Abstract
The class E genome of human cytomegalovirus (HCMV) contains long and short segments that invert due to recombination between flanking inverted repeats, causing the genome to isomerize into four distinct isomers. To determine if isomerization is important for HCMV replication, one copy of each repeat was deleted. The resulting virus replicated in cultured human fibroblasts with only a slight growth impairment. Restriction and Southern analyses confirmed that its genome is locked in the prototypic arrangement and unable to isomerize. We conclude that efficient replication of HCMV in fibroblasts does not require (i) the ability to undergo genome isomerization, (ii) genes that lie partially within the deleted repeats, or (iii) diploidy of genes that lie wholly within repeats. The simple genomic structure of this virus should facilitate studies of genome circularization, latency or persistence, and concatemer packaging as such studies are hindered by the complexities imposed by isomerization.
Keywords: genome isomerization, cytomegalovirus, segment inversion, class E genome
Introduction
Herpesviruses have large (150-kb to 235-kb) double-stranded linear DNA genomes that are categorized into six classes designated A–F based on structural complexities related to the presence of direct and inverted repeats (Roizman and Pellett, 2001). Herpes simplex virus type 1 (HSV-1), herpes simplex virus type 2, and human cytomegalovirus (HCMV) have class E genomes that feature a long segment (L) comprised of unique long (UL) sequences bordered by inverted ab and b'a' repeats, a short segment (S) comprised of unique short (US) sequences bordered by a'c' and ca inverted repeats, and an L/S junction comprised of b'a'c' sequences (Fig. 1A). In a process termed isomerization, the L and S segments independently invert, forming four genomic isomers designated prototype (P), inverted L (IL), inverted S (IS), and inverted L and S (ILS) (Delius and Clements, 1976; Hayward et al., 1975; Kilpatrick and Huang, 1977; LaFemina and Hayward, 1980; Sheldrick and Berthelot, 1975; Skare and Summers, 1977; Wilkie, 1976).
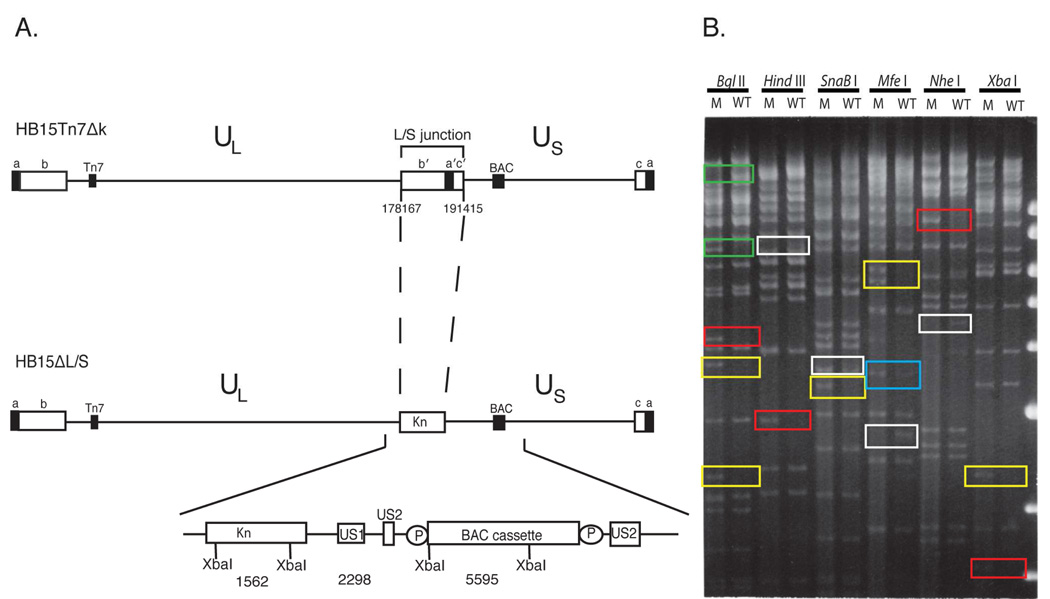
Figure 1.
Deletion of the L/S junction from the HCMV genome. (A) The P genome isomer of parental virus HB15Tn7Δk is shown with ab and b′a′ repeats flanking UL and a′c′ and ca repeats flanking US, the attTn7 site (Tn7) inserted in UL, and the BAC origin cassette (BAC) inserted in US. Below is shown the HB15ΔL/S genome in which Kn replaces b'a'c'. Sequences deleted from HB15Tn7Δk are indicated using nucleotide numbers that correspond to the AD169 genome sequence (Chee et al., 1990). The Kn - BAC origin region is expanded below to show the predicted sizes (bp) of diagnostic XbaI fragments, direct duplications of US2 sequences that allow excision of the BAC origin by homologous recombination, and LoxP sites (P) flanking the BAC origin. (B) Restriction analyses of HB15ΔL/S and HB15Tn7Δk. Virion DNAs from HB15ΔL/S (M) and HB15Tn7Δk (WT) were digested with the indicated restriction enzymes. The resulting DNA fragments were separated on 0.7% agarose and visualized with ethidium bromide and UV light. Colored boxes indicate fragments that are lost or created in HB15ΔL/S by deletion of b'a'c' (green), fragments that are unique to HB15ΔL/S by virtue of the Kn insertion (red), fragments that indicate retention of the BAC cassette in HB15ΔL/S (yellow), IS isomer S-terminal fragments missing from HB15ΔL/S (white), and a P isomer S-terminal fragment that is half-molar in HB15Tn7Δk and equi-molar in HB15ΔL/S (blue). The positions and sizes (kb) of DNA size markers are shown on the right.
