Summary
Generation of neurons in the vertebrate central nervous system requires complex transcriptional regulatory network and signaling processes in polarized neuroepithelial progenitor cells. Here we demonstrate that neurogenesis in the Xenopus neural plate in vivo and mammalian neural progenitors in vitro involves intrinsic antagonistic activities of the polarity proteins PAR-1 and aPKC. Furthermore, we show that Mind bomb (Mib), a ubiquitin ligase that promotes Notch ligand trafficking and activity, is a crucial molecular substrate for PAR-1. The phosphorylation of Mib by PAR-1 results in Mib degradation, repression of Notch signaling and stimulation of neuronal differentiation. These observations suggest a conserved mechanism for neuronal fate determination that might operate during asymmetric divisions of polarized neural progenitor cells.
Keywords: PAR-1, aPKC, Notch, Mind bomb, neurogenesis, progenitor, Xenopus
Introduction
Vertebrate neurogenesis relies on the complex transcriptional control of proneural gene expression leading to the establishment of neuronal progenitor domains during embryonic development (Bally-Cuif and Hammerschmidt, 2003; Kintner, 2002; Lee et al., 1995; Ma et al., 1996). Individual neurons are selected within these domains by the Notch pathway, a major molecular pathway associated with cell fate decisions in a wide spectrum of tissues, from Drosophila midgut and sensory organ precursors to mammalian muscle and blood progenitors, and associated with human disease (Androutsellis-Theotokis et al., 2006; Artavanis-Tsakonas et al., 1999; Lai, 2004; Le Borgne and Schweisguth, 2003; Louvi and Artavanis-Tsakonas, 2006; Micchelli and Perrimon, 2006; Mizutani et al., 2007; Ohlstein and Spradling, 2007). The interaction of Notch with its ligands results in the release of the Notch intracellular domain (ICD), which translocates into the nucleus and associates with transcriptional cofactors to activate downstream targets repressing differentiation in the signal-receiving cell (Bray, 2006; Nichols et al., 2007). In the signal-sending cell, the recycling and functional activity of Notch ligands monoubiquitinated by the E3 ligases Mind bomb (Mib) and Neuralized is a key regulatory step for signaling (Chitnis, 2006; Nichols et al., 2007; Roegiers and Jan, 2004). At present, molecular mechanisms influencing the segregation of signal-sending and signal-receiving cells are not fully understood, although available evidence points to the importance of progenitor cell polarization (Knoblich, 2008; Roegiers and Jan, 2004).
Cell polarity is another critical parameter influencing the outcome of neurogenesis. Progenitor cell polarization and asymmetric division underlie cell fate decisions in C. elegans blastomeres (Guo and Kemphues, 1996), Drosophila neuroblasts and sensory organ precursors (Betschinger and Knoblich, 2004; Roegiers and Jan, 2004). In Drosophila sensory organ precursors, polarized segregation of Neuralized and Numb appears responsible for Notch signaling asymmetry and subsequent cell fate determination (Knoblich, 2008; Le Borgne and Schweisguth, 2003; Roegiers and Jan, 2004). Although progenitor cell polarization has been also observed in vertebrate ectoderm and the developing central nervous system (Chalmers et al., 2003; Gotz and Huttner, 2005; Knoblich, 2008; Lechler and Fuchs, 2005; Ossipova et al., 2007), the significance of cell polarization for vertebrate neurogenesis and the molecular mechanisms involved remain to be clarified (Chenn and McConnell, 1995; Gotz and Huttner, 2005; Lake and Sokol, 2009; Noctor et al., 2004; Sanada and Tsai, 2005; Shen et al., 2006; Shen et al., 2002).
Atypical protein kinase C (aPKC) (Macara, 2004; Rolls et al., 2003; Wodarz and Huttner, 2003) and its molecular substrate PAR-1 (Benton and St Johnston, 2003; Drewes et al., 1997; Kemphues, 2000; Pellettieri and Seydoux, 2002; Tomancak et al., 2000) function antagonistically in cell polarity and play key roles in early development (Ossipova et al., 2007; Plusa et al., 2005). The phosphorylation of PAR-1 by aPKC leads to the segregation of aPKC and PAR-1 to opposite cellular poles and is critical for apical-basal cell polarity (Hurov et al., 2004; Suzuki et al., 2004). In this study we report that PAR-1 and aPKC act in opposite ways to regulate neurogenesis in both Xenopus embryos and mammalian neural progenitor cells. We next identify Mib as a critical phosphorylation target of PAR-1, linking the effect of PAR-1 on neurogenesis to the activity of the Notch ligand Dll1 in the signal-sending cell. This phosphorylation of Mib leads to the decrease in its levels, resulting in PAR-1-mediated stimulation of neurogenesis that is consistent with the neurogenic phenotype of Mib loss-of-function mutants in different models (Itoh et al., 2003; Koo et al., 2005; Lai et al., 2005). These observations suggest that PAR-1 promotes neuronal cell fate by inhibiting Notch signaling via Mib destabilization.
Results
PAR-1 and aPKC influence neurogenesis in Xenopus embryos
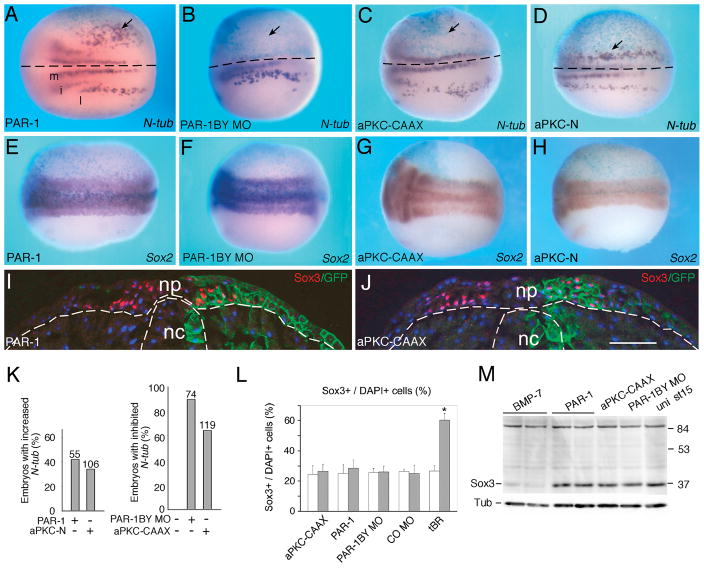
To study a function for apical-basal polarity proteins for neuronal fate determination in the vertebrate brain and spinal cord, we examined effects of the polarity kinase PAR-1 and its regulatory kinase aPKC (Goldstein and Macara, 2007; Hurov et al., 2004; Suzuki et al., 2004) on primary neurogenesis in Xenopus embryos (Fig. 1, Fig. S1). Overexpressed PAR-1A/MARK3 (later referred to as PAR-1) increased the number of N-tubulin-positive primary neurons in 42% of injected embryos (n=55) (Figs. 1A, K). The same effect was observed with PAR-1B/MARK2 (data not shown). Conversely, embryos injected with two previously described PAR-1 morpholino oligonucleotides (MO), which specifically deplete PAR-1B/MARK2 protein (Kusakabe and Nishida, 2004; Ossipova et al., 2005)(Fig. S1A), but not a control MO (COMO), revealed a significant reduction in primary neuron number in the majority of injected embryos (90.5%, n=74; Fig. 1B, K, and data not shown). Of note, PAR-1BY MO did not have any effects on Wnt/β-catenin signaling (Ossipova et al., 2005), indicating that the function of PAR-1 in neurogenesis is not mediated by the Wnt pathway. By contrast, aPKC-CAAX, an activated membrane-targeted form of aPKC (Ossipova et al., 2007), suppressed neurogenesis (63%, n=119), whereas aPKC-N, a dominant interfering form of aPKC, increased the number of primary neurons (38%, n=106; Fig. 1C, D). In support of these observations, in situ hybridization for a different neuronal gene, Hox11L2 (Patterson and Krieg, 1999), revealed enlarged clusters of primary sensory neurons in PAR-1 RNA-expressing embryos, while a kinase-dead form of PAR-1 had an inhibitory effect (Fig. S1B, C, F), consistent with its dominant negative activity (Sun et al., 2001). Other neuronal markers, including Dll1 (Delta-like 1 or Delta1)(Chitnis et al., 1995) and Xaml-1 (Tracey et al., 1998), were also sensitive to the modulation of PAR-1 and aPKC functions (Fig. S1D-F and data not shown). These results were confirmed in embryo sections (Fig. S1G–L). Our observations demonstrate that PAR-1 and aPKC modulate neurogenesis in the opposite manner, consistent with the negative regulation of PAR-1 by aPKC during the establishment of apical-basal polarity.
