Abstract
Obesity is a public health crisis in The United States. Targeting preadipocyte to adipocyte conversion may be an effective approach to regulate adipose mass. Using differential screening we identified Fstl1, a secreted glycoprotein with roles in immunomodulation, cell growth, cardioprotection, and vascularization, as a “preadipokine”. Fstl1 is highly expressed in 3T3-L1 preadipocytes and dramatically downregulated early in their differentiation to adipocytes. Northern blot analysis of murine tissues reveals white adipose tissue (WAT), lung and heart as primary sites of Fstl1 transcript expression. In WAT, Fstl1 transcript is restricted to the preadipocyte-containing stromal-vascular cell population. Time course studies in multiple adipogenesis models reveal downregulation of Fstl1 is a hallmark of white and brown adipocyte conversion. By Western blot, we show culture media of 3T3-L1 preadipocytes contains high levels of Fstl1 protein that rapidly decline in adipocyte conversion. Moreover, we observe a correlation between preadipocyte phenotype and Fstl1 expression in that TNFα-mediated dedifferentiation of 3T3-L1 adipocytes is accompanied by re-expression of Fstl1 transcript and protein. Treatment of 3T3-L1 preadipocytes with a panel of 18 hormones and other agents revealed the demethylating agent 5-aza-cytidine decreases Fstl1 transcript and protein levels by ~90%. Furthermore, of 10 additional preadipocyte-expressed genes analyzed we find Pref-1, Col1A1, Sca-1/Ly6a, Lox and Thbs2, are also downregulated by 5-aza-cytidine. Using luciferase reporter constructs containing 791 or 3922 bp of the Fstl1 5’-flanking region, we determine negative transcriptional regulation by Kruppel-like factor 15. Together, our data suggest downregulation of Fstl1 expression may be an important feature of preadipocyte to adipocyte conversion.
Keywords: Preadipocyte, adipogenesis, differentiation, obesity, Fstl1, Tsc-36, secreted factor, adipose tissue
INTRODUCTION
Obesity can lead to multiple of health problems including non-insulin dependent diabetes mellitus, hypertension, cardiac infarction and some types of cancers (Flier, 2004; Ntambi and Young-Cheul, 2000; Spiegelman and Flier, 2001). Dysregulation of multiple aspects of adipose tissue and adipocyte function is a hallmark of obesity and its co-morbidities (Flier, 2004; Greenberg and Obin, 2006; Lazar, 2005; Qatanani and Lazar, 2007; Spiegelman and Flier, 2001). White adipose tissue (WAT) is a unique organ that regulates the balance between energy storage and release to maintain energy homeostasis in accord with nutritional status (Zechner et al., 2009). WAT consists of a combination of adipocytes, small blood vessels, nerve tissue, fibroblasts, preadipocytes, stem cells, and other undefined cell types (Geloen et al., 1989; Zuk et al., 2002). WAT is now recognized as a dynamic secretory endocrine organ (Ailhaud, 2006; Breitling, 2009; Waki and Tontonoz, 2007). A number of soluble factors and cytokines including leptin, adiponectin, resistin and TNFα are synthesized and secreted by white adipocytes (Maeda et al., 2002; Spiegelman and Flier, 1996; Steppan et al., 2001). Recent studies in humans indicate that for a given individual the number of adipocytes within WAT is largely constant during adulthood (Spalding et al., 2008). However these studies also revealed that adipocytes within WAT turnover at a rate of ~10% per year (Spalding et al., 2008) and are presumably maintained at constant numbers via ongoing differentiation of preadipocytes to adipocytes.
Adipogenesis is the process of the formation of new adipocytes from preadipocyte precursors, and is thus important for the understanding and control of WAT function and obesity (Lefterova and Lazar, 2009). To date, this process has been studied primarily using in vitro cell culture models. 3T3-L1 preadipocytes, developed several decades ago by Green and colleagues, are the best characterized model of in vitro adipogenesis and are extensively utilized for studies of the molecular mechanisms of adipogenesis (Green and Kehinde, 1974; Green and Kehinde, 1975; Green and Meuth, 1974). 3T3-L1 preadipocytes were cloned from Swiss-3T3 cells, a cell type originally derived from disaggregated late-stage mouse embryo (Green and Kehinde, 1974). While 3T3-L1 preadipocytes exhibit a low degree of spontaneous adipogenesis, their standard in vitro differentiation involves a 2 d treatment of post-confluent cells with the adipogenic agents dexamethasone (Dex) and methylisobutylxanthine (Mix), which greatly enhances extent and uniformity of differentiation (Rubin et al., 1978). The adipogenic transcription factors C/EBPs (CCAAT/enhancer binding proteins) and PPARγ (peroxisome proliferator-activated receptor-gamma) play key roles in the transcriptional cascade governing adipogenesis (Gregoire, 2001; Gregoire et al., 1998; Lazar, 2005; Lefterova and Lazar, 2009; Rajala and Scherer, 2003; Rosen et al., 2002; Rosen and Spiegelman, 2001). The nature of PPARγ as a master regulator in adipogenesis has been documented in numerous studies, including the observation that forced expression of PPARγ, in the presence of a PPARγ ligand, stimulates adipogenesis in fibroblast cell lines (Hu et al., 1995; Lazar, 2005; Shao and Lazar, 1997; Tontonoz et al., 1994). C/EBPβ and C/EBPδ transcription factors are rapidly induced following exposure to adipogenic agents to impact expression of PPARγ and C/EBPα (Rosen and Spiegelman, 2001; Tontonoz et al., 1994) and C/EBPα cooperates with PPARγ to induce a maximal adipogenic response (Hu et al., 1995). The interplay of an expanding and complex network of various types of regulatory signals ultimately leads to the gene expression profile and phenotype of mature lipid-laden terminally-differentiated adipocyte. In many cases, expression of genes that define the mature white adipocyte are not readily detected at the preadipocyte stage, for example those functioning in aspects of insulin signaling, lipid metabolism, lipid droplet function, and numerous adipokines (Gregoire, 2001; Gregoire et al., 1998; Rajala and Scherer, 2003).
A number of large-scale microarray profiling studies of transcriptional changes during 3T3-L1 in vitro adipogenesis have documented upwards of hundreds of genes whose expression is dramatically increased or decreased (Burton et al., 2002; Burton and McGehee, 2004; Burton et al., 2004; Guo and Liao, 2000; Ross et al., 2002; Ross et al., 2000; Soukas et al., 2000; Soukas et al., 2001). Many studies have characterized expression and function of genes whose expression is absent/low in preadipocytes and dramatically upregulated in adipocyte conversion (Gregoire, 2001; Gregoire et al., 1998; Lefterova and Lazar, 2009; Rajala and Scherer, 2003; Rosen and Spiegelman, 2001). In marked contrast, only a few genes with highly selective expression in preadipocytes vs. mature adipocytes have been studied in any detail (Harp, 2004). These include the transmembrane protein Pref-1 (preadipocyte factor-1), which is active as a processed soluble factor (Smas and Sul, 1993) , the secreted factor Wnt10b (Ross et al., 2000), GATA-2 and GATA-3 transcription factors (Tong et al., 2000). Each of these acts as important preadipocyte-expressed inhibitory regulators of adipogenesis, as demonstrated by in vivo and in vitro studies. Pref-1 transcript is highly expressed in 3T3-L1 preadipocytes and absent in adipocytes (Smas and Sul, 1993) and is also present, albeit at markedly lower levels (Soukas et al., 2001), in the preadipocyte-containing stromal-vascular cell population in vivo (Zhou et al., 1999). Constitutive expression or addition of soluble Pref-1 inhibits 3T3-L1 adipocyte conversion (Smas et al., 1997; Smas and Sul, 1993). Pref-1-null mice exhibit obesity (Moon et al., 2002) and Pref-1 null mouse embryo fibroblasts exhibit enhanced differentiation to adipocytes (Kim et al., 2007b; Moon et al., 2002). In the canonical Wnt signaling pathway, Wnts bind to transmembrane Frizzled family receptors leading to stabilization of soluble β-catenin. β-catenin translocates to the nucleus to complex with TCF/LEF transcription factor leading to expression of genes inhibitory for adipogenesis (Prestwich and Macdougald, 2007). Overexpression of Wnts, for example Wnt10b, or stabilizing free cytosolic β-catenin blocks adipogenesis (Ross et al., 2000). GATA-2 and GATA-3 transcription factors are downregulated during adipocyte differentiation and their constitutive expression traps cells at the preadipocyte stage (Tong et al., 2000). A recent study suggests that certain preadipocyte-expressed genes may not only act to block adipogenesis when constitutively expressed in preadipocytes but, under some circumstances, can modulate the phenotype of mature adipocytes. In this case, knockdown of PPARγ in concert with ectopic expression of GATA-3 in 3T3-L1 adipocytes resulted in mature adipocytes taking on aspects of a dedifferentiated transcriptional profile (Schupp et al., 2009). In addition to the long-standing use of in vitro adipogenesis models, new investigations have made significant contributions towards the study of preadipocyte populations in vivo (Park et al., 2008). Until recently, such studies were severely limited due to lack of molecular markers and other tools for their identification and study. Cell lineage tracing approaches developed by Graff and coworkers demonstrated that white preadipocyte progenitor cells reside in the vasculature of adipose tissue (Tang et al., 2008). Friedman and colleagues reported that a subpopulation of early adipocyte progenitor cells, obtained from murine WAT and demonstrating Lin(−):CD29(+):CD34(+):Sca-1(+):CD24(+) surface marker expression, are capable of de novo formation of WAT upon injection into residual fat pads of lipodystrophic mice (Rodeheffer et al., 2008). However the process of emergence of functionally distinct preadipocytes from mesenchymal and/or other stem cell populations, in both the in vitro and in vivo setting, remains obscure. A further elucidation of the molecular definition of preadipocytes is essential to a full understanding of adipogenesis. Additionally, the identification of new genes preferentially down-regulated upon preadipocyte to adipocyte conversion may uncover novel and important regulatory roles for preadipocyte-enriched genes.
The follistatin-like-1 (Fstl1) gene, alternately named FRP (Zwijsen et al., 1994), Flik (Patel et al., 1996), Occ1 (Tochitani et al., 2001) and TSC-36 (Shibanuma et al., 1993), encodes a secreted glycoprotein of ~37 kDa. The Fstl1 cDNA was first isolated in a differential screening for TGF-β1-inducible genes in the MC3T3-E1 murine osteoblastic cell line (Shibanuma et al., 1993). The presence of an extracellular calcium-binding type region places Fstl1 in the BM-40/SPARC/osteonectin protein family (Hambrock et al., 2004; Shibanuma et al., 1993). BM-40/SPARC/osteonectin proteins function as regulators of interactions of cells with the extracellular milieu during development, in response to injury, and in other settings (Bradshaw and Sage, 2001). However, Fstl1 lacks a key structural/functional characteristic of the BM-40/SPARC/osteonectin protein family in that its extracellular calcium-binding domain is nonfunctional (Hambrock et al., 2004) indicating Fstl1 may have distinct functional features. The presence of a follistatin-like domain module that lies between the Fstl1 signal sequence and the extracellular calcium-binding domain region renders Fstl1 distantly-related to the activin and bone morphogenetic protein (BMP) antagonist follistatin (Hambrock et al., 2004; Shibanuma et al., 1993). Follistatin binds and sequesters activin (Chen et al., 2006); however binding of Fstl1 with activin has not been demonstrated. Although there are three other genes that are “follistatin-like” (Fstl3, Fstl4, and Fstl5), these have only weak and limited sequence relationship with Fstl1. Fstl4 and Fstl5 are essentially uncharacterized. Fstl3 has been demonstrated to bind and neutralize TGF-β family ligands, including activin and myostatin (Sidis et al., 2006). Fstl3 null mice exhibit a set of metabolic phenotypes including enhanced insulin sensitivity and reduced visceral fat (Mukherjee et al., 2007).
Initial reports supported a putative tumor suppressor function for Fstl1. For example, Fstl1 transcript expression was decreased in a panel of cancer cells (Mashimo et al., 1997) and following oncogenic transformation of fibroblasts (Johnston et al., 2000; Shibanuma et al., 1993). Ectopic expression of Fstl1 was reported to diminish growth of cancer cells (Sumitomo et al., 2000) and to decrease invasiveness of oncogenically-transformed cells (Johnston et al., 2000). In the last several years, functional roles for Fstl1 have been described in respect to BMP4 signals in embryonic development (Esterberg et al., 2008), revascularization after ischemic injury (Liu et al., 2006; Ouchi et al., 2008), myocyte cell survival (Oshima et al., 2008), immunomodulation (Kawabata et al., 2004; Le Luduec et al., 2008), cytokine regulation (Clutter et al., 2009; Miyamae et al., 2006), and cancer (Chan et al., 2009; Reddy et al., 2008; Trojan et al., 2005).
Information on Fstl1 in adipogenesis is limited to that included in a single report on in in vitro vs. in vivo adipogenesis transcriptional profiles using high density microarrays. In this study by Friedman and coworkers, Fstl1 was among the set of genes categorized as both highly enriched in 3T3-L1 preadipocytes vs. 3T3-L1 adipocytes and in the preadipocyte-containing SVF vs. adipocytes purified from WAT in vivo (Soukas et al., 2001). To our knowledge, this observation was not further followed up in respect to Fstl1, nor have any other reports addressed the expression, regulation, and potential role of Fstl1 in the adipocyte lineage. Through differential hybridization screening of nylon DNA arrays we identified Fstl1 transcript to be highly expressed in preadipocytes and to dramatically decrease during adipogenesis, becoming nearly undetectable in cultures of differentiated adipocytes and we report on the further characterization the expression and regulation of Fstl1 in preadipocytes and in adipogenesis.
