Abstract
This study describes a temporal profile of gene expression from normal human fetal testes and ovaries. Gonads from 34 fetuses between 9 wk and 20 wk of gestation were obtained from the Department of Pathology and the Birth Defects Research Laboratory at the University of Washington. Relative transcript levels were determined using the Affymetrix Human Genome U133A Plus 2.0 arrays. Sex determination occurs in the human gonad at ∼6 wk of gestation with development of the testis driven by expression of SRY. In this study, SRY transcript was present and elevated at 9 wk of gestation in the testis but was absent in the ovary. The transcript levels of other testis-specific factors SOX9 and AMH and the steroidogenic genes CYP17A1, CYP11A1, STAR, and HSD17B3 were all significantly higher in the testis. In contrast, transcripts known to be involved in meiosis, including STRA8, SPO11, SYCP3, TEX11, TEX14, and STAG3, showed highest expression in the fetal ovary beginning at Week 12. These gene expression profiles will be a resource for understanding and defining normal gonad development and provide the opportunity to dissect abnormal development.
Keywords: early development, embryo, ovary, testis
Global gene expression arrays demonstrate changes in gene expression in 34 human fetal ovary and testis samples between ∼9 and ∼19 weeks gestation.
INTRODUCTION
The development and function of the mammalian gonad have been studied for decades. The genetic control of gonadogenesis has been elucidated predominantly by examining single genes or small groups of genes. Sexual differentiation occurs in the human at around 6 wk of gestation [1] and begins ∼11.5 days postcoitum (dpc) in the mouse [2]. The critical role of the Sry gene in mammalian sexual differentiation was demonstrated by a series of classic experiments in the mouse [3–5]. At roughly the same developmental period, cells undergoing testicular differentiation as marked by Sry expression are also expressing Sox9 [6]. Furthermore, transgenic mice (XX) expressing Sox9 (Tg[Wt1-Sox9]92Asc) develop normal testicular cells [7], confirming the importance of Sox9 in testicular development. Other genes are also postulated to be involved in the process of sex determination, and these include Wt1 [8], Sf1 [9], Lhx9 [10], Nr0b1 (previously known as Dax1) [11], Dmrt1 [12], and Amh [13]. In a recent murine gonadal microarray study [14], genes known to be involved in sex determination such as Sry and Sox9 were highly expressed in the early male embryonic gonad compared with the female.
After differentiation of the testis, the gonad becomes involved in steroidogenesis, which appears to be gonadotropin independent [15]. In the investigation of global gene expression in the embryonic murine gonad [14], it was shown that the expression of transcripts encoding steroidogenic enzymes such as Star, Cyp17a1, Cyp11a1, and Hsd17b3 was increased in the latter stages of embryonic testicular development. In contrast, these changes were not present in the embryonic mouse ovary.
It has been long accepted that the “default” outcome of fetal gonadal differentiation is formation of the fetal ovary. However, ovarian changes in the fetal gonad are present early [16, 17], and it is now clear that expression of several genes, including Wnt4 [18] and Foxl2 [19], is critical to fetal ovarian development. Among the myriad genes known to be involved in early ovarian development and function [20] are Gdf9 [21], Nobox [22], Sohlh1, and Lhx8 [23]. The sheer number of genetic events that occur in the early ovary has caused investigators to question the passive nature of fetal ovarian development.
Most studies [24–28] of the human fetal ovary have centered on events occurring in meiosis I. These studies have focused on the pairing behavior of chromosomes during meiosis I in chromosomally normal and abnormal fetal oocytes. Critical to this process is the synaptonemal complex, a meiosis-specific structure that facilitates synapsis and meiotic recombination. Recently, it has been possible to directly visualize the synaptonemal complex and recombination proteins reliably in the human fetal oocyte [24–27]. To date, studies [28, 29] have revealed that the temporal appearance of recombination proteins in meiosis seems to be different in meiosis in the human fetal ovary and adult testis.
