Abstract
Mice that are ets variant gene 5 (ETV5) null (Etv5−/−) undergo the first wave of spermatogenesis but lose all spermatogonial stem cells (SSCs) during this time. The SSC loss in Etv5−/− mice begins during the neonatal period, suggesting a role for ETV5 in SSC self-renewal during this period. Herein, we show that Etv5 mRNA was present in perinatal mouse testis and that ETV5 was expressed in fetal Sertoli cells and by germ cells and Sertoli cells during the neonatal period. Transplantation of Etv5−/− germ cells failed to establish spermatogenesis in W/Wv mice testes, indicating that germ cell ETV5 has a key role in establishment or self-renewal of transplanted SSCs. The SSC self-renewal is stimulated by glial cell-derived neurotrophic factor (GDNF) acting through the RET/GDNF family receptor alpha 1 (GFRA1) receptor complex in SSCs. Immunohistochemistry, quantitative PCR, and laser capture microdissection revealed decreased RET mRNA and protein expression in spermatogonia of neonatal Etv5−/− mice by Postnatal Days 4–8, indicating that disrupted GDNF/RET/GFRA1 signaling may occur before initial spermatogonial stem/progenitor cell decrease. Etv5−/− spermatogonia had reduced proliferation in vivo and in vitro. Decreased cell proliferation may cause the observed decreases in the number of type A spermatogonia (Postnatal Day 17) and daily sperm production (Postnatal Day 30) in Etv5−/− mice, indicating quantitative impairments in the first wave of spermatogenesis. In conclusion, ETV5 is expressed beginning in fetal Sertoli cells and can potentially have effects on neonatal Sertoli cells and germ cells. In addition, ETV5 has critical effects on neonatal spermatogonial proliferation, which may involve impaired signaling through the RET receptor.
Keywords: ERM, first wave, GDNF, GFRA1, Sertoli cells, spermatogenesis, spermatogonial stem cells, testis
Neonatal expression of ETV5 in mouse testis is essential for spermatogonial stem cell maintenance and normal spermatogenesis.
INTRODUCTION
The regulation of spermatogonial stem cell (SSC) maintenance, self-renewal, and differentiation is critical for normal spermatogenesis and has significant clinical applicability in understanding some types of infertility. These cells also have the potential to generate somatic tissues [1, 2] that can potentially be used therapeutically, further emphasizing the importance of understanding SSC regulation.
During spermatogenesis, germ cell production is maintained by the self-renewal and differentiation of SSCs. These cells are located along the basement membrane of the seminiferous tubule and are surrounded by Sertoli cells, the somatic cells of the seminiferous epithelium. Together with the basement membrane, Sertoli cells form the stem cell niche by providing physical support for the SSCs and produce factors essential for SSC maintenance [3–5].
Recent investigations using mice with targeted disruption of the transcription factor ets variant gene 5 (ETV5; also known as ets-related molecule, or ERM) have shown that this molecule is involved in testicular SSC maintenance [6]. ETV5 knockout (Etv5−/−) mice undergo a first wave of spermatogenesis during juvenile life that appears normal. However, by the end of the first week of neonatal life, Etv5−/− mice have decreased glial cell-derived neurotrophic factor (GDNF) family receptor alpha 1 (GFRA1)-positive spermatogonia compared with controls, indicating reductions in spermatogonial stem/progenitor cells [7]. This decrease progresses during juvenile life, eventually resulting in total loss of SSCs and a Sertoli cell-only phenotype [6] in the adult seminiferous tubules. Although the first wave of spermatogenesis in Etv5−/− mice appears to be morphologically normal [6], a thorough analysis assessing the efficiency of this process has not been performed.
In recent years, valuable insights have been gained into factors that regulate SSC maintenance and self-renewal. GDNF, a distant member of the transforming growth factor β family, is secreted by Sertoli cells. GDNF signals through the receptor tyrosine kinase RET present on spermatogonial stem/progenitor cells [8] and requires a ligand-specific coreceptor, GFRA1, which is anchored to the plasma membrane [9–11]. RET signaling causes the secondary activation of important signaling pathways essential for SSC self-renewal [12, 13]. Loss of any of the components of the GDNF/RET/GFRA1 signaling pathway results in loss of SSCs [8, 14]. These in vivo studies were corroborated by in vitro data demonstrating the importance of this pathway for SSC self-renewal [12, 15].
Sertoli cells of Etv5−/− mice express GDNF at a level equal to that of wild-type (WT) mice [6], indicating that ETV5 does not directly or indirectly regulate GDNF. However, the phenotypic similarities between mice lacking ETV5 or GNDF suggest the possibility of overlap or commonality in their downstream mechanisms of action.
Analysis of expression of ETV5 mRNA and protein indicated that ETV5 was strongly expressed in Sertoli cells beginning at 3–4 wk onward and persisting in adults [6]. Subsequent work [16, 17] indicated that ETV5 was also produced by SSCs. We have recently determined that, although initial development of the Etv5−/− testis is normal with no differences in the number of GFRA1-positive spermatogonia at Postnatal Day 4 compared with that in WT testes, all spermatogonial stem/progenitor cells are lost by age 5 wk [7]. The initiation of SSC loss in Etv5−/− mice by Postnatal Week 1 strongly suggests that ETV5 is produced during fetal and neonatal periods and has an important role in establishment and proliferation of SSCs during this time frame.
The objectives of this study were to investigate the expression and mechanism of action of ETV5 in neonatal WT testes. Our results indicate that ETV5 expression begins in fetal Sertoli cells and neonatal gonocytes of the germ cell lineage. Furthermore, loss of ETV5 results in decreased RET expression with possible inhibition of downstream GDNF/RET/GFRA1 signaling. This may be one of the mechanisms by which loss of ETV5 disrupts SSC self-renewal, leading to a quantitative decrease in the number of germ cells during the first wave of spermatogenesis and ultimate loss of all SSCs.