RESULTS
Identification of Fstl1 as a Robust Preadipocyte Signature Gene
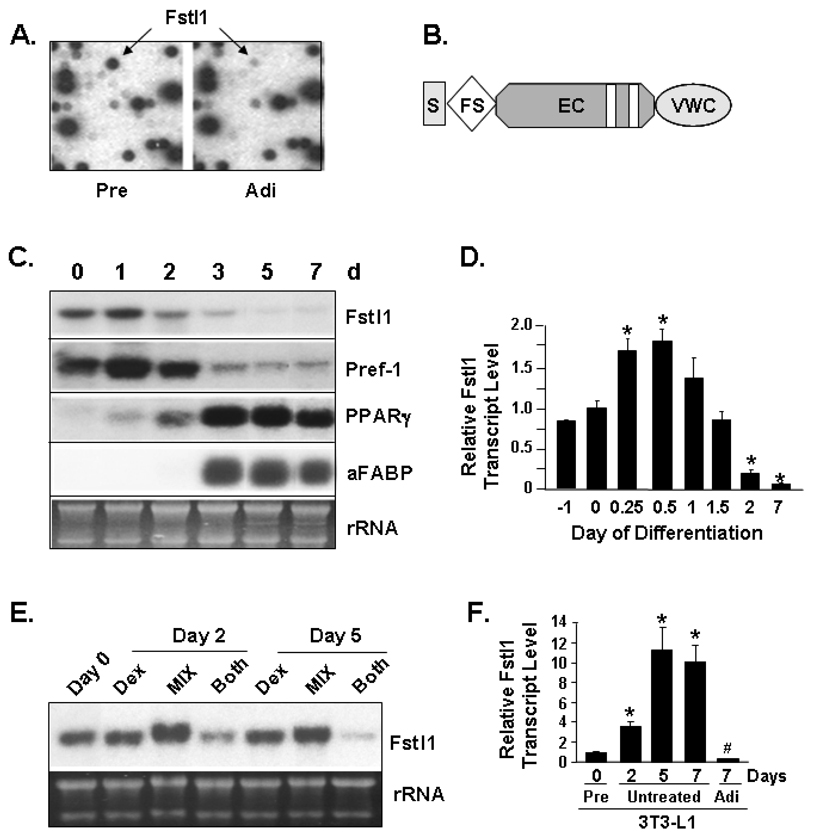
To identify and characterize distinct gene expression patterns that may define preadipocytes, we undertook DNA array analysis of ~5,000 murine genes utilizing filter arrays. Differential screening was conducted via hybridization with reverse-transcribed cDNA probes prepared from 3T3-L1 preadipocyte or 3T3-L1 adipocyte total RNA. Fstl1 demonstrated a very high degree of differential transcript enrichment in preadipocytes vs. adipocytes. A portion of the arrays showing the differential signal for Fstl1 is shown in Figure 1A. Figure 1B depicts the structural features found in the 306 amino acid Fstl1 protein. These are an N-terminal signal sequence, a single follistatin-like domain module showing distant homology to follistatin, a region of homology with the extracellular calcium binding domain of the BM40/SPARC/osteonectin family, and a von Willebrand type C homology region. To further assess expression of Fstl1 transcript during adipocyte differentiation, we examined a time course of in vitro adipocyte conversion of murine 3T3-L1 preadipocytes using Northern blot analysis. For 3T3-L1 differentiation, adipogenesis is induced by a standard 2 d treatment with Dex and Mix; a high degree of adipocyte conversion usually occurs by 5–7 d. As shown in Figure 1C, downregulation of Fstl1 transcript is first noted at 2 d, prior to the removal of adipogenic inducing agents. It is further decreased at 3 d and is markedly reduced upon full differentiation to mature adipocytes at 5–7 d. Here, Pref-1 was utilized as a preadipocyte marker transcript and PPARγ and adipocyte fatty acid binding protein (aFABP) transcripts as adipocyte markers. As shown, the downregulation of Fstl1 transcript at 2 d is inversely related to the appearance of transcripts for these adipocyte-specific genes.
Figure 1. Fstl1 Transcript Expression During Adipocyte Conversion of 3T3-L1 Preadipocytes.
A. Initial differential screening identification of Fstl1 as a novel gene enriched in 3T3-L1 preadipocytes vs. 3T3-L1 adipocytes. Shown is a portion of the DNA filter array indicating Fstl1 signal (arrows) in 3T3-L1 preadipocytes (Pre) and minimal signal in 3T3-L1 adipocytes (Adi). B. Structural motifs of the Fstl1 protein. S, signal sequence; FS, follistatin-like module; EC, extracellular calcium binding homology region with vertical bars indicating EF-hand type motifs; VWC, von Willebrand type C homology region. C. Regulation of Fstl1 transcript in 3T3-L1 adipogenesis. 3T3-L1 cells were harvested as preadipocytes before induction of adipogenesis (0) and at indicated time points in days (d) post-induction of adipocyte differentiation. Northern blot analysis was performed with murine Fstl1, Pref-1, PPARγ and aFABP cDNA probes. D. Q-PCR analysis of Fstl1 transcript expression in 3T3-L1 adipogenesis. Signal was corrected against Gapdh level. * indicates p<0.01 compared to d 0 sample. Value in 0 d was set to 1. E. Fstl1 transcript regulation by adipogenic cocktail components. Post-confluent 3T3-L1 preadipocytes were treated with 10% FBS supplemented with 1 µM Dex, 0.5 mM Mix, or both Mix and Dex for 48 h. Cultures were either harvested at 48 h or maintained through d 5 in 10% FBS and then harvested. Northern blot was performed using murine Fstl1 cDNA probes. For C and E, the EtBr staining of rRNA is shown as a gel loading control. F. Q-PCR analysis of Fstl1 transcript in post-confluent 3T3-L1 preadipocytes. Cells were harvested at 0 d, or at 7 d post-standard adipogenic induction (first and last lanes). Cells not treated for adipogenic induction (untreated) were harvested at indicated days following the 0 d time point. *, # indicates p<0.001 in regard to increased or decreased Fstl1 transcript level relative to 0 d, respectively.
To examine expression of Fstl1 in very early 3T3-L1 adipogenesis, we conducted time course studies focusing on the first two days. Q-PCR analysis in Figure 1D shows that during the first 1.5 d of adipogenic induction a transient increase of Fstl1 transcript is noted, peaking at ~ 2-fold at 0.25–0.5 d post-adipogenic induction. This is followed by a ~ 5-fold diminution of Fstl1 transcript expression between 1.5 d and 2 d and a ~12-fold decrease of Fstl1 transcript by 7 d. While we have not investigated the function nor mechanism of the early transient increase in Fstl1 transcript, its increase is coincident with the clonal expansion phase of in vitro adipogenesis as post confluent preadipocytes undergo several rapid rounds of the cell cycle (Tang et al., 2003a; Tang et al., 2003b). We next examined the regulation of Fstl1 transcript in response to treatment of 3T3-L1 preadipocytes with the individual Dex and Mix components of the adipogenic cocktail. While the response of preadipocytes to Dex/Mix induced adipogenesis are likely complex and combinatorial, members of the C/EBP family of pro-adipogenic transcription factors were the first identified direct targets for Dex and Mix action (Cao et al., 1991; Umek et al., 1991; Yeh et al., 1995). Upon adipogenic induction, Dex stimulated C/EBPδ gene expression and MIX stimulated C/EBPβ expression (Cao et al., 1991; Umek et al., 1991; Yeh et al., 1995). These early responses feed into subsequent transcriptional cascades including upregulation of C/EBPα (Cao et al., 1991; Umek et al., 1991; Yeh et al., 1995). Downregulation of Fstl1 transcript requires the combined actions of the Dex/ Mix adipogenic cocktail and is not regulated by either individual component (Figure 1E).
Lastly we further confirmed that the decrease we observed in Fstl1 transcript level was dependent on the adipogenic program and not merely due to the consequence of prolonged in vitro culture of 3T3-L1 preadipocytes, irrespective of whether or not they were treated with adipogenic agents (Figure 1F). For this, cultures of 3T3-L1 preadipocytes at 2 d post-confluence, the standard point for induction of adipogenesis, were either changed into Dex/Mix adipogenic media for 2 d or subject only to media change with standard growth media. RNA was analyzed from 0 d cultures and from preadipocyte cultures at 2 d, 5 d, and 7 d; these latter 3 samples had not been exposed to adipogenic treatment. We also analyzed RNA from cultures at 7 d post-adipogenic induction for comparison; these evidenced the typical morphology of mature lipid-filled adipocytes (data not shown). As is shown in Figure 1F, we observed the anticipated downregulation of Fstl1 transcript in 7 d adipocytes vs. 0 d preadipocytes (first and last columns, respectively). However, contrary to this downregulation as cells undergo adipogenic conversion, we found an ~10-fold increase in Fstl1 transcript level in parallel cultures of untreated 3T3-L1 preadipocytes (i.e. those not subject to adipogenic hormone treatment) were maintained in culture through 7 d. While we do not yet know what role this increase in Fstl1 transcript level may play in post-confluent 3T3-L1 preadipocytes, together our data clearly show that decreased levels of Fstl1 transcript are specific to the adipogenic differentiation program. It is not merely a result of prolonged maintenance of cultures of post-confluent, presumably growth-arrested, 3T3-L1 preadipocytes.
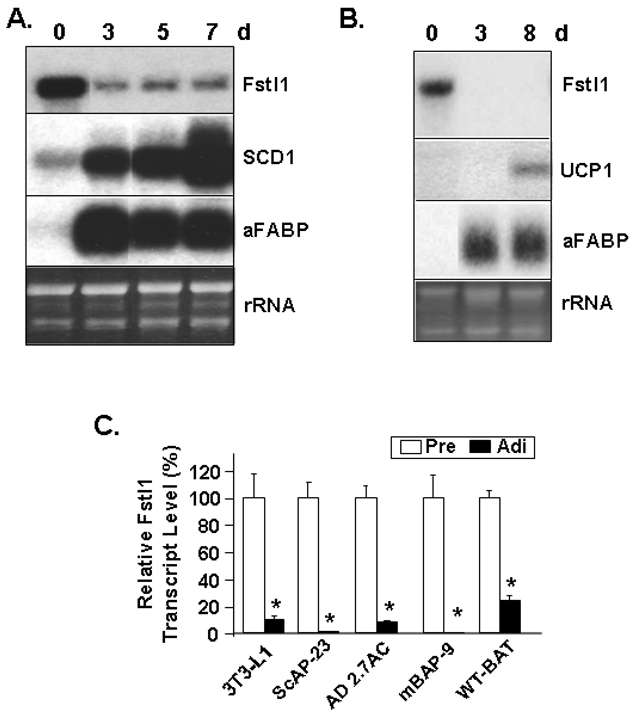
We extended study of differentiation-dependent downregulation of Fstl1 transcript to additional models of in vitro adipogenesis. The Northern blot in Figure 2A shows marked downregulation of Fstl1 transcript also accompanies adipogenic conversion of ScAP-23 cells, a new white preadipocyte cell line generated in our laboratory from murine subcutaneous WAT (Kim et al., 2007a) . Mammals also have brown adipose tissue (BAT) which serves primarily to dissipate energy as opposed to storing it (Lowell and Flier, 1997). Brown adipocytes are present in adult humans and have an emerging role in human metabolism and obesity (Cinti, 2006). Whereas it was initially thought that these brown adipocytes were found only interspersed within select other tissues, it has recently been reported that BAT can be present as a distinct tissue entity in some adult humans (Cypess et al., 2009). The Northern blot in Figure 2B shows that, similar to white adipogenesis, downregulation of Fstl1 transcript level is also a hallmark of brown adipogenesis. This was examined using an established brown adipocyte cell line (WT-BAT) established by Kahn and coworkers from BAT of neonatal mice (Klein et al., 2002). The expression of the brown adipocyte-specific transcript uncoupling protein 1 (UCP1) at day 8 is a marker of brown adipocyte conversion. Figure 2C is Q-PCR data for levels of Fstl1 transcript in preadipocytes and mature 7 d adipocytes in five in vitro models of adipogenesis. The Ad2.7AC cell line is derived from bone marrow stromal cells and is committed to an adipocyte lineage (Lecka-Czernik et al., 1999). The mBAP-9 preadipocyte cell line has recently been generated in this laboratory from BAT of 3 wk-old mice (manuscript in preparation). Fstl1 is a differentiation-dependent downregulated transcript in each of these cell culture models of in vitro adipogenesis.
Figure 2. Fstl1 Transcript Expression is Decreased in Multiple in vitro Models of White and Brown in vitro Adipogenesis.
A. Fstl1 downregulation during ScAP-23 differentiation. Cells were harvested as preadipocytes prior to induction of adipogenesis at day (d) 0 and at indicated days post-induction of adipogenic differentiation. Northern blot shows transcript level for Fstl1, SCD1 and aFABP. B. Fstl1 transcript expression is decreased in brown adipogenesis. WT-BAT preadipocytes were harvested at confluence (0 d) and adipocytes harvested at day 3 and day 8 post-induction of differentiation. Northern blot analysis was performed with murine Fstl1, UCP1, and aFABP probes. For panels A and B, EtBr staining of rRNA is shown as a gel loading control. C. Q-PCR analysis of Fstl1 transcript levels in preadipocytes (Pre) and adipocytes (Adi) of 3T3-L1, ScAP-23, AD2.7AC, mBAP-9, WT-BAT cell lines. Value in the respective preadipocyte sample was set to 1 and * indicates p<0.001 for Adi vs. Pre.
Fstl1 Transcript Expression in WAT is Restricted to the Preadipocyte-Containing Stromal-Vascular Fraction
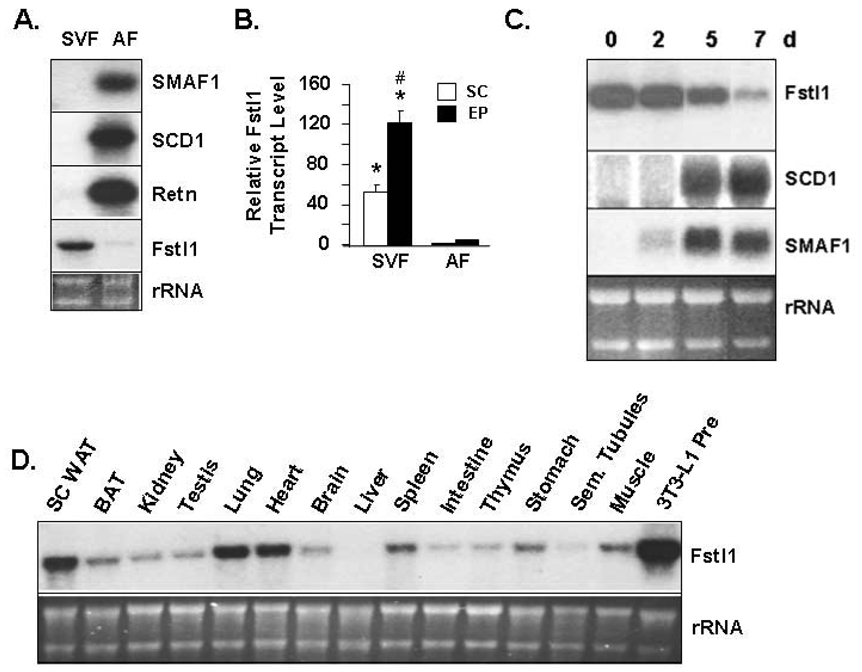
A microarray analysis by Soukas and coworkers revealed distinct transcriptional profiles for 3T3-L1 in vitro vs. in vivo murine adipogenesis (Soukas et al., 2001); in that case comparison of the preadipocyte-containing stromal vascular fraction (SVF) and adipocyte fraction of WAT served as a proxy for in vivo white adipogenesis. Soukas and coworkers reported that a large number of genes are highly differentially expressed for in vitro vs. in vivo sources (Soukas et al., 2001). Of interest, Fstl1 was among the various genes listed in their microarray data to be enriched in preadipocytes vs. adipocytes both in vitro and in vivo; suggestive of importance in the adipocyte lineage. To investigate Fstl1 transcript regulation during adipogenesis in a model that represents the process of adipogenesis in vivo we used collagenase digestion of murine WAT to prepare purified adipocytes and the preadipocyte-containing SVF. The Northern blot in Figure 3A illustrates that Fstl1 transcript was not detected in the adipocyte fraction of murine SC WAT, a cell population that does not contain preadipocytes, but is readily detected in the preadipocyte-containing SVF. For this study, stearoyl-CoA desaturase-1 (SCD1) small adipocyte factor 1 (SMAF1) and resistin (Retn) transcripts were used as adipocyte markers. While our data does not exclude the possibility that other SVF cell types may also be a source of Fst1l expression within WAT, we have found that the SVF and each of the preadipocyte cell lines examined in Figure 2C express from ~500 to ~3000 times higher levels (p<0.01) of Fstl1 transcript level than do cells of the murine macrophage line RAW264.7 (data not shown). This suggests that macrophages within WAT are likely not a source of Fstl1 transcript. This view is also supported by the fact that we do not observe any consistent degree of increase in Fstl1 transcript level in obese (ob/ob) WAT compared with that for wild type mice (data not shown). Obese mice evidence dramatically enhanced macrophage numbers within their WAT, which results in the detection of markedly increased levels for macrophage-expressed transcripts therein (Cancello et al., 2006; Kamei et al., 2006; Permana et al., 2006; Weisberg et al., 2003). If macrophages made a significant contribution to WAT level of Fstl1 transcript expression, we would expect to observe a dramatically increased Fstl1 transcript level in obese WAT.
