. doi: 10.1074/jbc.M501171200. [DOI] [PubMed] [Google Scholar]
- 42.Banham JE, Timmel CR, Abbott RJ, Lea SM, Jeschke G. The characterization of weak protein-protein interactions: evidence from DEER for the trimerization of a von Willebrand Factor A domain in solution. Angew Chem Int Ed Engl. 2006;45:1058–1061. doi: 10.1002/anie.200503720. [DOI] [PubMed] [Google Scholar]
- 43.Wu HY, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, Sen KI, Janakiraman V, Seshagiri S, Gerfen GJ, Girvin ME, Backer JM. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110 alpha and are disrupted in oncogenic p85 mutants. P Natl Acad Sci USA. 2009;106:20258–20263. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shekar SC, Wu H, Fu Z, Yip SC, Cahill SM, Girvin ME, Backer JM. Mechanism of constitutive PI 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J. Biol. Chem. 2005 doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 45.Alexander N, Bortolus M, Al-Mestarihi A, McHaourab H, Meiler J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16:181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.