Figure 1. PAR-1 and aPKC regulate neurogenesis in Xenopus embryos and mammalian neural progenitors.
Four-cell embryos were unilaterally injected with LacZ RNA (50–100 pg), indicated RNAs or MOs and subjected to wholemount in situ hybridization with N-tubulin (A–D) or Sox2 (E–H) anti-sense probes at neurula stages. The injected area is identified by β-galactosidase activity (light blue). Dorsal views are shown, anterior is to the left, the injected side is at the top. Arrows point to altered gene expression at the injected side. (A) Three stripes of primary neurons (medial, m, intermediate, i, lateral, l) are indicated. (A, B) PAR-1 RNA (300 pg, doses are per embryo) increased (A), whereas PAR-1BY MO (5 to 10 ng) decreased (B) N-tubulin positive cells on the injected side. (C, D) aPKC-CAAX RNA (30 to 100 pg) inhibited (C), and aPKC-N RNA (4 ng) expanded (D) N-tubulin-positive cells. (A–D) Dotted lines indicate midline. (E–H) Modulation of PAR-1 and aPKC does not influence the pan-neural marker Sox2 (doses are as in A–D). (I, J) Phenotypically active doses of PAR-1 (I, 300 pg) and aPKC-CAAX (J, 100 pg) RNAs do not alter the number of Sox3-positive progenitor cells. GFP-CAAX RNA was used as a lineage tracer. Stage 15 embryos were cryosectioned and immunostained with anti-Sox3C antibodies (red) and GFP-antibodies (green). (I, J) Dotted lines demarcate neural plate (np) and notochord (nc). Scale bar, 100 μm. (K) Summary of data combined for several independent experiments presented in A–D. Frequency of embryos with a change in N-tubulin-positive cells is shown for each experimental group. Numbers of embryos per group are indicated above bars. (L) Sox3+ progenitor analysis in cryosections. Summary of data for embryos injected with aPKC-CAAX (100 pg), PAR-1 (300 pg), tBR (1 ng) RNAs and PAR-1BY MO, COMO (5 to 10 ng). tBR RNA strongly upregulated the number of Sox3-positive progenitors. Mean ratios of Sox3+/DAPI+ cells +/− s. d. are shown. At least 8 embryos were used in each experimental group. *, p<0.01, indicates significant difference from uninjected cells. M, Lack of effect of PAR-1 and aPKC manipulation on Sox3 protein levels in neurula stage (st 15) embryos (doses are as in A–D). BMP7 RNA (500 pg) dramatically reduced Sox3 protein levels.
To evaluate the number of neural progenitors and the size of the neural plate in the manipulated embryos, we studied the expression of pan-neural markers, such as Sox2 and Sox3 (Mizuseki et al., 1998; Pevny and Placzek, 2005), using in situ hybridization and immunohistochemistry. No significant effects on the Sox2 expression domain (Fig. 1E–H) and the number of Sox3-positive progenitors (Fig. 1I, J, L and data not shown) were detected in stage 15/16 embryos, injected with the same phenotypically active doses of PAR-1, aPKC-CAAX, PKC-N RNAs (n>30 for each group) and PAR-1BY MO or COMO (n > 80 for each group). Moreover, western analysis did not reveal any alteration in Sox3 protein levels (Fig. 1M). By contrast, manipulating BMP signaling by injection of BMP7 (Fig. 1M) or a truncated BMP receptor (tBR, Fig. 1L) readily affected Sox3 protein levels and progenitor number. Furthermore, we did not observe significant changes in mitotic cell number, when measured by phospho-histone 3 staining in embryo sections (Fig. S2A–F). Nevertheless, the same PAR-1 RNA coexpressed with the neuralizing factor Chordin activated the N-tubulin gene in ectodermal explants (Fig. S1M), further supporting our data obtained by in situ hybridization. To assess the location of ectopic neurons more precisely, we performed double in situ hybridization for N-tubulin positive cells and the pan-neural marker Sox2 or the epidermal keratin XK70. These experiments show that overexpressed PAR-1 expands the existing N-tubulin stripes within the neurogenic territory, which extends into the XK70-positive area, adjacent to the Sox2-positive domain (Fig. S1N, O). No ectopic neuronal differentiation was detected in response to PAR-1 in ventral ectoderm (data not shown), indicating that PAR-1 activity is only manifested within the normal neurogenic territory, and, therefore, differs from the neuralizing influence of Chordin and other BMP antagonists. Although we cannot rule out a possibility that PAR-1 and aPKC influence progenitor proliferation at early stages, our results suggest that PAR-1 influences neuronal differentiation rather than progenitor pool expansion.
PAR-1 and aPKC control mammalian neural progenitor differentiation
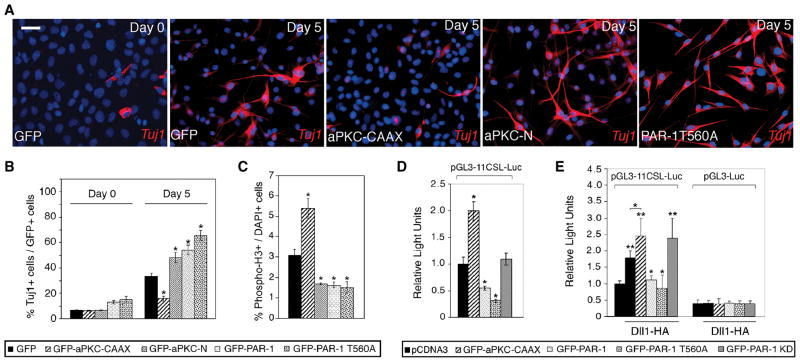
The observed changes in cell fate may result from intrinsic activities of these polarity proteins or be an indirect consequence of disrupted cytoskeletal organization and lack of tissue integrity (Ghosh et al., 2008; Kim and Walsh, 2007). To address this question, we used mouse cerebellar multipotent progenitor C17.2 cells, which do not exhibit epithelial morphology and are capable of neuronal differentiation (Snyder et al., 1992). These cells were infected with lentiviruses expressing GFP-tagged aPKC and PAR-1 proteins (Fig. 2A, B, S3A, B) and allowed to differentiate at low serum conditions. After five days of differentiation, frequency of neuron-specific tubulin TuJ1 staining in the control GFP-virus-infected (or uninfected) cells increased to 31.9 %, while the population of cells expressing the progenitor marker Nestin was reduced to 9.2 % (Fig. 2A, B and data not shown). Many cells expressing aPKC-CAAX retained Nestin expression (31.5 %) and did not differentiate into neurons (Fig. S3C, D). By contrast, overexpression of aPKC-N, wild-type PAR-1 or PAR-1T560A with a mutation in the aPKC phosphorylation site (Ossipova et al., 2007) caused extensive morphological differentiation, revealed by the formation of extended bipolar processes, and high frequency of TuJ1 staining (47.3 %, 52 % and 63.3 % of cells, respectively, Fig. 2A, B and data not shown). Together, these results demonstrate that PAR-1 enhances, whereas aPKC suppresses neuronal differentiation of C17.2 cells, and extend the data obtained in Xenopus embryos to the mammalian system. Of note, in contrast to what we observed in Xenopus embryos, PAR-1 and PKC-N decreased, whereas aPKC-CAAX increased C17.2 cell proliferation, when measured by phospho-histone 3 staining (Fig. 2C, S3E) and assessing cell growth in vitro (Fig. S3F). These findings indicate that the observed increase in TuJ1-positive neurons stimulated by PAR-1 is due to enhanced cell differentiation in the absence of cell proliferation, rather than the expansion of the progenitor pool. Our experiments also suggest that the observed effects of PAR-1 and aPKC on neurogenesis do not require epithelial tissue organization and likely represent intrinsic signaling properties of PAR-1 and aPKC.
