Abstract
Generation of adenovirus-based vectors through homologous recombination within Escherichia coli cells is one of the most efficient strategies. A common challenge associated with this method is the formation of colonies containing self-ligated shuttle plasmid. To improve homologous recombination, a new pAdEasy-1-bearing competent cell line was constructed so that it no longer requires co-transformation with two plasmids and can generate more recombinant colonies (ninefold). New and efficient approaches were also tested to block shuttle plasmid self-ligation by a combined treatment of the plasmid with Taq DNA polymerase and calf intestine phosphatase (CIP) or blocking the formation of self-ligated plasmid-containing colonies by subcloning a suicide gene, ccdB, into the plasmid construct. Present experimental data show that these modifications are effective in eliminating self-ligated plasmid-containing colony background and offer greater simplicity, faster experimental progress, and higher efficiency in performing homologous recombination within E. coli cells, which could facilitate the production of high-titer infectious viral particles.
Keywords: Recombinant adenovirus-based vectors, Homologous recombination, Shuttle plasmid self-ligation, Transfection
1. Introduction
Viruses are natural “professional” vehicles for transferring foreign nucleic acids into host cells, making them useful vectors for gene therapy (Kay et al., 2001). Recombinant adenoviruses received early attention as vehicles to transfer genes into the respiratory epithelium of patients being treated for diseases such as cystic fibrosis (Rosenfeld et al., 1992; Rich et al., 1993; Zabner et al., 1994; Johnson et al., 1995). Quickly it was realized that adenovirus-based vectors were a versatile gene delivery system, capable of highly efficient heterologous gene delivery into a broad range of cell types, both quiescent and dividing (Brody and Crystal, 1994; Kovesdi et al., 1997; Hitt et al., 1997; Krasnykh et al., 1998; Benihoud et al., 1999; Kay et al., 2001). Adenovirus-based vectors do not integrate into the host cell genome, avoiding insertional mutagenesis within target cells. Capsid modifications of these vectors are produced easily, allowing retargeting of their tropism to different tissues (Dmitriev et al., 2002; Belousova et al., 2002; Gaden et al., 2004; Alba et al., 2005; Hesse et al., 2007). Furthermore, high-titered vector stocks can be prepared readily, which is a prerequisite for efficient in vivo gene delivery. Finally, new scale-up adenovirus production systems have been developed, facilitating their use in human clinical trials (Garnier et al., 1994; Côté et al., 1998; Lee et al., 2003; Cortin et al., 2004).
Decades of studying adenovirus biology have led to a clear understanding of the viral life cycle, genomic organization, and functions of the majority of viral proteins. Adenoviruses are medium-sized (80–100 nm), nonenveloped, icosahedral, complex, double-stranded DNA viruses that induce a cytopathic reproductive cycle within a large variety of somatic cells. To date, 51 immuno-logically distinct human adenovirus serotypes have been identified. Among them, serotypes 2 (Ad2) and 5 (Ad5) have been used extensively to construct replication-defective gene-transfer vectors for mammalian cells. The genetic information of Ad2 and Ad5 is compacted within linear, 36 kb long, double-stranded DNA genomes, with both strands coding for transcripts, nearly all of which are spliced heavily. Conventional adenoviral transcription units are referred to as early (E1–E4) and late, depending on their temporal expression to the onset of viral DNA replication (Horwitz, 1996; Shenk, 1996).
The high density and complexity of the adenoviral genome poses problems for recombinant manipulation, usually restricting manipulation to certain regions. In most recombinant adenoviral vectors, transgenes are introduced in place of E1 and E3. The E1 gene is essential for the assembly of infectious virus particles and is supplied in trans by packaging cell lines such as 293 (Graham et al., 1977) and 911 (Fallaux et al., 1996). Deletions within E1 render the virus defective in replication and production of infectious viral particles within target cells. The E3 gene encodes proteins involved in evading host immunity. Accordingly, vector viruses retain most of the viral genome within an approximately 30 kb DNA molecule (Crouzet et al., 1997; He et al., 1998).
Several approaches have been used to generate recombinant adenoviruses. The first method involves direct ligation of adenoviral DNA fragments to restriction endonuclease fragments containing the gene of interest (Ballay et al., 1985; Rosenfeld et al., 1991). In vitro manipulation is technically challenging due to the relatively large vector genome (36 kb), lack of suitable unique restriction sites and low efficiency of large DNA fragment ligation (Becker et al., 1994; Ketner et al., 1994). The second approach, which is used more commonly, involves cloning a gene of interest into a shuttle vector and then transferring the gene into the adenovirus genome by means of homologous recombination within an adenovirus packaging cell line (Ballay et al., 1985; Becker et al., 1994). The resulting desired recombinants are identified by screening individual plaques generated in a lawn of packaging cells (Becker et al., 1994). This approach has proven to be extremely useful, but the low efficiency of homologous recombination, the need for repeated rounds of plaque purification, and the long durations required for completion of the viral production process have hampered the widespread use of adenovirus-based vectors.
More recently, a third approach has been developed that takes advantage of the highly efficient homologous recombination machinery present in Escherichia coli to generate recombinant adenovirus through a double-recombination step between a co-transformed adenoviral backbone plasmid vector and a shuttle plasmid vector carrying a gene of interest (Ketner et al., 1994; Chartier et al., 1996; Crouzet et al., 1997; He et al., 1998). This approach obviated the enzymatic manipulation and ligation steps involved in generating the adenoviral recombinants by previous methods. However, generation of self-ligated shuttle plasmids following recombination in E. coli cells produces undesirable to background colonies, which can lower the efficiency for selecting recombinant adenoviral plasmids. Thus, new approaches are needed to improve homologous recombination and selection for recombinant adenoviral colonies.