MATERIALS AND METHODS
Animal Care and Husbandry
129Sv/Ev WT mice were bred and maintained in our mouse colony as previously described [7]. Etv5−/− mice on a 129Sv/Ev background [6] were produced by breeding Etv5+/− mice, and the pups were genotyped as described earlier [7]. All were housed at 25°C with a 12L:12D photoperiod and were given water and a standard rodent diet ad libitum. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Illinois and conducted in accord with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
RNA Isolation and Real-Time PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocols. The concentration and purity of RNA were estimated using a Gene Quant Pro spectrophotometer (Amersham Biosciences, Uppsala, Sweden) before analytical procedures were performed. First-strand cDNA was synthesized from total RNA (1 μg) using Superscript RT and random primers (Invitrogen, Carlsbad, CA). Real-time PCR was performed with the ABI Prism 7000 Sequence detection system using validated ABI Taqman gene expression assays (Applied Biosystems, Foster City, CA). The expression value of each gene was normalized to the amount of an internal control gene (18S ribosomal RNA) cDNA to calculate a relative amount of RNA in each sample. The expression value of each gene in a WT control was arbitrarily defined as 1 U. All assays were carried out in triplicate for each target mRNA, and the normalized expression values for all control and treated samples were averaged. A relative quantitative fold change was determined using the ρρCt method (ABI Chemistry Guide No. 4330019). ABI Taqman gene expression assays used for specific transcripts were Mm00465816_m1 (Etv5) and Mm00436304_m1 (Ret).
Immunohistochemistry for ETV5, RET, and MKI67
Testes were fixed in 10% (v/v) neutral buffered formalin. After fixation, tissues were embedded in paraffin, sectioned at 5 μm, and mounted on glass slides. Tissue sections were deparaffinized in xylene, rehydrated through a graded series of ethanol, and washed with distilled water. Endogenous peroxidase activity was blocked, and antigen retrieval was performed as described previously [18].
For ETV5 immunohistochemistry, sections were blocked with 10% normal goat serum for 30 min, followed by incubation with the primary antibody (ETV5 3H7 hamster monoclonal clone 7; Dr. K. Murphy, Washington University, St. Louis, MO) at 4°C overnight, rinsed in PBS, and incubated with goat anti-hamster biotinylated secondary antibody at room temperature for 60 min. Slides were rinsed again and incubated for 30 min with avidin-biotin-peroxidase complex staining system (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) and then stained with 3,3′-diaminobenzidine and counterstained with hematoxylin (Sigma-Aldrich, St. Louis, MO).
For RET immunohistochemistry, the slides were washed with PBS and incubated for 30 min in 10% (v/v) normal rabbit serum. After serum block, the sections were incubated for 1.5 h with anti-RET (1:100, goat polyclonal; Neuromics, Bloomington, MN). The sections were washed with PBS and incubated for 1 h with biotinylated anti-goat antibody (Vector Laboratories). Labeling was visualized by incubation with horseradish peroxidase-streptavidin (Zymed Laboratories, San Francisco, CA) for 30 min, followed by immersion in 3-amino-9-ethylcarbazole solution (Zymed Laboratories); slides were then counterstained with hematoxylin and mounted.
Immunohistochemistry for MKI67 was performed using a mouse anti-human monoclonal IgG to human MKI67 (1:1000; BD Transduction Laboratories, Lexington, KY) and biotinylated anti-mouse antibody (Vector Laboratories) as described previously [18]. Microscopic images were obtained digitally using a ProgResC3 camera (Jenoptik, Jena, Germany) and compiled using Adobe Photoshop (Adobe Systems, San Jose, CA).
Determination of Spermatogonial Proliferation In Vivo
To assess early spermatogonial proliferation in WT and Etv5−/− mice, 8-day-old mice testes (n = 4 for WT and n = 3 for Etv5−/−) were embedded in paraffin, and 2–4 pairs of serial 3-μm sections were stained for RET, which is expressed at high levels in spermatogonial stem/progenitor cells [8, 19], and for a cell proliferation marker, MKI67. Following immunostaining, tissues were counterstained with Mayer hematoxylin. The percentage of proliferating type A spermatogonia was determined as the number of cells per testis serially costained with MKI67 and RET divided by the total number of RET-positive cells (56–100 cells were counted per animal).
Enrichment of Spermatogonial Stem/Progenitor Cells
For each experiment, spermatogonial stem/progenitor cells were isolated from the testes of 4- to 5-day-old WT and Etv5−/− mice (n = 4 for each group). Testes were decapsulated, and Leydig cells and peritubular myoid cells were eliminated by a two-step enzymatic digestion [12]. Briefly, testes were suspended in Dulbecco modified Eagle medium (DMEM)/F12 media containing collagenase (1.5 mg/ml) and DNase (1 μg/ml) and then incubated at 33°C for 10 min in a shaking water bath at 100 cycles/min. After three washes in DMEM/F12, seminiferous cord fragments, devoid of Leydig cells and other interstitial cells, were incubated in this medium containing collagenase (1.5 mg/ml), hyaluronidase (1.5 mg/ml), trypsin (0.5 mg/ml), and DNase (1 μg/ml) for 20 min under the conditions already described. All tissue culture reagents were obtained from Hyclone/Fisher Scientific, Pittsburgh, PA, and enzymes were from Sigma-Aldrich. Dispersed cells (Sertoli cells, peritubular myoid cells, and germ cells) were washed twice and then incubated in DMEM/F12 containing 10% fetal calf serum (FCS) for 4 h at 33°C for differential plating [12, 20]. Differential plating was done by seeding the mixed population at a concentration of 1 × 105 cells/cm2 in FCS-coated 35-mm-diameter culture dishes or 24-well plates (both from Invitrogen). Sertoli cells and myoid cells attached to the culture plates. The spermatogonia remained in suspension and were collected and washed in DMEM/F12 before culturing. Purity of the spermatogonial population was evaluated by staining for VASA and was 70%–80%.