Genes known to be involved in meiosis are highly expressed in the mouse ovary, including Sycp1, Msh4 and Msh5, and Dmc1 [14]. Other genes associated with meiosis [30, 31] include Spo11 [32, 33], Stag3 [34], Sycp3 [35], Tex11 [36], and Tex14. Notably, a 100-fold increase of Stra8 was demonstrated in the fetal ovary coincident with the onset of meiosis (14.5 dpc) in the mouse array [14]. Expression of Stra8 is specific to the fetal ovary and the postnatal testis, and its role in mammalian meiosis has initiated further studies [37–40] of testicular gonadogenesis. Elimination of the gene for murine Stra8 resulted in a developmental block at the level of premeiotic replication and meiosis [41].
Examination of gene expression in the fetal ovary and testis of the human is critical to our understanding of testicular and ovarian gonadogenesis. Comparison of the human data with other mammalian models may further this understanding. However, access to sufficient and quality human fetal tissue has made it challenging to characterize events such as sex determination, ovarian meiosis, and gonadotropin-independent steroidogenesis in this species. Thus, this study provides a valuable resource by outlining the profiles of gene expression that occur in midgestation in the human ovary and testis.
As the number of unique global expression databases from various species increases, comparisons of conserved gene expression profiles can be made. This comparison will offer insight into novel species differences and generate a higher level of confidence in specific targets of various therapies that will likely be applicable to higher organisms such as humans. The development of the mammalian fetal gonad has been studied for more than half a century, but global molecular examination of this tissue has not been performed, to our knowledge. Examining patterns of gene expression at a genomic level during human fetal gonad development is necessary to better understand the processes that result in either a male or female phenotype, and this data set is certain to provide us with a platform to explore many issues in mammalian gonadal development and function.
MATERIALS AND METHODS
Tissue Collection and RNA Preparation
Gonads from human fetuses between 9 wk and 20 wk of gestation were obtained from the Department of Pathology and the Birth Defects Research Laboratory at the University of Washington. All samples were derived from elective abortions. Fetuses with known or suspected genetic disorders were excluded from consideration. Although karyotypic information was not generally available, we also excluded all cases known to involve chromosome abnormalities. Fetal sexing was done on the basis of morphology, and in all cases the sex assignment was confirmed on the arrays by appearance of known sex-specific markers described herein. All procedures were approved by the University of Washington and Washington State University institutional review boards, and informed consent was obtained from all study participants.
The limited availability of human fetal tissue precludes biological replicates for each time point, and this study attempted to obtain an even distribution of ovary and testis samples across the gestational ages that were available. Tissue was collected into ≥10 volumes of RNAlater and stored at 2–5°C until RNA extraction. Total RNA was extracted from the tissue following the protocol outlined in the Qiagen (Studio City, CA) RNeasy Protect Mini/Midi Kit. The RNA was quantified by NanoDrop spectrophotometry (Thermo Fisher Scientific Inc., Wilmington, DE) and evaluated for quality using the Agilent Technologies (Palo Alto, CA) 2100 Bioanalyzer.
Microarray Processing
Fifty nanograms of total RNA from each of the tissue samples was used to create the target for the microarray using the Ovation Biotin RNA Amplification and Labeling System by NuGEN (San Carlos, CA). The labeled cDNA was fragmented, hybridized to Human Genome U133A Plus 2.0 arrays (Affymetrix, Santa Clara, CA), and stained in accord with the manufacturer's standard protocol. The arrays were washed using the Affymetrix GeneChip Fluidics Station 400 and scanned using a GeneArray Scanner 2500A (Agilent Technologies). All reactions and microarray hybridization procedures were performed in the Laboratory for Biotechnology and Bioanalysis I at Washington State University as described previously for murine studies [14].
Quantitative RT-PCR
A two-step real-time RT-PCR was utilized to measure the expression of STRA8 [38–40]. Each RNA sample was analyzed in triplicate. STRA8 primers amplify a 126-base pair (bp) product (5′-CCTCAAAGTGGCAGGTTCTGAA-3′ and 5′-TCCTCTAAGCTGCTTGCATGC-3′); control ACTB primers amplify a 70-bp product (5′-GCGAGAAGATGACCCAGATCA-3′ and 5′-CACAGCCTGGATAGCAACGTAC-3′). Expression of STRA8 was normalized to ACTB expression. A baseline expression level was determined by calculating the mean expression level (values are mean ± SD throughout) of the testis samples across the time course (10.37 ± 0.73; n = 6). The expression level of each ovary sample (n = 8) was then calculated relative to this baseline.