Figure 2. PAR-1 and aPKC regulate neurogenesis in mammalian neural progenitors.
C17.2 cells infected with lentiviruses expressing GFP or different aPKC and PAR-1 constructs were allowed to differentiate for five days. (A) Representative images of cells stained for TuJ1 (red) and DAPI (blue) before and after differentiation. Neuronal differentiation is revealed by bipolar morphology of cells, containing long processes, and TuJ1 staining. PAR-1, PAR-1T560A and aPKC-N stimulated, whereas aPKC-CAAX suppressed neuronal differentiation. Scale bar, 20 μm. (B) Summary of experiments shown in (A). *, p<0.01, indicates significant differences from the control GFP group. (C) Phospho-histone H3 positive cells in C17.2 cultures expressing indicated lentiviral constructs. PAR-1, PAR-1T560A and aPKC-N decreased, whereas aPKC-CAAX increased the number of mitotic cells. Data are from three independent experiments expressed as ratio of PH3+/DAPI+ cells. *, p<0.01 indicates significant differences from the control GFP group. (D) aPKC and PAR-1 regulate Notch reporter activity in neural progenitor cells. C17.2 cells were transfected with aPKC and PAR-1 constructs as indicated, and the pGL3-11CSL-Luc reporter and harvested 24 hrs later for luciferase activity measurement. Relative luciferase activity was normalized to Renilla enzyme activity. (B–E) Data are presented as the means ± s. d. of three independent experiments. *, p<0.05. (E) PAR-1 inhibits Dll1 activity in signal-sending cells. HEK293T cells were transfected with Dll1 and aPKC or PAR-1 constructs, then cocultured for 24 hrs with NIH3T3 cells expressing Notch and pGL3-11CSL-Luc reporter. pGL3-Luc is a control reporter. Each group contained triplicate samples. **, p<0.01, or *, p<0.05, indicate significant differences from the control- or the Dll1-transfected group, respectively.
Regulation of Notch signaling in signal sending cells
As the Notch pathway is a major regulator of neurogenesis, we hypothesized that the observed effects of PAR-1 and aPKC may be mediated by Notch signaling and measured Notch transcriptional activity in C17.2 cells cotransfected with various aPKC and PAR-1 constructs, using a luciferase reporter containing multimerized CSL-binding sites. We observed that the reporter was activated by aPKC-CAAX and inhibited by wild-type PAR-1 and PAR-1T560A, whereas PAR-1KD did not have a significant effect (Fig. 2D). Thus, our results link the antagonistic effects of PAR-1 and aPKC on neurogenesis to Notch pathway regulation, consistent with other studies implicating polarity proteins in Notch signaling (Bayraktar et al., 2006; Smith et al., 2007).
Since PAR-1 modulated Dll1, but not Notch ICD, signaling in Xenopus ectoderm (Ossipova et al., 2007), we hypothesized that PAR-1 regulates Dll1, rather than Notch, function. We therefore asked whether PAR-1 is active in signal-sending cells expressing Dll1, which are cocultured with the population of signal-receiving cells expressing a Notch reporter (Itoh et al., 2003; Lindsell et al., 1995)(Fig. 2E). Both PAR-1 and PAR-1T560A repressed Dll1-dependent reporter activity, while aPKC-CAAX and PAR-1KD had a modest enhancing effect, consistent with the view that PAR-1 inhibits Dll1 activity in signal-sending cells.
PAR-1 phosphorylates Mib and regulates its expression levels in vitro and in vivo
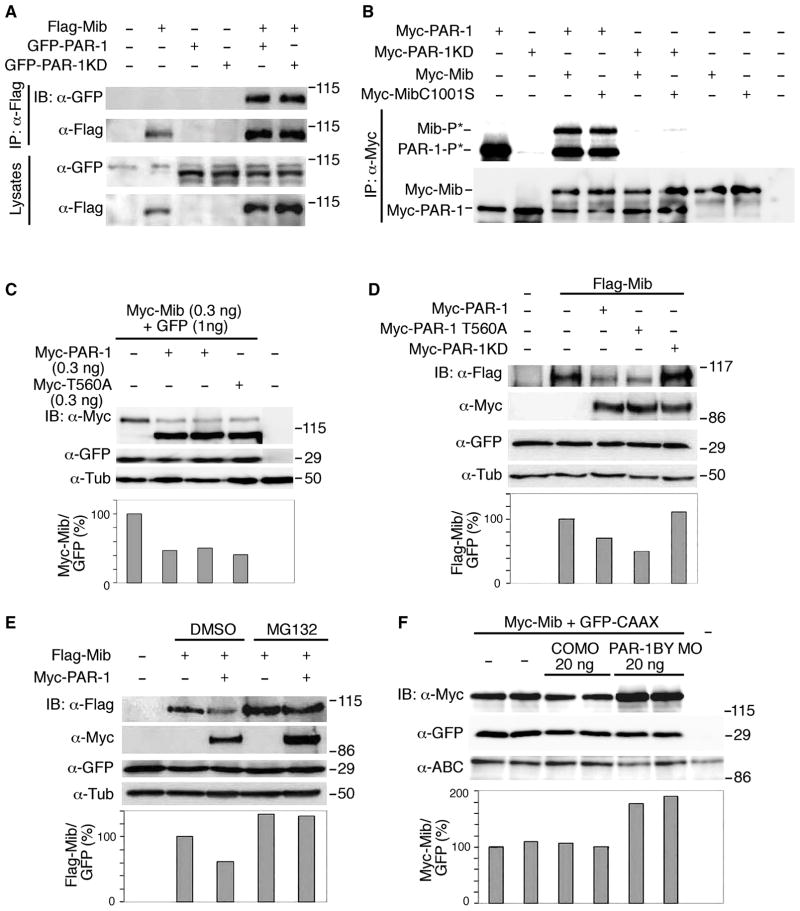
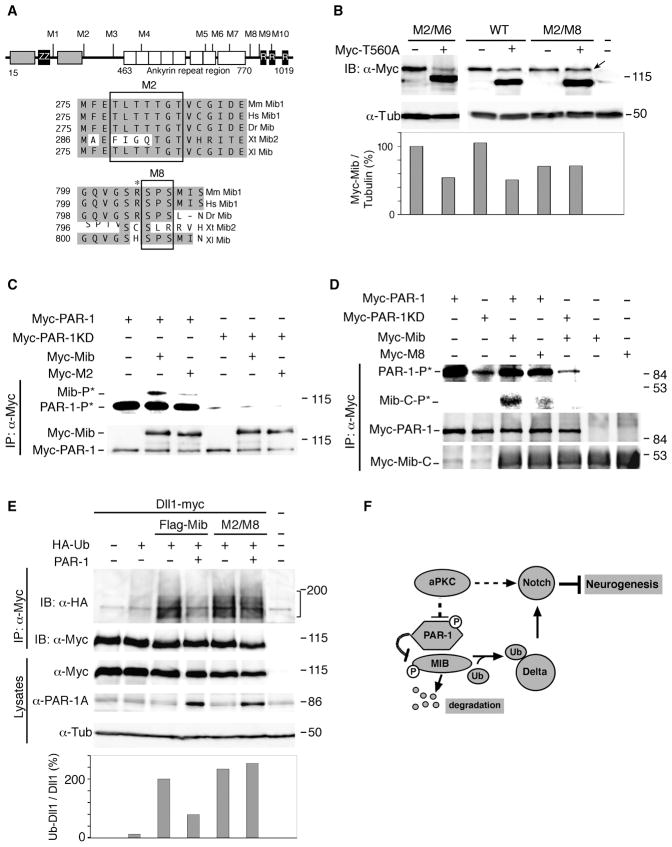
We next wanted to test the hypothesis that PAR-1 regulates Dll1 by phosphorylating E3-ubiquitin ligases, such as Neuralized or Mib. Supporting this possibility, a mass-spectrometry-based approach identified a mammalian PAR-1 homolog among proteins associated with Mib (Choe et al., 2007). The physical interaction of PAR-1 and Mib was confirmed by protein co-precipitation from cell lysates (Fig. 3A). Similar results were obtained using PAR-1 and Mib constructs with different epitope tags (data not shown). An immune complex kinase assay revealed that PAR-1, but not PAR-1KD, phosphorylated Mib and MibC1001S, an E3 ubiquitin ligase mutant (Itoh et al., 2003)(Fig. 3B), identifying Mib as an in vitro substrate for PAR-1.