In this paper several new approaches were tested and evaluated to improve current conditions for preparing recombinant adenoviral vectors through homologous recombination. First, a modified strain of competent BJ5183 cells harboring adenoviral backbone DNA was generated to enhance homologous recombination through chemical transformation. Secondly, the linearized and dephosphorylated shuttle plasmid was treated with Taq DNA polymerase prior to homologous recombination within E. coli cells to ensure correct recombination. Lastly, a bacterial suicide gene, ccdB (Bernard and Couturier, 1992; Bernard et al., 1993, 1994), was subcloned into the backbone of the shuttle plasmid to prevent shuttle plasmid self-ligation. Experimental tests demonstrated that these modifications were effective in generating valid recombinant adenoviral plasmids with nearly zero self-ligated shuttle-plasmid background and producing high-titered infectious adenoviral vectors in 293T packaging cells.
2. Materials and methods
2.1. Plasmid construction
AdEasy™ Adenoviral Vector Systems were purchased from Stratagene (La Jolla, CA, Cat. No. 240009). To construct a shuttle plasmid that expressed GFP as a reporter, a GFP open reading frame (ORF) was amplified by polymerase chain reaction (PCR) from pcDNA3.1 CT-GFP (Invitrogen, Carlsbad, CA, Cat. No. 482001) with a primer pair of 5′-gagatatcagtcgaggctgatcagcg-3′ and 5′-gtaatacgactcactatag-3′ in a 25-μL reaction volume. PCR product was cloned into a TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA, Cat. No. KNM2000-01) by following the manufacturer's instructions. The GFP gene was released subsequently from the resultant pCR2.1-GFP plasmid by digestion with EcoR V (NEB, Beverly, MA, Cat. No. R0195S) and then ligated into EcoR V-linearized pShuttle-CMV plasmid DNA. Insertion of GFP in pShuttle-CMV was verified by colony-PCR using primers 5′-gaagtgaaatctgaataattttgt-3′ and 5′-gtaatacgactcactatag-3′ and DNA sequencing. The resultant pShuttle-CMV-GFP plasmid was transfected into 293T cells and GFP expression was evaluated using an inverted fluorescent microscope at 24 h post-transfection.
To construct a pShuttle-CMV-GFP-ccdB plasmid, a ccdB expression cassette was amplified by PCR from pSilent-R1/R2/ccdB (a kind gift from Dr. Shaobin Zhong, Department of Plant Pathology, North Dakota State University) with primers 5′-gctagtttaaacgacctgcagactggct-3′ and 5′-gcatcaagaacagaagtatgtc-3′ . In this PCR amplification, Deep Vent DNA polymerase (NEB, Beverly, MA, Cat. No. M0258S) was used and the PCR product was directly ligated into pShuttle-CMV-GFP DNA that had been linearized with Pme I (NEB, Cat. No. R0560S). Ligates were then transformed into a ccdB-survival E. coli strain, DB3.1 (Invitrogen, Carlsbad, CA, Cat. No. 11782-018). Insertion of the ccdB gene was verified by PCR under the same conditions as its amplification and DNA sequencing. The killing of BJ5183 cells by ccdB gene products was verified by transforming the plasmid DNA into BJ5183 competent cells. Transformations with the ccdB-containing plasmid clones were evaluated by colony formation.
2.2. DNA treatments by Pme I, calf intestine phosphatase (CIP) and Taq DNA polymerase
Approximately 100 μg of pShuttle-CMV-GFP DNA was linearized with Pme I and then separated by 0.8% agarose gel electrophoresis, followed by gel extraction (QIAquick Gel Extraction Kit, Qiagen, Valencia, CA, Cat. No. 28706). Recovered DNA was quantified by OD absorption using a UV spectrophotometer (Beckman DU 800). Approximately 20 μg of recovered DNA was dephosphorylated with CIP (NEB, Beverly, MA, Cat. No. M0290S) and then precipitated with EtOH and dissolved in 50 μL ddH2O. Twenty-five microliters of dissolved DNA was included in a 50-μL PCR master mix with 1.0 unit Taq DNA polymerase, and incubated at 72 °C for 30 min. After incubation, treated DNA was precipitated with EtOH, dissolved in 25 μL ddH2O and used for transformation of competent E. coli or self-ligation tests. One hundred nanograms of plasmid DNA treated as specified was included in a 10-μL ligation mix with 1 Unit T4 DNA ligase (Promega, Madison, WI, Cat. No. M1801) and incubated at 16 °C for 9 h. After self-ligation, 2 μL of the ligation mix was used to transform 200 μL plasmid-free BJ5183 competent cells.
2.3. Mammalian cell culture
293T cells were obtained from ATCC and grown with DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT, Cat. No. SV30014.03), 100 U/mL penicillin, 100 μg/mL streptomycin sulfate, and 4 mM l-glutamine (Sigma–Aldrich St. Louis, MO, Cat. No. G1146) at 37 °C in a humidified 5.0% CO2 incubator. Cells were split at a 1:6 ratio every 3–4 days.
2.4. Competent cell preparation and transformation
Highly-competent BJ5183 E. coli cells, with or without pAdEasy-1, were prepared according to a modified protocol as previously described (Hanahan, 1983). Treated bacteria cells were used immediately for transformation or kept at –80 °C for long-term storage by addition of 15% glycerol (Sigma–Aldrich, St. Louis, MO, Cat. No. G5516).
To transform competent cells with plasmid or ligation mix, 10–200 ng DNA was mixed with E. coli cells and incubated on ice for 30–60 min. Cells were then heat-shocked at 42 °C for exactly 90 s and then incubated on ice for 10 min. Eight hundred micro-liters of SOC was added to each heat-shocked tube and incubated at 37 °C for 60 min. Appropriate amounts of the transformation mixture were plated onto LB plates with the corresponding antibiotic for selection and incubated overnight. To test the competence of each preparation, 50 pg plasmid DNA was used per tube to avoid saturating the competent cells, followed by transformation as described. The transformation efficiency was calculated according to the following equation: colony forming units/μg DNA = [(# of colonies/pg DNA transformed) × (106 pg/μg)]/% plated of total transformation.