Data Analysis
Analysis of data was conducted within the R statistical computing environment (www.R-project.org). Visual inspection of 49 scanned images and output from the “affyQCreport” package of BioConductor (www.bioconductor.org) revealed that 34 of 49 arrays were within acceptable limits. The 15 arrays that were unacceptable had significant degradation as determined by poor 3′/5′ ratios of affy control probe sets present on each array. For the remaining 34 arrays, probe sets were filtered on the following: probe sets not classified as present in any array were eliminated, and two separate normalizations of the data (MAS5 and RMA) were then performed. The background intensity values for each normalization routine were determined using probe sets labeled absent in all microarrays, and only those probe sets above the 99th percentile of the background were retained for further analysis.
To determine differential expression, pairwise comparisons were performed using the limma package of BioConductor. Significance was set at α = .05 with a Benjamini and Hochberg correction for a false discovery rate. Differential expression of a given probe was determined using only the RMA normalized data set after application of the previously described filters. For the array data, raw microarray data (CEL files) were imported into the statistical computing environment R using the BioConductor package affy, checked for quality, and subsequently preprocessed using the RMA algorithm from the affy package. In addition to R, GeneSpring (www.agilent.com/chem/genespring) software was utilized to facilitate the study of this large data set. Pathway and annotated cluster analysis was performed by using the functional annotation tools of DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov) [42]. Probe identifications ascertained in the microarray analysis as belonging to either pattern one or pattern two in one or both tissues were uploaded to DAVID Bioinformatics Resources, and their biological process annotation was defined by using the functional annotation tools. Only biological process gene ontology (GO) terms defined as significantly overrepresented by DAVID Bioinformatics Resources were included for further analysis.
Data from the RT-PCR studies were analyzed first using paired t-test to determine whether ovarian STRA8 within each gestational age differed in ovarian and testicular samples. ANOVA was used to determine the influence of gestational age on ovarian STRA8 expression. Post hoc analysis of the significant effect of gestational age in the ovarian samples was completed using Tukey test.
RESULTS
The data from 34 samples (17 ovary samples and 17 testis samples) that passed quality control parameters can be accessed through the National Center for Biotechnology Information via the Gene Expression Omnibus data repository (http://www.ncbi.nih.gov/geo/) accession numbers GSM387005–GSM387038. Each time point was represented by one sample except in the ovary, where there were three samples at 9.6 wk, two samples at 13.6 wk, and two samples at 16.9 wk. In the testis, there were two samples at 11 wk, two samples at 13.9 wk, and two samples at 16.1 wk. The average raw signal was used in the figures herein.
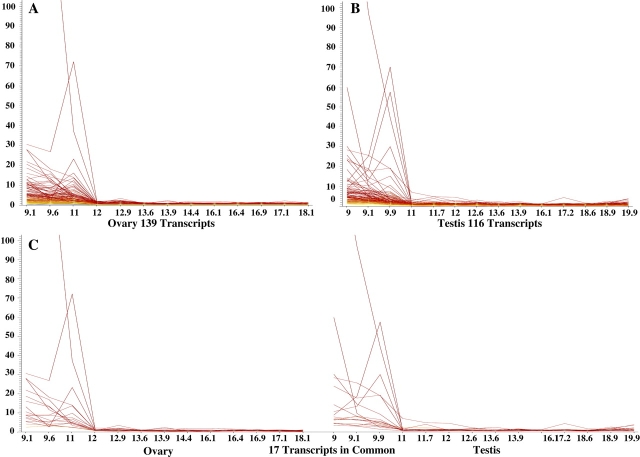
The Human Genome U133A Plus 2.0 array represents 54 613 probe sets. One approach to deal with a data set this large is to determine if there are any patterns of expression visible across the time course in each tissue. To accomplish this, a K-means clustering analysis was performed within GeneSpring. Next, it was important to examine the individual transcripts within those expression patterns and to determine which genes were significantly and differentially expressed. The definition used to meet the criterion of being “differentially expressed” was that the raw signal was ≥50 for at least one time point, the transcript level was increased or decreased at least 2-fold during the time course, and the changes were statistically significant (P < 0.05). Two similar patterns of expression were observed in each tissue (Figs. 1 and 2). Pattern one represents probe sets that were present at an elevated level at 9 wk of gestation and then decreased at least 2-fold throughout the remainder of the time course (Fig. 1, A–C). In the ovary, 139 probe sets exhibited this pattern of expression (Fig. 1A). In the testis, 116 probe sets exhibited a similar pattern (Fig. 1B). Of all the probe sets in pattern one, 17 were expressed in both the ovary and testis (Fig. 1C).