Figure 3. PAR-1 phosphorylates Mib and regulates its protein levels.
(A) PAR-1 interacts with Mib. Levels of Flag-Mib, GFP-PAR-1 and GFP-PAR-1KD were assessed in HEK293T cell lysates 18 hrs after transfection with indicated plasmids, followed by immunoprecipitation (IP) with anti-Flag beads and immunoblotting (IB) analysis. (B) Phosphorylation of Mib and MibC1001S by PAR-1 in the immune complex kinase assay. Autoradiography (top) demonstrates the autophosphorylation of PAR-1 and reveals its kinase activity towards Mib and Mib C1001S. No significant kinase activity is detectable in lysates containing PAR-1KD. Anti-Myc antibodies (bottom) detect similar protein levels in immunoprecipitates. (C) PAR-1 downregulates Mib in Xenopus embryos. Coexpressed GFP is not decreased by PAR-1. Western analysis of lysates of embryos (stage 10.5) that were injected with Myc-Mib (top band) and Myc-PAR-1 and GFP RNAs. Lower panel, a ratio of Myc-Mib/GFP levels; α-tubulin controls loading.
(D) Flag-Mib levels are downregulated by myc-PAR-1 in HEK293T cells. Western analysis of cell lysates prepared 48 hrs after cotransfection of cells with indicated plasmids and pEGFP-C3. Lower panel, a ratio of Flag-Mib/GFP levels. (E) PAR-1 stimulates Mib degradation by the proteasome pathway. Transfected HEK293T cells were treated with 10 μM MG132 or DMSO (control) for 6 hrs. MG132 stabilizes Mib and rescues PAR-1 effects. Lower panel, a ratio of Flag-Mib/GFP levels. (F) Endogenous PAR-1 downregulates Mib in vivo. Embryos were injected at the two-cell stage with indicated MOs, Myc-Mib (0.3 ng) and GFP-CAAX (0.45 ng) RNAs. Western analysis of stage 10.5 embryo lysates with indicated antibodies. PAR-1 MO, but not control MO (CO MO) upregulates Mib at gastrula stage, but has no effect on GFP-CAAX or β-catenin levels. Lower panel, a ratio of Myc-Mib/GFP levels.
We next examined the effect of PAR-1 on Mib in vivo. PAR-1 or PAR-1T560A downregulated Mib levels in lysates of embryos and cultured cells, whereas PAR-1KD had no effect (Fig. 3C, D, Fig. S4A, B). PAR-1 did not have a significant effect on endogenous α-tubulin or exogenous coexpressed GFP (Fig. 3C–E). Levels of the E3 ligase-deficient Mib mutant remained unaffected (Fig. S4B), indicating that the E3 ligase activity of Mib is necessary for the PAR-1 effect. Moreover, PAR-1 did not decreased Mib protein levels in cultured cells and Xenopus embryos treated with the proteasome inhibitor MG132, suggesting that PAR-1 promotes Mib degradation via the proteasome pathway (Fig. 3E and data not shown).
To assess whether endogenous PAR-1 functions to downregulate Mib, we examined Mib levels in Xenopus embryos depleted of PAR-1. Lysates of embryos injected with a PAR-1BY MO revealed a specific increase in Mib protein levels (Figs. 3F and S4C). A similar result was obtained with a second PAR-1BY MO with a different nucleotide sequence, but not with a control MO (data not shown). Together, these observations support a model, in which PAR-1 phosphorylates Mib to stimulate its proteasome-dependent degradation.
PAR-1 inhibits Dll1 ubiquitination and signaling activity, but does not influence a Mib-independent form of Dll1
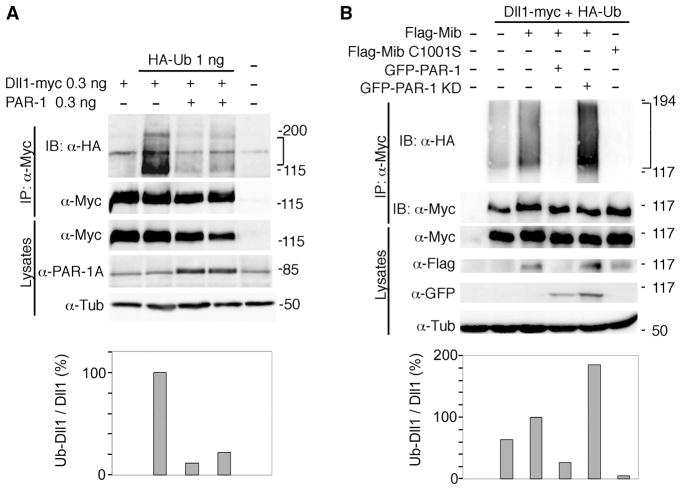
Since Mib is a known major regulator of Dll1 ubiquitination and signaling activity, our results predict that PAR-1 should influence Dll1 ubiquitination. Consistent with this prediction, ubiquitination of Dll1 in both Xenopus embryos and mammalian cells was inhibited by PAR-1, but not PAR-1KD (Fig. 4A, B). These results support the model, in which PAR-1 inhibits Dll1 signaling by antagonizing Mib function.
Figure 4. Regulation of Dll1 ubiquitination by PAR-1.
(A) Dll1 ubiquitination is inhibited by PAR-1 in Xenopus ectoderm. Embryos were injected with RNAs at indicated doses per embryo. At stages 10–11, embryo lysates were collected for immunoprecipitation (IP) of Dll1-myc with anti-Myc antibodies and immunoblotted with anti-HA or anti-Myc to visualize ubiquitinated (brackets) or total Dll1, respectively. (B) Mib-dependent Dll1 ubiquitination is inhibited by PAR-1 in HEK-293T cells. Cells were transfected with Dll1-myc, Flag-Mib or Flag-Mib C1001S, HA-ubiquitin and PAR-1 DNAs as indicated. Ubiquitinated Dll1 (brackets) was precipitated from lysates of cells 48 hr after transfection with anti-Myc antibodies and detected with anti-HA antibodies. Lower panels: ratio of ubiquitinated (HA-tagged) Dll1 to total (myc-tagged) Dll1 in immunoprecipitates.
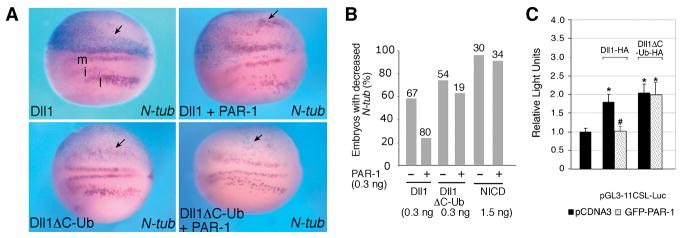
As Dll1 signaling activity depends on the ubiquitination of its intracellular domain by Mib, we fused Dll1 to the ubiquitin moiety to generate a Mib-independent construct (Dll1ΔC-Ub)(Itoh et al., 2003). We next examined PAR-1 effects on the ability of Dll1 and Dll1ΔC-Ub to promote Notch signaling and inhibit neuronal differentiation. PAR-1 rescued neurogenesis downregulated by Dll1, but not Notch ICD (Fig. 5A, B, S5), supporting the idea that PAR-1 functions at the level of Dll1, upstream of Notch receptor activation. By contrast, embryos expressing Dll1ΔC-Ub showed reduced number of neurons even in the presence of PAR-1 RNA (Fig. 5A, B). Our observations were further supported in the Dll1-dependent co-culture assay by measuring Notch reporter activity in mammalian cells. Whereas both Dll1 and Dll1ΔC-Ub activated the reporter to comparable levels, PAR-1 suppressed Dll1, but not Dll1ΔC-Ub responses (Fig. 5C). These experiments demonstrate that PAR-1 inhibits Dll1 activity both in Xenopus ectoderm and mammalian cells in a Mib-dependent manner.