2.5. Homologous recombination in E. coli cells
To carry out recombination, highly-competent BJ5183 cells carrying the pAdEasy-1 plasmid were prepared as described. pShuttle-CMV-GFP plasmid DNA was linearized by Pme I, dephosphorylated by CIP, and treated further by Taq DNA polymerase as described. For plasmid pShuttle-CMV-GFP-ccdB, DNA was either used in supercoiled form, or linearized by Pme I before transformation. After these treatments, 1.0 μg of treated shuttle plasmid DNA was added into each 200 μL aliquot of competent BJ5183 cells, transformed as described, plated on LB plates containing 50 mg/L kanamycin, and then incubated at 37 °C for 24–36 h. Well-isolated colonies were picked and inoculated in 3.0 mL LB medium with 50 mg/L kanamycin for small-scale plasmid extraction. Extracted plasmids were dissolved in 200 μl TE buffer, 5.0 μl of which were used for primary identification through agarose gel electrophoresis. Self-ligated shuttle plasmid could be identified and discarded due to huge difference in molecular size between the shuttle plasmid and recombinant adenoviral plasmid. Recombinant adenoviral plasmids were further distinguished from pAdEasy-1 and stabilized by another round of transformation into a plasmid-stable E. coli strain (XL-10) and selection with 50 mg/L kanamycin.
2.6. Transfection into 293T cells and infectious adenoviral generation
Transfection of recombinant adenoviral DNA into 293T cells was performed with a dextran and polybrene enhanced calcium phosphate transfection method as described previously (Wu and Lu, 2007). Cultures were incubated at 37 °C in a 5.0% CO2 incubator for 8 h and then media was replaced with 10 mL warm DMEM media with 10% FBS per flask. Conditioned medium was replaced with fresh medium every other day until 10–12 days post-transfection.
2.7. Viral stock preparation and amplification in 293T cells
To prepare primary viral-vector stocks, growth medium was discarded from transfected cells at day 10–12 post-transfection, leaving approximately 2.0 mL per flask. Flasks were subjected to four rounds of freeze–thaw cycles between an –80 °C freezer and a 37 °C water bath. At the last cycle, the cells were collected into a 15-mL centrifuge tube and centrifuged at 5000 rpm for 15 min. The supernatant was collected and aliquots of 0.5 mL/tube were stored at –80 °C. Titers of the viral stock were determined as described in the following “vector titration” section. To amplify titers of the primary viral stock, 293T cells were seeded 24 h before infection, allowing the formation of an approximately 70% cell monolayer the next day. At the time of infection, the medium was discarded and the cell monolayer was infected with 0.5 mL primary viral stock at 37 °C for 1.0 h. After infection, cells were washed twice with PBS and maintained in DMEM supplemented with 5% FBS. Infection of 293T cells and amplification of the vector virus was monitored by GFP expression using an inverted fluorescent microscope. Infected 293T cells were harvested when maximum cytopathic effect (CPE) appeared and then viral stocks were prepared as described above.
2.8. Vector titration
Titration of viral vectors generated in transfected 293T cells was performed by a limiting dilution method described previously (Wu and Lu, 2007). Infected cultures were kept at 37 °C in a CO2 incubator for 3 days. Vector titers were determined by counting the number of GFP-positive cells at the end-point dilution as follows: Vector titer (IU/mL) = # of GFP+ cells × 5 (IF) × DF, where IU = infectious unit, IF = inoculum factor, and DF = dilution factor.
3. Results
3.1. Blocking shuttle vector self-ligation by Taq DNA polymerase treatment
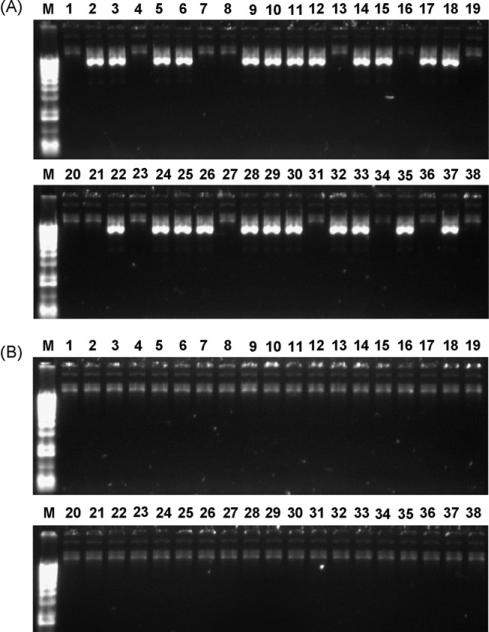
As shown in Table 1, transformation of E. coli cells with ligation mix containing Pme I-linearized self-ligated plasmid DNA generated more than 1000 colonies. Additional CIP or Taq DNA polymerase treatments lowered the number of colonies to less than 10. It was notable that when both CIP and Taq DNA polymerase treatments were combined, no kanamycin-resistant colonies were observed. When treated DNA was used for recombinant adenovirus generation in E. coli cells, CIP-based treatment resulted in up to 60% of the colonies containing self-ligated shuttle plasmid DNA (Fig. 1A), while the combined treatments of CIP and Taq DNA polymerase induced no colonies with self-ligated shuttle plasmid DNA (Fig. 1B).
Table 1.
Effect of selected treatments on colony formation of transformed competent BJ5183 cells with self-ligated shuttle vector plasmid DNA
| Plasmid DNA treatment | Number of self-ligated colonies |
|---|---|
| Pme I | 1.3 ± 0.4 × 103 |
| Pme I + CIP | 7.25 ± 2.5 |
| Pme I + Taq | 2.0 ± 1.4 |
| Pme I + CIP + Taq | 0.0 |
Pme I: shuttle vector plasmid DNA was linearized by Pme I; CIP: shuttle vector plasmid DNA was dephosphorylated by calf intestinal phosphatase (CIP); Taq: shuttle vector plasmid DNA was treated with one unit of Taq DNA polymerase at 72 °C for 30 min. 100 ng of each treated plasmid DNA was used in a ligation test and transformed into plasmid-free competent BJ5183 cells. Transformed cells were plated on LB plates with 50 mg/L kanamycin. The numbers of colonies formed were calculated.
Fig. 1.