Culture of Spermatogonial Stem/Progenitor Cells
After enrichment, single-cell suspensions from WT and Etv5−/− testes were plated in microtiter wells (1 or 2 × 104 cells/well) and cultured in triplicate in minimal media (DMEM/F12 with 10% Nu serum; Fisher Scientific, Pittsburgh, PA) alone or in this media with 50 ng/ml rat GDNF (R & D Systems, Minneapolis, MN), or with 50 ng/ml rat GDNF, 300 ng/ml rat GFRA1-Fc fusion protein (R & D Systems), and 1 ng/ml human FGF2 (BD Biosciences/Pharmingen, San Diego, CA) [15]. To eliminate residual adherent somatic cells, after 3 days of culture spermatogonia were gently flushed, carefully removed from the wells, and replated in new wells containing fresh media and growth factors. To allow controlled conditions, no feeder layers were used [12]. The spermatogonia were cultured for another 3 days and then observed in phase-contrast microscopy, and the number of colonies (i.e., groups of >10 cells) was counted [12]. The results were expressed as the number of colonies obtained per 2 × 104 cells seeded and represented the means of triplicate repetitions (five separate experiments).
Donor Cell Preparation
Testes were harvested from 5-day-old Etv5−/− or WT littermate pups. Testes were decapsulated and digested in DMEM containing 0.05 mg/ml DNase I, 0.25 mg/ml trypsin, and 0.5 mg/ml collagenase type IV (all from Sigma-Aldrich) for 30 min at 37°C with pipette mixing every 10 min. Cells were allowed to settle for 10 min on ice, supernatant was removed, and fresh digestion media were added. Cells were incubated at 37°C for 15 min with pipette mixing every 5 min and were centrifuged at 500 × g for 4 min; the supernatant was removed, and the pellet was resuspended in DMEM containing 0.3 mg/ml soybean trypsin inhibitor (Sigma-Aldrich). Cells were centrifuged and washed thrice in DMEM before counting and resuspension in DMEM at 8.6 × 106 cells/ml. Cells were kept on ice until transplanted. One part trypan blue (0.04%) was added to nine parts cell suspension before each transplant to aid visualization.
Testicular Transplants
Recipient mice were 54-day-old W/Wv mice (Jackson Laboratories, Bar Harbor, ME). W/Wv mice have mutations in both Kit alleles. Endogenous germ cells are thus unable to respond to stem cell factor produced by Sertoli cells, and these mice lack spermatogenesis, despite having normal Sertoli cells [21]. This lack of endogenous spermatogenesis allows the W/Wv to serve as hosts for germ cell transplantation, as any spermatogenesis observed is due to transplanted germ cells [22]. Recipient mice were anesthetized, and approximately 7 μl of a single-cell suspension of WT (n = 6 testes) or Etv5−/− (n = 10 testes) was infused into the seminiferous tubules of each testis via the efferent ductules [21]. Sixty days after transplantation, testes were perfusion fixed with cold 10% (v/v) neutral buffered formalin and then embedded in paraffin. Blocks were serially sectioned at 5 μm with every 10–15 sections evaluated (30–40 sections/half testis). All tubules within a section were evaluated. Slides were stained with periodic acid-Schiff (PAS)-hematoxylin and evaluated for the presence or absence of spermatogenesis.
Tissue Collection and Cryopreservation for Laser Capture Microdissection
Testes from 5-day-old WT and Etv5−/− mice (n = 6 for each) were embedded in embedding compound (Tissue-Tek, Torrance, CA) and stored at −80°C. Frozen tissue sections (7 μm) were made in a cryostat, and the slides were stored at −80°C until immunohistochemistry.
GCNA1 Staining of Germ Cells for Laser Capture Microdissection
We used rat monoclonal anti-germ cell nuclear antigen 1 (GCNA1), kindly provided by Dr. George Enders (University of Kansas Medical Center, Kansas City, KS), as a marker for microdissection of spermatogonia from 5-day-old neonatal testes [23]. GCNA1 was used because of its high specificity, which allowed for a short incubation time during the staining process. Testes sections were thawed for 30 sec and stained with GCNA1 antibody for 3 min, immediately followed by incubation with Alexa Fluor 488 anti-rat secondary antibody (Invitrogen) for 30 sec. Sequential dehydration was performed (70% ethanol for 30 sec, 95% ethanol for 30 sec, and 100% ethanol for 1 min), followed by placement in histological grade xylene for 1 min [24]. Tissue sections were then immediately transferred for laser capture microdissection (LCM).
LCM and RNA Extraction
The LCM was performed at the core facilities of the Institute for Genomic Biology at the University of Illinois at Urbana-Champaign. Microdissection and capture of spermatogonia were done on an Arcturus Veritas microdissection instrument (MDS Analytical Technologies, Sunnyvale, CA). Approximately 100 cells were collected from every sample. The parameters used for LCM were 5-μm spot size, 50-mW power, and 10-msec pulse duration. Total RNA from the captured spermatogonia was isolated with the PicoPure RNA Isolation kit (Arcturus Veritas; MDS Analytical Technologies) according to the manufacturer's instructions. RNA quantity was measured by using the NanoDrop system (NanoDrop Technologies, Wilmington, DE). Real-time PCR was performed as already described.
Enumeration of Type A Spermatogonia and Measurement of Daily Sperm Production
For type A spermatogonial counts, the testes were embedded in glycol methacrylate, and sections were stained with PAS-hematoxylin. Type A spermatogonia were identified as spermatogonia if they had one flattened surface resting on the basal lamina and no heterochromatin. Sertoli cells were identified by their characteristic tripartite nucleolus. The numbers of type A spermatogonia and Sertoli cells in 80–100 seminiferous tubules in 17-day-old WT and Etv5−/− testes (n = 3) were counted, and data are presented as the number of type A spermatogonia/Sertoli cells. For daily sperm production (DSP), WT and Etv5−/− testes (n = 4) were homogenized for 3 min in 10 ml of physiological saline containing 0.05% (v/v) Triton X-100 (Sigma-Aldrich) using a Semi-Micro Waring blender (Waring, Torrington, CT). Two hundred microliters of homogenate was diluted with 300 μl of saline and 500 μl of 4% trypan blue; the stained spermatids were counted using a hematocytometer, and DSP was estimated as described previously [25].
Statistical Analysis
Data were analyzed using Student t-test and are presented as the mean ± SEM except for the LCM experiment, in which paired t-test was used. Differences were considered significant at P < 0.05. Statistical analysis was performed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA) or Microsoft Excel (Microsoft, Redmond, WA).