Figure 3. Fstl1 Transcript Expression in Cellular Fractions of WAT and in Adipogenesis of Primary Preadipocytes.
A. Fstl1 transcript in stromal vascular fraction (SVF) and adipocyte fraction (AF) of murine WAT. Northern blot shows transcript level for SMAF1, SCD1, Retn and Fstl1. B. Q- PCR assessment of Fstl1 transcript level in cell fractions of WAT. cDNA derived from SVF or adipocyte fraction (AF) of subcutaneous (SC) or epididymal (EP) WAT was used for Q-PCR analysis of Fstl1 transcript. Note: the third column represents SC tissue but the white color of the bar cannot be discerned due to low column height. The level in SC AF was set to 1. * indicates p<0.001 for SC SVF vs. SC AF and for EP SVF vs. EP AF. # indicates p<0.001 for EP SVF vs. SC SVF. C. Fstl1 transcript downregulation during adipogenic conversion of rat primary white preadipocytes. RNA was collected from preadipocytes (0) and at indicated times post-induction of adipocyte differentiation and analyzed by Northern blot using murine Fstl1, SCD1 and SMAF1 cDNA probes. D. Fstl1 transcript level in a panel of adult murine tissues. 5 µg of total RNA from indicated tissues were analyzed by Northern blot using murine Fstl1 cDNA probe. For A, C and D, EtBr staining of rRNA is shown as a gel loading control.
It is well-established that increased intra-abdominal WAT is linked to a higher risk of diabetes and cardiovascular disease compared to subcutaneous adipose tissue (SC), suggestive of functional heterogeneity of adipocytes (Gesta et al., 2007). We used Q-PCR, Figure 3B, to compare Fstl1 transcript expression in intra-abdominal epididymal (EP) WAT and in subcutaneous (SC) WAT. As expected, for SC and EP WAT, Fstl1 transcript is enriched over 50-fold in the SVF vs. the adipocyte fraction. Additionally, the Fstl1 transcript level in EP SVF is approximately 2-fold higher than that in SC SVF, indicating Fstl1 appears to demonstrate a degree of depot-dependent differential expression (Figure 3B). We next examined regulation of Fstl1 transcript following culture of the preadipocyte-containing SVF cells harvested from rat WAT and during differentiation of these primary white preadipocytes to adipocytes, Figure 3C. We utilized rat cells for this study as we typically observe a more uniform and a higher percentage of adipocyte differentiation with primary cultures of rat SVF cells compared with murine SVF cells. Dramatic downregulation of Fstl1 transcript occurred as a result of differentiation of primary white preadipocytes to adipocytes (Figure 3C). As was noted for the established preadipocyte cell lines, presented in Figure 1 and Figure 2, the primary cell data indicates that downregulation of Fstl1 transcript also inversely correlates with emergence of the adipocyte phenotype, indicated by expression of the adipocyte marker transcripts SCD1 and SMAF1. The Northern blot in Figure 3D shows that SC WAT is a primary site of Fstl1 transcript expression, with prominent expression also noted for lung and heart. Markedly lower levels of Fstl1 transcript were detected most other tissues examined except for liver, which appears negative. While our studies are the first to report Fstl1 transcript expression levels in WAT, data on other murine tissues shown in Figure 3D is largely consistent with published reports (Adams et al., 2007; Liu et al., 2006; Mashimo et al., 1997).
Expression and Regulation of Fstl1 Protein in Adipogenesis
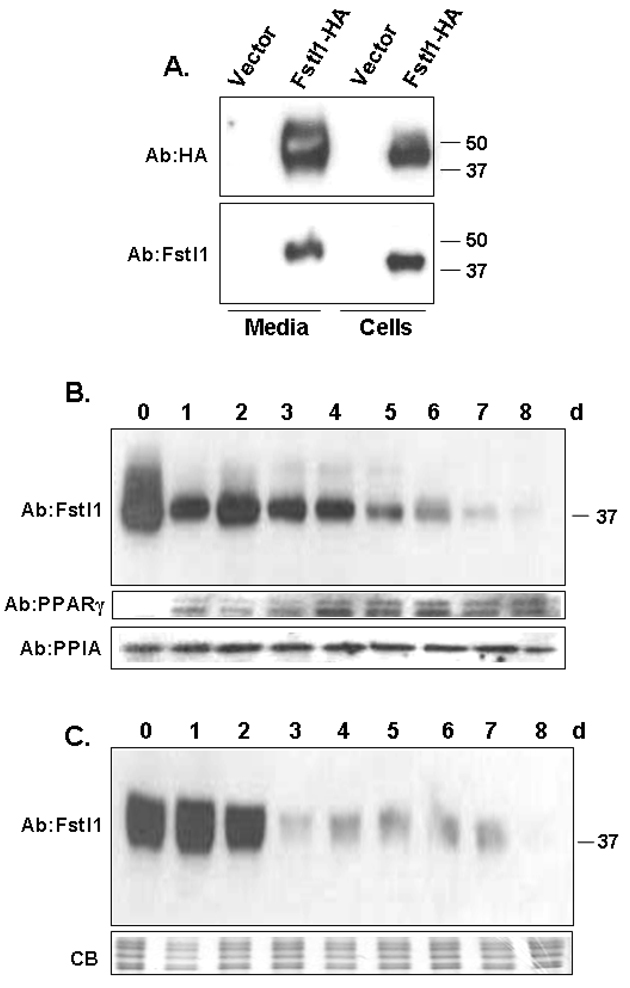
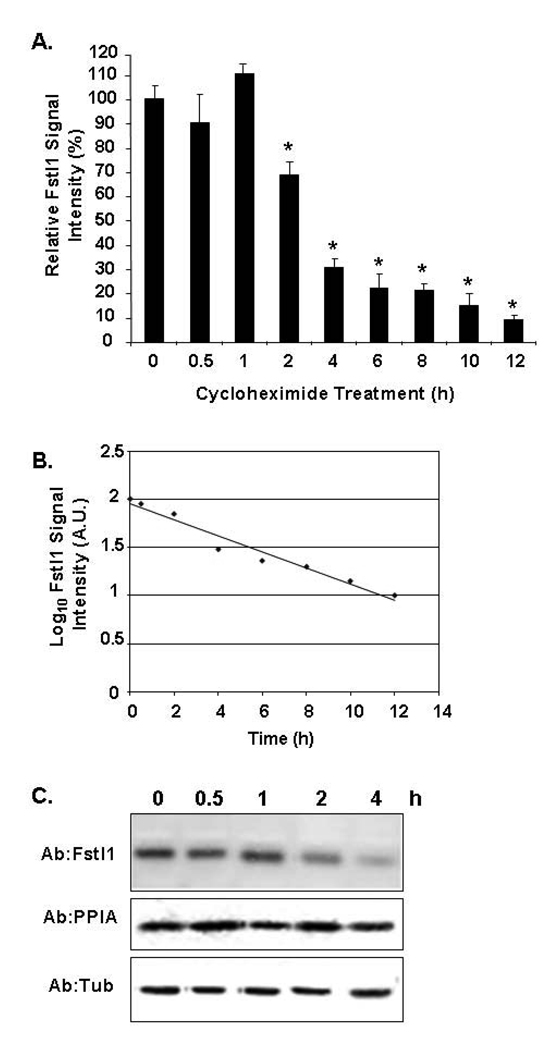
To begin studies on the Fstl1 protein, we first characterized a commercially available Fstl1 antibody utilizing transfection of an HA-tagged Fstl1 expression construct into COS cells, followed by analysis of lysate and culture media by Western blot. The top panel of Figure 4A indicates successful detection of the Fstl1-HA protein in media and cells using anti-HA antibody, consistent with the secreted nature of Fstl1 protein. The lower panel of Figure 4A indicates that anti-mouse Fstl1 antibody readily detects ectopically expressed Fstl1-HA protein in both COS cell lysates and culture media. The molecular mass found for secreted Fstl1 in media appears larger than that found in cell lysates, consistent with a previous report on glycosylation of Fstl1 (Hambrock et al., 2004). No background or cross-reactive species are evident for vector-only transfectants, confirming the utility of the Fstl1 antibody for Western blot analysis. To determine Fstl1 protein expression during adipogenesis, 3T3-L1 cell lysates and culture media were collected prior to induction of adipogenesis as confluent preadipocytes (0 d), or at daily times post-induction of differentiation. An anti-PPARγ antibody was employed as a marker for adipocyte differentiation. Endogenous Fstl1 protein level in 3T3-L1 cell lysates was downregulated at 1 d post treatment with adipogenic agents and markedly decreased levels were noted at 5–6 d. Fstl1 protein was all but absent by 7–8 d post-adipogenic induction, Figure 4B. Figure 4C illustrates that Fstl1 protein is secreted into culture media by 3T3-L1 preadipocytes, defining Fstl1 as a new “preadipokine”. Levels of Fstl1 protein secreted into media declined dramatically by 3 d. For both Figures 4B and 4C, we chose to present blot data of a longer film exposure time in order to be fully informative in respect to the lower levels of Fstl1 protein expressed at the later time points. As a result earlier time points appear somewhat overexposed. To determine protein half-life for secreted Fstl1, 3T3-L1 preadipocytes were cultured with cycloheximide. Media was collected at subsequent time points and assessed for endogenous Fstl1 protein by Western blot analysis using anti-Fstl1 antibody. The graph in Figure 5A shows quantitation of Western blot signals and a semi-log plot of the results is presented in Figure 5B. These data indicate that the protein half-life of preadipocyte-secreted Fstl1 is ~3 h. A representative Western blot for Fstl1 protein for the 0–4 h time points of the cycloheximide study are shown in Figure 5C; a minimal exposure time for digital image acquisition was used such that signal differences at each of the time points are maximally evident. Fstl1 protein has been demonstrated to be a target for MMP-2 mediated proteolytic degradation to generate multiple smaller protein species (Dean et al., 2007) and active MMP-2 is present in 3T3-L1 preadipocytes (Croissandeau et al., 2002). However, in our studies to date of preadipocyte-secreted Fstl1, we have not observed shorter Fstl1 species indicative of proteolysis.
Figure 4. Expression and Secretion of Fstl1 Protein During 3T3-L1 Adipogenesis.
A. Assessment of Fstl1 antibody. Vector only or an Fstl1-HA expression construct was transiently transfected into COS cells. Culture media and cell lysates were analyzed by Western blot using anti-HA (top panel) or anti-Fstl1 (lower panel) primary antibodies. B, C. Fstl1 downregulation in 3T3-L1 cell lysates (B) and culture media (C) during adipocyte differentiation. Cells and culture media were harvested from confluent 3T3-L1 preadipocytes prior to induction of adipogenesis (day 0) or daily post-induction of differentiation. Western blot was performed using anti-Fstl1 primary antibody. For B (lower two panels), the membrane was re-processed using anti-PPARγ primary antibody as a differentiation marker and anti-PPIA (peptidylprolyl isomerase A) primary antibody as a loading control. For panel C (lower panel), the gel was stained with Coomassie Blue (CB) to assess evenness of sample loading. For Fstl1 blots in Figures B and C, a longer exposure is presented to best illustrate signal at the later time points.
Figure 5. Determination of Protein Half-Life for Endogenous Preadipocyte-Secreted Fstl1.
3T3-L1 preadipocytes were treated with 5 µg/ml of cycloheximide and harvested at indicated time points in hours (h) post-treatment. A. Quantitation of Western blot signal for preadipocyte-secreted Fstl1 in triplicate time point samples. B. Semi-log plot of decrease of preadipocyte-secreted Fstl1 protein level over time in hours (h). C. Western blot for preadipocyte-secreted Fstl1 protein in representative of samples for the 0–4 h time points. β tubulin and PPIA are shown as controls. A shorter exposure is presented in order to best visualize Fstl1 signals at each intermediate time point.