Generation of recombinant adenoviral plasmids in bacterial cells. (A) DNA from pShuttle-CMV-GFP was linearized by Pme I, dephosphorylated, and used for transformation of competent BJ5183 E. coli cells. Plasmid DNA prepared from inoculated colonies, as described in Section 2, were analyzed in supercoiled form by 0.4% agarose gel electrophoresis and ethidium bromide staining. Lane M, Fisher 1 kb DNA ladder (Cat. No. BP2553-100); lanes 1–38, plasmid DNA from different colonies. Based on the difference in migration rates, colonies in lanes 2, 3, 5, 6, 9–12, 14, 15, 17, 18, 22, 24–26, 28–30, 32, 33, 35, 37 contained self-ligated shuttle plasmid DNA; the remaining 15 out of 38 colonies potentially contained valid recombinant aden-oviral plasmids. (B) Analysis of plasmid DNA from post-recombination colonies. In addition to the treatments in (A), pShuttle-CMV-GFP DNA was treated with Taq DNA polymerase as described in Section 2. Based on the migration rates, none of the 38 colonies contained self-ligated shuttle plasmid DNA.
3.2. Homologous recombination in chemically competent BJ5183 E. coli cells carrying pAdEasy-1
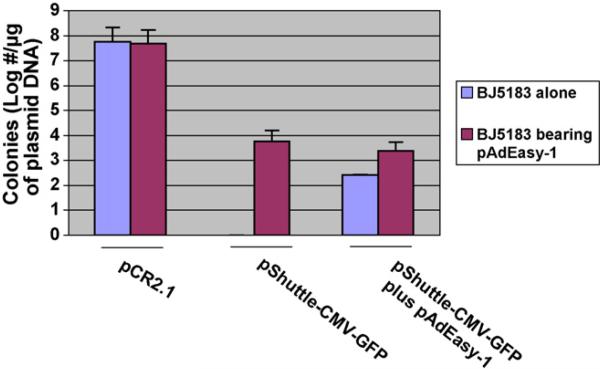
To simplify homologous recombination, a new strain of BJ5183 E. coli strain harboring pAdeasy-1 was constructed. These newly generated BJ5183 E. coli cells possessed similar transformation competence to plasmid-free cells. As shown in Fig. 2, transformation of these cells with 1 μg of pCR2.1 resulted in the formation of 4.9 × 107 and 5.8 × 107 kanamycin-resistant colonies on LB plates, respectively. In addition, transformation of these two types of competent cells with pShuttle-CMV-GFP plasmid DNA generated zero kanamycin-resistant colonies for plasmid-free competent cells as compared to 5.9 × 103 for the pAdEasy-1-carrying competent cells. Furthermore, co-transformation of these two competent cells with both pAdEasy-1 and pShuttle-CMV-GFP plasmid DNA resulted in nine times more colonies in pAdEasy-1-carrying competent cells than the control plasmid-free counterpart (2400 vs. 260). These experimental data indicate that the generation of pAdEasy-1-carrying competent cells no longer requires co-transformation of the shuttle plasmid and pAdEasy-1, thus, simplifying the transformation procedure, as well as largely enhancing the transformation efficiency.
Fig. 2.
Comparison of transformation and recombination efficiencies of competent cells made from E. coli BJ5183 and BJ5183 carrying pAdEasy-1. The competence of two types of cells was compared by transformation with 25 ng pCR2.1 plasmid and plating on kanamycin LB plates. The number of colonies was calculated as described in Section 2. Recombination efficiencies were compared by either transforming pAdEasy-1-carrying competent BJ5183 E. coli cells with 1.0 μg Pme I-linearized, CIP and Taq DNA polymerase treated pShuttle-CMV-GFP plasmid DNA alone, or co-transforming plasmid-free competent cells with 500 ng of treated pShuttle-CMV-GFP and 500 ng of supercoiled pAdEasy-1 plasmid DNA. Numbers of colonies were counted as described. Data was obtained from representative experiments performed in triplicate.
3.3. Adaptation of the ccdB gene into the shuttle vector backbone
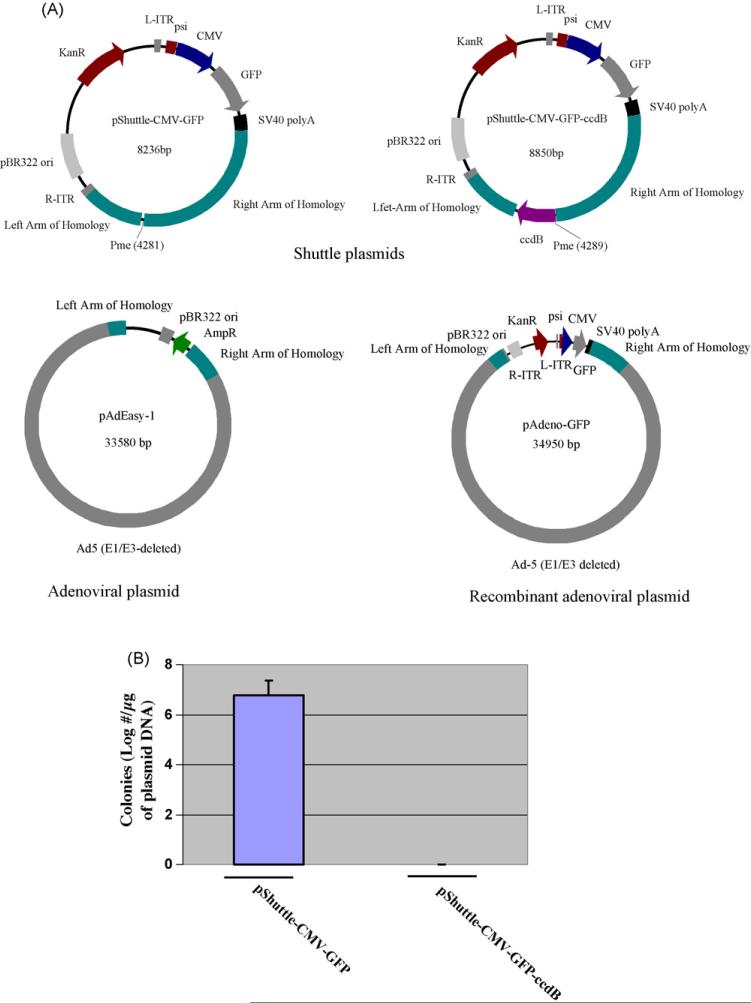
To block shuttle plasmid self-ligation, a ccdB expression cassette was incorporated into the Pme I restriction site of the pShuttle-CMV-GFP plasmid (Fig. 3A). As constructed, the resultant ccdB-carrying shuttle plasmid regained the Pme I restriction site, and thus, could still be linearized by Pme I in the same manner as the unmodified shuttle plasmid. The ccdB expression cassette was subcloned into the shuttle plasmid in such a way that it will be maintained if the shuttle plasmid DNA is self-ligated, but lost if a double-arm homologous recombination takes place between the two homologous arms of the shuttle plasmid and pAdEasy-1 (Fig. 3B).
