Abstract
Expression profiling of post-mortem human brain tissue has been widely used to study molecular changes associated with neuropsychiatric diseases as well as normal processes such as aging. Changes in expression associated with factors such as age, gender or postmortem interval are often more pronounced than changes associated with disease. Therefore in addition to being of interest in their own right, careful consideration of these effects are important in the interpretation of disease studies. We performed a large meta-analysis of genome-wide expression studies of normal human cortex to more fully catalogue the effects of age, gender, postmortem interval and brain pH, yielding a “meta-signature” of gene expression changes for each factor. We validated our results by showing a significant overlap with independent gene lists extracted from the literature. Importantly, meta-analysis identifies genes which are not significant in any individual study. Finally, we show that many schizophrenia candidate genes appear in the meta-signatures, reinforcing the idea that studies must be carefully controlled for interactions between these factors and disease. In addition to the inherent value of the meta-signatures, our results provide critical information for future studies of disease effects in the human brain.
Introduction
Many studies have concerned genome-wide expression analysis of human postmortem brain tissue with an aim to identify changes in expression associated with neuropsychiatric disease (Mirnics et al., 2006). Human brain tissue presents a particular challenge for the analysis of gene expression due to variability between individuals and tissue heterogeneity, making the detection of small expression changes difficult. It is routine to match samples across conditions and check for confounding effects of gender, age and other factors. This is not always easy, as postmortem brain tissue is a limited resource and often sample sizes are small. Another common method of reducing the effects of these factors involves adjustment during data analysis. These methods include stratification of samples or implementation of statistical techniques based on observed covariate distributions in the compared populations. However, many studies are underpowered to detect genes so affected. This greatly complicates the detection of molecular changes associated with neuropsychiatric disorders such as schizophrenia and bipolar disorder (Tomita et al., 2004).
It is therefore important to understand the effects of factors such as age, gender, brain pH and postmortem interval (PMI) on gene expression, both to control for confounding sources of variability when seeking disease effects, and as a means of elucidating biologically interesting patterns due to the factors themselves. A number of studies have examined expression differences associated with age (Lu et al., 2004, Erraji-Benchekroun et al., 2005), sex (Galfalvy et al., 2003, Vawter et al., 2004, Reinius et al., 2008) and brain pH (Li et al., 2004, Mexal et al., 2006). Because of small samples sizes and the presence of noise, our knowledge of gene expression changes associated with these factors is likely to be incomplete.
One approach to detecting weak patterns is to use meta-analysis. In a meta-analysis, the results from multiple studies are statistically pooled to provide an overall estimate of significance of an effect. While meta-analysis has been increasingly used in the study of gene expression data (Rhodes et al., 2004, Cahan et al., 2007, Borozan et al., 2008), to our knowledge only a few studies have done so with post-mortem human brain data (Elashoff et al., 2007, Choi et al., 2008, de Magalhaes et al., 2009).
In this paper we present a large cross-laboratory meta-analysis of human postmortem brain data, integrating expression data from multiple studies, rather than a simple comparative analysis of published gene lists. Our primary focus in this study is to examine gene expression changes in normal brain with respect to four factors: age, gender, post-mortem interval (PMI) and brain pH, for which data sets from control subjects are publicly available. While many studies treat these factors as a nuisance and attempt to limit their range or control for them, we show that considerable variability in gene expression exists due to these factors. Our results provide new information on gene expression changes attributable to these factors.
Experimental Procedures
Data Collection
Genome-wide expression data sets were selected on the basis of public availability, inclusion of normal subjects, use of neocortical tissue, and the availability sample characteristic data. Details on each of the eleven datasets, including the source citation, can be found in Table 1. Sources include the Stanley Medical Research Institute (SMRI), the Harvard Brain Bank, and the Gene Expression Omnibus (GEO). GEO studies were identified by extensive manual and keyword searches. While the SMRI has additional data sets, these represent repeated runs of the samples from the same subjects, so we arbitrarily selected one data set to represent each of the two SMRI brain collections. Sample characteristics for the normal subjects within each dataset were collected (see Supplementary Table 1 for a summary). Datasets consisted of single-channel intensity data generated from various Affymetrix platforms and one dataset from the Illuimina HumanRef-8 BeadArray. For the majority of the datasets, we obtained pre-processed data in which the expression levels were summarized, log transformed and normalized by using the ‘rma’ function in the R bioconductor ‘affy’ package (RDevelopmentCoreTeam, 2005). Where possible, we obtained the raw data (.cel files) for the remaining datasets and reprocessed it using the ‘rma’ function. For the studies in which the raw data was not available, we used the data in its given format.
Table 1.