RESULTS
ETV5 Expression During Testicular Development
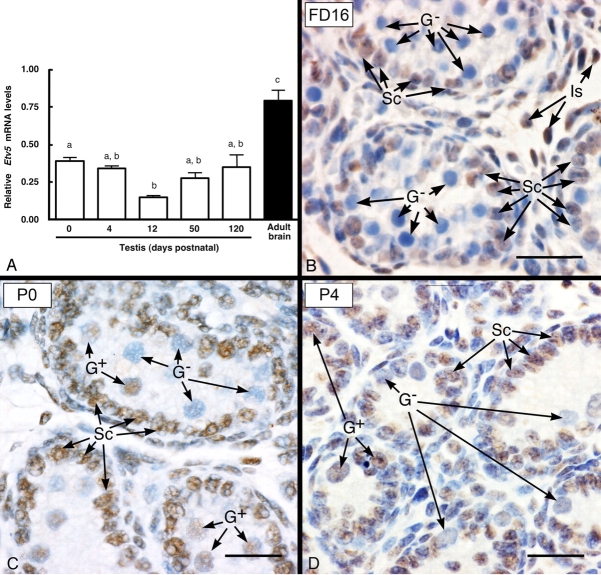
Etv5 mRNA was expressed in WT testis at the earliest age examined (birth), and the level was comparable to that in adult (120-day-old) testis and approximately half of that in brain, which have the highest known expression of Etv5 mRNA expression in mice (Fig. 1A). The finding of Etv5 mRNA in neonatal testis led us to investigate ETV5 expression by immunohistochemistry during late fetal and early neonatal life. At Fetal Day 16, ETV5 expression was detected only in Sertoli cells of seminiferous cords, and gonocytes were negative for ETV5 expression (Fig. 1B). At Postnatal Day 0, Sertoli cells and a subpopulation of gonocytes stained positive for ETV5 (Fig. 1C), indicating ETV5 expression by both cell types at this age. Similar ETV5 staining in Sertoli cells and a subpopulation of gonocytes/spermatogonia was seen at Postnatal Day 4 (Fig. 1D). The number of gonocytes/spermatogonia positive for ETV5 ranged from 50%–65% at these neonatal time points.
FIG. 1.
Temporal and cellular expression of ETV5 in WT testes. The expression of Etv5 mRNA by real-time PCR in neonatal testes (A) was approximately half of that in adult (120-day-old) brain, which has the highest Etv5 mRNA level in mice, and is comparable to that in adult testis. Results are the mean ± SEM. Values with different lowercase letters are significantly different from each other (P < 0.05); n = 4 for each time point except the adult brain sample. B–D) Immunohistochemistry for ETV5 in WT testis at Fetal Day 16 (FD16) and Postnatal Days (P) 0 and 4. B) At FD16, ETV5 expression in the seminiferous cords was limited to Sertoli cell nuclei (Sc), whereas the gonocytes (G−) were negative. Some interstitial cells (Is) were also positive. C and D) In contrast to fetal testis, the pattern of expression was different at P0 and P4, when Sertoli cells and some gonocyte nuclei (G+) showed positive staining, but other gonocytes were negative (G−). Bar = 25 μm.
Etv5−/− Germ Cells Fail To Establish Normal Spermatogenesis after Transplantation
ETV5 expression in gonocytes and in spermatogonia at Postnatal Day 0 and Postnatal Day 4 suggests that this transcription factor could be necessary in the germ cells for their self-renewal. To test this hypothesis, we transplanted 5-day-old WT or Etv5−/− SSCs into 10 W/Wv testes. Etv5−/− SSCs failed to establish normal spermatogenesis after 60 days in any of the W/Wv testes transplanted. In contrast, WT SSCs established full spermatogenesis (Fig. 2) in W/Wv hosts (five of six testes transplanted).
FIG. 2.
Etv5−/− germ cells fail to initiate spermatogenesis after transplantation into W/Wv testes. Establishment of spermatogenesis (Sp) was observed in W/Wv testes 60 days after transplantation with WT germ cells (A) but not with Etv5−/− germ cells (B). Bar = 25 μm.
RET Expression Is Reduced in Etv5−/− Testes and Microdissected Spermatogonia
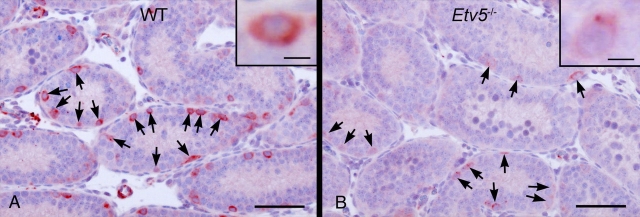
Expression of Ret mRNA and protein was compared in neonatal WT and Etv5−/− testes. Ret mRNA expression in Etv5−/− testes was decreased to 60% of that in WT testes at Postnatal Day 4 (Fig. 3). Progressive reductions were seen until Postnatal Day 24, when Ret mRNA expression was only 10% of that seen in age-matched WT controls (Fig. 3). Real-time PCR analysis on microdissected spermatogonia from Postnatal Day 5 Etv5−/− and WT testes also showed decreased levels of Ret mRNA (Fig. 4), indicating that the decrease in RET represented decreased expression in individual cells rather than simply a decrease in spermatogonial stem/progenitor cell numbers. Consistent with real-time PCR analysis, immunostaining showed a decrease in RET expression in Etv5−/− compared with WT spermatogonia at Postnatal Days 8 (Fig. 5) and 12 (data not shown).
FIG. 3.
Ret mRNA is reduced in neonatal Etv5−/− testes. Results are presented as the mean ± SEM and are given as the percentage of WT controls at each age (n = 4) for WT and Etv5−/− testes at all ages. Values with different lowercase letters are significantly different (P < 0.05) compared with WT controls.
FIG. 4.
GCNA1-stained microdissected spermatogonia from 5-day-old Etv5−/− testes have decreased levels of Ret mRNA relative to those of WT controls (n = 6). Data are presented as the mean ± SEM. *Significantly different at P < 0.05.
FIG. 5.
RET immunohistochemistry in WT and Etv5−/− testes (n = 4). The intensity of RET staining per spermatogonia is markedly reduced in Etv5−/− testis (B), although the numbers of cells staining for RET (arrows) are comparable to those of the WT testis (A). Insets are higher magnification of spermatogonia showing differences in staining intensity. Bar = 50 μm (10 μm in insets).