Figure 5. PAR-1 does not affect signaling of a Mib-independent form of Dll1.
(A, B) Embryos were injected with indicated RNAs and processed for in situ hybridization with N-tubulin probe as described in Fig. 1. RNAs were injected at 0.3 ng. Dorsal views are shown, with anterior to the left, the injected side is up (light blue). Arrows point to changes in N-tubulin expression. (A) PAR-1 suppressed the inhibitory effect of untagged Dll1 on neurogenesis, but had no effect on the activity of Dll1ΔC-Ub.
(B) Quantification of PAR-1 effects on Dll1 and Notch signaling. Numbers of embryos per group are indicated above bars. Data are pooled from several independent experiments. RNA doses are indicated per embryo.
(C) PAR-1 does not affect Notch reporter activation by Dll1ΔC-Ub. Signal-sending cells were transfected with PAR-1, Dll1 or Dll1ΔC-Ub constructs, and cocultured with signal-receiving cells that were transfected with Notch and pGL3-11CSL-Luc, as described in Fig. 2D. *, p<0.01, and #, p<0.01, indicate significant differences from control- or the Dll1-transfected group, respectively. Results are presented as the means ± s. d. of three independent experiments, each carried out with triplicate samples per group.
Specific phosphorylation sites in Mib are required for PAR-1-dependent changes in Dll1 ubiquitination
To assess the functional significance of Mib phosphorylation by PAR-1, we wanted to identify sites in Mib that are phosphorylated by PAR-1. We generated a series of mutated Mib proteins with substitutions in the putative serine/threonine phosphorylation motifs (Fig. 6A), and assessed their levels in the absence or presence of PAR-1 in Xenopus embryo lysates. Whereas the majority of Mib mutant proteins remained sensitive to PAR-1-mediated degradation, the levels of the M2/M8 Mib construct, carrying mutations in the M2 and M8 sites, were not altered by exogenous PAR-1 (Fig. 6B), indicating that these sites are critical for Mib regulation. In vitro kinase assays using wild-type and mutated Mib constructs confirmed that these sites are subject to phosphorylation by PAR-1 (Fig. 6C, D). The phosphorylation analysis of S804/S806 residues was carried out in the Mib construct with deleted amino-terminal phosphorylation sites. Similarly to other PAR-1 sites identified in Dsh and PAR-3 (Sun et al., 2001, Benton and St. Johnston, 2003), the S804 has an immediately adjacent upstream basic residue (Fig. 6A). Importantly, these phosphorylation sites were conserved among vertebrate Mib1, but not Mib2 homologs (Fig. 6A), suggesting functional significance.
Figure 6. PAR-1 influences Dll1 ubiquitination by phosphorylating Mib.
(A) Putative PAR-1 phosphorylation sites in the Mib protein. The HERC2 domain (grey box), zinc finger regions, ZZ-type (ZZ) and ring-type (R), and the ankyrin repeat regions are shown. Alignment of the M2 and M8 Mib region that are critical for PAR-1-dependent degradation. Asterisk marks positively charged residue upstream of S804/S806. Sequence accession numbers: Mm Mib1, NM_144860; Hs Mib1, NP_065825; Dr Mib, AF537301; Xt Mib1, BC167461, Xt Mib2, NP_001123444; Xl Mib, NP_001085805. (B) Mib M2/M8 is insensitive to downregulation by PAR-1. Embryos were injected with indicated RNAs at the two-cell stage for western analysis at stage 10.5. Mib (top band) and PAR-1T560A (bottom band) are detected with anti-Myc antibodies. α-tubulin is a control for loading. (C, D) Decreased phosphorylation of Mib proteins with mutated M2 (C) and M8 (D) sites as compared to wild-type Mib. Arrow points to lack of decrease in M2/M8 levels in presence of PAR-1. (C) M2 mutations are in the context of the full-length protein. (D) Phosphorylation of Mib-M8 mutant is analyzed in the context of a carboxy-terminal fragment of Mib (amino acids 691–1031) which migrates faster than PAR-1. Carboxy-terminal fragments of the wild-type Mib or M8 mutants are compared. The details of the immune complex kinase assay are as in Fig. 3B legend. (E) M8* Mib promotes Dll1 ubiquitination in a PAR-1-independent manner. Embryos were injected with Dll1-myc (0.3 ng), HA-Ub (1 ng), PAR-1 (0.4 ng), Flag-Mib or Flag-M8* Mib (0.3 ng) RNAs as indicated. Dll1 ubiquitination was analyzed as described in Fig. 4A. Compared to Fig. 4A, image was acquired at a different exposure time, since Delta is strongly ubiquitinated by Flag-Mib. Lower panel shows ratios of ubiquitinated (HA-tagged) Dll1 to total (myc-tagged) Dll1 in immunoprecipitates. (F) A model for regulation of neurogenesis by PAR-1 and aPKC. aPKC promotes Notch signaling and negatively regulates PAR-1 activity in neural progenitors. PAR-1 phosphorylates Mib leading to its degradation, suppressed Dll1 monoubiquitination, inhibited Notch signaling and promoted neurogenesis.
We next analyzed whether PAR-1 can inhibit the activity of the M2/M8 mutant in the Dll1 ubiquitination assay. PAR-1 inhibited Dll1 ubiquitination by the wild-type Mib, but not by the M2/M8 Mib (Fig. 6E). This finding suggests that the inhibitory effect of PAR-1 on Delta ubiquitination involves Mib phosphorylation.
Discussion
An important question of developmental biology is how cell and tissue polarity influences cell fate specification during embryogenesis. Our experiments reveal that PAR-1 promotes, whereas aPKC suppresses neuronal production in Xenopus neural plate and mammalian neural progenitor cultures. Although aPKC is known to phosphorylate the Notch inhibitor Numb, which acts as cell fate determinant in Drosophila (Le Borgne et al., 2005), the functional relevance of this process for Notch signaling is not fully clear (Nishimura and Kaibuchi, 2007; Smith et al., 2007). In this work, we demonstrate that Mib, an E3 ubiquitin ligase required for Dll1 function, is phosphorylated by PAR-1 and subsequently degraded. This leads to inhibition of Dll1-Notch signaling and stimulation of neuronal differentiation. These results functionally connect polarity proteins to Notch-mediated cell fate decisions and provide a mechanism for the PAR-1 function in neurogenesis.
Although PAR-1 was implicated in Notch signaling in Drosophila and Xenopus (Bayraktar et al., 2006; Ossipova et al., 2007), no biochemical mechanism underlying these effects has been demonstrated. Our study identifies Mib as a critical molecular substrate of PAR-1. In response to PAR-1-mediated phosphorylation, Mib appears to undergo proteasome-mediated self-degradation, which requires Mib enzymatic activity and may be due to polyubiquitination. Once Mib is degraded, it can no longer monoubiquitinate Dll1, resulting in inhibited Notch signaling. Despite the identification of a number of potential substrates of PAR-1, including Tau, MAP2, MAP4, Dishevelled, PAR-3 and Oskar (Benton and St Johnston, 2003; Drewes et al., 1997; Drewes et al., 1995; Riechmann et al., 2002; Sun et al., 2001), Mib appears to be relevant to the function of PAR-1 in neurogenesis. First, Mib levels are increased in embryos depleted of PAR-1, indicating post-transcriptional regulation of Mib by endogenous PAR-1. Second, whereas PAR-1 inhibited Dll1 ubiquitination in a Mib-dependent manner, no effect on Dll1 was observed in the presence of the M2/M8 Mib mutant with substitutions in putative PAR-1 phosphorylation sites. Third, PAR-1 readily suppressed Dll1 activity, but it did not affect the Dll1-ubiquitin fusion construct, which promotes Notch signaling in a Mib-independent manner. Taken together, these findings suggest that PAR-1 inhibits Delta ubiquitination and signaling activity by phosphorylating Mib and thereby causing its degradation.