FIG. 1.
Probe sets with at least 2-fold increase in signal intensity from 9 to 11 wk of gestation to 12 to 18 wk of gestation. These changes were statistically significant (P < 0.05). This pattern was observed in 139 ovary probe sets (A) and in 116 testis probe sets (B). C) There were 17 probe sets that shared this pattern of expression in both the ovary and testis. The vertical axis represents normalized intensity, and the horizontal axis represents weeks of gestation.
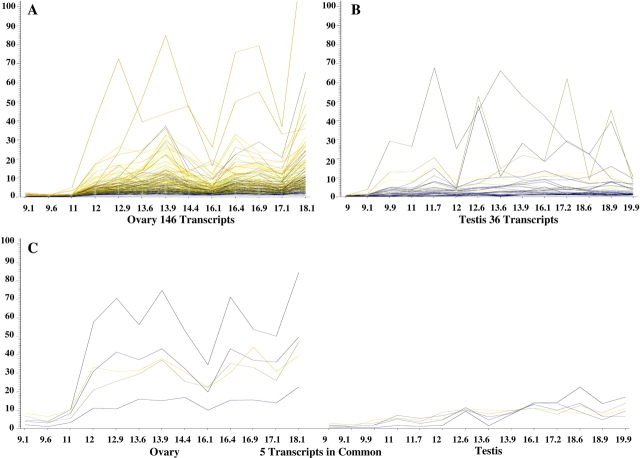
FIG. 2.
Probe sets with at least 2-fold increase in signal intensity from 9 to 11 wk of gestation to 12 to 18 wk of gestation. These changes were statistically significant (P < 0.05). This pattern was seen in 146 ovary probe sets (A) and in 36 testis probe sets (B). C) There were five probe sets that shared this pattern of expression in both the ovary and testis. The vertical axis represents normalized intensity, and the horizontal axis represents weeks of gestation.
Analysis of the probe sets using DAVID Bioinformatics Resources was performed. Of 139 probe sets representing pattern one in the ovary, 92 probe sets were associated with significantly overrepresented GO terms in a biological process. The majority of these ovarian transcripts had functions associated with development (n = 44) and cell communication (n = 38). Of 116 probe sets representing pattern one in the testis, 76 were associated with significantly overrepresented GO terms in a biological process. The majority of these testicular transcripts had functions associated with a developmental process (n = 25) and localization (n = 25), specifically transport (n = 21). Of 17 probe sets shared between the two tissues, 15 probe sets were associated with significantly overrepresented GO terms, with the most common process being multicellular organismal development (n = 6).
A second expression pattern was evident from the clustering analysis. This pattern depicted probe sets that had low levels of expression early in the developmental time course and then increased significantly (Fig. 2, A–C). In the fetal ovary, 146 probe sets were present at low levels at 9.1–11 wk of gestation; after 12 wk, they increased at least 2-fold and were elevated for the remainder of the time course (Fig. 2A). Analysis of these probe sets with DAVID Bioinformatics Resources revealed that the majority of these genes fell into the following four categories of function: biopolymer metabolic process (n = 39), including RNA biosynthetic processing; transcription (n = 22); reproduction (n = 14), specifically gamete generation; and cell cycle processes (n = 14) such as meiosis.