Human postmortem brain datasets included in meta-analysis
| Dataset | Reference | Description | Microarray Platform | Brain region(s) | No. of normal Subjects | |
|---|---|---|---|---|---|---|
| A | Stanley Chen | n/a | Schizophrenia, Bipolar, depression | HG-U133A/B (RMA) | DLPFC | 13 |
| B | GSE1572 | Lu et al. (2004) | Aging study | HG-U95vA (RMA) | Frontal lobe | 30 |
| C | GSE2164 | Vawter et al. (2004) | Gender differences in expression | HG-U95vA (RMA) | DLPFC | 10 |
| D | GSE3790 | Hodges et al. (2006) | Huntington’s | HG-U133A/B (MAS 5.0) | Frontal cortex | 36 |
| E | GSE11882 | Berchtold et al. (2008) | Gender and aging | HG-U133 Plus 2.0 (GC-RMA) | Superior Frontal Gyrus | 47 |
| F | GSE11512 | Somel et al. (2009) | Transcriptional neoteny | HG-U133 Plus2.0 (RMA) | Frontal cortex | 15 |
| G | GSE8919 | Myers et al. (2007) | Cortical gene expression | Illumina Sentrix BeadChip (Illumina Software) | Cerebral cortex, temporal lobe, frontal lobe, parietal lobe | 193 |
| H | Stanley Kato | Iwamoto et al. (2005) | Schizophrenia, Bipolar | HG-U133A (RMA) | DLPFC | 34 |
| I | GSE13162 | Chen-Plotkin et al. (2008) | Frontotemporal lobar degeneration | HG-U133A (RMA) | Frontal cortex | 8 |
| J | GSE5390 | Lockstone et al. (2007) | Down Syndrome | HG-U133A (RMA) | DLPFC | 8 |
| K | McLean PFC | n/a | Schizophrenia, Bipolar | HG-U133A (RMA) | PFC | 27 |
Regression Analysis
Gene expression values for each probe, in each dataset, were modeled as a function of each of the factors (age, sex, pH, and PMI). P-values were computed using one-sided tests, preformed independently for the two alternative null hypotheses (i.e. gene expression does not increase with the confounding variable and gene expression does not decrease with the confounding variable). To make a fair comparison across studies, we re-annotated probe sequences for each array and mapped them to the corresponding GenBank gene using the Gemma database (http://www.chibi.ubc.ca/Gemma/). Probes which were annotated to more than one gene were removed from consideration. For cases in which multiple probes mapped to a single gene, we combined p-values by retaining only the minimum p-value. Analyses were conducted in R (RDevelopmentCoreTeam, 2005); code is available at http://chibi.ubc.ca/~mmistry/.
Meta-analysis of Differential Expression
The following meta-analysis was carried out for each of the four confounding factors, and each hypothesis independently. We computed a summary statistic S across n studies for each gene t using Fisher’s method (Fisher, 1948), which has been used previously in other microarray meta-analyses (Rhodes et al., 2002, Hess and Iyer, 2007)
where pi is the regression p-value in the ith experiment. A given gene was included in the analysis given if it was measured in at least three datasets and the particular sample characteristic was reported. A p-value for S(t) is computed by observing that, under the null hypothesis of uniform p-values within each study, S(t) has a χ2 distribution with 2n degrees of freedom. The meta-analysis p-values for each signature were processed with the R ‘qvalue’ package to control the false discovery rate, yielding a q-value measure for each gene (Storey and Tibshirani, 2003).
Validation analysis
We extracted genes lists from the expression analysis literature for age (Erraji-Benchekroun et al., 2005), gender (Galfalvy et al., 2003) and brain pH (Mexal et al., 2006). Each set consisted of a list of probes (Affymetrix probe sets) differentially expressed in human postmortem brain as reported in their respective studies, which were then split based on direction of change. Each probe was mapped to its corresponding gene using Gemma. Genes were removed if they were not included in our meta-analysis. Details on each of the validation sets can be found in Supplementary Table 1. Agreement of the meta-signature ranking with the validation sets was performed using receiver operating characteristic (ROC) curve analysis.
Each meta-signature was further analyzed for functional enrichment of Gene Ontology (GO) terms (Ashburner et al., 2000), using the ‘over-representation analysis’ (ORA) method in ErmineJ (Lee et al., 2005). ORA evaluates the genes that meet a specified selection criterion (meta-q < 0.001) and determines if there are gene sets which are statistically over-represented. Probabilities are computed using the binomial approximation to the hypergeometric distribution then corrected for multiple testing using the Benjamini-Hochberg procedure.
Results
Meta-analysis of differential expression
We assessed the global levels of gene expression across datasets by assigning rank values to each gene based on its mean expression value within a dataset. While we observe variation between studies, there still emerges a clear pattern of genes which are consistently strongly or weakly expressed in the brain (data not shown) supporting the feasibility of comparing studies to one another.