Despite significant morphological differences in neurulation between amphibians and other vertebrates, the molecular mechanism that connects PAR-1 and aPKC to neurogenesis appears be conserved in Xenopus and mammalian cells. We note that aPKC and PAR-1 regulated neurogenesis in C17.2 progenitor cells that lack epithelial morphology. Xenopus neural plate consists of the superficial epithelial layer and the deep cell layer, lacking clear epithelial structure. Primary neurons are known to originate mainly if not exclusively from the deep cell layer (Chalmers et al., 2002; Hartenstein, 1989). Whereas we show that PAR-1 inhibits Notch signaling during neurogenesis, it is currently unclear in which cell layer this inhibition takes place. We have observed that PAR-1 expands all neurons within the neurogenic domain (data not shown), but does not appear to induce neurons in non-neural ectoderm when expressed in ventral blastomeres (Fig. 1, S1 and data not shown). Given the dynamic pattern of Sox2 expression, the localization of some PAR-1 induced neurons outside the Sox2-expressing domain does not contradict this conclusion. Previous work demonstrated a similar localization of sensory neurons outside of the Sox2-positive area (Hardcastle and Papolopulu, 2000), which may be due to downregulation of Sox2 in the lateral neural plate or migration of sensory neurons away from the neural plate. Separation of individual cell layers and lineage analysis of individual neuronal progenitors would be required to definitively establish the origin of ectopic neurons forming in response to PAR-1.
Besides stimulating neuronal differentiation, PAR-1 may also affect tissue integrity and promote migration of cells within the ectoderm. PAR-1 is known to be required for different morphogenetic processes, including convergent extension in Xenopus (Kusakabe and Nishida, 2004; Ossipova et al., 2005) and border cell migration in Drosophila (McDonald et al., 2008). While the relevance of these effects to neurogenesis remains to be established, we note that the loss of Strabismus/Vangl2, another protein involved in epithelial polarity and morphogenesis, has been shown to influence neuronal differentiation in mouse embryos (Lake and Sokol, 2009). Future studies are warranted to determine how the epithelial structure of each ectodermal layer is affected by aPKC and PAR-1 and which layer of ectoderm is critical for the observed changes in neurogenesis.
Our results are consistent with a model, in which the apical PAR protein complex acts to maintain the progenitor cell state (Chia et al., 2008; Costa et al., 2008; Lee et al., 2006; Rolls et al., 2003), whereas the basolaterally localized PAR-1 stimulates progenitor differentiation by interfering with Notch signaling (Fig. 6F). In agreement with this idea, inactivation of Cdc42, a protein required for apical aPKC complex activity, leads to increased neuron production in the mouse cortex (Cappello et al., 2006). Further supporting this hypothesis, the Notch pathway has been shown to maintain the progenitor state in multiple cell lineages including neural progenitors (Androutsellis-Theotokis et al., 2006; Fre et al., 2005; Louvi and Artavanis-Tsakonas, 2006; Mizutani et al., 2007). Since we did not see a change in the number of proliferating progenitors in Xenopus embryos, PAR-1 appears to function by promoting progenitor differentiation. Nevertheless, we cannot exclude the possibility that it influences progenitor proliferation at earlier developmental stages, since many primary neurons are already born by the early neurula stage (Hartenstein, 1989; Lamborghini, 1980). Also, the observed increase in the number of primary neurons may be insufficient to detect a change in the total number of Sox2 and Sox3 positive progenitors. Alternatively, PAR-1 may affect neurogenesis by altering the balance between neuronal and glial progenitors, after cells become committed to the neural fate.
The apical-basal polarity and the orientation of the mitotic spindle have been linked to daughter cell fate determination during division of Drosophila neuroblasts, sensory organ precursors and mammalian neuroepithelial cells (Gotz and Huttner, 2005; Kaltschmidt et al., 2000). Asymmetric cell division of neuroepithelial progenitors has been also described in Xenopus and zebrafish embryos (Chalmers et al. 2003; Geldmacher-Voss et al., 2003; Tawk et al., 2007). Thus, besides an effect on Notch-mediated process of lateral inhibition, aPKC and PAR-1 may function to unequally distribute Notch pathway components such as Mib or Neuralized in neuroepithelial progenitors. Whereas the subcellular localization of Mib in dividing neuroepithelial progenitors is currently unknown, its unequal segregation may result in asymmetric Notch signaling, as has been demonstrated for Numb and Neuralized in Drosophila sensory organ precursors (Knoblich, 2008; Le Borgne and Schweisguth, 2003; Roegiers and Jan, 2004). Further analysis of endogenous protein distribution is needed to establish the relevance of the observed mechanism to cell fate determination in polarized stem/progenitor cells undergoing asymmetric division.
Experimental procedures
Embryo culture and microinjections, in situ hybridization and lineage tracing
Fertilization and embryo culture have been performed as described (Itoh et al., 2005). DNA constructs, RNA synthesis, morpholino oligonucleotides, RT-PCR and mutagenesis are described in detail in Supplementary on-line materials. Embryos were microinjected in 1/3 x MMR, 2 % Ficoll-400 (Pharmacia) in the animal pole with 10 nl of a solution containing 0.1 – 2 ng of RNA per blastomere at the two to four-cell stage, and cultured in 0.1 x MMR until desired stages. In loss-of-function experiments, PAR-1BY MO and PAR-1BX MO (Ossipova et al., 2005) or control MO were injected at 5 –10 ng per blastomere. Of note, PAR-1BY MOs did not have an effect on Wnt/β-catenin signaling.
In situ hybridization and XGal staining were carried out using standard techniques (Harland, 1991) with the following anti-sense probes: N-tubulin (Oschwald et al., 1991), Sox2 (Mizuseki et al., 1998), XDll1 (Chitnis et al., 1995), Hox11L2 (Patterson and Krieg, 1999), Xaml (Tracey et al., 1998), epidermal type I keratin XK70 (Winkles et al., 1985). For double in situ hybridization, probes were synthesized using digoxygenin- or fluorescein RNA Labeling Mix (Roche) and detected with corresponding antibodies (Roche). Magenta phosphate, Fast Red and NBT/BCIP (Roche) were used for chromogenic reactions. For 10–12 μm sections, embryos were embedded in coldwater fish gelatin/sucrose mixture as described (Fagotto and Gumbiner, 1994) and cryosectioned using the Leica cryostat CM3050. Images were digitally acquired on a Zeiss Axiophot microscope. Quantification of results is presented as a percent of embryos with a conspicuous phenotypic change. RNA injected embryos usually revealed a range of phenotype severity.
Immunoprecipitation, western analysis and kinase assay
Immunoprecipitation (IP) and Western blotting were carried out with cell and embryo lysates essentially as described (Gloy et al., 2002; Itoh et al., 1998). Ubiquitination assay in cultured cells was as described (Itoh et al., 2003) with the following modifications. HEK-293T cells were transiently transfected with 0.4 μg total of expression plasmid DNA per well in 6-well plates using Effectene (Qiagen). Cells were collected 48 hr after transfection, lysed in 300 μl of lysis buffer (Gloy et al., 2002) containing protease inhibitor cocktail (Roche Diagnostics) and analyzed by immunoblotting or IP/immunoblotting. For IP, the 200 μl lysates were cleared by centrifugation at 10,000 g at 4°C for 5 min to remove cellular debris and incubated with 20 μl of anti-myc antiboby (9E10) overnight at 4°C. The immunoprecipitates were incubated with protein A-agarose beads (12.5 μl of 50 % suspension) for 1 hr at 4°C, washed 3 times with ice-cold lysis buffer and resuspended in a sample buffer. One-fourth of the material was analyzed by 8% SDS PAGE. The equivalent of 0.5 embryo per lane was loaded for analysis of embryo lysates, and the equivalent of 10–15 embryos per lane was loaded for analysis of immunoprecipitated proteins. Peroxidase activity was detected by enhanced chemiluminescence as described (Itoh et al., 1998). Protein detection was performed using Fujifilm Image Reader LAS-3000 System and software (FujiFilm). Quantification of band intensities by densitometry was carried out using NIH Image J software, using a representative of at least three independent experiments.