Etv5−/− Spermatogonia Have Decreased Proliferation In Vitro
We investigated the ability of Etv5−/− spermatogonia to proliferate in vitro in the presence of GDNF, FGF2, and the soluble receptor GFRA1. The ability of neonatal Etv5−/− spermatogonia to produce colonies was significantly reduced compared with than of WT germ cells (Fig. 6) at the same age. In addition to a significantly lower colony number, the size of these colonies was greatly decreased in Etv5−/− cultures. The WT germ cell proliferation frequently resulted in large colonies (Fig. 7A), which were never observed in Etv5−/− germ cell cultures (Fig. 7B). Decreased proliferation in Etv5−/− vs. WT germ cell cultures was more pronounced in groups exposed to GDNF or GDNF+GFRA1+FGF2, likely a result of the more robust WT germ cell proliferation under these conditions.
FIG. 6.
Etv5−/− spermatogonia showed decreased proliferation in vitro. The WT and Etv5−/− spermatogonia were cultured for 6 days in various combinations of GDNF, GFRA1-Fc fusion protein, and FGF2, and the number of proliferating colonies was counted. Data are expressed as the mean ± SEM. *Significant difference between the WT control and corresponding Etv5−/− culture (P < 0.05).
FIG. 7.
Neonatal Etv5−/− (B) spermatogonia produce markedly smaller colonies than WT (A) cultures. Cells were cultured in the presence of GDNF, GFRA1-Fc fusion protein, and FGF2. Bar = 25 μm.
Proliferation of Etv5−/− Spermatogonia In Vivo Is Decreased
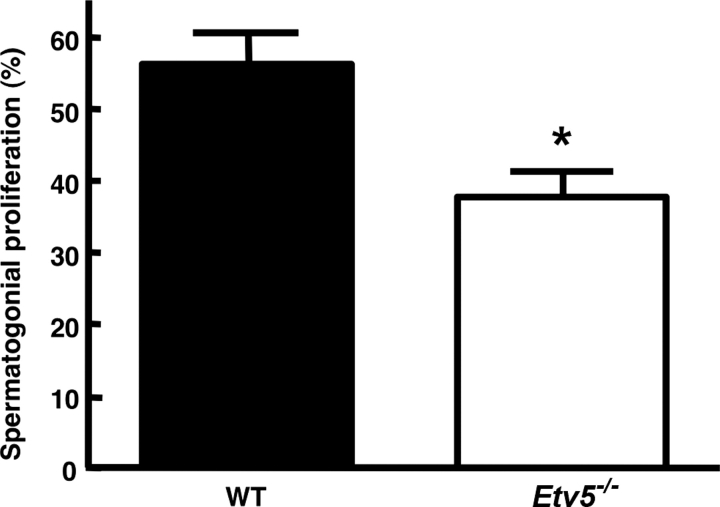
We evaluated the proliferation of Etv5−/− spermatogonial stem/progenitor cells in Postnatal Day 8 testes. Consistent with the in vitro data, proliferation was significantly reduced (Fig. 8), again indicating that ETV5 is necessary for normal spermatogonial proliferation.
FIG. 8.
Spermatogonial stem/progenitor cells from Etv5−/− testes have decreased proliferation in vivo at Postnatal Day 8. Data are shown as the percentage of RET-positive spermatogonia that are proliferating as determined by RET and MKI67 immunostaining in serial Postnatal Day 8 WT (n = 4) and Etv5−/− (n = 3) testis sections. Results are the mean ± SEM. *Significant difference (P < 0.05).
Etv5−/− Testes Have Decreased Type A Spermatogonia and DSP
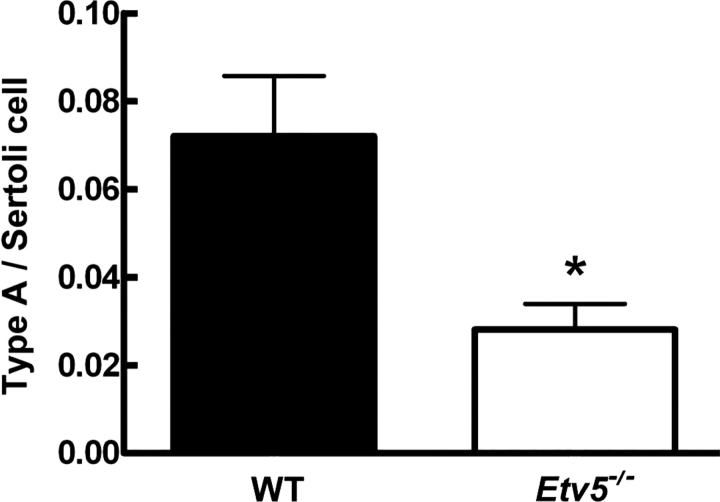
The number of type A spermatogonia was significantly reduced in Etv5−/− mice at Postnatal Day 17 (Fig. 9). Similarly. the DSP was significantly decreased by 60% in Etv5−/− mice at Postnatal Day 30 (Supplemental Fig. S1 available at www.biolreprod.org), indicating that reduced proliferation of Etv5−/− spermatogonial stem/progenitor cells results in decreased numbers of cells in later stages of spermatogenesis during the first wave.
FIG. 9.
At Postnatal Day 17, the number of type A spermatogonia was significantly reduced in Etv5−/− testes relative to that in WT testes (n = 3 for both groups). *Significant difference (P < 0.05).
DISCUSSION
The ets proteins have roles in many biological processes, including cell proliferation, differentiation, apoptosis, immune response and oncogenic transformation, lung development, and epithelial-mesenchymal interactions [26–28]. In the murine lung, ETV5 has been localized in type I and type II epithelial cells and has been shown to regulate surfactant protein C and caveolin 1 [29, 30]. In mouse ovary, ETV5 is expressed in granulosa cells and cumulus cells and up-regulates the expression of cyclooxygenase 2, a rate-limiting enzyme for prostaglandin synthesis [31]. ETV5 is expressed by subsynaptic nuclei at the neuromuscular junction and has a role in synapse assembly and function [32]. ETV5 has been implicated in the progression and metastasis of human breast and endometrial carcinomas [33].