Re-emergence of Fstl1 Expression Tracks with TNFα-Induced Adipocyte Dedifferentiation
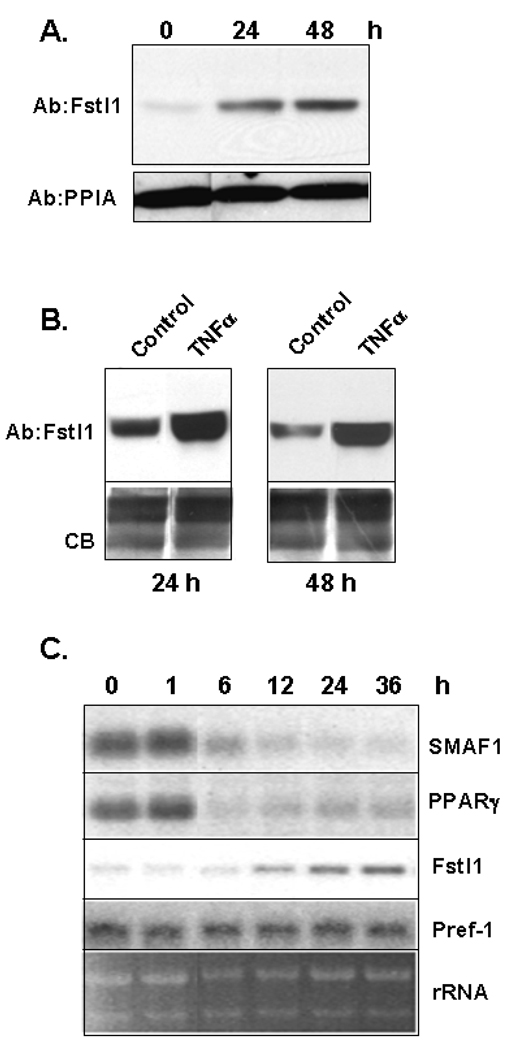
It is well known that TNFα is a contributing cause of the insulin resistance seen in obesity and obesity-linked diabetes (Hotamisligil, 2006). Genome-scale transcriptional profiling has revealed that TNFα treatment of 3T3-L1 adipocytes results in suppression of a large number of adipocyte-specific genes while concomitantly activating many preadipocyte genes (Ruan et al., 2002a). TNFα is also a major regulator of WAT gene expression in vivo (Ruan and Lodish, 2003; Ruan et al., 2002b). Western blot analysis in Figure 6A and Figure 6B indicates TNFα treatment of 3T3-L1 adipocytes results in a marked upregulation of Fstl1 protein level both in cell lysates and in media. The Northern blot in Figure 6C shows that treatment of 3T3-L1 adipocytes with TNFα results in re-expression of Fstl1 transcript over the 36 h time period examined. Decreased SMAF-1 and PPARγ transcript expression indicate de-differentiation of adipocytes, as anticipated. These observations of re-emergence of Fstl1 expression upon TNFα-mediated adipocyte dedifferentiation indicate that Fstl1 transcript and protein expression closely tracks with, and is inversely related to, the adipocyte phenotype. As was previously reported by others (Xing et al., 1997), transcript for the preadipocyte gene Pref-1 remains largely constant with TNFα treatment.
Figure 6. Regulation of Fstl1 Protein and Transcript Expression by TNFα.
A and B. Fstl1 protein level in cell lysates (A) and in media (B) of 3T3-L1 adipocytes (time 0 and control, respectively) or following treatment with 10 ng/ml TNFα for 24 h or 48 h. For panel A, samples were processed for Western blot using either Fstl1 antibody or for loading control, anti-PPIA antibody. For panel B, the gel was stained with Coomassie Blue (CB) to assess even sample loading. C. Fstl1 transcript level in 3T3-L1 adipocytes during treatment with TNFα. 3T3-L1 adipocytes were treated with 10 ng/ml TNFα and RNA samples were collected prior to treatment (0) and at indicated times in hours (h) thereafter. Northern blot was performed using murine SMAF1, PPARγ, Fstl1 and Pref-1 cDNA probes. EtBr staining for rRNA is shown as a gel loading control.
Preadipocyte Expression of Fstl1 is Decreased by 5-Aza-CR
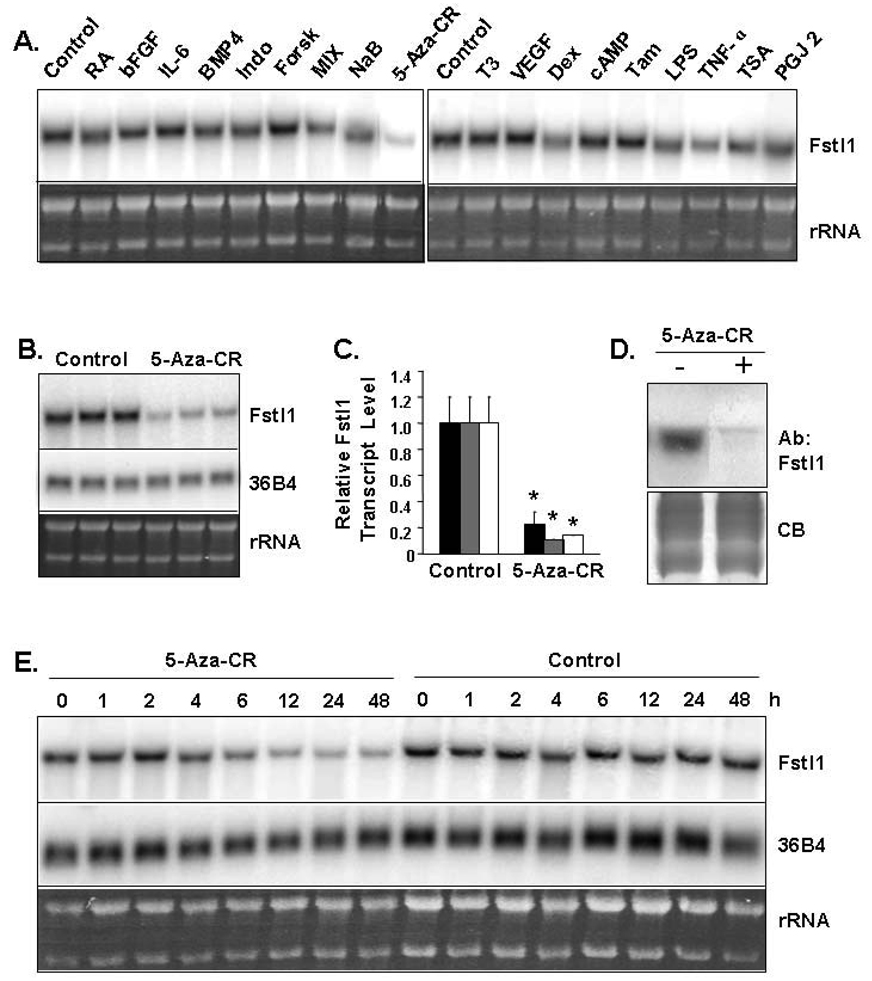
To investigate signals that may regulate Fstl1 transcript expression, 3T3-L1 preadipocytes were individually treated with 18 hormones and other agents; the majority of these have roles in adipogenesis and/or adipocyte function. Figure 7A shows Northern blot analysis of 3T3-L1 preadipocytes following 24 h agent treatment. Of all the agents tested, 5-aza-CR was observed to have the most dramatic effect on modulation of Fstl1 transcript level. The incorporation of 5-aza-CR, an analogue of cytosine, into DNA leads to inhibition of methyltransferase activity to result in DNA demethylation. Demethlyation of a gene is nearly exclusively reported to correlate with its activation (Christman, 2002; Jones, 1985). The decrease in Fstl1 transcript level by 5-aza-CR was confirmed on triplicate samples presented in the Figure 7B Northern blot. To quantitative these effects we used Q-PCR; this was assessed employing three separate internal standards, Gapdh, 36B4 and 18S (Figure 7C). We find that, on average, 5-aza-CR resulted in an 89% decrease in Fstl1 transcript level in 3T3-L1 preadipocytes. Figure 7D shows that protein level of Fstl1 in media of 3T3-L1 preadipocytes is markedly decreased after a 24 treatment with 5-aza-CR. We also examined the time course of Fstl1 transcript downregulation by 5-aza-CR in 3T3-L1 preadipocytes. Figure 7E indicates that Fst1l transcript levels are decreased by 6 h of treatment and further decreased at 12–48 h. The transcript level of a control transcript 36B4 remained relatively unchanged through 48 h.
Figure 7. Levels of Fstl Transcript and Protein are Decreased in 3T3-L1 Preadipocytes by 5-Aza-CR Treatment.
A. Confluent 3T3-L1 preadipocytes were either untreated (control) or treated with the indicated agents for 24 h. RNA samples were collected and analyzed by Northern blot using murine Fstl1 cDNA probe. The abbreviations used are: RA, retinoic acid; b-FGF, basic fibroblast growth factor; IL-6, interleukin-6; BMP4, bone morphogenetic protein 4; Indo, indomethicin; Forsk, forskolin; Mix, methylisobutylxanthine; NaB, sodium butyrate; 5-aza-CR, 5-aza-cytidine; Dex, dexamethasone; Tam, tamoxifen; T3, triiodothyronine; VEGF, vascular endothelial cell growth factor; cAMP, cyclic adenosine monophosphate; LPS, lipopolysaccharide; TSA, trichostatin A. B. Triplicate samples for control untreated or 5-aza-CR treated 3T3-L1 preadipocytes. Northern blot was performed using murine Fstl1 cDNA probe. C. Q- PCR assessment of Fstl1 transcript level in control and 5-aza-CR treated 3T3-L1 preadipocytes. Fstl1 transcript signal was corrected against three internal standards (Gapdh, black bar; 36B4, gray bar; and 18S, white bar). The transcript level for control samples was set to a value of 1. * indicates p<0.001 for treated samples vs. respective controls. D. Secreted Fstl1 protein expression by 3T3-L1 preadipocytes is decreased by 5-aza-CR treatment. 3T3-L1 preadipocytes were treated with 1 mM 5-aza-CR or untreated (control) for 24 h. Culture media was collected for Western blot analysis by using anti-Fstl1 antibody. The gel was stained with Coomassie Blue (CB) to assess evenness of sample loading. E. Regulation of Fstl1 transcript expression during 5-aza-CR treatment of 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were treated with 1 mM 5-aza-CR or untreated (control) for indicated time in hours (h). Northern blot was performed using a murine Fstl1 probe or a 36B4 probe as control. For panels A, B and E, EtBr staining of rRNA is shown as a gel loading control.
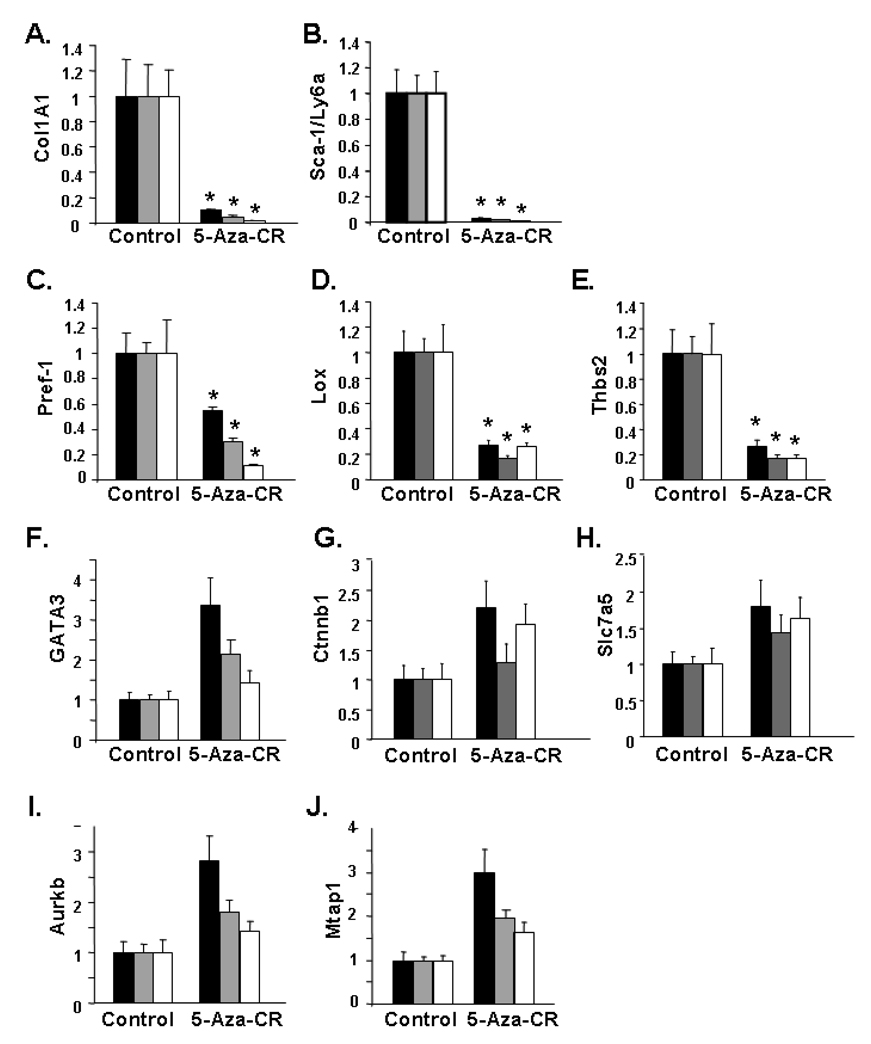
It has been demonstrated that for cultures of Swiss 3T3 and C3H10T1/2 mouse cells treated with 5-aza-CR, three mesenchymal-derived phenotypes, characterized as muscle cells, adipocytes and chondrocytes emerge (Pinney and Emerson, 1989; Taylor and Jones, 1979). Based on regulation of Fstl1 expression by 5-aza-CR, we hypothesize that Fstl1 may be involved in cell fate determination in the preadipocyte/mesenchymal lineage. To investigate whether the phenomenon of transcript downregulation by 5-aza-CR might be a common theme for genes that are specifically decreased during preadipocyte to adipocyte conversion, we carried out additional transcript analysis using Q-PCR. We have previously reported that downregulation of a subset of preadipocyte genes, which includes Lox, Thbs2, Ctnnb1, Slc7a5, Aurkb and Mtap1 occurs in both 3T3-L1 and ScAP-23 adipogenesis (Kim et al., 2007a). Other studies have indicated the specific expression/enrichment of Col1A1 (Weiner et al., 1989; Yi et al., 2001), Sca-1/Ly6a (Rodeheffer et al., 2008), Pref-1(Smas et al., 1997; Smas and Sul, 1993), and GATA-3 (Tong et al., 2000) in adipogenic precursors. Figures 8A–E reveals that transcript levels for a subset of the genes examined, namely Col1A1, Sca-1/Ly6a, Pref-1, Lox and Thbs2 are also downregulated by 5-aza-CR treatment of 3T3-L1 preadipocytes. Several of these genes (Col1A1, Pref-1, Thbs2), encode secreted/soluble factors (Bornstein et al., 2000; Smas et al., 1997; Weiner et al., 1989; Yi et al., 2001). Lox is known to be involved in extracellular matrix production (Lucero and Kagan, 2006), and Sca-1/Ly6a is a surface antigen (Rodeheffer et al., 2008). Transcript levels for the five other genes examined are very marginally increased (Figure 8F–J).
Figure 8. Assessment of Effects of 5-Aza-CR Treatment of 3T3-L1 Preadipocytes on Transcript Levels of 10 Preadipocyte-Selective Genes.
Q-PCR assessment of relative transcript level in untreated (control) and 5-aza-CR treated 3T3-L1 preadipocytes. Signal for each indicated transcript was corrected against three internal standards (Gapdh, black bar; 36B4, gray bar; and 18S, white bar). The transcript level for control samples was set to a value of 1. * indicates a significant decrease (p<0.005) for treated samples vs. their respective controls.