That isomerization occurs by homologous recombination between inverted repeats was first suggested by studies in which insertion of L/S sequences into the UL region of the HSV-1 genome gave rise to additional genome inversions (Mocarski, Post, and Roizman, 1980). Subsequent studies revealed that HSV-1 isomerization does not require specific inverted sequences (Chou and Roizman, 1985; Chou and Roizman, 1989; Davison and Wilkie, 1981; Davison and Wilkie, 1983; Dutch, Bianchi, and Lehman, 1995; Dutch et al., 1992; Dutch, Zemelman, and Lehman, 1994; Mocarski and Roizman, 1982a; Mocarski and Roizman, 1982b; Pogue-Geile, Lee, and Spear, 1985; Pogue-Geile and Spear, 1986; Sarisky and Weber, 1994; Smiley, Duncan, and Howes, 1990; Smiley, Fong, and Leung, 1981; Smiley, Lavery, and Howes, 1992; Varmuza and Smiley, 1985; Weber et al., 1988; Weber, Levine, and Glorioso, 1990) but does require replication by the highly recombinagenic HSV-1 DNA synthesis machinery (Dutch et al., 1992; Mocarski and Roizman, 1982a; Sarisky and Weber, 1994; Weber et al., 1988; Weber, Levine, and Glorioso, 1990).
The significance of genome isomerization is not known. It is not required for replication as many herpesviruses do not contain inverted repeats or invertible elements (Roizman and Pellett, 2001), and deletion of the majority of the b'a'c' region from the HSV-1 genome gives rise to replication competent mutants that do not undergo genome isomerization (Jenkins and Roizman, 1986; Poffenberger and Roizman, 1985; Poffenberger, Tabares, and Roizman, 1983). While such isomerically “frozen” mutants exhibit very modest growth impairments in vitro, they are profoundly attenuated in vivo, suggesting that either isomerization or diploidy of genes encoded by bac/b'a'c' sequences are important in vivo (Jenkins, Donoghue, and Martin, 1996; Jenkins and Martin, 1990).
To date, HSV-1 remains the only class E genome virus in which isomerically frozen b'a'c' deletion mutants have been characterized. Moreover, no mutants have yet been constructed in which the entire b'a'c' region is deleted (HSV-1 mutants retain small amounts of b’ and c’ sequences at the UL/b’ and c’/US boundaries) (Jenkins, Donoghue, and Martin, 1996). In this study we sought to determine if deletion of the entire b'a'c' region from the class E genome of human cytomegalovirus (HCMV) eliminates genome isomerization and how this impacts viral replication in vitro.
Results
Construction of HB15ΔL/S
The HB15Tn7Δk bacterial artificial chromosome (BAC) was derived from BAC HB15 (Hobom et al., 2000). It contains the HCMV genome (strain AD169) modified with a mini-attTn7 site in UL18 (Hahn et al., 2003a), a LoxP-flanked BAC origin cassette between US1 and US2, and a short duplication of US2 sequences that facilitates excision of the BAC cassette by homologous recombination (Fig. 1A) (Hobom et al., 2000). To construct an HCMV genome lacking the internal b'a'c' sequences, linear recombination in E. coli was used to replace the b'a'c' sequences in HB15Tn7Δk with a kanamycin-resistance (Kn) marker. Candidate clones were selected with kanamycin and screened using PCR assays spanning the predicted UL-Kn and Kn-US junctions (not shown). Two clones were further analyzed by XbaI restriction (not shown). As predicted, both clones contained novel 1.6- and 2.3-kb XbaI fragments indicative of correct insertion of the Kn marker (Fig. 1A). Clone 2 was designated HB15ΔL/S.
HB15ΔL/S BAC DNA was transfected into human MRC-5 fibroblasts to determine if it could reconstitute a replication competent virus. Viral cytopathic effect was detected 15 days post transfection and subsequent serial passage did not suggest an obvious growth defect (not shown).
Restriction analysis of HB15ΔL/S virion DNA
To further confirm the genomic structure of HB15ΔL/S and rule out possible unforeseen rearrangements or deletions, restriction fragment patterns for HB15ΔL/S and HB15Tn7Δk virion DNAs were compared using a panel of restriction enzymes (Fig. 1B). Most of the observed restriction pattern differences were predicted by deletion of b'a'c' sequences (Fig. 1B, green boxes) or insertion of Kn (Fig. 1B, red boxes), or were consistent with retention of the BAC origin cassette in HB15ΔL/S and its excision from HB15Tn7Δk (Fig. 1B, yellow boxes). The latter results were not surprising given that excision is largely driven by selection against the packaging of over length genomes and even with the BAC cassette retained the HB15ΔL/S genome is under length, while that of HB15Tn7Δk would be significantly over length were the BAC cassette not excised.