We used linear regression to evaluate gene expression changes with respect to four factors (age, sex, brain pH, and PMI), within each dataset. We considered both directions of change (up- and down-regulation) for each factor, creating up to eight different scenarios for each dataset. Although the focus of this paper is to report on the results from the meta-analysis across datasets, we briefly summarized here the results from individual studies. The numbers of genes that show evidence change with each of the factors (q < 0.01) is given in Table 2. Not surprisingly, the datasets with smaller sample size showed fewer statistically significant changes associated with the factors. Overall the factors associated with the most robust differential expression were age and brain pH. Genes found to change in individual datasets but not from meta-analysis can be found in Supplementary Table 7 at http://chibi.ubc.ca/~mmistry/.
Table 2.
Significant genes (q<0.01) identified within each individual dataset
| Dataset | Age | Sex | pH | PMI | ||||
|---|---|---|---|---|---|---|---|---|
| Down | Up | Female | Male | Down | Up | Down | Up | |
| Stanley Chen | 0 | 0 | 2 | 7 | 0 | 0 | 0 | 0 |
| Stanley Kato | 0 | 0 | 1 | 10 | 703 | 0 | 0 | 0 |
| GSE1572 | 162 | 64 | 1 | 6 | n/a | n/a | 0 | 0 |
| GSE2164 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| GSE8919 | 153 | 133 | 6 | 19 | n/a | n/a | 2 | 278 |
| GSE11512 | 0 | 0 | 3 | 13 | 1 | 0 | 0 | 0 |
| GSE11882 | 428 | 0 | 2 | 14 | n/a | n/a | 1 | 0 |
| GSE13162 | 0 | 0 | 0 | 0 | n/a | n/a | 0 | 0 |
| GSE5390 | 1 | 1 | n/a | n/a | n/a | n/a | 0 | 0 |
| McLeanPFC | 0 | 0 | 1 | 5 | 0 | 0 | n/a | n/a |
| GSE3790 | 0 | 0 | 1 | 13 | n/a | n/a | n/a | n/a |
| ‘Union’ Signature | 689 | 198 | 10 | 33 | 704 | 0 | 2 | 278 |
| Meta-signature Overlap | 415 | 102 | 6 | 19 | 191 | 0 | 1 | 46 |
Q-values were calculated from regression p-values for each factor within each dataset. The number of genes reported here are significant at q < 0.01. The ‘union’ signature represents the union of unique genes identified for each factor across all datasets. The meta-signature overlap indicates the number of union signature genes overlapping with the corresponding meta-signature.
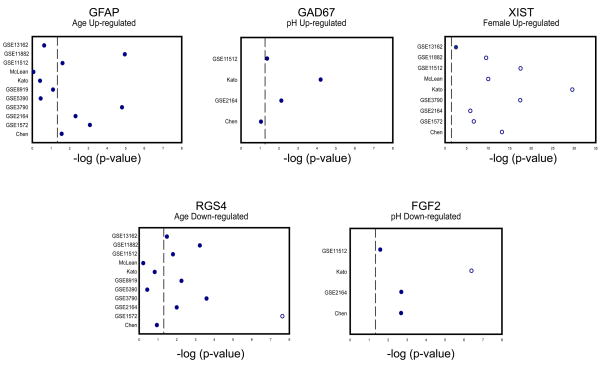
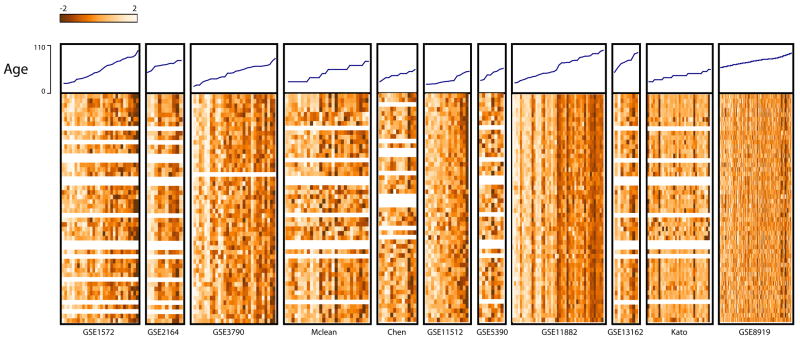
To examine changes in gene expression that were consistent across all datasets, or supported by evidence from multiple data sets, we implemented a cross-study meta-analysis approach (see Methods). The output of this analysis is eight meta-signatures (up or down for each of the four factors). The top ten genes from each meta-signature can be found in Table 7. To examine the results, we first extracted the corresponding p-values from each individual dataset and visualized them as (smoothed) plots in the order determined by the meta-signature order (Figure 1). We observe that genes that have good meta-q-values tended to have good p-values in multiple, but not necessarily all studies. We also demonstrate this using an alternative heat map visualization (Supplementary Figure 1). More detailed results are plotted for some example genes in Figure 2. These plots show that p-values for a given gene vary across individual datasets, and the meta-analysis can identify genes which show only weak or non-significant effects in some data sets. For example, in Figure 2, for age genes Gfap and Rgs4, we see weak changes in expression level (up and down, respectively), that are not significant after multiple test correction in those studies. On the other hand, we also find genes that show significant effects in most if not all studies (e.g., Xist, Figure 2). In Figure 3 we assembled the top 50 genes down-regulated with age, and plotted expression levels within each dataset with samples ordered by increasing age. For most of the studies, we observe a gradient across the dataset as expression decreases from high to low levels, illustrating that the meta-analysis recovers many genes which show fairly consistent trends across data sets.