Fig. 3.
CcdB gene adaptation to the adenovirus vector system and functional testing. (A) Schematic map of the ccdB gene cloned into shuttle-CMV-GFP. CcdB gene was inserted between the left and right homologous recombination arms of the shuttle vector plasmid so that it will be lost upon recombination, while if self-ligation occurs, it will block colony formation by eventually killing BJ5183 cells. (B) Functional test of cloned ccdB gene on BJ5183 cells. 25 ng of both pShuttle-CMV-GFP and pShuttle-CMV-GFP-ccdB plasmids were used to transform 200 μL of competent BJ5183 cells. The number of colonies formed per microgram of each plasmid DNA was determined as described in Section 2. Data was obtained from representative experiments performed in triplicate.
To test if the incorporation of the ccdB gene into the shuttle plasmid would prevent the formation of BJ5183 cell colonies carrying self-ligated shuttle plasmid DNA, two types of shuttle plasmids were used to transform plasmid-free competent BJ5183 cells. As expected, transformation with the pShuttle-CMVGFP plasmid into BJ5183 competent cells generated 6.8 × 107 kanamycin-resistant colonies from 1 μg of parental plasmid DNA, while transformation with the ccdB-carrying shuttle plasmid did not generate any kanamycin-resistant self-ligated colonies (Fig. 4). The incorporation of the ccdB gene expression cassette completely blocks the formation of self-ligated colonies in BJ5183 E. coli cells.
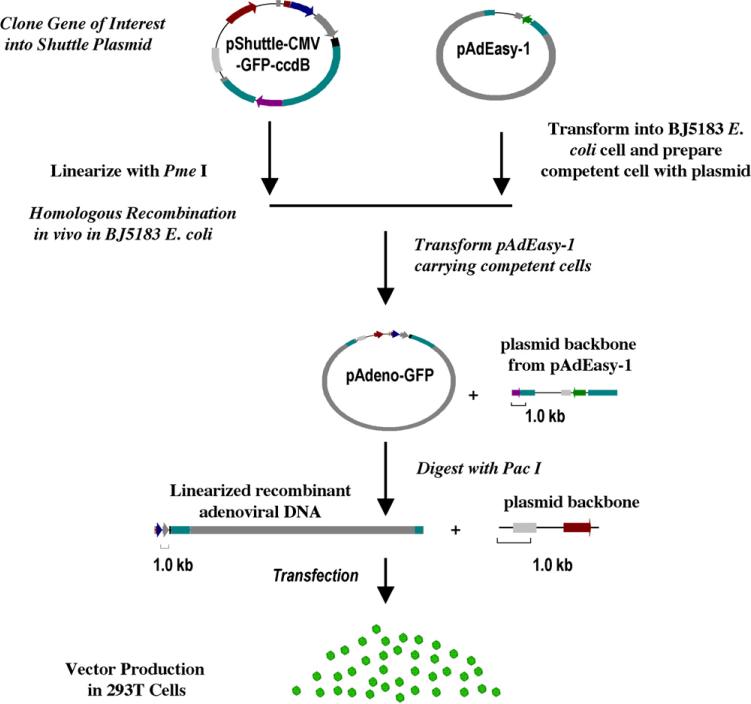
Fig. 4.
Schematic outline of the optimized recombinant adenovirus vector system through plasmid recombination in E. coli cells. First, the gene of interest is cloned into a shuttle vector, such as the illustrated pShuttle-CMV-GFP-ccdB as illustrated; Second, the resulting plasmid is digested with a restriction enzyme, Pme I(ccdB-containing plasmids can also be used in supercoiled form). For shuttle plasmids not containing the ccdB gene, linearized DNA is further treated with CIP and Taq DNA polymerase; Third, competent BJ5183 E. coli cells carrying pAdEasy-1 is transformed with linearized plasmid DNA; Fourth, the recombinant adenoviral plasmid is digested with Pac Ito expose the adenoviral inverted terminal repeats (ITR) and transfected into a 293T packaging cell line. Infectious recombinant adenoviral vector particles are obtained from the transfected cells and amplified by infecting fresh 293T cells.
3.4. Homologous recombination using the ccdB shuttle vector plasmid
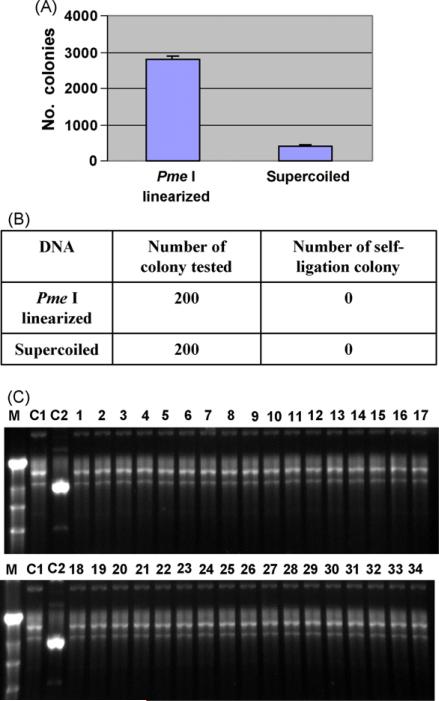
The shuttle plasmid DNA was prepared and used to transform competent BJ5183 cells carrying the pAdEasy-1 plasmid. As shown in Fig. 5A, transformation with Pme I-linearized shuttle vector DNA generated an average of 2780 kanamycin-resistant colonies, while transformation with supercoiled shuttle vector DNA generated an average of 412 kanamycin-resistant colonies. It was notable that transformation with supercoiled shuttle vector DNA also generated quite a lot of kanamycin-resistant colonies (Fig. 5A). To ensure that these are ccdB-negative colonies, a total of 200 transformed-cell colonies were tested by PCR for their retention of the ccdB gene as an indication of shuttle plasmid self-ligation. None of these colonies contained the ccdB gene expression cassette (Fig. 5B). Further verification by agarose gel electrophoresis showed no shuttle plasmid band, which is characteristic of self-ligated shuttle plasmids (Fig. 5C). These results indicate that all surviving colonies are generated by recombinant adenoviral plasmids and do not contain any self-ligated shuttle plasmid DNA.