ETV5 was previously shown to be necessary for maintenance of SSCs. Chen et al. [6] reported the total loss of adult germ cells in Etv5−/− mice following the first wave of spermatogenesis. In adult mice, ETV5 was initially reported in Sertoli cells but not in germ cells. However, Oatley et al. [16] subsequently reported Etv5 mRNA expression in SSCs following long-term culture and Etv5 mRNA and protein expression in spermatogonia in vivo at Postnatal Day 8 [17].
Our recent analysis revealed that, although GFRA1-positive spermatogonial numbers were initially similar in developing WT and Etv5−/− testes (Postnatal Day 4), their loss begins by the end of the first postnatal week [7]. These results indicated that ETV5 expression might be present and critical for SSC maintenance in Sertoli cells, germ cells, or both before Postnatal Day 8, the earliest age at which ETV5 expression has been documented [6, 17]. In the present study, we characterized the temporal and cellular expression of ETV5 during the perinatal period in the testis. Our results indicate that testicular ETV5 expression is present as early as late fetal life and that ETV5 shows a staggered ontogeny in different cell types of the testis.
ETV5 expression was present in Sertoli cells on Fetal Day 16, the earliest time point examined. In contrast, gonocytes were negative in Fetal Day 16 testis. At birth (Postnatal Day 0), Sertoli cells remained immunopositive for ETV5, but by this age a subpopulation of gonocytes was also positive. Real-time PCR data were consistent with immunohistochemistry showing significant amounts of Etv5 mRNA in whole testis and spermatogonial stem/progenitor cells (data not shown) from neonatal mice, although testicular Etv5 mRNA concentrations were lower in the neonatal period than in the adult period. Etv5 mRNA expression continues into the juvenile period, although by Postnatal Day 12 the expression levels are sharply reduced compared with those of neonatal testes. This is consistent with previous work in which ETV5 expression could not be detected in seminiferous epithelium of 2-wk-old mice containing the ETV5 gene linked to a β-galactosidase reporter [6]. The mechanisms responsible for the initial production of Sertoli cell ETV5 prenatally and the onset of germ cell ETV5 expression during the neonatal period, as well as the factors responsible for the decrease seen in the late preweaning period, are unclear.
Thus, it appears that germ cells and Sertoli cells could be important sites of ETV5 synthesis and function during the neonatal period of rapid testicular growth and establishment of the first wave of spermatogenesis. The production of ETV5 in fetal Sertoli cells suggests that this protein may be important even before birth in these cells. Although we confirmed that germ cells express ETV5, the exact spermatogonial population in which ETV5 is expressed remains to be determined. Our immunohistochemical data showing ETV5 expression at Postnatal Day 0 indicate that spermatogonial stem/progenitor cells likely express ETV5. In addition, we were unable to consistently find expression in germ cells of juvenile and adult testes, which could be due to ETV5 expression being limited to spermatogonial stem/progenitor cells, which are scarce. However, these data are inconsistent with a previous study [17] in which all spermatogonia lining the basement membrane were found to be positive in adult mouse testis, and more investigation is needed in this area. In addition to Sertoli cells and germ cells, Leydig cells were also ETV5 positive. The significance of ETV5 in Leydig cells is not yet apparent, and testosterone levels [6] were not found to be significantly different from those in the WT mice.
The expression of ETV5 in spermatogonial stem/progenitor cells raises the important question of whether ETV5 in these cells is essential for their maintenance or whether the loss of SSCs in Etv5−/− mice might totally be due to impaired function of Sertoli cells in these animals. We used the germ cell transplantation technique to directly determine whether ETV5 in SSCs is required for establishment of spermatogenesis in W/Wv mice testes, which lack endogenous spermatogenesis.
Our results show that qualitatively normal spermatogenesis was established when neonatal WT germ cells were transplanted into W/Wv mice testes. In contrast, transplanted neonatal Etv5−/− germ cells failed to produce spermatogenesis in any host mice. It is unclear from these results whether this was due to an inability of the grafted Etv5−/− germ cells to normally home into the SSC niche established by the Sertoli cells and/or an inability of the Etv5−/− germ cells to proliferate normally once they had entered the niche to establish spermatogenesis. However, these results clearly show that ETV5 is necessary for grafted germ cells to establish spermatogenesis in host animals, and the loss of ETV5 in neonatal germ cells may contribute to the loss of spermatogenesis in developing Etv5−/− testis.
Previous findings by Oatley et al. [17] have shown that a 60% knockdown in Etv5 mRNA by short interfering RNA technology led to decreased SSC colony formation in vitro. We estimated the proliferation of spermatogonial stem/progenitor cells in WT and Etv5−/− testes to determine whether proliferation of spermatogonia is obligatorily dependent on ETV5. Our analysis indicated that loss of ETV5 reduces but does not eliminate spermatogonial proliferation in vivo as assessed by expression of the proliferation marker MKI67. Similarly, the in vitro proliferative capacity of WT spermatogonial stem/progenitor cells was markedly greater than that of a similar population isolated from Etv5−/− testes under a variety of culture conditions. These data indicate that ETV5, although not obligatory for proliferation of spermatogonia, is essential for a normal rate of self-renewal in vivo and in vitro. Using TUNEL staining, we also confirmed (consistent with our previous report [6]) that apoptosis was not the cause of SSC loss in neonatal Etv5−/− testes (data not shown).
Proliferation of Etv5−/− spermatogonia in vitro was reduced even in the presence of high levels of GDNF, the major growth factor responsible for SSC proliferation and self-renewal in vitro [15, 34]. These results suggested a decreased ability of Etv5−/− spermatogonia to respond to GDNF stimulation and indicated that one or more components of the GDNF/RET/GFRA1 signaling pathway could be altered in Etv5−/− spermatogonia.