In the testis, 36 probe sets were expressed at a low level at 9 wk and then increased at least 2-fold after 12 wk (Fig. 2B). Twenty-one probes were associated with significantly overrepresented GO terms, most commonly a biosynthetic process (six probes) (such as steroidogenesis [three probes]). Of all the probe sets in this “low to high” pattern, only five were common to both the ovary and testis (Fig. 2C). Ontological analysis of these shared probe sets revealed functions associated with protein modification, development, and DNA metabolic processes. All GO output data, including descriptions of GO terms and pathways and P values in addition to complete lists of probe sets shown in Figures 1, 2, 4, and 5, are available on the Griswold Lab Microarray Data Site (http://www.wsu.edu/∼griswold/microarray/). Contact information is provided on the site, and inquiries about all of the Griswold array data are welcome.
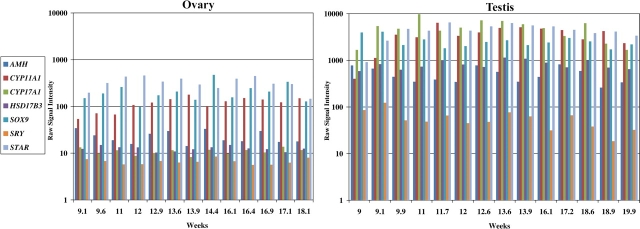
FIG. 4.
Ovarian and testicular expression profiles of transcripts known to be involved in sex determination, organogenesis, and steroidogenesis. The vertical axis represents raw signal intensity, and the horizontal axis represents weeks of gestation.
Known Biological Processes and Sex-Specific Expression
This study outlines the human fetal gonadal transcriptome during midgestation (∼9–20 wk). A number of known biological processes had either already occurred (sex determination at 6 wk) and were over, were about to begin (meiosis in the ovary at 11–12 wk), or began at sex determination and were still ongoing (organogenesis and steroidogenesis in the testis). The expression profiles of a few tissue-specific transcripts known to be involved in these processes and present on the human array are shown in Figures 4 and 5. Although sex determination was initiated ∼4 wk before the earliest time points in this study, the transcript level for SRY was still elevated in the testis at 9 wk (raw signal 123) before dropping to low levels by Week 11 (raw signal 48) (Fig. 4). SRY transcript levels were negligible in the ovary (raw signal 6). SOX9 transcript was ∼27-fold higher in the testis than the ovary and maintained a high level throughout testis development. Upon sex determination, the fetal testis begins producing antimüllerian hormone (AMH) and androgen. The transcript for AMH and a number of transcripts involved in steroidogenesis (CYP17A1, CYP11A1, STAR, and HSD17B3) were observed to be elevated in the testis throughout the time course (Fig. 4).
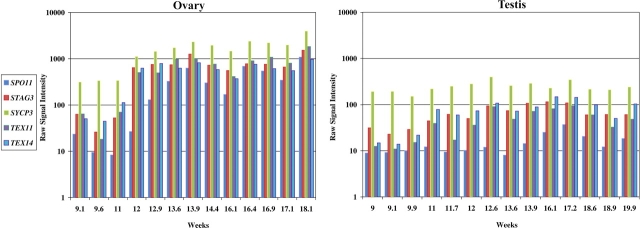
FIG. 5.
Ovarian and testicular expression profiles of transcripts known to be involved in meiosis. The vertical axis represents raw signal intensity, and the horizontal axis represents weeks of gestation.
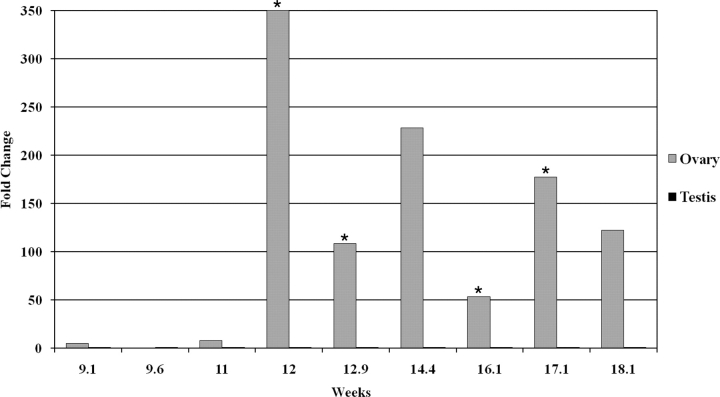
Meiosis is initiated at 11–12 wk in the human ovary, and STRA8 is expected to be a marker for the initiation of the meiotic program. Unfortunately, STRA8 is not present on the human array used in this study. Therefore, to identify the initiation of meiosis, STRA8 transcript levels were measured by quantitative RT-PCR in a number of fetal ovarian and testicular samples (Fig. 3). At all time points, STRA8 was expressed at a significantly higher level in the ovary (P < 0.01). Furthermore, ovarian gene expression varied significantly over the gestational period examined. STRA8 transcript levels were low in early gestation (9.1 wk and 11 wk) in the human ovary, but by 12 wk of gestation, STRA8 transcript was expressed at a level exceeding 50 times the expression in early gestation. This expression remained elevated during the remainder of the gestational period examined (P < 0.01). In addition to STRA8, a large number of transcripts related to meiosis (SPO11, SYCP3, STAG3, TEX11, and TEX14) were elevated after 12 wk in the ovary and remained elevated for the remainder of the period. These transcripts were expressed at significantly lower levels in the fetal testis (Figs. 3 and 5).