Functional Analysis of the Fstl1 Promoter Region
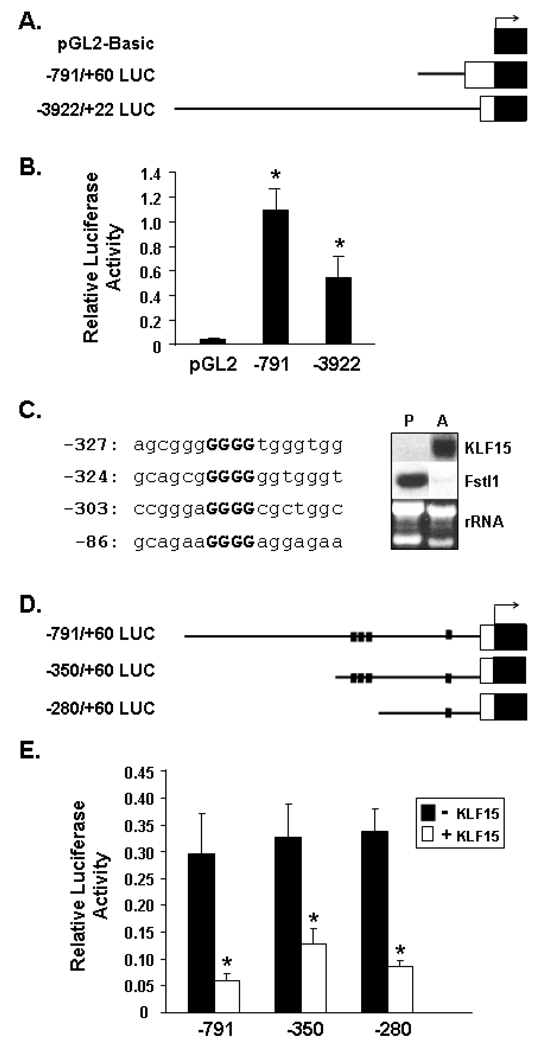
To date, there is no information regarding the transcriptional control of the Fstl1 gene promoter in any cell type, and very little is generally known on mechanisms of gene downregulation of preadipocyte-expressed genes during adipogenesis. To begin to characterize regulation of the Fstl1 promoter, two portions of the 5′ flanking regulatory region of the murine Fstl1 gene, encompassing −791/+60 bp and −3922/+22 bp, were used to generate luciferase reporter constructs, Figure 9A. 3T3-L1 preadipocytes were transfected with these Fstl1 luciferase reporter constructs or empty pGL2-basic vector. As shown in the graph of Figure 9B, luciferase activities of the Fstl1 luciferase promoter constructs are markedly elevated compared to pGL2-basic negative control.
Figure 9. Promoter Activity and KLF15 Regulation of the Murine Fstl1 5’ Flanking Region.
A. Schematic diagram of Fstl1-luciferase constructs. The solid line represents the Fstl1 promoter region, the white box the 5’ untranslated region of Fstl1, and the black box the luciferase reporter gene. B. Promoter activity of Fstl1 5’ flanking region in 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were transfected with the indicated luciferase reporter constructs (−791 or −3922) or empty pGL2-Basic vector (pGL2). An * indicates p<0.001 compared to empty pGL2-Basic vector. C. Left Panel: Sequence of putative KLF15 sites in the Fstl1 5’ flanking region. The consensus core motif of the inverted KLF sites are in bold. Numbers at left refer to the most 5’ position of the sequence relative to the Fstl1 transcription start site. Right Panel: Northern blot analysis of KLF15 and Fstl1 transcripts in 3T3-L1 preadipocytes (P) and 7 d adipocytes (A), with rRNA shown as a loading control. D. Schematic diagram of Fstl1-luciferase constructs containing putative KLF15 binding sites. The diagram key is as for A, above. The small black squares indicate putative KLF15 binding sites. E. Regulation of Fstl1 promoter by KLF15 in preadipocytes. 3T3-L1 preadipocytes were transfected with luciferase reporter constructs containing 791 bp, 350 bp or 280 bp of the Fstl1 gene 5’ flanking region with or without cotransfection of a KLF15 expression construct. * indicates p<0.01 for (+) KLF15 co-transfection vs. (−) KLF co-transfection. Data represent the mean ± S.D. from a minimum of triplicate transfections.
We next explored possible mechanisms underlying differentiation-dependent downregulation of Fstl1 gene transcription. Unfortunately, attempts to compare Fstl1 promoter activity in 3T3-L1 preadipocytes vs. 3T3-L1 adipocytes were not fruitful due to inherent and well-known challenges of efficiently transfecting mature adipocytes. We also attempted to study differentiation-dependent transcriptional signals by assessing luciferase activity in 3T3-L1 stably transfected pooled populations of Fstl1 luciferase promoter constructs before and following their differentiation to adipocytes. We encountered a high degree of variation in such studies. We ascribe this to a reduced capacity 3T3-L1 preadipocytes to undergo effective adipocyte differentiation due to increased cell passaging needed in such studies, and perhaps also to integration site effects. We therefore turned to bioinformatic analysis of the sequence contained within the 791 bp fragment of the Fstl1 promoter to identify putative regulatory sites at which repressive transcription factors with a role in adipogenesis might function. Sequence analysis with MatInspector software identified four consensus inverted Kruppel-like factor (KLF) binding sites. Three of these sites are closely grouped at the −280 to −350 bp region and a fourth is located between −70 and −86 (Figures 9C and 9D).
KLFs are DNA-binding transcriptional regulators containing Cys2/His2 zinc fingers that play diverse roles during differentiation and development (Black et al., 2001; Dang et al., 2000). Studies over the last several years have indicated a regulatory role for several KLFs in adipogenesis (Banerjee et al., 2003; Birsoy et al., 2008; Li et al., 2005; Mori et al., 2005; Oishi et al., 2005; Wu et al., 2005). A number of KLFs, such as KLF15, can function in both transcriptional activation and transcriptional repression in an apparently context-dependent manner. KLF15 expression is upregulated during adipocyte differentiation (Mori et al., 2005), which we also find (left panel of Figure 9C), and KLF15 and C/EBPα act synergistically to increase the activity of the PPARγ gene promoter in 3T3-L1 (Mori et al., 2005). Overexpression of KLF15 induces adipocyte maturation and activates expression of the gene for the insulin-responsive glucose transporter, GLUT4, in adipocytes (Gray et al., 2002). On the other hand, KLF15 can also function in transcriptional repression; two genes transcriptionally repressed by KLF15 in adipocytes were recently identified (Nagare et al., 2009). We hypothesized that KLF15 might play a role in repression of Fstl1 gene expression in adipogenesis. We determined transcriptional regulation of the Fstl1 promoter by KLF15 employing co-transfection of Fstl1 luciferase constructs and a KLF15 expression construct in 3T3-L1 preadipocytes. In addition to the −791/+60 Fstl1 reporter construct, we generated a −350/+60 and a −280/+60 construct, the latter containing a single putative KLF15 binding site (Figures 9C and 9D). As illustrated in Figure 9E, relative luciferase activities of all three promoter constructs, −280/+60 LUC, −350/+60 LUC and −791/+60 LUC are decreased by KLF15. These data suggest that differential expression of Fstl1 transcript in preadipocytes vs. adipocytes may involve transcriptional repression by KLF15.
DISCUSSION
Our data clearly demonstrate that Fstl1 is expressed in preadipocytes and that dramatic downregulation of Fstl1 transcript and protein level is a robust hallmark of adipogenesis. Furthermore, Fstl1 transcript is re-expressed upon TNFα treatment of 3T3-L1 adipocytes. TNFα has been recognized as a link between obesity and insulin resistance and treatment of mature 3T3-L1 adipocytes with TNFα results in a degree of morphological, biochemical, and transcriptional dedifferentiation to a preadipocyte-like phenotype (Ruan et al., 2002a; Ruan and Lodish, 2003; Ruan et al., 2002b). Fstl1 evidences a tight link to the preadipocyte phenotype as its expression closely tracks with TNFα-induced 3T3-L1 adipocyte dedifferentiation. We find that Fstl1 is highly expressed in the media of cultured 3T3-L1 preadipocytes, typically detecting robust Fstl1 protein signal on Western blots using 5 µl or less of straight unconcentrated culture media. If Fstl1 has effects on adipogenic differentiation, or in regard to adipose tissue function, we postulate that it would most likely exert these as an autocrine or paracrine signal and perhaps through effects on cell matrix. Whether a specific receptor molecule exists for Fstl1 is not known nor have candidates been reported. Fstl1 was initially discovered as a TGF-β1-stimulated transcript in osteoblasts (Shibanuma et al., 1993). TGF-β1 has significant effects on ECM-related proteins through the induction of proteinase inhibitors and the suppression of proteinases that degrade matrix proteins (Shibanuma et al., 1993). In vitro and in vivo studies have shown that matrix remodeling is essential for adipogenesis and to also play key regulatory roles in WAT expansion in obesity (Lilla et al., 2002; Rupnick et al., 2002). Members of the BM-40/osteonectin/SPARC protein family have been demonstrated to contribute to the organization of the extracellular matrix via growth factor modulation, cell adhesion inhibition, proteinase inhibition and other effects (Bradshaw and Sage, 2001). It is possible that Fstl1 may affect ECM remodeling during adipogenesis. To date our attempts at using shRNA-mediated knockdown to address function of Fstl1 in the adipocyte lineage have been ineffective in achieving significant reduction of Fstl1 protein level in 3T3-L1 preadipocytes. This may be attributable to the very high level of Fstl1 transcript and protein expressed by these cells.
In WAT, adipocyte precursor cells have been recently described to reside in the vasculature (Tang et al., 2008). Interestingly, a comprehensive in situ hybridization study of Fstl1 transcript expression in murine embryogenesis indicates particular enrichment of Fstl1 transcript in the vasculature (Adams et al., 2007). For example, in the developing gut Fstl1 expression is high in the sub-epithelial vascular presumptive lamina propria (Adams et al., 2007). In the developing lung, high levels of Fstl1 expression are localized to mesenchymal cells in the vasculature developing around alveoli (Adams et al., 2007). At mid-gestation stages of murine embryogenesis, Fstl1 is strongly expressed in the developing somites and in the mesenchymal component of other tissues but is largely excluded from epithelium (Adams et al., 2007). The possibility that Fstl1 is a marker of mesenchymal cells, particularly those with stem cell characteristics, is further supported by the observation that intestinal mesenchmymal stem cell lines and putative intestinal mesenchymal stem cells in vivo display dramatically elevated expression of Fstl1 compared to that of various other cell lines or surrounding cell types, respectively (Geske et al., 2008). Fstl1 transcript has also been reported to be expressed in C2C12 myoblasts and decreased during their myogenic conversion, albeit apparently not to the same all-or-nothing extent we observe for adipogenesis (Rosenberg et al., 2006). Together, these observations raise the notion that downregulation of Fstl1 transcript level may be a conserved event in the differentiation of mesenchymal stem-like type cell precursors to their respective terminally-differentiated phenotypes. The adipose stromal compartment of adult tissue contains putative stem cell populations that can be induced to differentiate in vitro to osteogenic, adipogenic, myogenic, neuronal and chondrogenic lineage (Zuk et al., 2002). Precursor mesenchymal/stromal cell types (i.e. Swiss 3T3 and C3H10T1/2 cell lines) undergo differentiation to muscle, cartilage and fat cells after treatment with 5-aza-CR (Taylor and Jones, 1979). Of the agents tested we find that 5-aza-CR led to the most dramatic alteration of Fstl1 transcript level in 3T3-L1 preadipocytes, with a ~90% downregulation that was also reflected at the protein level. Demethylation is nearly exclusively reported to enhance transcription of cognate genes (Christman, 2002; Jones, 1985). We therefore surmise that the decreased level of Fstl1 transcript in 3T3-L1 preadipocytes by 5-aza-CR may be an indirect result of the demethylation-mediated activation of a currently unidentified regulatory gene. This gene would in turn repress gene transcription of select preadipocyte genes such as Fstl1. This may be one step in a regulatory cascade whereby 5-aza-CR treatment induces mesenchymal stem-like cells to differentiate along distinct lineages.
Studies on transcriptional mechanisms of gene activation in adipogenesis are frequently reported. In marked contrast, very few studies to date have attempted to assess mechanisms of selective gene expression in preadipocytes vs. adipocytes. A very recent report utilizing microarray analysis of KLF15-overexpressing 3T3-L1 adipocytes reported that, of ~6,000 genes examined, 247 were downregulated in KLF15-expressing adipocytes compared to control adipocytes (Nagare et al., 2009). Combining this transcriptional profiling data with a that from a ChIP-on-Chip study resulted in the identification of two genes, adrenomedullin (Adm) and adenine phosphoribosyl transferase (Aprt), that were negatively regulated by direct promoter binding of KLF15 (Nagare et al., 2009). This observation and our data herein suggest that in adipocytes KLF15 may serve to actively suppress a subset of preadipocyte-expressed genes, and it also reinforces the notion that KLF15 has both activating and repressive roles in the adipocyte lineage. However, unlike the near all-or-nothing expression of Fstl1 transcript in 3T3-L1 preadipocytes vs. differentiated adipocytes, adrenomedullin transcript expression decreases by only approximately two-thirds during adipogenesis of 3T3-L1 preadipocytes to adipocytes (Nagare et al., 2009). In addition to the transcriptional effects of KLF15, additional genetic and epigenetic mechanisms may function to initiate downregulation of Fstl1 and other preadipocyte genes during adipogenesis and act to ensure their persistent repression in mature adipocytes.
While the role of Fstl1 in myogenesis is also not known, an interesting mechanism for decreased of Fstl1 transcript during myogenesis has been uncovered. The onset of myogenesis is accompanied by enhanced expression of MyoD, a well-established master regulator of muscle differentiation. Use of an in vitro estrogen-mediated activation of a MyoD-ER fusion protein to identify MyoD-regulated transcripts revealed negative regulation of Fstl1 transcript by MyoD (Bergstrom et al., 2002; Rosenberg et al., 2006). It was determined that MyoD enhances transcription of miR-206, one of several identified muscle-specific microRNAs. miR-206 in turn targets Fstl1 transcript for degradation via interaction at the Fstl1 3’ untranslated region (Rosenberg et al., 2006). As miR-206 is muscle specific, it would appear that this particular microRNA mechanism would not be applicable to downregulation of Fstl1expresssion in adipogenesis. To date, several microRNAs have been investigated in detail in regard to adipogenesis, most notably miR-103, miR-143 and miR-let-7 (Sun et al., 2009; Takanabe et al., 2008; Xie et al., 2009). Analysis with TargetScan software fails to identify binding sites for these within the Fstl1 transcript. However, it is possible that other microRNAs expressed and/or regulated in the adipocyte lineage could be involved. Curiously, the 3’ untranslated region of the human Fstl1 transcript itself encodes a microRNA, mir-198, one of the few microRNAs present in a protein-coding transcript. However mir-198 does not appear conserved in murine (Lewis et al., 2005). Production of mir-198 may lead to degradation of the Fstl1 transcript, although this has not been formally examined. The only functional role for mir-198 to date is restriction of HIV-1 replication in monocytes by a mechanism that apparently involves repression of cyclin T1 (Sung and Rice, 2009). The function of mir-198 within the adipocyte lineage has not been examined. Of interest, however, in a study of expression level of 155 different micro RNAs in human omental and subcutaneous adipose tissue, miR-198 was unique in that its level of expression showed the highest degree of correlation with mean omental adipocyte diameter. An inverse correlation was noted wherein smaller omental adipocytes expressed higher levels of mir-198, and vice versa (Kloting et al., 2009).