Fig. 5.
Generation of recombinant adenovirus plasmids using the ccdB-containing shuttle plasmid. (A) Competent BJ5183 E. coli cells were transformed using 1.0 μg Pme I-linearized DNA from pShuttle-CMV-GFP-ccdB plasmid, or supercoiled DNA from of the same plasmid, and numbers of recombinant colonies were counted as described in Section 2. Data was obtained from representative experiments performed in triplicate. (B) Detection of shuttle plasmid self-ligation by PCR using primers specific for the amplification of the ccdB gene cassette. Two hundred colonies transformed with linearized and supercoiled shuttle plasmid DNA were tested and none of them were found to contain the ccdB gene. (C) Detection for shuttle plasmid self-ligation by agarose gel migration. As in Fig. 1(A and B), none of the colonies contained self-ligated shuttle plasmid DNA based on the migration rates. Lane M, Fisher λ DNA-Hind III digest DNA ladder (Cat. No. BP2556-200); lane C1, control plasmid DNA from pAdEasy-1; lane C2, control plasmid DNA from pShuttle-CMV-GFP-ccdB; lanes 1–17, plasmid DNA from colonies generated by transforming Pme I linearized pShuttle-CMV-GFP-ccdB DNA; lanes 18–34, plasmid DNA from colonies generated by transforming supercoiled pShuttle-CMV-GFP-ccdB DNA.
3.5. Verification of recombinant plasmids
After primary identification by agarose gel electrophoresis, plasmid DNA from representative colonies were transformed into E. coli strain XL-10, and selected as described. Plasmid DNA derived from individual kanamycin-resistant colonies, along with control DNA from pAdEasy-1, pShuttle-CMV-GFP and pShuttle-CMV-GFP-ccdB, were digested with Pac I and analyzed by agarose gel electrophoresis. As shown in Fig. 6A, recombinant plasmids obtained through this optimized system showed the correct patterns of restriction bands. Moreover, there was no difference among recombinant plasmids derived from the ccdB gene-adapted shuttle plasmid and those from the unmodified plasmids. This was further confirmed by DNA transfection and production of infectious viral particles using 293T packaging cells as described.
Fig. 6.
Recombinant adenoviral plasmid transfection and infectious virus generation in HEK 293T cells. (A) Representative Pac I digestion of DNA from plasmids pShuttle-CMVGFP, pShuttle-CMV-GFP-ccdB, pAdEasy-1, and recombinant adenoviral plasmids from recombination using DNA from either pShuttle-CMV-GFP or pShuttle-CMV-GFP-ccdB. Lane M1, Fisher 1 kb DNA ladder; lane M2, Fisher λ DNA-Hind III digest DNA ladder; lane 1, pShuttle-CMV-GFP; lane 2, pShuttle-CMV-GFP-ccdB; lane 3, pAdEasy-1; lane 4, recombinant adenoviral plasmid from recombination using DNA from pShuttle-CMV-GFP; lane 5, recombinant adenoviral plasmid from recombination using DNA from pShuttle-CMV-GFP-ccdB. Digested DNA was analyzed by 0.4% agarose gel electrophoresis. Based on the migrations, characteristic bands of correct size were observed for each plasmid, and the recombinant adenoviral plasmids derived from each shuttle plasmid showed the same characteristic bands (a 4.5-kb band from bacterial plasmid backbone and a 31.8-kb band from recombinant adenoviral DNA). (B) DNA from recombinant adenoviral plasmids were digested with Pac I, precipitated with EtOH, and transfected into 293T cells as described in Section 2. Representative photomicrographs taken on day 10 post-transfection demonstrate the formation of infection loci within the transfected cell monolayer. (C) Primary viral stock was amplified by infecting fresh 293T cell monolayer. A 50% confluent cell monolayer was infected at an MOI of 10 and representative photomicrographs were taken on day 2 post-infection. (D) Consecutive amplification of viral stock in 293T cells. When the stock titers were relatively low (less than 108 IU/mL), each amplification process gradually boosted the titer about 10 fold. Data was obtained from representative experiments performed in triplicate. FL, fluorescent light; NL, normal light.
3.6. Adenoviral DNA transfection and vector production
Pac I-digested recombinant plasmid DNA was transfected into 293T cells and robust GFP expression was observed within 24 h. Viral vector production continued to increase for 3–4 days post-transfection. Beyond this time period, GFP expression appeared to decrease in most of the transfected cells. However, in some transfected cells, infectious adenovirus particles were generated and able to infect and replicate within adjacent cells. This process continued and eventually led to the formation of infection loci within the transfected 293T cell monolayer 5–10 days post-transfection (Fig. 6B). This appeared to be a typical phenomenon documented by others in literature (He et al., 1998).
This study also confirmed that adenoviral-vector titers from the initial transfection could be significantly increased by repeated viral infection of fresh 293T cells. As shown in Fig. 6C, 100% GFP-positive cells were obtained on day 2 post-infection by infecting a 50% confluent 293T cell monolayer at an MOI of 10. In addition to GFP expression, infected 293T cells also showed the characteristic CPE associated with adenovirus infection (cell roundup). Following several rounds of consecutive infection and amplification, high-titer vector stocks up to 4.5 × 108 IU/mL were prepared without vector concentration (Fig. 6D).