The following antibodies were used: anti-HA and anti-myc monoclonal antibodies are hybridoma supernatants of 9E10 and 12CA5 cells (Roche), anti-myc monoclonal antibodies 9E10 (Roche), anti-β-catenin 8E7 (Millipore, USA), anti-tubulin (BioGenex), anti-GFP monoclonal antibodies (Santa Cruz Biotechnology), anti-GFP JL8 monoclonal antibodies (Clontech), anti-Flag M2 (Sigma), anti-tubulin B512 (Sigma), anti-PAR-1 (Ossipova et al., 2005). Immune complex kinase assays were as described in (Ossipova et al., 2005) using in vitro synthesized myc-tagged proteins in TNT system (Promega) and Molecular Dynamics PhosphorImager. The following DNA templates were used: pCS2-Myc-PAR-1, pCS2-Myc-PAR-1 kinase dead, pCS3-Myc-Mib, pCS3-Myc-MibC1001S, pCS3-Myc-M2, pCS3-Myc-M8 (691–1031).
Cell culture, MG132 treatment, and Notch reporter assay
HEK-293T and NIH3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gemini-Bioscience) and penicillin/streptomycin (Sigma). C17.2 progenitor cells were cultured in DMEM containing 10 % FBS, 5 % horse serum (Snyder et al., 1992; Zhang et al., 2006). For differentiation assays, C17.2 cells were seeded on coverslips in 12-well plates (1×105 per well) and shifted the next day to DMEM containing 2 % FBS. For MG132 treatment, the inhibitor was added at 10 μm to HEK293T cell culture medium for 6 hrs before harvesting the cells. Staining with anti-Nestin (R401, DSHB) or anti-TuJ1 (Covance) mouse monoclonal antibodies was carried out in the beginning (day 0) and at the end (day 5) of differentiation.
For Notch reporter assays, 5×104 C17.2 cells were plated in 12-well plates for transfection the next day using linear polyethylenimine (M.W. 25,000, Polysciences). For transfection, each well received 0.25 μg of pGL3-11CSL-Luc (a gift of J. Kitajewski), 0.02 μg of pTK-Renilla-Luc (Promega) and 0.5 μg of pCS2-Flag-aPKC-CAAX, pCDNA3.1-aPKC-N, pCS2-Myc-PAR-1, pCS2-Myc-PAR-1T560A or pCS2-Myc-PAR-1KD were used. Specificity of pGL-11CSL-Luc responses to Notch ICD was established in separate experiments (Ezan et al., data not shown). Total DNA amount was adjusted to 1 μg by supplementing pCDNA3 DNA. For the co-culture assay, 293T cells were transfected in 6-well plates at 80 % confluence with 2 μg pCS2-Dll1-myc or empty pCDNA3 together with aPKC-CAAX and PAR-1 constructs and NIH3T3 cells were transfected in 24-well plates at 50 % confluence with 0.3 μg of pGL3-11CSL-Luc, 0.005 μg of TK-Renilla-Luc, 0.2 μg of pBOS-Notch1 and 0.5 μg of pCDNA3. One day after transfection, HEK293T cells were added on to NIH3T3 transfected cells, co-cultured for an additional 24 h and harvested in 100 μl of lysis buffer (Promega) for luciferase activity measurement. Firefly and Renilla luciferase activities were determined using the Dual Luciferase assay system (Promega) and the Veritas microplate luminometer (Turner Biosystems). Normalized results are expressed as the means ± s. d. of three independent experiments performed with triplicate samples. Statistical analysis was performed using two-tailed paired Student’s t-test.
Immunocytochemistry of Xenopus embryos and cultured cells
Indirect immunofluorescence analysis on Xenopus embryo cryosections was performed as described previously (Ossipova et al., 2007; Fagotto and Gumbiner, 1994). Embryos were cryosectioned at 10μm using the Leica cryostat CM3050. The following antibodies were used: anti-XSox3c (Zhang et al., 2003, 1:500), anti-GFP (B2, Santa-Cruz 1:200), anti-phospho-histone H3 (Cell Signaling, 1:300). Images were digitally acquired on a Zeiss Axiophot microscope at 200x magnification. Aphidicolin treatment (Sigma) was performed as described (Hardcastle and Papalopulu, 2000). Representative sections of 5–10 embryos per experimental group are shown.
C17.2 cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with PBS, 0.2% Triton X-100, blocked with PBS-5% goat serum, and probed with anti-β-tubulin III (TuJ1, Covance; 1:500) or anti-Nestin (R401, DSHB, 1:50) and Cy3-conjugated goat anti-mouse antibody (Jackson ImmunoResearch; 1:100). Coverslips were mounted in Vectashield/DAPI (Vector Laboratories). The number of labeled cells (at least 50 cells per field) was determined under a fluorescence microscope (AxioImager, Zeiss) with Axiovision imaging software (Zeiss). The ratio of TuJ1- or Nestin-positive cells to GFP-expressing cells for each field is presented. Results are expressed as the means ± s. d. of five random fields per sample with triplicate samples per group from three to five independent experiments in parallel cultures.
Lentiviral vector construction, lentivirus production and transduction
Self-inactivating lentiviral vector pVVPW, pVVPW-IRES-eGFP, the envelope plasmid pMD.G and the packaging plasmid pCMV R8.2 were kindly provided by Luca Gusella (Battini et al., 2006). GFP-tagged versions of aPKC-CAAX, aPKC-N, PAR-1 and PAR-1T560A were generated from cDNAs encoding rat PKC zeta, accession number NM_022507 (Parkinson et al., 2004), and Xenopus PAR-1A, accession number AF509737, and PAR-1T560A (Ossipova et al., 2005; Ossipova et al., 2007) by Pfu-mediated PCR. GFP-tagged aPKC-CAAX and aPKC-N were subcloned in frame into pXT7-GFP vector (Itoh et al., 2000) as Eco RI inserts. PAR-1 and PAR-1T560A were subcloned as Bgl II/Sal I inserts. To generate GFP–tagged versions in pVVPW, aPKC-CAAX and aPKC-N were amplified using pXT7-eGFP constructs as templates and inserted into the Nhe I and Eco RI sites of VVPW. GFP–tagged PAR-1 and PAR-1T560A in pVVPW were subcloned as Nhe I/Not I inserts. Plasmids were verified by sequencing. Primer oligonucleotide sequences and further construction details are available upon request.
Lentiviruses were produced by transient transfection as described (Battini et al., 2006), with several modifications. Briefly, 3×106 HEK293T cells were plated into 10 cm2 plate and adherent cells were transfected 24 h later using 150 μl of linear polyethylenimine (1 mg/ml, Polysciences) with a total of 16 μg of plasmid DNA: 8 μg of the lentiviral vector, 6 μg of pCMV R8.2, and 2 μg of pMD.G. Cell culture medium was replaced 24 hours after transfection with medium supplemented with 4 mM sodium butyrate. Culture supernatant were collected at 48–96 h after transfection, filtered and concentrated by ultracentrifugation (50,000 g, for 2.5 h at 4°C). The viral pellet was resuspended in complete culture medium and aliquots were stored at −80°C. Viral titers estimated from GFP expression in HEK293T cells transduced with serial dilutions of concentrated lentiviruses, ranged from 106 to 108 transducing units per milliliter. For transduction, cells were seeded in 12-well plates (1×105 per well) and shifted the following day to virus-containing medium in the presence of 8 μg/ml polybrene (Fluka). Viruses were removed after 24 h of incubation, and cells were expanded for differentiation, western analysis and immunocytochemistry. More than 90% of cells expressed GFP at least 7 days after transduction as assessed by flow cytometry, western analysis and immunofluorescence.