BMP4 is one important signaling pathway involved in the differentiation of mesenchymal lineages (Bowers et al., 2006; Bowers and Lane, 2007; Butterwith et al., 1996; Otto et al., 2007; Tang et al., 2004). For example, treatment of multipotential murine CH310T1/2 mesenchymal cells with BMP4 primes these cells for greatly enhanced adipocyte conversion upon subsequent exposure to adipogenic inducing agents (Butterwith et al., 1996; Tang et al., 2004). We did not, however, observe regulation of Fstl1 transcript by BMP4 treatment of 3T3-L1 preadipocytes. Nonetheless, a possible link between Fstl1 and BMP4 signaling in regard to adipogenesis remains to be more fully explored. This is particularly so in light of reports in zebrafish and chick development indicating that Fstl1 may impact BMP4 signaling pathways (Amthor et al., 1996; Esterberg et al., 2008; Towers et al., 1999). In zebrafish, BMP4 signaling acts in early development to establish mesodermal cell identity. Transcripts for Fstl1 orthologs are expressed in zebrafish axial and paraxial mesoderm during late gastrulation and segmentation stages (Esterberg et al., 2008). In this model, morpholino-based knockdown of Fstl1 orthologs enlarged the tailbud domain and led to increased expression of markers indicative of enhanced BMP4 signaling (Esterberg et al., 2008). Follistatin can act by directly binding to and inhibiting function of the TGF-β superfamily member activin (Chen et al., 2006). To our knowledge, whether Fstl1 binds activin or functions by an analogous mechanism has not been reported nor explored. This may be unlikely given the low degree of sequence conservation of the follistatin-like module of Fstl1 with that of follistatin, and the fact that follistatin has three such modules in tandem vs. the single follistatin-like domain of Fstl1. Additional studies indicate that the expression of Fstl1 in embryogenesis may also involve interplay of the hedgehog signaling pathway (Amthor et al., 1996; Towers et al., 1999), which has a described regulatory role in adipogenesis (Cousin et al., 2007). It remains to be determined if and where in development and differentiation of adipocytes Fstl1 fits in regard to the complex cell signaling networks mediated by BMP4 and sonic hedgehog.
As cells transition from preadipocytes to terminally differentiated adipocytes, a major phenotypic alteration is cessation of cell proliferation due to cell cycle withdrawal (Gregoire, 2001; Gregoire et al., 1998). This suggests a positive correlation of Fstl1 expression with cell growth/proliferation. However a number of studies, particularly in cancer cells, have reported that Fstl1 is anti-proliferative and suggested a tumor suppressor role for Fstl1. Fstl1 transcript level was observed to be decreased in cancer vs. normal cell lines and to decrease following experimental transformation of cells with select oncogenes (Mashimo et al., 1997; Shibanuma et al., 1993; Sumitomo et al., 2000). Ectopic expression of Fstl1 in cancer cells or transformed fibroblasts reduced proliferation and/or invasiveness (Johnston et al., 2000; Sumitomo et al., 2000). A number of other studies support an anti-proliferative role for Fstl1. This includes inhibited proliferation of human lung cancer cells (Sumitomo et al., 2000), vascular smooth muscle cells (Liu et al., 2006), a rheumatoid synovial cell line (E11), Ovca420 ovarian (Chan et al., 2009), and AN3CA endometrial cancer cells (Chan et al., 2009). In the latter instance, ectopic expression of Fstl1 enhanced apoptosis, and reduced cell migration and invasion (Chan et al., 2009). These observations on the anti-proliferative effects of Fstl1 are consistent with aforementioned studies of zebrafish morphants that reported enhanced proliferation and tissue expansion upon knockdown of Fstl1 orthologs (Esterberg et al., 2008). Overall, these data suggest that Fstl1 might function during embryogenesis and in cancer cells to inhibit cell growth and/or migration. In contrast to the anti-proliferative role described for Fstl1 in cultured cells, studies reported to date in human clinical cancer samples do not reveal a consistent pattern of Fstl1 deregulation indicative of tumor suppressor function. For example, Fstl1 transcript is downregulated in primary clear cell renal cell carcinoma (ccRCC) compared with adjacent normal kidney tissue (Tan et al., 2008). It is also decreased in metatstatic ccRCC vs. primary ccRCC tumors (Tan et al., 2008) and in majority of ovarian and endometrial cancers (Chan et al., 2009). On the other hand, Fstl1 is reported up-regulated in primary and secondary glioblastoma multiforma with normal glial cells negative for Fstl1 expression (Reddy et al., 2008). Analysis of prostate cancers revealed that overexpression of Fstl1 was associated with higher metastatic potential (Nagare et al., 2009). Our observations of high expression of Fstl1 in preadipocytes and its marked decrease in early adipogenesis, a time coincident with their cell cycle withdrawal, and our observed transient increase in Fstl1 transcript coincident with the clonal expansion period of adipogenesis, appears consistent with a pro-proliferative role for Fstl1 in the adipocyte lineage.
Fstl1 has an emerging role as a signaling molecule in immunity. As such, preadipocyte expression of Fstl1 within WAT may contribute to the complex interplay between WAT and the immune system. Reports to date on the role of Fstl1 in immune response, however, appear contradictory in nature in that both anti- and - pro- inflammatory functions have been described. In respect to the former case, Kawabata and colleagues reported that treatment with recombinant Fstl1 ameliorates joint inflammation in a mouse model of arthritis leading to decreased expression of disease–promoting genes such as c-fos and IL-6 (Kawabata et al., 2004). A second study on rheumatoid arthritis reported recombinant Fstl1 expression downregulates synovial production of MMP-1, MMP-3 and prostaglandin E2 (Tanaka et al., 2003). Fstl1 is overexpressed during both the induction and maintenance phase of heart allograft tolerance with ectopic Fstl1 expression in this model reducing production of several pro-inflammatory cytokines and prolonging allograft survival (Le Luduec et al., 2008). On the other hand, various data support a pro-inflammatory function for Fstl1. In apparent contrast to the report of Kawataba, Miyamae and colleagues reported Fstl1 to be increased in a collagen-induced murine model of arthritis (Miyamae et al., 2006). They found that Fstl1 exacerbates arthritis when delivered by gene transfer, apparently by enhancing T-cell IFNγ production, and that neutralization of Fstl1 inhibited arthritis and diminishes IFNγ and CXCL10 levels in arthritic joints (Miyamae et al., 2006). Fstl-1 transfection into macrophages and fibroblasts increased levels of IL-1β, TNFα, and IL-6 (Miyamae et al., 2006). It has also been reported that Fstl1 and/or its autoantibodies are overexpressed in synovial tissues from rheumatoid arthritis patients (Ehara et al., 2004; Tanaka et al., 1998). Circulating Fstl1 has recently been proposed as an inflammatory serum biomarker in humans and increased serum levels reported in acute coronary syndrome (Widera et al., 2009). It remains to be determined whether expression of Fstl1 by preadipocytes plays a paracrine signaling role in the regulation of cytokine production by adipocytes or macrophages within WAT.
Given the fact that myocytes and preadipocytes have the same mesodermal origin, recent reports of Fstl1 function and mechansism(s) in muscle may have implications for the role of Fstl1 in the adipocyte lineage. Fstl1 transcript was found to be upregulated in hearts of transgenic mice with cardiac-specific ectopic Akt expression. These observations led to the discovery that Fstl1 is secreted by neonatal rat ventricular cardiomyocytes (Oshima et al., 2008). Ectopically expressed Fstl1 protects these cells from hypoxia-induced apoptosis and protects myocardial tissue from effects of ischemic stress (Oshima et al., 2008). Ectopic expression of Fstl1 in cardiomyocytes in vitro increased levels of phosphorylated Akt and enhanced ERK phosphorylation, two key protective signals for myocardium (Oshima et al., 2008). In gastrocnemius muscle Fstl1 is upregulated by ischemia to promote revascularization (Ouchi et al., 2008). In vitro studies of human umbilical vein endothelium cells revealed Fstl1 facilitates endothelial network formation, promotes cell migration and suppresses apoptosis (Ouchi et al., 2008). The initial stage in formation of the “primitive organs”, which are described to precede the appearance of bona fide adipose tissue during development, is formation and expansion of dense capillary networks. Enhancement of blood supply via vascularization within WAT is also a key step that is required for the expansion of adipose mass in obesity and to prevent hypoxia therein (Rupnick et al., 2002). It is possible that Fstl1 functions within WAT to promote or maintain vascularization and/or as a preadipocyte-secreted cell survival factor.
In conclusion, reports of the last few years have illuminated Fstl1 expression and action in muscle, immune response, embryonic development and cancer, indicating a complex role(s) for Fstl1 therein. We postulate that preadipocyte-expressed Fst1l may exert similar pleiotropic actions in adipogenesis and adipose tissue function.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
3T3-L1 cells (American Type Culture Collection, Manassas, VA) were maintained in DMEM with 10% calf serum (CS). For differentiation, cells were treated at 2 d post-confluence with DMEM supplemented with 10% fetal bovine serum (FBS), 0.5 mM 3-isobutyl-1-methylxanthine (Mix), and 1 µM dexamethasone (Dex) for 48 h. After induction, cultures were maintained in 10% FBS medium. ScAP-23 cells were maintained in DMEM containing 10% CS and passaged prior to reaching confluence. For adipocyte differentiation, preconfluent ScAP-23 preadipocytes were cultured in DMEM containing 10% FBS in the presence of the adipogenic inducers 0.5 mM Mix, 1 µM Dex, 17 nM insulin, and 0.2 mM indomethacin for 70 h. Agents were then removed and cultures maintained in DMEM containing 10% FBS and 17 nM insulin for an additional 4 d. The AD2.7AC cell line (Lecka-Czernik et al., 1999)was obtained from Dr. B. Lecka-Czernik (University of Toledo College of Medicine, Toledo, OH). Their culture and adipogenic induction is as for ScAP-23. For culture and differentiation of primary white adipocytes, WAT collected from male C57BL/6 mice or Sprague-Dawley rats was digested with 1 mg/ml of type I collagenase for 40 min at 37°C with shaking. Following digestion, material was filtered through a 300 micron pore size nylon mesh (Sefar America Inc., Depew, NY) and filtrate centrifuged at 2,000 rpm for 5 min. The floating adipocyte fraction was removed and the pellet of preadipocyte-containing stromal-vascular fraction (SVF) cells was resuspended in DMEM containing 10% FBS and plated. Upon confluence cells were either harvested or subjected to culture in differentiation media that consisted of DMEM containing 10% FBS, 0.5 µM Dex, 0.25 mM Mix, and 17 nM insulin for 3 d. After this, media was removed and cell cultures were maintained in DMEM containing 10% FBS and 17 nM insulin for an additional 4 d. For differentiation of a brown preadipocyte cell line (termed herein WT-BAT) obtained from C.R. Kahn (Joslin Diabetes Foundation, Harvard Medical School, Boston, MA), cells were cultured to confluence in DMEM supplemented with 10% FBS, 20 nM insulin and 1 nM triiodothyronine (T3) (Klein et al., 2002). Confluent cells were incubated in differentiation medium that contained 0.5 mM Mix, 0.5 µM Dex and 0.125 mM indomethacin for 2 d. After induction, cells were maintained in medium supplemented with 20 nM insulin and 1 nM T3 for an additional 4 d. The culture and adipogenic induction of mBAP-9 cells is as for WT-BAT. A manuscript more fully describing derivation and characterization of the mBAP-9 cell line is in preparation. COS cells and NIH-3T3 cells were cultured in DMEM supplemented with 10% FBS.
For treatment with TNFα, 3T3-L1 adipocytes were incubated with 10 ng/ml TNFα for the indicated times. For assessment of Fstl1 regulation by components of the adipogenic cocktail, post-confluent 3T3-L1 preadipocytes were treated with 1 µM Dex or 0.5 mM Mix or the combination of Mix and Dex for 48 h. For assessment of regulation of Fstl1 transcript by a panel of various other agents, confluent 3T3-L1 preadipocytes were maintained in fresh DMEM with 10% CS for 24 h as control or treated for 24 h with either 10 nM retinoic acid; 2 ng/ml basic fibroblast growth factor (b-FGF); 10 ng/ml interleukin 6 (IL-6); 100 ng/ml bone morphogenetic protein 4 (BMP4); 200 µM indomethacin; 10 µM forskolin; 0.5 mM Mix; 10 nM sodium butyrate; 1 mM 5-azacytidine (5-aza-CR), 10 nM T3 (triiodothyronine); 100 ng/ml VEGF (vascular endothelial cell growth factor); 1 µM dexamethasone; 1 nM dibutyryl cAMP (cyclic adenosine monophosphate); 5 µM tamoxifen; 100 ng/ml LPS (lipopolysaccharide); 10 ng/ml TNFα; 1 µsM TSA (trichostatin A); 5 µg/ml PPARγ ligand 15-deoxy-Δ12, 14-prostaglandin J2. For 5-aza-CR analysis, 3T3-L1 preadipocytes were treated with 1 mM 5-aza-CR for the indicated times.