4. Discussion
In this study, several attempts were taken to address the problems related to the self-ligation of shuttle plasmid, as well as the low recombination efficiency, within E. coli cells, through an initial transformation of BJ5183 cells with pAdEasy-1, growth selection with ampicillin, and preparation of chemically competent E. coli cells carrying the pAdEasy-1 plasmid. As a result, recombinant adenoviral plasmids were generated with high efficiency and zero background. The competence of the newly constructed pAdEasy-1-carrying cells was determined to be comparable to that of the plasmid-free counterpart cells. Instead of co-transformation with two plasmids, the plasmid-carrying competent cells mediated efficient generation of recombinant adenoviral plasmids by a single plasmid transformation. It should be noted that by homologous recombination within E. coli cells carrying the pAdEasy-1 plasmid, kanamycin-resistant colonies carry actually both the recombinant adenoviral plasmid and pAdEasy-1. This potential problem for preparing pure target plasmids can be solved by transforming the plasmid mix into another plasmid-stabilizing E. coli strain, such as DH5α or XL-10, and selecting with kanamycin. New kanamycin-resistant colonies will carry only the recombinant adenoviral plasmid.
To address the issue of self-ligated shuttle plasmid production when preparing recombinant adenoviral plasmids, current approaches include linearization and dephosphorylation of the shuttle plasmid prior to co-transformation with pAdEasy-1. However, these treatments are not sufficient enough to block self-ligation. Experimental results show that the percentage of colonies carrying self-ligated shuttle plasmids can reach as high as 60% (Fig. 1). To promote more effective blockage of plasmid self-ligation, an additional treatment of shuttle plasmid DNA with Taq DNA polymerase was introduced following linearization and dephosphorylation. Experimental data obtained demonstrated that the additional Taq DNA treatment blocked the formation of self-ligated shuttle plasmid completely. Through these treatments, the terminal transferase activity of Taq DNA polymerases is known to add a3′-A overhang to each end of the DNA fragment (Clark, 1988), while CIP treatment creates ends that do not possess a 5′ phosphate group. Thus, this combined treatment creates a condition that makes the self-ligation of shuttle plasmid DNA essentially impossible. Taq DNA polymerase mediated addition of a 3′-A overhang to linearized DNA ends is an well-documented molecular biology technique that has been very useful and widely employed in TA vector-based cloning and sequencing. In addition, this step of Taq DNA treatment was determined to cause no adverse impact on transformation. When the treated shuttle plasmid DNA was used to transform pAdEasy-1-carrying competent cells or co-transform plasmid-free competent cells with pAdEasy-1 DNA, over 5.9 × 103 and 2.6 × 102 recombinant colonies were generated, respectively. These data also argue for improved transformation efficiencies of competent cells carrying pAdEasy-1 for recombinant adenoviral plasmid generation.
In addition, another strategy was tested for knocking out shuttle plasmid self-ligation background by introducing a ccdB expression cassette between the two homologous recombination arms of the shuttle plasmid. Since ccdB was positioned this way, it will be lost upon successful recombination between the shuttle plasmid and pAdEasy-1, allowing colony formation of E. coli carrying the desired recombinant plasmid. Self-ligated shuttle plasmids will retain an intact ccdB gene, and the accumulation of CcdB protein will eventually kill E. coli cells and prevent them from forming colonies on LB plates. As demonstrated, transformation with pShuttle-CMV-GFP generated 6.8 × 107 colonies per microgram plasmid, while the ccdB containing plasmid, pShuttle-CMV-GFP-ccdB, did not generate any colonies. Furthermore, it was confirmed that transformation with shuttle plasmid DNA containing the ccdB expression cassette led to high efficiency generation of colonies carrying the desired recombinant plasmid. These findings together indicate that the ccdB gene can be used effectively for eliminating shuttle plasmid self-ligation in BJ5183 cells. Another advantage of using the ccdB gene to block shuttle plasmid self-ligation is that it works when transformed in supercoiled form. Theoretically, this holds an important implication because it broadens the range of heterologous genes that can be cloned into adenoviral vectors. The original system is dependent on the linearization of shuttle plasmid DNA carrying target gene by Pme I to reduce background. As a consequence, any gene sequence that carries Pme I restriction sites will not be introduced into recombinant adenoviral plasmids since it will be disrupted by Pme I digestion. It should be noted that no change has been introduced to the homologous arm sequences. Therefore, the described modifications to the adenoviral shuttle construct will not introduce any sequence mismatching during the adenoviral homologous recombination event.
In summary, these new modifications make the generation of recombinant adenoviral vectors easy, straightforward, efficient, and most importantly, reduces the background from self-ligated shuttle vector plasmid-containing colonies to zero. These modifications have proven to be efficient and rapid in creating recombinant adenoviral vectors expressing heterologous genes, and would be especially valuable for creating a large number of recombinant adenoviral vectors with different genes. These modifications will have significant implications in using adenovirus-based vectors for gene therapy research and clinical applications.
Acknowledgements
We would like to thank Jarred Yasuhara-Bell for his comments on manuscript preparation and Shaobin Zhong for plasmid pSilent-R1/R2/ccdB. This research was supported in part by grants from the Hawaii Community Foundation (20070438) and the National Institutes of Health (S11NS043499 and G12RR003061).