GDNF is produced by Sertoli cells and then acts through the RET/GFRA1 receptor complex on SSCs to stimulate SSC self-renewal. GDNF mRNA production was comparable in Sertoli cells from WT and Etv5−/− testes, indicating that impaired GDNF production was not the cause of SSC loss in Etv5−/− testes [6]. This is consistent with our in vitro data showing impaired proliferation of spermatogonial stem/progenitor cells from Etv5−/− testes even in the presence of high concentrations of GDNF and GFRA1. In addition, no differences in staining intensity of GFRA1 were observed between WT and Etv5−/− spermatogonial stem/progenitor cells by Schlesser et al. [7] in seminiferous tubule whole mounts, suggesting that GFRA1 is also not directly influenced by ETV5.
In striking contrast to the lack of effect on GDNF and GFRA1 in Etv5−/− testes, Ret mRNA expression was sharply reduced in Etv5−/− testes. This may represent a key role in overall decreased self-renewal and proliferation of Etv5−/− spermatogonial stem/progenitor cells and would explain the inability of these cells to respond normally to GDNF. Like GDNF, RET is essential for SSC self-renewal in the testis, and Ret−/− testes lose all SSCs [8]. Ret mRNA was decreased in Etv5−/− testes compared with WT testes as early as Postnatal Day 4, and these decreases become more pronounced with age.
Spermatogonial stem/progenitor cell loss in Etv5−/− testes begins during the first postnatal week [7]. Therefore, it is essential to establish whether the observed decreases in Ret mRNA postnatally in Etv5−/− testes do indeed reflect decreased Ret mRNA expression in individual spermatogonial stem/progenitor cells and not merely a decrease in spermatogonial numbers.
To differentiate between these two possibilities, we used LCM to obtain RNA samples from individual spermatogonia, and we examined RET expression by immunohistochemistry. Although GCNA1 does not specifically label spermatogonial stem/progenitor cells, it allowed us to obtain an enriched population of neonatal spermatogonia (Postnatal Day 5). Our LCM results indicate that Ret mRNA expression is decreased in RNA samples obtained from Etv5−/− spermatogonia. This suggests that the decreased Ret mRNA in the Etv5−/− testes results from decreased expression of Ret mRNA per spermatogonium. This conclusion is supported by RET immunohistochemistry. There was a marked decrease in staining intensity of RET-positive spermatogonia by Postnatal Day 8 in Etv5−/− testes. These data suggest that RET expression in spermatogonia may be directly or indirectly regulated by ETV5.
This conclusion is also consistent with analysis of the patterns of the decreases in RET expression vs. the decrease in spermatogonial stem/progenitor cell numbers. For example, Ret mRNA was down 40% at Postnatal Day 4, although spermatogonial stem/progenitor cell numbers in WT and Etv5−/− testes are equal at this age [7]. Similarly, at Postnatal Day 12, spermatogonial stem/progenitor cell numbers in the Etv5−/− testes were down 37%, but Ret mRNA was down 75%. This suggests that, although there is a progressive loss of spermatogonial stem/progenitor cell numbers postnatally in the Etv5−/− testes, the decrease in Ret mRNA precedes the onset of spermatogonial stem/progenitor cell loss and the decrease in Ret mRNA at later ages is greater than the magnitude of spermatogonial stem/progenitor cell loss. This is supported by the LCM data presented herein that Ret mRNA per spermatogonium is significantly reduced during the neonatal period. Similarly, immunohistochemistry indicates decreased RET expression per spermatogonium. Decreased RET expression could result in impaired GDNF/RET/GFRA1 signaling, lead to the loss of SSCs in Etv5−/− mice, and have a major role in the eventual Sertoli cell-only phenotype of these animals.
There is indirect evidence that Ret gene expression could be under direct control of ETV5. The murine RET promoter (GenBank accession No. AY255629) contains multiple ETS-binding motifs (ggaa) [35]. ETV5 may also regulate expression of germ cell RET indirectly by influencing expression of other genes. Another possibility is that RET is regulated in a paracrine manner by Sertoli cell factors induced by ETV5 in these cells. Indeed, Batourina et al. [36] have shown that RET expression in ureteric bud is regulated by retinoic acid-induced expression of an unknown gene or genes in surrounding stromal cells.
During the first wave of spermatogenesis, Sertoli cells and germ cells are actively dividing, as this early expansion of Sertoli cells establishes the maximum testis size and adult sperm production [37]. Although initial results showed the presence of first-wave spermatogenesis in Etv5−/− testes [6], the sharply decreased proliferation of spermatogonial stem/progenitor cells from Etv5−/− testes in vitro and in vivo implies that there would be decreased numbers of SSCs and subsequently later stages of spermatogenesis in the Etv5−/− testes. To directly test this hypothesis, we quantitated type A spermatogonia and DSP during the first wave of spermatogenesis. Our results indicate that there were decreases in type A spermatogonia accompanied by decreased DSP during juvenile life. Thus, Etv5−/− testes have a quantitatively impaired first wave of spermatogenesis due to decreased proliferation of the spermatogonial stem/progenitor cells and subsequent decreases in more differentiated progeny. Whether impaired germ cell proliferation at stages later than spermatogonial stem/progenitor cells could contribute to this decreased number of differentiated germ cells during the first wave is not directly addressed by the present data but could also be a factor.
In conclusion, ETV5 expression begins in fetal Sertoli cells and is expressed in Sertoli cells and spermatogonia during perinatal development. Germ cell ETV5 is essential for establishment of ongoing spermatogenesis. Proliferation was reduced in Etv5−/− germ cells in vitro and in vivo, and the first wave of spermatogenesis is quantitatively reduced. This appears to result from impairment in the GDNF/RET/GFRA1 signaling pathway as a result of decreased RET expression. These results provide mechanistic insights into the role of ETV5 in neonatal SSC self-renewal and establishment of overall spermatogenesis.
Supplementary Material
Acknowledgments
We are grateful to Dr. G. Enders for providing anti-GCNA1 antibody. We thank Dr. Raju S. Mantena, University of Illinois, for assistance with RET immunostaining.
Footnotes
1Supported in part by the Billie A. Field Endowment, University of Illinois (P.S.C.), subproject CIG-05-111 (R.A.H.) provided by CICCR of CONRAD, Eastern Virginia Medical School; NIH grant HD44543 (M.C.H.) and the Howard Hughes Medical Institute (K.M.M.). Work at the University of Illinois was conducted in a facility constructed with support from Research Facilities Improvement Program Grant No. C06 RR16515 from the National Center for Research Resources, National Institutes of Health.