RNA Preparation, Northern Blot Analysis, DNA Array Hybridization and Q-PCR
Total RNA was extracted with TriZol Reagent (Invitrogen Corp) according to manufacturer instruction. For Fstl1 expression in murine tissues, 8 wk old C57BL/6 male mice were utilized, with all animal treatments conducted with approval of the University of Toledo College of Medicine Institutional Animal Care and Use Committee. Fractionation of whole adipose tissue was performed by digestion with 1 mg/ml of type I collagenase for 40 min with shaking at 37°C. Following digestion, material was filtered through a 300 micron pore size nylon mesh (Sefar America Inc., Depew, NY) and filtrate centrifuged at 2,000 rpm for 5 min. Floating adipocyte fraction was collected as adipocyte fraction (AF) and the pellet was collected as SVF. For array analysis, duplicate mouse ResGen GeneFilter Arrays (Invitrogen Corp.) containing 5,184 genes, were processed according to manufacturer's instructions. Filters were hybridized with [α-33P] dATP labeled cDNA probes synthesized from 8 µg of total RNA from 3T3-L1 preadipocytes and adipocytes. For Northern blot analyses, 5 µg of total RNA was electrophoresed in 1% agarose formaldehyde gels in MOPS buffer and transferred to Hybond-N nylon membrane (GE Healthcare, Piscataway, NJ). Hybridizations were performed under high stringency conditions in ExpressHyb (BD Biosciences Clontech), according to the manufacturer's directions. Probes were labeled with [α-32P] dATP using a Megaprime DNA labeling system (Amersham Biosciences). After washing, membranes were exposed at −80°C to Kodak Biomax film with a Kodak Biomax intensifying screen.
For Q-PCR analysis, total RNA was subject to purification with an RNeasy RNA purification kit with DNase I treatment (Qiagen Corp., Valencia, CA) and 5 µg used for first strand cDNA synthesis with SuperScript II RNase H-reverse transcriptase (Invitrogen Corp.) and an oligo(dT)-22 primer. Q-PCR was conducted with an ABI 7500 Q-PCR System. Target cDNA levels were analyzed by SYBR green-based Q-PCR in 25 µl reactions containing 1X SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 100 nM each forward and reverse primers, and 10 ng of cDNA. Q-PCR primers were designed to span introns and sequences are available on request. Transcript expression was normalized against Gapdh transcript level except when otherwise stated. In all cases the same amount of input RNA/cDNA was used in side-by-side comparisons. The cycle threshold value was generated using ABI PRISM 7500 SDS software version 1.2 and exported to an Excel spreadsheet to calculate fold differences. Statistical analyses were conducted using single factor ANOVA.
Expression Construct for HA-Tagged Fstl1, Transfection and Western Blot Analysis
An Fstl1 CDNA clone was obtained from ATCC (I.M.A.G.E. ID: 2647002). PCR with the following primers (5′ GCC AAG CTT CCG GAC CCG AGC ACG ATG TGG3’) and (5′ GGC GTC GAC TTA AAG AGC GTA ATC TGG AAC ATC GTA TGG GTA GAT CTC TTT GGT GTT CAC CTT3’) was used to generate the coding region of murine Fstl1 with a C-terminal HA tag. Primers introduced restriction sites (5’ Hind III and 3’ Sal I, underlined) and Hind III/Sal I restricted PCR product was purified and inserted into Hind III and Sal I sites of pcDNA3.1 to produce the expression construct, Fstl1-HA-pcDNA.
For transfection, COS cells were seeded at 6×105 cells/100 mm dish. 10 µg of Fstl1-HA-pcDNA or pcDNA empty vector was transfected using the DEAE-dextran method. For Western blot analysis culture media and cell lysates were collected at 48 h post-transfection. Media was centrifuged at 5,000 rpm for 3 min and cells harvested by lysis in TNN (+) buffer (10 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM EDTA, supplemented with a protease inhibitor cocktail). Lysates were incubated on ice for 30 min with intermittent vortexing, supernatant collected via centrifugation, and protein content determined (Bio-Rad Laboratories). To examine Fstl1 protein expression in medium during 3T3-L1 cell differentiation, culture medium was harvested daily from the same well of cells in a 6-well plate, with medium changed 24 h pre-harvest. For assessment of TNFα regulation of Fstl1 in 3T3-L1 adipocytes, cells were incubated with or without 10 ng/ml TNFα for indicated times. To determine Fstl1 protein half life, 3T3-L1 preadipocytes were treated with 5 µg/ml of cycloheximide and harvested at indicated time points. For Fstl1 protein regulation by 5-aza-CR treatment, 3T3-L1 preadipocytes were treated with or without 1 mM 5-aza-CR for 24 h and culture media harvested. For Western blot, proteins were separated in a 10% SDS-PAGE gels under reducing conditions and electroblotted onto polyvinylidene fluoride (PVDF) membrane (Millipore Corp.). Membranes were blocked 1 h in 5% nonfat milk/0.5% Tween 20 in PBS and then incubated with primary antibody for 1 h, followed by three 10 min washes. Anti-mouse Fstl1 antibody was purchased from R&D Systems, PPIA and tubulin antibodies were from Santa Cruz Biotechnology. Secondary antibody was incubated for 30 min followed by three 10 min washes. All washes were in 0.5% Tween 20 in PBS. Signal was detected by ECL Plus enhanced chemiluminescence (GE Healthcare) and by exposure to X-ray film except for quantitative studies of protein half-life for which digital data was acquired using a Kodak Digital Image Station.
Luciferase Constructs and Reporter Assay
To prepare luciferase reporter constructs containing regions of the mouse Fstl1 promoter, murine liver genomic DNA was subject to PCR with primers designed based on murine Fstl1 genomic flanking region sequence available through the Ensembl database (www.ensembl.org). For the −280/+60 promoter construct, PCR was conducted with a promoter proximal primer (5’ GGC CTC GAG TGG CGG CAG CGA GTT AGA GGC) that included an Xho I site (underlined) spanning through 60-base pair (bp) of the Fstl1 transcript in combination with a promoter distal primer (5’ GGC ACG CGT TGA TTT CTG TGA TTT CCC CCG C) including an Mlu I site (underlined). For the −350/+60 promoter construct, PCR was conducted with the same promoter proximal primer as for the −280/+60 promoter construct and a promoter distal primer (5’ GGC ACG CGT CGA GTT CCTTCT GTT ACC CAC) including an Mlu I site (underlined). For the −791/+60 promoter construct, PCR was conducted with a promoter proximal primer (5′ GGC AGA TCT TGG CGG CAG CGA GTT AGA GGC 3′) including a Bgl II site (underlined) and a promoter-distal primer (5′ GGC GGT ACC GAA AGA GAT TGG GGA TCC ACAC 3′) including a Kpn I site (underlined). For the −3922/+22 promoter construct, PCR was conducted with a promoter proximal primer (5’ GGC ACG CGT TTA TCA CCA GGC TCC GAG GAA G 3’) including an Mlu I site (underlined), and a promoter distal primer (5’ CAG CCG GTA CCA ATG CAC ACG CAT TGC CAC) including Kpn I site (underlined). The resulting PCR products were ligated into the corresponding restriction sites of the pGL2-Basic vector (Promega Corp, Madison, WI). Promoter constructs were confirmed by complete sequencing.
To assess transcriptional response of the murine Fstl1 promoter region, Fstl1 promoter-luciferase constructs were transfected into 3T3-L1 preadipocytes, either singly or in combination with a KLF15 expression construct, using Lipofectamine 2000 (Invitrogen Corp). Empty vector pDNA3.1 was included as needed to maintain equal mass of DNA per transfection and all transfections included a pRLNull Renilla luciferase construct (Promega Corp.) as an internal control. Firefly and Renilla luciferase activity was determined at 48 h post-transfection using a Promega dual luciferase kit and a Turner Industries luminometer (Promega Corp.), with values corrected against Renilla luciferase signals. Statistical analyses were conducted using single factor ANOVA.
AKNOWLEDGEMENTS
We are grateful to Dr. Ji Young Kim (University of Toledo College of Medicine) for provision of select RNA samples. We thank Dr. Beata Lecka-Czernik (University of Toledo College of Medicine) for providing AD2.7AC cells and Dr. C.R. Kahn (Harvard Medical School) for providing the brown preadipocyte cell line.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams D, Larman B, Oxburgh L. Developmental expression of mouse Follistatin-like 1 (Fstl1): Dynamic regulation during organogenesis of the kidney and lung. Gene Expr Patterns. 2007;7:491–500. doi: 10.1016/j.modgep.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailhaud G. Adipose tissue as a secretory organ: from adipogenesis to the metabolic syndrome. C R Biol. 2006;329:570–577. doi: 10.1016/j.crvi.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Amthor H, Connolly D, Patel K, Brand-Saberi B, Wilkinson DG, Cooke J, Christ B. The expression and regulation of follistatin and a follistatin-like gene during avian somite compartmentalization and myogenesis. Dev Biol. 1996;178:343–362. doi: 10.1006/dbio.1996.0223. [DOI] [PubMed] [Google Scholar]
- Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Kruppel-like factor KLF2 inhibits per oxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Bornstein P, Kyriakides TR, Yang Z, Armstrong LC, Birk DE. Thrombospondin 2 modulates collagen fibrillogenesis and angiogenesis. J Investig Dermatol Symp Proc. 2000;5:61–66. doi: 10.1046/j.1087-0024.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- Bowers RR, Kim JW, Otto TC, Lane MD. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc Natl Acad Sci U S A. 2006;103:13022–13027. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers RR, Lane MD. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle. 2007;6:385–389. doi: 10.4161/cc.6.4.3804. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R. Robust signaling networks of the adipose secretome. Trends Endocrinol Metab. 2009;20:1–7. doi: 10.1016/j.tem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Burton GR, Guan Y, Nagarajan R, McGehee RE., Jr Microarray analysis of gene expression during early adipocyte differentiation. Gene. 2002;293:21–31. doi: 10.1016/s0378-1119(02)00726-6. [DOI] [PubMed] [Google Scholar]
- Burton GR, McGehee RE., Jr Identification of candidate genes involved in the regulation of adipocyte differentiation using microarray-based gene expression profiling. Nutrition. 2004;20:109–114. doi: 10.1016/j.nut.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Burton GR, Nagarajan R, Peterson CA, McGehee RE., Jr Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–185. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Butterwith SC, Wilkie RS, Clinton M. Treatment of pluripotential C3H 10T1/2 fibroblasts with bone morphogenetic protein-4 induces adipocyte commitment. Biochem Soc Trans. 1996;24:163S. doi: 10.1042/bst024163s. [DOI] [PubMed] [Google Scholar]
- Cancello R, Tordjman J, Poitou C, Guilhem G, Bouillot JL, Hugol D, Coussieu C, Basdevant A, Bar Hen A, Bedossa P, Guerre-Millo M, Clement K. Increased infiltration of macrophages in omental adipose tissue is associated with marked hepatic lesions in morbid human obesity. Diabetes. 2006;55:1554–1561. doi: 10.2337/db06-0133. [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Chan QK, Ngan HY, Ip PP, Liu VW, Xue WC, Cheung AN. Tumor suppressor effect of follistatin-like 1 in ovarian and endometrial carcinogenesis: a differential expression and functional analysis. Carcinogenesis. 2009;30:114–121. doi: 10.1093/carcin/bgn215. [DOI] [PubMed] [Google Scholar]
- Chen YG, Wang Q, Lin SL, Chang CD, Chuang J, Ying SY. Activin signaling and its role in regulation of cell proliferation, apoptosis, and carcinogenesis. Exp Biol Med (Maywood) 2006;231:534–544. doi: 10.1177/153537020623100507. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Cinti S. The role of brown adipose tissue in human obesity. Nutr Metab Cardiovasc Dis. 2006;16:569–574. doi: 10.1016/j.numecd.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Clutter SD, Wilson DC, Marinov AD, Hirsch R. Follistatin-like protein 1 promotes arthritis by up-regulating IFN-gamma. J Immunol. 2009;182:234–239. doi: 10.4049/jimmunol.182.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin W, Fontaine C, Dani C, Peraldi P. Hedgehog and adipogenesis: fat and fiction. Biochimie. 2007;89:1447–1453. doi: 10.1016/j.biochi.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Croissandeau G, Chretien M, Mbikay M. Involvement of matrix metalloproteinases in the adipose conversion of 3T3-L1 preadipocytes. Biochem J. 2002;364:739–746. doi: 10.1042/BJ20011158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol Cell Biol. 2007;27:8454–8465. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara Y, Sakurai D, Tsuchiya N, Nakano K, Tanaka Y, Yamaguchi A, Tokunaga K. Follistatin-related protein gene (FRP) is expressed in the synovial tissues of rheumatoid arthritis, but its polymorphisms are not associated with genetic susceptibility. Clin Exp Rheumatol. 2004;22:707–712. [PubMed] [Google Scholar]
- Esterberg R, Delalande JM, Fritz A. Tailbud-derived Bmp4 drives proliferation and inhibits maturation of zebrafish chordamesoderm. Development. 2008;135:3891–3901. doi: 10.1242/dev.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- Geloen A, Roy PE, Bukowiecki LJ. Regression of white adipose tissue in diabetic rats. Am J Physiol. 1989;257:E547–E553. doi: 10.1152/ajpendo.1989.257.4.E547. [DOI] [PubMed] [Google Scholar]
- Geske MJ, Zhang X, Patel KK, Ornitz DM, Stappenbeck TS. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. Sublines of mouse 3T3 cells that accumulate lipid. Cell. 1974;1:113–116. [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Obin MS. Obesity and the role of adipose tissue in inflammation and metabolism. Am J Clin Nutr. 2006;83:461S–465S. doi: 10.1093/ajcn/83.2.461S. [DOI] [PubMed] [Google Scholar]
- Gregoire FM. Adipocyte differentiation: from fibroblast to endocrine cell. Exp Biol Med (Maywood) 2001;226:997–1002. doi: 10.1177/153537020122601106. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- Guo X, Liao K. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene. 2000;251:45–53. doi: 10.1016/s0378-1119(00)00192-x. [DOI] [PubMed] [Google Scholar]
- Hambrock HO, Kaufmann B, Muller S, Hanisch FG, Nose K, Paulsson M, Maurer P, Hartmann U. Structural characterization of TSC-36/Flik: analysis of two charge isoforms. J Biol Chem. 2004;279:11727–11735. doi: 10.1074/jbc.M309318200. [DOI] [PubMed] [Google Scholar]
- Harp JB. New insights into inhibitors of adipogenesis. Curr Opin Lipidol. 2004;15:303–307. doi: 10.1097/00041433-200406000-00010. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hu E, Tontonoz P, Spiegelman BM. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc Natl Acad Sci U S A. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston IM, Spence HJ, Winnie JN, McGarry L, Vass JK, Meagher L, Stapleton G, Ozanne BW. Regulation of a multigenic invasion programme by the transcription factor, AP-1: re-expression of a down-regulated gene, TSC-36, inhibits invasion. Oncogene. 2000;19:5348–5358. doi: 10.1038/sj.onc.1203927. [DOI] [PubMed] [Google Scholar]