The remaining differences could be attributed to a failure of the S segment of HB15ΔL/S to invert. In HB15Tn7Δk, terminal restriction fragments can differ in size according to the orientations of the L or S segments if the restriction site nearest the genome end falls within UL or US. Thus, L or S segment inversion can be detected by the presence of terminal fragments unique to IL or IS genomes. Table 1 shows predicted terminal fragment sizes for the enzymes used here. MfeI and XbaI cut within the b repeat and therefore L terminal fragments from P and IL genomes are identical, while for the remaining enzymes the L terminal fragments were too large to resolve. Hence, this analysis was unable to evaluate L segment inversion. However, several S-terminal fragments unique to IS genomes were easily resolved and in each case were present in HB15Tn7Δk DNA but absent from HB15ΔL/S DNA (Fig. 1B, white boxes), indicating that the S segment of HB15ΔL/S does not invert. Failure of the S segment to invert further predicts that S terminal fragments from the P orientation should be equi-molar in HB15ΔL/S since all genomes should have this S end, while in HB15Tn7Δk the same fragments should be half-molar since half the genomes have a different S end. This was clearly the case for the 3.5-kb S-terminal MfeI fragment (Fig. 1B, blue box).
Table 1.
Sizes* (kb) of Terminal and Junction restriction fragments for the four genomic isomers
| L-terminal | S-terminal | Junction | ||||||
|---|---|---|---|---|---|---|---|---|
| P | IL | P | IS | P | IL | IS | ILS | |
| BglII | 31.1 | 18.0 | 12.7 | 17.7 | 35.2 | 48.3 | 30.2 | 43.3 |
| HindIII | 17.2 | 12.6 | 17.7 | 7.0 | 19.0 | 23.6 | 29.7 | 34.4 |
| SnaBI | 11.4 | 11.5 | 14.5 | 3.6 | 14.5 | 14.5 | 25.4 | 25.3 |
| MfeI | 6.5 | 6.5 | 3.5 | 2.6 | 8.6 | 8.6 | 9.4 | 9.4 |
| NheI | 12.0 | 21.1 | 21.6 | 4.5 | 25.0 | 15.9 | 42.2 | 33.0 |
| XbaI | 8.0 | 8.0 | 11.2 | 11.5 | 18.9 | 18.9 | 18.6 | 18.6 |
Junctions and L-termini can contain one to several a sequences, whereas S-termini contain predominantly zero or one a sequence (Tamashiro et al., 1984; Tamashiro and Spector, 1986). Sizes given are for fragments that contain one a sequence.
Southern analysis of HB15ΔL/S virion DNA
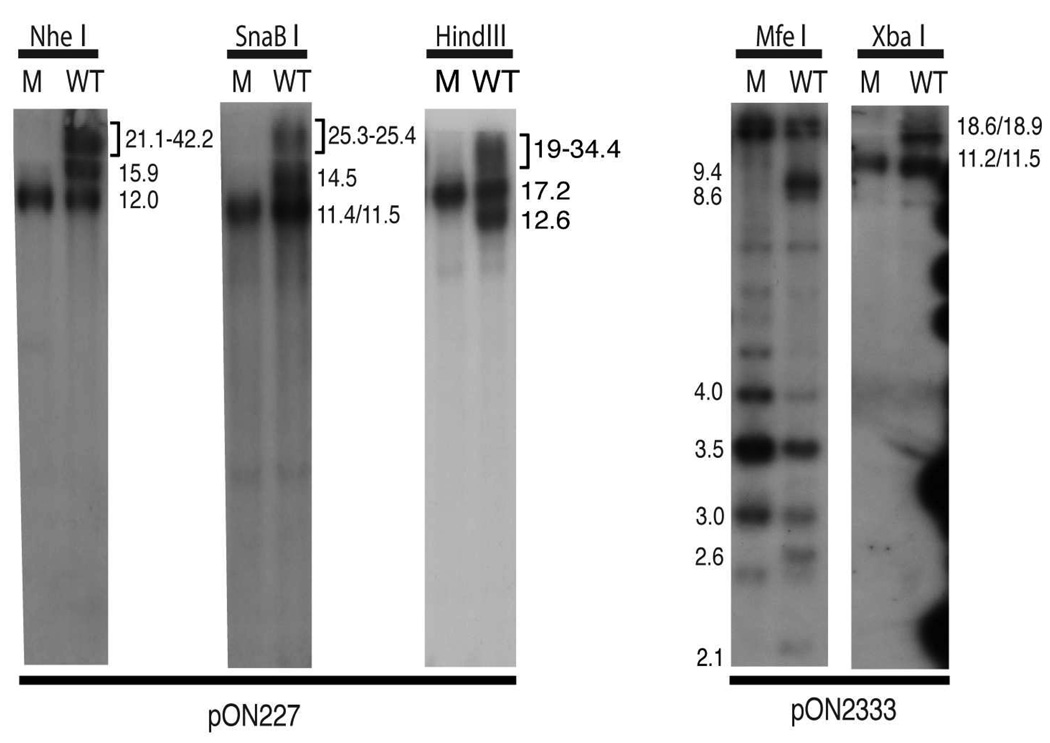
The agarose-separated fragments shown in Fig. 1B were transferred to a nylon membrane and hybridized with probes pON1101 to detect fragments internal to the BAC origin, pON227 to detect restriction fragments from the L end of the genome, and pON2333 to detect fragments from the S end of the genome. The latter two probes are predicted to hybridize to junction fragments in addition to terminal fragments. The predicted fragment sizes for each enzyme are given in Table 1.