REFERENCES
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G.Pluripotency of spermatogonial stem cells from adult mouse testis. Nature 2006; 440: 1199–1203. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, Toyoshima M, Niwa O, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell 2004; 119: 1001–1012. [DOI] [PubMed] [Google Scholar]
- Griswold MD.The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol 1998; 9: 411–416. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ohmura M, Ohbo K.The niche for spermatogonial stem cells in the mammalian testis. Int J Hematol 2005; 82: 381–388. [DOI] [PubMed] [Google Scholar]
- Hofmann MC.Gdnf signaling pathways within the mammalian spermatogonial stem cell niche. Mol Cell Endocrinol 2008; 288: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005; 436: 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesser HN, Simon L, Hofmann MC, Murphy KM, Murphy T, Hess RA, Cooke PS.Effects of ETV5 (ets variant gene 5) on testis and body growth, time course of spermatogonial stem cell loss, and fertility in mice. Biol Reprod 2008; 78: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J.Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod 2006; 74: 314–321. [DOI] [PubMed] [Google Scholar]
- Jing S, Wen D, Yu Y, Holst PL, Luo Y, Fang M, Tamir R, Antonio L, Hu Z, Cupples R, Louis JC, Hu S, et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell 1996; 85: 1113–1124. [DOI] [PubMed] [Google Scholar]
- Treanor JJ, Goodman L, de Sauvage F, Stone DM, Poulsen KT, Beck CD, Gray C, Armanini MP, Pollock RA, Hefti F, Phillips HS, Goddard A, et al. Characterization of a multicomponent receptor for GDNF. Nature 1996; 382: 80–83. [DOI] [PubMed] [Google Scholar]
- He Z, Jiang J, Kokkinaki M, Golestaneh N, Hofmann MC, Dym M.Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells 2008; 26: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braydich-Stolle L, Kostereva N, Dym M, Hofmann MC.Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol 2007; 304: 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Kimura T, Nakano T, Ogura A, Shinohara T.Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 2007; 134: 1853–1859. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489–1493. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL.Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A 2004; 101: 16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL.Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A 2006; 103: 9524–9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL.Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan S, Simon L, Meling DD, Cyr DG, Gutstein DE, Fishman GI, Guillou F, Cooke PS.Proliferation of adult Sertoli cells following conditional knockout of the Gap junctional protein GJA1 (connexin 43) in mice. Biol Reprod 2007; 76: 804–812. [DOI] [PubMed] [Google Scholar]
- Aponte PM, van Bragt MP, de Rooij DG, van Pelt AM.Spermatogonial stem cells: characteristics and experimental possibilities. APMIS 2005; 113: 727–742. [DOI] [PubMed] [Google Scholar]
- Dirami G, Ravindranath N, Pursel V, Dym M.Effects of stem cell factor and granulocyte macrophage-colony stimulating factor on survival of porcine type A spermatogonia cultured in KSOM. Biol Reprod 1999; 61: 225–230. [DOI] [PubMed] [Google Scholar]
- Ohta H, Tohda A, Nishimune Y.Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol Reprod 2003; 69: 1815–1821. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Arechaga JM, Avarbock MR, Brinster RL.Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol 1997; 41: 111–122. [PubMed] [Google Scholar]
- Enders GC, May JJ., IIDevelopmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 1994; 163: 331–340. [DOI] [PubMed] [Google Scholar]
- Murakami H, Liotta L, Star RA.IF-LCM: laser capture microdissection of immunofluorescently defined cells for mRNA analysis rapid communication. Kidney Int 2000; 58: 1346–1353. [DOI] [PubMed] [Google Scholar]
- Cooke PS, Porcelli J, Hess RA.Induction of increased testis growth and sperm production in adult rats by neonatal administration of the goitrogen propylthiouracil (PTU): the critical period. Biol Reprod 1992; 46: 146–154. [DOI] [PubMed] [Google Scholar]
- Sementchenko VI, Watson DK.Ets target genes: past, present and future. Oncogene 2000; 19: 6533–6548. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD.The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2: 827–837. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T.Molecular biology of the Ets family of transcription factors. Gene 2003; 303: 11–34. [DOI] [PubMed] [Google Scholar]
- Lin S, Perl AK, Shannon JM.Erm/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. J Biol Chem 2006; 281: 16716–16726. [DOI] [PubMed] [Google Scholar]
- Kathuria H, Cao Y, Hinds A, Ramirez MI, Williams MC.ERM is expressed by alveolar epithelial cells in adult mouse lung and regulates caveolin-1 transcription in mouse lung epithelial cell lines. J Cell Biochem 2007; 102: 13–27. [DOI] [PubMed] [Google Scholar]
- Eo J, Han K, Murphy KM, Song H, Lim HJ.Etv5, an ETS transcription factor, is expressed in granulosa and cumulus cells and serves as a transcriptional regulator of the cyclooxygenase-2. J Endocrinol 2008; 198: 281–290. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Huber RM, Ladle DR, Murphy K, Arber S.ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron 2007; 55: 726–740. [DOI] [PubMed] [Google Scholar]
- Monge M, Colas E, Doll A, Gonzalez M, Gil-Moreno A, Planaguma J, Quiles M, Arbos MA, Garcia A, Castellvi J, Llaurado M, Rigau M, et al. ERM/ETV5 up-regulation plays a role during myometrial infiltration through matrix metalloproteinase-2 activation in endometrial cancer. Cancer Res 2007; 67: 6753–6759. [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Nolan C, Dym M, Hofmann MC.Role of glial cell line-derived neurotrophic factor in germ-line stem cell fate. Ann N Y Acad Sci 2005; 1061: 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K.The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta 2006; 1766: 79–87. [DOI] [PubMed] [Google Scholar]
- Batourina E, Gim S, Bello N, Shy M, Clagett-Dame M, Srinivas S, Costantini F, Mendelsohn C.Vitamin A controls epithelial/mesenchymal interactions through Ret expression. Nat Genet 2001; 27: 74–78. [DOI] [PubMed] [Google Scholar]
- Sharpe RM, McKinnell C, Kivlin C, Fisher JS.Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003; 125: 769–784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.