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- Alba R, Bosch A, Chillon M. Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. 2005;12:S18–S27. doi: 10.1038/sj.gt.3302612. [DOI] [PubMed] [Google Scholar]
- Ballay A, Levrero M, Buendia MA, Tiollais P, Perricaudet M. In vitro and in vivo synthesis of the hepatitis B virus surface antigen and of the receptor for polymerized human serum albumin from recombinant human adenoviruses. EMBO J. 1985;4(13B):3861–3865. doi: 10.1002/j.1460-2075.1985.tb04158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TC, Noel RJ, Coats WS, Gomez-Foix AM, Alam T, Gerard RD, Newgard CB. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- Belousova N, Krendelchtchikova V, Curiel DT, Krasnykh V. Modulation of adenovirus vector tropism via incorporation of polypeptide ligands into the fiber protein. J. Virol. 2002;76(17):8621–8631. doi: 10.1128/JVI.76.17.8621-8631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr. Opin. Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA–topoisomerase II complex. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- Bernard P, Kezdy KE, Melderen LV, Steyaert J, Wyns L, Pato ML, Higgins PN, Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 1993;234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- Bernard P, Gabant P, Bahassi EM, Couturier M. Positive selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Brody SL, Crystal RG. Adenovirus-mediated in vivo gene transfer. Ann. NY Acad. Sci. 1994;716:90–101. doi: 10.1111/j.1749-6632.1994.tb21705.x. [DOI] [PubMed] [Google Scholar]
- Chartier C, Degryse M, Gantzer M, Dieterie A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM. Novel non-templated nucleotide addition reactions catalyzed by prokaryotic and eukaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortin V, Thibault J, Jacob D, Garnier A. High-titer adenovirus vector production in 293S cell perfusion culture. Biotechnol. Prog. 2004;20(3):858–863. doi: 10.1021/bp034237l. [DOI] [PubMed] [Google Scholar]
- Côté J, Garnier A, Massie B, Kamen A. Serum-free production of recombinant proteins and adenoviral vectors by 293SF-3F6 cells. Biotechnol. Bioeng. 1998;59(5):567–575. [PubMed] [Google Scholar]
- Crouzet J, Naudin L, Orsini C, Vigne E, Ferrero L, Le Roux A, Benoit P, Latta M, Torrent C, Branellec D, Denèfle P, Mayaux JF, Perricaudet M, Yeh P. Recombinational construction in Escherichia coli of infectious adenoviral genomes. PNAS. 1997;94:1414–1419. doi: 10.1073/pnas.94.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev IP, Kashentseva EA, Curiel DT. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J. Virol. 2002;76(14):6893–6899. doi: 10.1128/JVI.76.14.6893-6899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, Van Ormondt H, Hoeben RC, Van Der Eb AJ. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 1996;7(2):215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- Gaden F, Franqueville L, Magnusson MK, Hong SS, Merten MD, Lindholm L, Boulanger P. Gene transduction and cell entry pathway of fiber-modified adenovirus type 5 vectors carrying novel endocytic peptide ligands selected on human tracheal glandular cells. J. Virol. 2004;78(13):7227–7247. doi: 10.1128/JVI.78.13.7227-7247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier A, Côté J, Nadeau I, Kamen A, Massie B. Scale-up of the adenovirus expression system for the production of recombinant protein in human 293S cells. Cytotechnology. 1994;15(1–3):145–155. doi: 10.1007/BF00762389. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russel WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa TL, Yu J, Kinzle WK, Vogelstein B. A simplified system for generating recombinant adenoviruses. PNAS. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse A, Kosmides D, Kontermann RE, Nettelbeck DM. Tropism modification of adenovirus vectors by peptide ligand insertion into various positions of the adenovirus serotype 41 short-fiber knob domain. J. Virol. 2007;81(6):2688–2699. doi: 10.1128/JVI.02722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt MM, Addison CL, Graham FL. Human adenovirus vectors for gene transfer into mammalian cells. Adv. Pharmacol. 1997;40:137–206. doi: 10.1016/s1054-3589(08)60140-4. [DOI] [PubMed] [Google Scholar]
- Horwitz MS. Adenoviruses. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Fields Virology. Lippincott; Philadelphia: 1996. pp. 2149–2171. [Google Scholar]
- Johnson LG, Boyles SE, Wilson J, Boucher RC. Normalization of raised sodium absorption and raised calcium-mediated chloride secretion by adenovirus-mediated expression of cystic fibrosis transmembrane conductance regulator in primary human cystic fibrosis airway epithelial cells. J. Clin. Invest. 1995;95:1377–1382. doi: 10.1172/JCI117789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay MA, Glorioso J, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Efficient manipulation of the human adenovirus genome as an infectious yeast artificial chromosome clone. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdi I, Brough DE, Bruder JT, Wickham TJ. Adenovirus vectors for gene transfer. Curr. Opin. Biotechnol. 1997;8:583–589. doi: 10.1016/s0958-1669(97)80033-x. [DOI] [PubMed] [Google Scholar]
- Krasnykh V, Dmitriev I, Mikheeva G, Miller CR, Belousova N, Curiel DT. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 1998;72(3):1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Yap MG, Hu WS, Wong KT. Low-glutamine fed-batch cultures of 293-HEK serum-free suspension cells for adenovirus production. Biotechnol. Prog. 2003;19(2):501–509. doi: 10.1021/bp025638o. [DOI] [PubMed] [Google Scholar]
- Rich DP, Couture LA, Cardoza LM, Guiggio VM, Armentano D, Espino PC, Hehir K, Welsh MJ, Smith AE, Gregory RJ. Development and analysis of recombinant adenoviruses for gene therapy of cystic fibrosis. Hum. Gene Ther. 1993;4:461–476. doi: 10.1089/hum.1993.4.4-461. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MA, Siegfried W, Yoshimura K, Yoneyama K, Fukayama M, Stier LF, Paakko PK, Gilardi P, Stratford-Perricaudet LD, Perricaudet M, Jallat S, Pavirani A, Lecocq J-P, Crystal RG. Adenovirus-mediated transfer of a recombinant alpha 1-antitrypsin gene to the lung epithelium in vivo. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MA, Yoshimura K, Trapnell BC, Yoneyama K, Rosenthal ER, Dalemans W, Fukayama M, Bargon J, Stier LE, Stratford-Perricaudet L, Perricaudet M, Guggino WB, Pavirani A, Lecocq J, Crystal RG. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68:143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- Shenk T. Adenoviridae and their replication. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Fields Virology. Lippincott; Philadelphia: 1996. pp. 2111–2148. [Google Scholar]
- Wu C, Lu Y. Inclusion of high molecular weight dextran in calcium phosphate-mediated transfection significantly improves gene transfer efficiency. Cell Mol. Biol. 2007;53(4):67–74. [PMC free article] [PubMed] [Google Scholar]
- Zabner J, Couture LA, Smith AE, Welsh MJ. Correction of cAMP-stimulated fluid secretion in cystic fibrosis airway epithelia: efficiency of adenovirus-mediated gene transfer in vitro. Hum. Gene Ther. 1994;5:585–593. doi: 10.1089/hum.1994.5.5-585. [DOI] [PubMed] [Google Scholar]