The pON1101 probe failed to hybridize to HB15Tn7Δk DNA, confirming that the BAC origin had efficiently self-excised from the HB15Tn7Δk genome (not shown). For HB15ΔL/S DNA each restriction digest contained a fragment that hybridized to pON1101 and had a size consistent with retention of the BAC origin in HB15ΔL/S (not shown).
In HB15Tn7Δk DNA the pON227 probe detected the 12-kb NheI L-terminal fragment from P genomes as well as several larger hard to resolve fragments including the 21-kb L-terminal fragment from IL genomes and junction fragments from P, IL, IS, and ILS genomes (Fig. 2). Similar results were obtained for SnaBI fragments probed with pON227 and for MfeI and XbaI fragments probed with pON2333. In each case, all of the larger junction fragments were missing from HB15ΔL/S DNA (Fig. 2). The lack of P junctions was predicted due to the deletion, however, the lack of IL, IS, and ILS junctions clearly demonstrates that HB15ΔL/S fails to undergo both S and L segment inversion.
Figure 2.
Southern analyses of HB15ΔL/S and HB15Tn7Δk. DNA fragments from the gel shown in Fig. 1B were transferred to a nylon membrane and sequentially hybridized with probe pON227 to detect L-terminal fragments and junctions or with pON2333 to detect S-terminal fragments and junctions. The sizes (kb) of relevant restriction fragments are indicated.
In some digests terminal fragment size differences were sufficient to differentiate IL or IS genome ends from P genome ends. HindIII analysis provided clear evidence that the L segment of HB15ΔL/S does not invert as the 12.6-kb HindIII IL L-terminal fragment was clearly resolved in HB15Tn7Δk DNA but absent from HB15ΔL/S DNA (Fig. 2). Similarly, as noted above, HB15ΔL/S DNA lacked hybridizing fragments migrating in the region where the 21-kb NheI IL L-terminal fragment should migrate (Fig. 2). Due to the large sizes of these fragments the heterogeneity that occurs at both junctions and termini as a result of variable reiterations of the 0.5-kb a sequence (Tamashiro et al., 1984; Tamashiro and Spector, 1986) were not apparent. However, the MfeI S-terminal fragments were small enough to reveal variations in the number of terminal a sequences and hence these patterns contained additional fragments that formed ladders with ~0.5-kb increments. S-terminal MfeI fragments from P genomes (3.0-, 3.5-, and 4.0-kb fragments representing S-termini with zero, one, and two a sequences, respectively) were present on both genomes. However, the 2.1- (no a sequence) and 2.6-kb (one a sequence) fragments indicative of IS genomes were present in HB15Tn7Δk but absent from HB15ΔL/S DNA (Fig. 2), again confirming that HB15ΔL/S fails to undergo S segment inversion.