FIG. 3.
Quantitative RT-PCR of STRA8 message levels in human embryonic gonads. Depicted are fold changes normalized to ACTB expression. Welch t-test was used to determine the statistical significance between testis and ovary, and all samples had P < 0.01. Within each tissue, values noted by an asterisk were significantly different (P < 0.01) from the previous gestational age (ANOVA with Tukey post hoc test). A baseline expression level was determined by calculating the mean expression level of the testis samples across the time course (10.37 ± 0.73; n = 6). The expression level of each ovary sample was then calculated relative to this baseline.
DISCUSSION
The genetic control of mammalian gonadal development and sex determination continues to be extensively studied [43]. Global gene expression studies [14, 44–46] via microarray analysis have a critical role in furthering our understanding of gonadal function before and after birth. To our knowledge, this study represents the first report of global gene expression in the human fetal ovary and testis.
Two major patterns of expression were evident in the fetal ovary and testis. Several probe sets that were elevated early in the time course (∼9 wk) and then over the next few weeks dropped to low levels and remained at low levels throughout the remainder of the time course. Ontological classification of the transcripts in both tissues pointed toward involvement in cell differentiation and structure, which is not surprising because this period is just after sex determination and the gonads are rapidly increasing in size and changing morphology.
A second pattern of expression emerged showing probe sets that were low at Week 9 and increased significantly at Week 12 and remained high. This group was composed of transcripts that, in contrast to the first pattern, were tissue specific and were associated with known biological processes. Many of the ovarian transcripts were strongly associated with meiosis and related processes. A similar pattern was seen with the STRA8 expression profile (i.e., minimal ovarian expression until Week 12 and later). As expected, transcripts involved in meiosis were not expressed in the fetal testis.
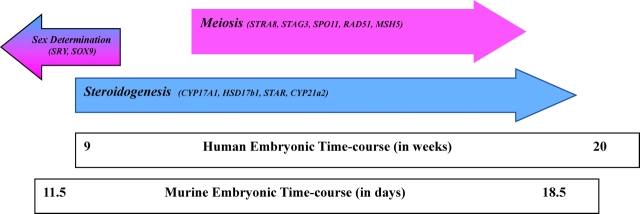
The second pattern of expression in the testis showed many of the probe sets to be involved in steroidogenesis. It is to be expected that the testis in midgestation is involved in steroidogenesis. This classification was further supported by independently examining the expression profiles of several transcripts known to be involved in steroidogenesis and by noting a similar elevated expression during the same period. Thus, the same biological process and the time frame of activity were identified by the following two independent approaches: 1) clustering and pattern analysis, followed by ontology analysis, and 2) plotting expression profiles of specific transcripts known to be involved in biological processes. With these results, it becomes fairly easy to overlay the timing of events occurring within the human fetal gonads with the murine model (Fig. 6). With the generation of global gene expression data sets comes the immediate desire to perform direct comparisons between species. Unfortunately, these comparisons are limited because of chip design, annotation, and availability. For example, although the Murine 430 2.0 affy array harbors 45 037 transcripts and the Human Genome U133A Plus 2.0 array represents 54 613 transcripts, only ∼24 181 transcripts are present on both chips. We were able to present a limited comparison of a few selected genes and common biological processes, but the lack of a more complete overlap between the mouse and human chips made a comprehensive comparison impossible (http://www.wsu.edu/∼griswold/microarray/).