- Jones PA. Altering gene expression with 5-azacytidine. Cell. 1985;40:485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- Kamei N, Tobe K, Suzuki R, Ohsugi M, Watanabe T, Kubota N, Ohtsuka-Kowatari N, Kumagai K, Sakamoto K, Kobayashi M, Yamauchi T, Ueki K, Oishi Y, Nishimura S, Manabe I, Hashimoto H, Ohnishi Y, Ogata H, Tokuyama K, Tsunoda M, Ide T, Murakami K, Nagai R, Kadowaki T. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J Biol Chem. 2006;281:26602–26614. doi: 10.1074/jbc.M601284200. [DOI] [PubMed] [Google Scholar]
- Kawabata D, Tanaka M, Fujii T, Umehara H, Fujita Y, Yoshifuji H, Mimori T, Ozaki S. Ameliorative effects of follistatin-related protein/TSC-36/FSTL1 on joint inflammation in a mouse model of arthritis. Arthritis Rheum. 2004;50:660–668. doi: 10.1002/art.20023. [DOI] [PubMed] [Google Scholar]
- Kim JY, Wu Y, Smas CM. Characterization of ScAP-23, a new cell line from murine subcutaneous adipose tissue, identifies genes for the molecular definition of preadipocytes. Physiol Genomics. 2007a;31:328–342. doi: 10.1152/physiolgenomics.00206.2006. [DOI] [PubMed] [Google Scholar]
- Kim KA, Kim JH, Wang Y, Sul HS. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007b;27:2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Fasshauer M, Klein HH, Benito M, Kahn CR. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. Bioessays. 2002;24:382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K, Stumvoll M, Bluher M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:e4699. doi: 10.1371/journal.pone.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA. PPAR gamma, 10 years later. Biochimie. 2005;87:9–13. doi: 10.1016/j.biochi.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Le Luduec JB, Condamine T, Louvet C, Thebault P, Heslan JM, Heslan M, Chiffoleau E, Cuturi MC. An immunomodulatory role for follistatin-like 1 in heart allograft transplantation. Am J Transplant. 2008;8:2297–2306. doi: 10.1111/j.1600-6143.2008.02398.x. [DOI] [PubMed] [Google Scholar]
- Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999;74:357–371. [PubMed] [Google Scholar]
- Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, Tan S, Friedman JM, Friedman SL, Walsh MJ. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- Lilla J, Stickens D, Werb Z. Metalloproteases and adipogenesis: a weighty subject. Am J Pathol. 2002;160:1551–1554. doi: 10.1016/S0002-9440(10)61100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang L, Wang W, Lin J, Han J, Sun H, Guo H, Sun R, Wu Q. TSC-36/FRP inhibits vascular smooth muscle cell proliferation and migration. Exp Mol Pathol. 2006;80:132–140. doi: 10.1016/j.yexmp.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Flier JS. Brown adipose tissue, beta 3-adrenergic receptors, and obesity. Annu Rev Med. 1997;48:307–316. doi: 10.1146/annurev.med.48.1.307. [DOI] [PubMed] [Google Scholar]
- Lucero HA, Kagan HM. Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol Life Sci. 2006;63:2304–2316. doi: 10.1007/s00018-006-6149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, Komuro R, Ouchi N, Kihara S, Tochino Y, Okutomi K, Horie M, Takeda S, Aoyama T, Funahashi T, Matsuzawa Y. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- Mashimo J, Maniwa R, Sugino H, Nose K. Decrease in the expression of a novel TGF beta1-inducible and ras-recision gene, TSC-36, in human cancer cells. Cancer Lett. 1997;113:213–219. doi: 10.1016/s0304-3835(97)04700-9. [DOI] [PubMed] [Google Scholar]
- Miyamae T, Marinov AD, Sowders D, Wilson DC, Devlin J, Boudreau R, Robbins P, Hirsch R. Follistatin-like protein-1 is a novel proinflammatory molecule. J Immunol. 2006;177:4758–4762. doi: 10.4049/jimmunol.177.7.4758. [DOI] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Sidis Y, Mahan A, Raher MJ, Xia Y, Rosen ED, Bloch KD, Thomas MK, Schneyer AL. FSTL3 deletion reveals roles for TGF-beta family ligands in glucose and fat homeostasis in adults. Proc Natl Acad Sci U S A. 2007;104:1348–1353. doi: 10.1073/pnas.0607966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagare T, Sakaue H, Takashima M, Takahashi K, Gomi H, Matsuki Y, Watanabe E, Hiramatsu R, Ogawa W, Kasuga M. The Kruppel-like factor KLF15 inhibits transcription of the adrenomedullin gene in adipocytes. Biochem Biophys Res Commun. 2009;379:98–103. doi: 10.1016/j.bbrc.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Young-Cheul K. Adipocyte differentiation and gene expression. J Nutr. 2000;130:3122S–3126S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Ouchi N, Sato K, Izumiya Y, Pimentel DR, Walsh K. Follistatin-like 1 is an Akt-regulated cardioprotective factor that is secreted by the heart. Circulation. 2008;117:3099–3108. doi: 10.1161/CIRCULATIONAHA.108.767673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TC, Bowers RR, Lane MD. BMP-4 treatment of C3H10T1/2 stem cells blocks expression of MMP-3 and MMP-13. Biochem Biophys Res Commun. 2007;353:1097–1104. doi: 10.1016/j.bbrc.2006.12.170. [DOI] [PubMed] [Google Scholar]
- Ouchi N, Oshima Y, Ohashi K, Higuchi A, Ikegami C, Izumiya Y, Walsh K. Follistatin-like 1, a secreted muscle protein, promotes endothelial cell function and revascularization in ischemic tissue through a nitric-oxide synthase-dependent mechanism. J Biol Chem. 2008;283:32802–32811. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KW, Halperin DS, Tontonoz P. Before they were fat: adipocyte progenitors. Cell Metab. 2008;8:454–457. doi: 10.1016/j.cmet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Patel K, Connolly DJ, Amthor H, Nose K, Cooke J. Cloning and early dorsal axial expression of Flik, a chick follistatin-related gene: evidence for involvement in dorsalization/neural induction. Dev Biol. 1996;178:327–342. doi: 10.1006/dbio.1996.0222. [DOI] [PubMed] [Google Scholar]
- Permana PA, Menge C, Reaven PD. Macrophage-secreted factors induce adipocyte inflammation and insulin resistance. Biochem Biophys Res Commun. 2006;341:507–514. doi: 10.1016/j.bbrc.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Pinney DF, Emerson CP., Jr 10T1/2 cells: an in vitro model for molecular genetic analysis of mesodermal determination and differentiation. Environ Health Perspect. 1989;80:221–227. doi: 10.1289/ehp.8980221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwich TC, Macdougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19:612–627. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qatanani M, Lazar MA. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes Dev. 2007;21:1443–1455. doi: 10.1101/gad.1550907. [DOI] [PubMed] [Google Scholar]
- Rajala MW, Scherer PE. Minireview: The adipocyte--at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003;144:3765–3773. doi: 10.1210/en.2003-0580. [DOI] [PubMed] [Google Scholar]
- Reddy SP, Britto R, Vinnakota K, Aparna H, Sreepathi HK, Thota B, Kumari A, Shilpa BM, Vrinda M, Umesh S, Samuel C, Shetty M, Tandon A, Pandey P, Hegde S, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V, Kondaiah P, Somasundaram K, Rao MR. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res. 2008;14:2978–2987. doi: 10.1158/1078-0432.CCR-07-4821. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Erickson RL, Gerin I, DeRose PM, Bajnok L, Longo KA, Misek DE, Kuick R, Hanash SM, Atkins KB, Andresen SM, Nebb HI, Madsen L, Kristiansen K, MacDougald OA. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver × receptor alpha in adipocyte metabolism. Mol Cell Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes. 2002a;51:1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- Ruan H, Lodish HF. Insulin resistance in adipose tissue: direct and indirect effects of tumor necrosis factor-alpha. Cytokine Growth Factor Rev. 2003;14:447–455. doi: 10.1016/s1359-6101(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Ruan H, Miles PD, Ladd CM, Ross K, Golub TR, Olefsky JM, Lodish HF. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-alpha: implications for insulin resistance. Diabetes. 2002b;51:3176–3188. doi: 10.2337/diabetes.51.11.3176. [DOI] [PubMed] [Google Scholar]
- Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem. 1978;253:7570–7578. [PubMed] [Google Scholar]
- Rupnick MA, Panigrahy D, Zhang CY, Dallabrida SM, Lowell BB, Langer R, Folkman MJ. Adipose tissue mass can be regulated through the vasculature. Proc Natl Acad Sci U S A. 2002;99:10730–10735. doi: 10.1073/pnas.162349799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp M, Cristancho AG, Lefterova MI, Hanniman EA, Briggs ER, Steger DJ, Qatanani M, Curtin JC, Schug J, Ochsner SA, McKenna NJ, Lazar MA. Re-expression of GATA2 Cooperates with Peroxisome Proliferator-activated Receptor-{gamma} Depletion to Revert the Adipocyte Phenotype. J Biol Chem. 2009;284:9458–9464. doi: 10.1074/jbc.M809498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Lazar MA. Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation. J Biol Chem. 1997;272:21473–21478. doi: 10.1074/jbc.272.34.21473. [DOI] [PubMed] [Google Scholar]
- Shibanuma M, Mashimo J, Mita A, Kuroki T, Nose K. Cloning from a mouse osteoblastic cell line of a set of transforming-growth-factor-beta 1-regulated genes, one of which seems to encode a follistatin-related polypeptide. Eur J Biochem. 1993;217:13–19. doi: 10.1111/j.1432-1033.1993.tb18212.x. [DOI] [PubMed] [Google Scholar]
- Sidis Y, Mukherjee A, Keutmann H, Delbaere A, Sadatsuki M, Schneyer A. Biological activity of follistatin isoforms and follistatin-like-3 is dependent on differential cell surface binding and specificity for activin, myostatin, and bone morphogenetic proteins. Endocrinology. 2006;147:3586–3597. doi: 10.1210/en.2006-0089. [DOI] [PubMed] [Google Scholar]
- Smas CM, Chen L, Sul HS. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol Cell Biol. 1997;17:977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–980. [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Naslund E, Britton T, Concha H, Hassan M, Ryden M, Frisen J, Arner P. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- Sumitomo K, Kurisaki A, Yamakawa N, Tsuchida K, Shimizu E, Sone S, Sugino H. Expression of a TGF-beta1 inducible gene, TSC-36, causes growth inhibition in human lung cancer cell lines. Cancer Lett. 2000;155:37–46. doi: 10.1016/s0304-3835(00)00407-9. [DOI] [PubMed] [Google Scholar]
- Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 Regulates 3T3-L1 Adipogenesis. Mol Endocrinol. 2009 doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H, Kita T, Satoh N, Shimatsu A, Hasegawa K. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376:728–732. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- Tan X, Zhai Y, Chang W, Hou J, He S, Lin L, Yu Y, Xu D, Xiao J, Ma L, Wang G, Cao T, Cao G. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123:1080–1088. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ozaki S, Kawabata D, Kishimura M, Osakada F, Okubo M, Murakami M, Nakao K, Mimori T. Potential preventive effects of follistatin-related protein/TSC-36 on joint destruction and antagonistic modulation of its autoantibodies in rheumatoid arthritis. Int Immunol. 2003;15:71–77. doi: 10.1093/intimm/dxg005. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ozaki S, Osakada F, Mori K, Okubo M, Nakao K. Cloning of follistatin-related protein as a novel autoantigen in systemic rheumatic diseases. Int Immunol. 1998;10:1305–1314. doi: 10.1093/intimm/10.9.1305. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci U S A. 2003a;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci U S A. 2003b;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101:9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Tochitani S, Liang F, Watakabe A, Hashikawa T, Yamamori T. The occ1 gene is preferentially expressed in the primary visual cortex in an activity-dependent manner: a pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex. Eur J Neurosci. 2001;13:297–307. doi: 10.1046/j.0953-816x.2000.01390.x. [DOI] [PubMed] [Google Scholar]
- Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Towers P, Patel K, Withington S, Isaac A, Cooke J. Flik, a chick follistatin-related gene, functions in gastrular dorsalisation/neural induction and in subsequent maintenance of midline Sonic hedgehog signalling. Dev Biol. 1999;214:298–317. doi: 10.1006/dbio.1999.9421. [DOI] [PubMed] [Google Scholar]
- Trojan L, Schaaf A, Steidler A, Haak M, Thalmann G, Knoll T, Gretz N, Alken P, Michel MS. Identification of metastasis-associated genes in prostate cancer by genetic profiling of human prostate cancer cell lines. Anticancer Res. 2005;25:183–191. [PubMed] [Google Scholar]
- Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- Waki H, Tontonoz P. Endocrine functions of adipose tissue. Annu Rev Pathol. 2007;2:31–56. doi: 10.1146/annurev.pathol.2.010506.091859. [DOI] [PubMed] [Google Scholar]
- Weiner FR, Shah A, Smith PJ, Rubin CS, Zern MA. Regulation of collagen gene expression in 3T3-L1 cells. Effects of adipocyte differentiation and tumor necrosis factor alpha. Biochemistry. 1989;28:4094–4099. doi: 10.1021/bi00435a070. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widera C, Horn-Wichmann R, Kempf T, Bethmann K, Fiedler B, Sharma S, Lichtinghagen R, Leitolf H, Ivandic B, Katus HA, Giannitsis E, Wollert KC. Circulating Concentrations of Follistatin-Like 1 in Healthy Individuals and Patients with Acute Coronary Syndrome as Assessed by an Immunoluminometric Sandwich Assay. Clin Chem. 2009 doi: 10.1373/clinchem.2009.129411. [DOI] [PubMed] [Google Scholar]
- Wu J, Srinivasan SV, Neumann JC, Lingrel JB. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry. 2005;44:11098–11105. doi: 10.1021/bi050166i. [DOI] [PubMed] [Google Scholar]
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Northrop JP, Grove JR, Kilpatrick KE, Su JL, Ringold GM. TNF alpha-mediated inhibition and reversal of adipocyte differentiation is accompanied by suppressed expression of PPARgamma without effects on Pref-1 expression. Endocrinology. 1997;138:2776–2783. doi: 10.1210/endo.138.7.5242. [DOI] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- Yi T, Choi HM, Park RW, Sohn KY, Kim IS. Transcriptional repression of type I procollagen genes during adipocyte differentiation. Exp Mol Med. 2001;33:269–275. doi: 10.1038/emm.2001.44. [DOI] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- Zhou YT, Wang ZW, Higa M, Newgard CB, Unger RH. Reversing adipocyte differentiation: implications for treatment of obesity. Proc Natl Acad Sci U S A. 1999;96:2391–2395. doi: 10.1073/pnas.96.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen A, Blockx H, Van Arnhem W, Willems J, Fransen L, Devos K, Raymackers J, Van de Voorde A, Slegers H. Characterization of a rat C6 glioma-secreted follistatin-related protein (FRP). Cloning and sequence of the human homologue. Eur J Biochem. 1994;225:937–946. doi: 10.1111/j.1432-1033.1994.0937b.x. [DOI] [PubMed] [Google Scholar]