HB15ΔL/S replicates with a slight growth impairment relative to HB15Tn7Δk
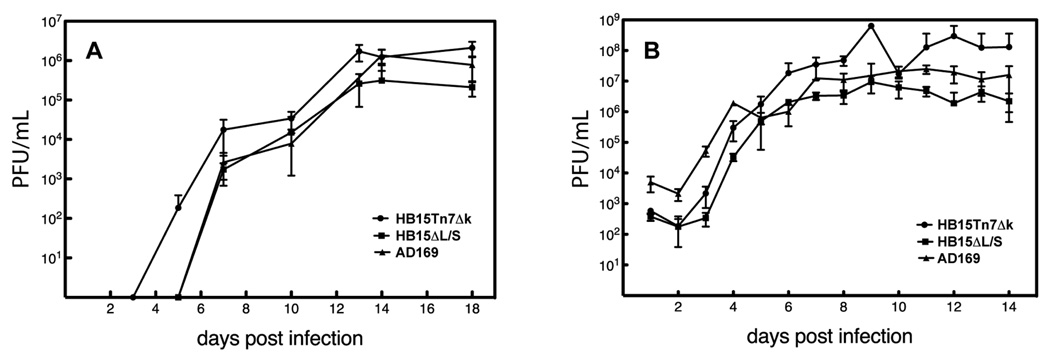
To determine if failure to isomerize or deletion of the internal L/S junction has an impact on viral replication, single step (MOI=4) and multistep (MOI=0.01) growth curves were performed with HB15ΔL/S, its parental virus HB15Tn7Δk, and AD169 (the strain from which HB15Tn7Δk was derived). Interestingly, HB15Tn7Δk replicated to slightly higher titers than AD169, suggesting that our AD169 stock, which as was obtained from ATCC in 1980, may have diverged from the stock that was used to construct HB15Tn7Δk (Hobom et al., 2000). While this comparison serves as a measure of intrastrain variation, the valid comparison is between HB15ΔL/S and its parental, HB15Tn7Δk. Under both MOI conditions HB15ΔL/S achieved titers approximately 1–1.5 logs lower than those of HB15Tn7Δk and at low MOI exhibited somewhat delayed kinetics (Fig. 3). While these differences could be attributable to intrastrain variation (as suggested by the range between HB15Tn7Δk and AD169), it should be noted that while the AD169 and HB15Tn7Δk stocks are distantly related, the HB15ΔL/S and HB15Tn7Δk are not; both were amplified through limited passage after reconstitution from their respective BAC DNAs. Thus, it is likely that, similar to HSV-1 isomerically frozen mutants, deletion of HCMV b’a’c’ sequences results in modest decreases in viral replicative fitness.
Figure 3.
Growth curves of virus strain AD169 and BAC-derived viruses HB15ΔL/S and HB15Tn7Δk. Carefully matched inocula of each virus were used to infect MRC-5 cells at MOIs of 0.01 (A) or 4 (B) and titers of infectious virus in the culture supernatants were determined on the days indicated.
Discussion
Structurally complex genomes able to form multiple genome variants are a hallmark of the Herpesviridae. Isomerization, the process of segment flipping due to recombination between inverted repeats, is common in viruses of the alpha subfamily; arrays of reiterated direct repeats that vary in length depending on the number of repeat copies are a common feature of genomes from the gamma subfamily; whereas in the beta subfamily direct terminal repeats that are sometimes reiterated but often single copy are common. Unique among betaherpesviruses is the highly complex class E genome of HCMV.
Early on the class E genome structure of HSV-1 and the phenomenon of isomerization was believed to be integral to and perhaps demonstrative of a unique mechanism of DNA replication. This idea was disproved when it was found that HSV-1 mutants deleted of their internal repeats do not isomerize yet are replication competent in vitro (Jenkins and Roizman, 1986; Poffenberger and Roizman, 1985; Poffenberger, Tabares, and Roizman, 1983). Moreover, that a virus that normally does not isomerize can gain the ability to isomerize simply by artificial creation of inverted repeats (McVoy and Ramnarain, 2000) further suggested that there is nothing unique about the replication mechanisms or enzymatic machinery of viruses that isomerize versus those that do not – probably any herpesvirus genome will isomerize during replication if it contains inverted repeats. Thus, isomerization appears to have arisen by serendipity, a consequence of accidental acquisition of inverted repeats and a replication mechanism that is highly recombinagenic by nature.
Even so, inverted repeats have been retained in the genomes of many herpesviruses and this suggests a replicative advantage in vivo, if not in vitro. Indeed, isomerization-defective HSV-1 mutants have been shown to be attenuated in vivo (Jenkins, Donoghue, and Martin, 1996; Jenkins and Martin, 1990), but this could be an effect of gene dosage rather than an advantage conferred by isomerization, since by necessity these studies compared wild type HSV-1, in which genes within the repeats are diploid, to isomerization-defective mutants in which these genes are haploid. Thus, inverted repeats may serve as a stable way to double the dosage of certain genes. Alternatively, isomerization and/or the presence of an internal cleavage site may be important in certain species, tissues, or cell types. Although noninverting mutants of HSV-1 replicate well in cell culture, they failed to replicate locally in corneal or brain tissues of mice following direct inoculation (Jenkins, Donoghue, and Martin, 1996). Similarly, acquisition of a class E genome by pseudorabies virus provides a growth advantage in chicken cells and in chickens and a disadvantage in rabbit cells and in mice (Reilly et al., 1991). The growth defect in rabbit cells was linked to inefficient genome packaging associated with pronounced alterations in the structure of concatemeric DNA (Rall et al., 1991).
Here, we sought to ascertain whether HCMV would tolerate deletion of its b'a'c' sequences, whether this would result in an isomerization-defective genome, and whether this would impact the efficiency of viral replication in vitro. The results clearly demonstrate that like HSV-1, deletion of the b'a'c' sequences results in an isomerization-defective HCMV that replicates with nearly wild type efficiency, at least in fibroblast cells. This is the first class E genome in which the b’a’c’ region has been fully deleted and the first characterization of an isomerically frozen class E mutant other than HSV-1. These results are similar to those obtained with HSV-1 mutants, in that HB15ΔL/S is replication competent but exhibits a modest growth impairment relative to its parental virus. As for the HSV-1 frozen mutants, the growth defect could either derive from changes in gene expression or the inability to isomerize.