FIG. 6.
Three embryonic processes mapped to the murine time course (in days) and to the human time course (in weeks). Depicted are sex determination in the male and female (blue or pink), meiosis in the female (pink), and steroidogenesis in the male (blue). Transcripts previously reported to be involved in each process and also found in this study are included.
The creation and availability of global gene expression data sets are becoming more and more common. Improvements in technology have alleviated the need for the copious amount of RNA that was originally required to complete such a study. Instead, obtaining access and availability to the actual organism providing the tissue may now be one of the most limiting factors. The tissue used in this study was made available as it was collected in a clinical setting. As such, the time course of this study was driven by what was available, making it impossible to obtain the biological “duplicates” and “triplicates” that would normally constitute the sample set. The lack of biological replicates for all samples, in addition to the innate biological variation observed with higher mammals such as humans, ultimately leads to a data set that requires an analysis of trends of gene expression over time rather than the drawing of conclusions from one or two time points. Early array data were typically confirmed using RT-PCR of select transcripts. However, with advances in array procedures and chip design, close scrutiny of the countless public databases by independent researchers has revealed these data to be extremely representative and in agreement with RT-PCR. Although these data have proven to be dependable, investigators who choose to utilize these data need to assume responsibility for additional work.
The availability of a time course to monitor global gene expression during the development of the embryonic testis and ovary is an important step in understanding the molecular and physiological changes necessary to elicit two distinct and normal reproductive tissues. Analysis of the human time course and comparison with the mouse database produced both confirmation and elaboration of previously known events occurring during embryonic gonad development, including sex determination, meiosis in the ovary, and steroidogenesis in the testis.
Acknowledgments
The authors would like to thank Derek Pouchnik and the Laboratory for Bioanalysis and Biotechnology I for GeneChip processing; and Peter Nelson, Fred Hutchinson Cancer Research Center (Seattle, WA), for assistance with the RNA quantification and quality assessment.
Footnotes
1Supported by HD 10808, U54 HD 42454 from NICHD and NIH R01 HD21341.
REFERENCES
- Ostrer H. Sex Determination. Philadelphia:: Lippincott Williams & Wilkins;; 1996. [Google Scholar]
- Byskov AG.Differentiation of mammalian embryonic gonad. Physiol Rev 1986; 66: 71–117. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R.A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990; 346: 245–250. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R.Male development of chromosomally female mice transgenic for Sry. Nature 1991; 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R.Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 1990; 348: 450–452. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R.Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet 1996; 14: 62–68. [DOI] [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, de Rooij DG, Schedl A.Sox9 induces testis development in XX transgenic mice. Nat Genet 2001; 28: 216–217. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Englert C.The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev 2002; 16: 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL.A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994; 77: 481–490. [DOI] [PubMed] [Google Scholar]
- Birk OS, Casiano DE, Wassif CA, Cogliati T, Zhao L, Zhao Y, Grinberg A, Huang S, Kreidberg JA, Parker KL, Porter FD, Westphal H.The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 2000; 403: 909–913. [DOI] [PubMed] [Google Scholar]
- Meeks JJ, Crawford SE, Russell TA, Morohashi K, Weiss J, Jameson JL.Dax1 regulates testis cord organization during gonadal differentiation. Development 2003; 130: 1029–1036. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D.Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev 2000; 14: 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA.Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell 1994; 77: 651–661. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD.Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod 2005; 72: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gehani F, Zhang FP, Pakarinen P, Rannikko A, Huhtaniemi I.Gonadotropin-independent regulation of steroidogenesis in the fetal rat testis. Biol Reprod 1998; 58: 116–123. [DOI] [PubMed] [Google Scholar]
- Di Carlo AD, Travia G, De Felici M.The meiotic specific synaptonemal complex protein SCP3 is expressed by female and male primordial germ cells of the mouse embryo. Int J Dev Biol 2000; 44: 241–244. [PubMed] [Google Scholar]