Supplementary Material
Acknowledgments
We thank A. Chitnis, E. Fuchs, P. Gergen, V. Joukov, C. Kintner, J. Kitajewski, P. Krieg, Q. Ma, Z. Pan, G. Thomsen, G. Weinmaster for plasmids, E. Snyder for C17.2 cells, L. Gusella and A. Terskikh for help with lentiviral preparations, M. Klymkowsky for the Sox3 antibody, R. Cagan, M. Rendl and A. Sproul for reading the manuscript, D. Weinstein, T. Haremaki and members of the Sokol laboratory for discussions. We especially thank J. Green for his support and discussions at the early stages of this project. This work was supported by the NIH (S. S.), the Grant Lavoisier and the Philippe Foundation fellowship (J. E).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Hammerschmidt M. Induction and patterning of neuronal development, and its connection to cell cycle control. Curr Opin Neurobiol. 2003;13:16–25. doi: 10.1016/s0959-4388(03)00015-1. [DOI] [PubMed] [Google Scholar]
- Battini L, Fedorova E, Macip S, Li X, Wilson PD, Gusella GL. Stable knockdown of polycystin-1 confers integrin-alpha2beta1-mediated anoikis resistance. J Am Soc Nephrol. 2006;17:3049–3058. doi: 10.1681/ASN.2006030234. [DOI] [PubMed] [Google Scholar]
- Bayraktar J, Zygmunt D, Carthew RW. Par-1 kinase establishes cell polarity and functions in Notch signaling in the Drosophila embryo. J Cell Sci. 2006;119:711–721. doi: 10.1242/jcs.02789. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development (Cambridge, England) 2003;130:2657–2668. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A. Why is delta endocytosis required for effective activation of notch? Dev Dyn. 2006;235:886–894. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, Tsai LH, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development (Cambridge, England) 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, Mandelkow EM, Mandelkow E. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem. 1995;270:7679–7688. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Gumbiner BM. Beta-catenin localization during Xenopus embryogenesis: accumulation at tissue and somite boundaries. Development (Cambridge, England) 1994;120:3667–3679. doi: 10.1242/dev.120.12.3667. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development (Cambridge, England) 2003;130:3767–3780. doi: 10.1242/dev.00603. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Marquardt T, Thaler JP, Carter N, Andrews SE, Pfaff SL, Hunter T. Instructive role of aPKCzeta subcellular localization in the assembly of adherens junctions in neural progenitors. Proc Natl Acad Sci U S A. 2008;105:335–340. doi: 10.1073/pnas.0705713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloy J, Hikasa H, Sokol SY. Frodo interacts with Dishevelled to transduce Wnt signals. Nat Cell Biol. 2002;4:351–357. doi: 10.1038/ncb784. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Papalopulu N. Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27(XIC1) and imparting a neural fate. Development (Cambridge, England) 2000;127:1303–1314. doi: 10.1242/dev.127.6.1303. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. In: Kay BK, Peng HB, editors. Methods Cell Biol. San Diego: Academic Press Inc; 1991. pp. 685–695. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3:399–411. doi: 10.1016/0896-6273(89)90200-6. [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Itoh K, Antipova A, Ratcliffe MJ, Sokol S. Interaction of dishevelled and Xenopus axin-related protein is required for wnt signal transduction. Mol Cell Biol. 2000;20:2228–2238. doi: 10.1128/mcb.20.6.2228-2238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/beta-catenin signaling. J Biol. 2005;4:3. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Krupnik VE, Sokol SY. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta-catenin. Curr Biol. 1998;8:591–594. doi: 10.1016/s0960-9822(98)70229-5. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- Kim S, Walsh CA. Numb, neurogenesis and epithelial polarity. Nat Neurosci. 2007;10:812–813. doi: 10.1038/nn0707-812. [DOI] [PubMed] [Google Scholar]
- Kintner C. Neurogenesis in embryos and in adult neural stem cells. J Neurosci. 2002;22:639–643. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development (Cambridge, England) 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Nishida E. The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. Embo J. 2004;23:4190–4201. doi: 10.1038/sj.emboj.7600381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development (Cambridge, England) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development (Cambridge, England) 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Lake BB, Sokol SY. Strabismus regulates asymmetric cell divisions and cell fate determination in the mouse brain. J Cell Biol. 2009 doi: 10.1083/jcb.200807073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamborghini JE. Rohon-beard cells and other large neurons in Xenopus embryos originate during gastrulation. J Comp Neurol. 1980;189:323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development (Cambridge, England) 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. NatRevNeurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Macara IG. Parsing the polarity code. Nat Rev Mol Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Khodyakova A, Aranjuez G, Dudley C, Montell DJ. PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr Biol. 2008;18:1659–1667. doi: 10.1016/j.cub.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Weinmaster G. Notch signaling--constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. NatNeurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Oschwald R, Richter K, Grunz H. Localization of a nervous system-specific class II beta-tubulin gene in Xenopus laevis embryos by whole-mount in situ hybridization. Int J Dev Biol. 1991;35:399–405. [PubMed] [Google Scholar]
- Ossipova O, Dhawan S, Sokol S, Green JB. Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev Cell. 2005;8:829–841. doi: 10.1016/j.devcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Tabler J, Green JB, Sokol SY. PAR1 specifies ciliated cells in vertebrate ectoderm downstream of aPKC. Development (Cambridge, England) 2007;134:4297–4306. doi: 10.1242/dev.009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson SJ, Good JA, Whelan RD, Whitehead P, Parker PJ. Identification of PKCzetaII: an endogenous inhibitor of cell polarity. Embo J. 2004;23:77–88. doi: 10.1038/sj.emboj.7600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KD, Krieg PA. Hox11-family genes XHox11 and XHox11L2 in xenopus: XHox11L2 expression is restricted to a subset of the primary sensory neurons. Dev Dyn. 1999;214:34–43. doi: 10.1002/(SICI)1097-0177(199901)214:1<34::AID-DVDY4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Seydoux G. Anterior-posterior polarity in C. elegans and Drosophila--PARallels and differences. Science. 2002;298:1946–1950. doi: 10.1126/science.1072162. [DOI] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Plusa B, Frankenberg S, Chalmers A, Hadjantonakis AK, Moore CA, Papalopulu N, Papaioannou VE, Glover DM, Zernicka-Goetz M. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118:505–515. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- Riechmann V, Gutierrez GJ, Filardo P, Nebreda AR, Ephrussi A. Par-1 regulates stability of the posterior determinant Oskar by phosphorylation. Nat Cell Biol. 2002;4:337–342. doi: 10.1038/ncb782. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. NatNeurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development (Cambridge, England) 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 2007;446:797–800. doi: 10.1038/nature05722. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Piano F, Riechmann V, Gunsalus KC, Kemphues KJ, Ephrussi A. A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat Cell Biol. 2000;2:458–460. doi: 10.1038/35017101. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Jr, Pepling ME, Horb ME, Thomsen GH, Gergen JP. A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development (Cambridge, England) 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- Winkles JA, Sargent TD, Parry DA, Jonas E, Dawid IB. Developmentally regulated cytokeratin gene in Xenopus laevis. Mol Cell Biol. 1985;5:2575–2581. doi: 10.1128/mcb.5.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Huttner WB. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech Dev. 2003;120:1297–1309. doi: 10.1016/j.mod.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development (Cambridge, England) 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yi MJ, Chen X, Cole F, Krauss RS, Kang JS. Cortical thinning and hydrocephalus in mice lacking the immunoglobulin superfamily member CDO. Mol Cell Biol. 2006;26:3764–3772. doi: 10.1128/MCB.26.10.3764-3772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.