That the HCMV genes encoded within the deleted b'a'c' sequences are dispensible was largely predictable from prior studies. The two haploid genes that are disrupted in HB15ΔL/S, IRL13 and IRS1, have been individually deleted without impact on fibroblast replication (Blankenship and Shenk, 2002; Dunn et al., 2003; Jones and Muzithras, 1992; Yu et al., 2002). The remaining genes, all within b’, are diploid in HB15Tn7Δk but haploid in HB15ΔL/S. However, it is important to note that these genes are not normally diploid. The strain AD169 genome has large 11-kb b repeats, whereas the b repeats of HCMV clinical strains are typically smaller than 1 kb. Current evidence suggests that during in vitro propagation of strain AD169 a ~10-kb region of UL encoding RL1–RL12 was duplicated, inverted, and inserted adjacent to b’, greatly expanding the b repeats and rendering RL1–RL12 diploid (Prichard et al., 2001). However, haploidy of these genes does not necessarily result in impaired growth. The varL variant of strain Towne has 60-bp b repeats, is haploid for all of the diploid b sequence genes of AD169 (Bradley et al., 2009), yet replicates in fibroblasts to titers comparable if not superior to those attained by strain AD169 (Hahn et al., 2003b). Even so, there remains a possibility that, like pseudorabies virus, a class E genome might provide HCMV with a replication advantage in certain cell types. For fibroblasts at least this appears not to be the case. While it would be of interest to evaluate replication efficiencies in other cell types such as epithelial and endothelial cells, the genetic background of HB15ΔL/S and HB15Tn7Δk does not permit efficient entry into such cell types due to a mutation in UL131 (Hahn et al., 2004; Wang and Shenk, 2005). Deletion of b'a'c' from a strain with broad-tropism will be needed to evaluate the importance of a class E genome in other cell types.
In recent years a possible evolutionary advantage has been proposed for retention of inverted or direct repeats, as such repeats may allow genomes to quickly regain an optimal genome length after acquiring or loosing sequences (Cui et al., 2008b). This was suggested partly by the observation that during passage in vitro both strain AD169 and the varS variant of strain Towne appear to have lost up to 15.5 kb from UL, yet genome length remained essentially unchanged due to compensatory enlargements of the b′ repeats (Prichard et al., 2001). Thus, in theory, loss or acquisition of sequences could be quickly compensated by modulating the length of inverted repeats.
There are several practical uses for HB15ΔL/S or similar deletion mutants. Deletion of the 13-kb b'a'c' region allows relatively large insertions of foreign sequences for vaccine vector or gene expression purposes. Moreover, HB15ΔL/S contains an attTn7 site that facilitates rapid site-specific insertion of foreign sequences by Tn7-mediated transposition (Hahn et al., 2003a). Also, because HB15ΔL/S virion DNA lacks internal junctions, circular or concatemeric DNA can be differentiated from virion DNA by the presence of terminal junctions. Assays based on detection of such junctions may be useful for in vitro studies of HCMV genome circularization, latency, or persistence. In our investigations HB15ΔL/S has been invaluable for probing the molecular mechanisms of DNA packaging inhibitors (Sauer and McVoy, unpublished data) and mutagenic analysis of HCMV DNA cleavage/packaging signals (Wang and McVoy, unpublished data) using a system analogous to one constructed in murine cytomegalovirus (Wang, Nixon, and McVoy, 2008). Such studies are intractable in the context of the isomerization-competent genome.
Materials and Methods
Cell and Virus culture
HCMV strain AD169 and BAC-derived viruses HB15Tn7Δk and HB15ΔL/S were propagated in MRC-5 human fetal lung fibroblasts (ATCC CCL-171) using Eagle’s minimum essential medium supplemented with 10% fetal bovine serum, 10,000 units/mL penicillin G, 10,000 µg/mL streptomycin sulfate, and 29.2 mg/mL L-glutamine (Gibco-Invitrogen).