- Peters H.Migration of gonocytes into the mammalian gonad and their differentiation. Philos Trans R Soc Lond B Biol Sci 1970; 259: 91–101. [DOI] [PubMed] [Google Scholar]
- Heikkila M, Prunskaite R, Naillat F, Itaranta P, Vuoristo J, Leppaluoto J, Peltoketo H, Vainio S.The partial female to male sex reversal in Wnt-4-deficient females involves induced expression of testosterone biosynthetic genes and testosterone production, and depends on androgen action. Endocrinology 2005; 146: 4016–4023. [DOI] [PubMed] [Google Scholar]
- Ottolenghi C, Omari S, Garcia-Ortiz JE, Uda M, Crisponi L, Forabosco A, Pilia G, Schlessinger D.Foxl2 is required for commitment to ovary differentiation. Hum Mol Genet 2005; 14: 2053–2062. [DOI] [PubMed] [Google Scholar]
- Pepling ME.From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 2006; 44: 622–632. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM.Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 1996; 383: 531–535. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM.NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 2004; 305: 1157–1159. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A.Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A 2006; 103: 8090–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EY, Chen YJ, Bonnet G, Gartler SM.An analysis of meiotic pairing in trisomy 21 oocytes using fluorescent in situ hybridization. Cytogenet Cell Genet 1998; 80: 48–53. [DOI] [PubMed] [Google Scholar]
- Cheng EY, Chen YJ, Gartler SM.Chromosome painting analysis of early oogenesis in human trisomy 18. Cytogenet Cell Genet 1995; 70: 205–210. [DOI] [PubMed] [Google Scholar]
- Cheng EY, Naluai-Cecchini T.FISHing for acrocentric associations between chromosomes 14 and 21 in human oogenesis. Am J Obstet Gynecol 2004; 190: 1781–1787. [DOI] [PubMed] [Google Scholar]
- Cheng YE, Gartler SM.A fluorescent in situ hybridization analysis of X chromosome pairing in early human female meiosis. Hum Genet 1994; 94: 389–394. [DOI] [PubMed] [Google Scholar]
- Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE.Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet 2005; 76: 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Judis L, Chan ER, Schwartz S, Seftel A, Lynn A.Cytological studies of meiotic recombination in human males. Cytogenet Genome Res 2004; 107: 249–255. [DOI] [PubMed] [Google Scholar]
- Svetlanov A, Cohen PE.Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp Cell Res 2004; 296: 71–79. [DOI] [PubMed] [Google Scholar]
- Vallente RU, Cheng EY, Hassold TJ.The synaptonemal complex and meiotic recombination in humans: new approaches to old questions. Chromosoma 2006; 115: 241–249. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S.Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 2000; 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD.The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Prieto I, Suja JA, Pezzi N, Kremer L, Martinez AC, Rufas JS, Barbero JL.Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol 2001; 3: 761–766. [DOI] [PubMed] [Google Scholar]
- Roig I, Liebe B, Egozcue J, Cabero L, Garcia M, Scherthan H.Female-specific features of recombinational double-stranded DNA repair in relation to synapsis and telomere dynamics in human oocytes. Chromosoma 2004; 113: 22–33. [DOI] [PubMed] [Google Scholar]
- Adelman CA, Petrini JH.ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 2008; 4: e1000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC.Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A 2008; 105: 14976–14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC.Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A 2006; 103: 2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD.Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 2008; 78: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD.Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC.In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 2006; 38: 1430–1434. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA.DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: eP3 [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P.Sex determination and gonadal development in mammals. Physiol Rev 2007; 87: 1–28. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD.The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319–330. [DOI] [PubMed] [Google Scholar]
- Herrera L, Ottolenghi C, Garcia-Ortiz JE, Pellegrini M, Manini F, Ko MS, Nagaraja R, Forabosco A, Schlessinger D.Mouse ovary developmental RNA and protein markers from gene expression profiling. Dev Biol 2005; 279: 271–290. [DOI] [PubMed] [Google Scholar]
- Kocabas AM, Crosby J, Ross PJ, Otu HH, Beyhan Z, Can H, Tam WL, Rosa GJ, Halgren RG, Lim B, Fernandez E, Cibelli JB.The transcriptome of human oocytes. Proc Natl Acad Sci U S A 2006; 103: 14027–14032. [DOI] [PMC free article] [PubMed] [Google Scholar]