Recombinant virus construction
BAC HB15Tn7Δk was derived from pHB15, an infectious BAC clone of the HCMV strain AD169 genome (Hobom et al., 2000), by insertion of a lacZα-mini-attTn7-Kn cassette at the UL18 locus and subsequent removal of Kn by methods described previously (Hahn et al., 2003a; Wang and McVoy, 2008). BAC HB15ΔL/S was constructed from HB15Tn7Δk using linear recombination to substitute Kn for HCMV nucleotides 178,167 to 191,415 (as numbered in GenBank accession #X17403 (Chee et al.)). A Kn-lacZ cassette from pYD-Tn1721 (a gift from Dong Yu) was PCR amplified using primers pYD1721-F (5′-CGGTCATATCTGTTTCCTGTA) and pYD1721-R (5′-AGCGGCCGCAGACTACAAGGA) and ligated into pGEM-T Easy (Promega) to make plasmid pMA228. Digestion with PacI and religation then removed lacZα to produce plasmid pMA236. Primers Forward ΔL/S (5′-CAGTTCATGTAAAAGTCGGTCTCGCCGTGTCCGGCCACGAAGAGGCTGCTCGGTCAT ATCTGTTTCCTGTA) and Reverse ΔL/S (5′-TGCGAGGGGATCATTTATGGGGTCACCGCGTTGTTCGCGAAACATGAACTAGCGGCCGCAGACTACAAGGA) were then used to PCR amplify Kn from pMA236 to produce a PCR product comprised of Kn flanked by HCMV homology sequences (underlined). Linear recombination between the PCR product and BAC HB15Tn7Δk was conducted as previously described (Wang and McVoy, 2008). Candidate clones were screened by PCR to identify clones with proper insertion of Kn (not shown). Two clones were further analyzed by XbaI restriction (not shown) and clone 2 was subsequently designated as HB15ΔL/S. Viruses HB15ΔL/S and HB15Tn7Δk were reconstituted by transfection of their respective BAC DNAs into MRC-5 cells as described previously (Hahn et al., 2003a).
DNA preparation, restriction, and Southern hybridization
Maxi-prep BAC DNAs were prepared using the Nucleobond BAC DNA purification kit (ClonTech) as previously described (Cui et al., 2008a). Virion DNAs were made by low MOI infection of confluent MRC-5 cultures and incubation until full cytopathic effect was reached. The culture media was clarified twice by centrifugation at 2100 rpm for ten minutes, then virions were pelleted at 25,000 rpm for 45 minutes at 4°C. Virion DNA was extracted from the pellets and ethanol precipitated as previously described (McVoy and Adler, 1994; McVoy, Nixon, and Adler, 1997). BAC or virion DNA samples were restricted overnight at 37 °C, electrophoretically separated on 0.7% agarose, and visualized by ethidium bromide staining and UV light. Separated fragments were transferred to Nytran nylon membranes (Schleicher & Schuell) and hybridized using [32]P-labeled probes derived from plasmids pON227, pON2333, or pON1101 as described previously (McVoy and Adler, 1994; McVoy and Ramnarain, 2000).
Viral growth curves
Frozen viral stocks were titered by limiting-dilution in 96-well plates as previously described (Cui et al., 2008a). Confluent MRC-5 cells in 75 cm2 flasks were infected with each virus at an MOI of 4 for single-step and 0.01 for multi-step growth curves. Cultures were washed with PBS 3.5 h post infection to remove unattached virus and fresh media was applied. Samples of culture media were removed daily for 18 days for single-step or every-other day for multi-step growth curves and titered by limiting-dilution as described above.
Acknowledgements
We thank Edward Mocarski for providing plasmids pON227, pON2333, and pON1101, and Dong Yu for plasmid pYD-Tn1721. This work was supported in part by NIH/NIAID grants R21AIO53768 and RO1AI046668 (to MM), NIH/NIAID fellowship 1F31A073209 (to AS), and DFG-HA 2233/2 (to GH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blankenship CA, Shenk T. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J Virol. 2002;76(23):12290–12299. doi: 10.1128/JVI.76.23.12290-12299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley AJ, Lurain NS, Ghazal P, Trivedi U, Cunningham C, Baluchova K, Gatherer D, Wilkinson GW, Dargan DJ, Davison AJ. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J Gen Virol. 2009 doi: 10.1099/vir.0.013250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MS, Bankier AT, Beck S, Sohni R, Brown CM, Cerny R, Horsnell T, Hutchinson CA, Kouzarides T, Marignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston K, Barrell BG. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. In: McDougall JK, editor. Cytomegaloviruses. Vol. 154. New York, N.Y: Springer-Verlag; 1990. pp. 125–169. [DOI] [PubMed] [Google Scholar]
- Chou J, Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Chou J, Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989;63(3):1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, McGregor A, Schleiss MR, McVoy MA. Cloning the complete guinea pig cytomegalovirus genome as an infectious bacterial artificial chromosome with excisable origin of replication. J Virol Methods. 2008a;149(2):231–239. doi: 10.1016/j.jviromet.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, McGregor A, Schleiss MR, Mcvoy MA. The Impact of Genome Length on Replication and Genome Stability of the Herpesvirus Guinea Pig Cytomegalovirus. Virology. 2008b doi: 10.1016/j.virol.2008.12.030. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Wilkie NM. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981;55(2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Wilkie NM. Inversion of the two segments of the herpes simplex virus genome in intertypic recombinants. J Gen Virol. 1983;64(Pt 1):1–18. doi: 10.1099/0022-1317-64-1-1. [DOI] [PubMed] [Google Scholar]
- Delius H, Clements JB. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003;100(24):14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE, Bianchi V, Lehman IR. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J Virol. 1995;69(5):3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE, Bruckner RC, Mocarski ES, Lehman IR. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992;66(1):277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE, Zemelman BV, Lehman IR. Herpes simplex virus type 1 recombination: the Uc-DR1 region is required for high-level a-sequence-mediated recombination. J Virol. 1994;68(6):3733–3741. doi: 10.1128/jvi.68.6.3733-3741.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G, Jarosch M, Wang JB, Berbes C, McVoy MA. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J Virol Methods. 2003a;107(2):185–194. doi: 10.1016/s0166-0934(02)00232-x. [DOI] [PubMed] [Google Scholar]
- Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78(18):10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G, Rose D, Wagner M, Rhiel S, McVoy MA. Cloning of the genomes of human cytomegalovirus strains Toledo, TownevarRIT3, and Towne long as BACs and site-directed mutagenesis using a PCR-based technique. Virology. 2003b;307(1):164–177. doi: 10.1016/s0042-6822(02)00061-2. [DOI] [PubMed] [Google Scholar]
- Hayward GS, Jacob RJ, Wadsworth SC, Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol. 2000;74(17):7720–7729. doi: 10.1128/jvi.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins FJ, Donoghue AM, Martin JR. Deletion of the Herpes simplex 1 internal repeat sequences affects pathogenicity in the mouse. Front Biosci. 1996;1:a59–a68. doi: 10.2741/a106. [DOI] [PubMed] [Google Scholar]
- Jenkins FJ, Martin JR. Role of the herpes simplex virus 1 internal repeat sequences in pathogenicity. Intervirology. 1990;31(2–4):129–138. doi: 10.1159/000150147. [DOI] [PubMed] [Google Scholar]
- Jenkins FJ, Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J Virol. 1986;59(2):494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Muzithras VP. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J Virol. 1992;66(4):2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick BA, Huang ES. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J Virol. 1977;24(1):261–276. doi: 10.1128/jvi.24.1.261-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina RL, Hayward GS. Structural organization of the DNA Molecules from human cytomegalovirus. In: Fields BN, Jaenisch R, editors. Animal Virus Genetics. New York: Academic Press; 1980. pp. 39–55. [Google Scholar]
- McVoy MA, Adler SP. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68(2):1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Nixon DE, Adler SP. Circularization and cleavage of guinea pig cytomegalovirus genomes. J Virol. 1997;71(6):4209–4217. doi: 10.1128/jvi.71.6.4209-4217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Ramnarain D. Machinery to support genome segment inversion exists in a herpesvirus which does not naturally contain invertible elements. J Virol. 2000;74(10):4882–4887. doi: 10.1128/jvi.74.10.4882-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Post LE, Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L–S junction into the genome causes additional genome inversions. Cell. 1980;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski ES, Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982a;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982b;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger KL, Tabares E, Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile KL, Lee GT, Spear PG. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile KL, Spear PG. Enhanced rate of conversion or recombination of markers within a region of unique sequence in the herpes simplex virus genome. J Virol. 1986;58(2):704–708. doi: 10.1128/jvi.58.2.704-708.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Penfold ME, Duke GM, Spaete RR, Kemble GW. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev Med Virol. 2001;11(3):191–200. doi: 10.1002/rmv.315. [DOI] [PubMed] [Google Scholar]
- Rall GF, Lu ZQ, Sugg N, Veach RA, Ben-Porat T. Acquisition of an additional internal cleavage site differentially affects the ability of pseudorabies virus to multiply in different host cells. J Virol. 1991;65(12):6604–6611. doi: 10.1128/jvi.65.12.6604-6611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly LM, Rall G, Lomniczi B, Mettenleiter TC, Kuperschmidt S, Ben-Porat T. The ability of pseudorabies virus to grow in different hosts is affected by the duplication and translocation of sequences from the left end of the genome to the UL-US junction. J Virol. 1991;65(11):5839–5847. doi: 10.1128/jvi.65.11.5839-5847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Pellett PE. The family of Herpesviridae : a brief introduction. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 2381–2397. [Google Scholar]
- Sarisky RT, Weber PC. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J Virol. 1994;68(1):34–47. doi: 10.1128/jvi.68.1.34-47.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J, Summers WC. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Smiley JR, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990;64(10):5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley JR, Fong BS, Leung WC. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Smiley JR, Lavery C, Howes M. The herpes simplex virus type 1 (HSV-1) a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992;66(12):7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro JC, Filpula D, Friedmann T, Spector DH. Structure of the heterogeneous L–S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984;52(2):541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro JC, Spector DH. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986;59(3):591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza SL, Smiley JR. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79(16):10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, McVoy MA. Mutagenesis of the murine cytomegalovirus M56 terminase gene. J Gen Virol. 2008;89(Pt 11):2864–2868. doi: 10.1099/vir.0.2008/003137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Nixon DE, McVoy MA. Definition of the minimal cis-acting sequences necessary for genome maturation of the herpesvirus murine cytomegalovirus. J Virol. 2008;82(5):2394–2404. doi: 10.1128/JVI.00063-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber PC, Challberg MD, Nelson NJ, Levine M, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber PC, Levine M, Glorioso JC. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie NM. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and XbaI. J Virol. 1976;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76(5):2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]