Table 7.
Top meta-signature genes
| Age Down-regulated | Age Up-regulated | ||
|---|---|---|---|
| OLFM1 | olfactomedin 1 | NEBL | nebulette |
| KCNF1 | potassium voltage-gated channel, subfamily F, member 1 | MED12 | mediator complex subunit 12 |
| RGS4 | regulator of G-protein signaling 4 | BCL2 | B-cell CLL/lymphoma 2 |
| PPP3CB | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform | GMPR | guanosine monophosphate reductase |
| ADCY2 | adenylate cyclase 2 (brain) | GFAP | glial fibrillary acidic protein |
| SVOP | SV2 related protein homolog (rat) | ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide |
| EFNB3 | ephrin-B3 | WWOX | WW domain containing oxidoreductase |
| ATP2B2 | ATPase, Ca++ transporting, plasma membrane 2 | PLEC1 | plectin 1, intermediate filament binding protein 500kDa |
| HPCA | hippocalcin | VCAN | versican |
| CALB1 | calbindin 1, 28kDa | AHCYL1 | S-adenosylhomocysteine hydrolase-like 1 |
| pH Down-regulated | pH Up-regulated | ||
| FGF2 | fibroblast growth factor 2 (basic) | SLC1A1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| AHCYL1 | S-adenosylhomocysteine hydrolase-like 1 | LARGE | like-glycosyltransferase |
| DTNA | dystrobrevin, alpha | C17orf81 | chromosome 17 open reading frame 81 |
| MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 | HISPPD2A | histidine acid phosphatase domain containing 2A |
| TJP1 | tight junction protein 1 (zona occludens 1) | PRKCD | protein kinase C, delta |
| S100A13 | S100 calcium binding protein A13 | DLG3 | discs, large homolog 3 (Drosophila) |
| RBBP6 | retinoblastoma binding protein 6 | KCNAB1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| GNG12 | guanine nucleotide binding protein (G protein), gamma 12 | SLC8A1 | solute carrier family 8 (sodium/calcium exchanger), member 1 |
| ANP32E | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | GABRA5 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| BAALC | brain and acute leukemia, cytoplasmic | RIT2 | Ras-like without CAAX 2 |
| Female Up-regulated | Male Up-regulated | ||
| XIST | X (inactive)-specific transcript (non-protein coding) | JARID1D | jumonji, AT rich interactive domain 1D |
| HDHD1A | haloacid dehalogenase-like hydrolase domain containing 1A | USP9Y | ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) |
| UTX | ubiquitously transcribed tetratricopeptide repeat, X chromosome | EIF1AY | eukaryotic translation initiation factor 1A, Y-linked |
| JARID1C | jumonji, AT rich interactive domain 1C | CYorf15B | chromosome Y open reading frame 15B |
| TSIX | XIST antisense RNA (non-protein coding) | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
| USP9X | ubiquitin specific peptidase 9, X-linked | UTY | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked |
| LOC554203 | alanyl-tRNA synthetase domain containing 1 pseudogene | RPS4Y1 | ribosomal protein S4, Y-linked 1 |
| STS | steroid sulfatase (microsomal), isozyme S | TTTY15 | testis-specific transcript, Y-linked 15 |
| ZFX | zinc finger protein, X-linked | CYorf15A | chromosome Y open reading frame 15A |
| PNPLA4 | patatin-like phospholipase domain containing 4 | TMSB4Y | thymosin beta 4, Y-linked |
| PMI Down-regulated | PMI Up-regulated | ||
| BRD8 | bromodomain containing 8 | GOSR2 | golgi SNAP receptor complex member 2 |
| RBM5 | RNA binding motif protein 5 | CYB5B | cytochrome b5 type B (outer mitochondrial membrane) |
| PUM2 | pumilio homolog 2 (Drosophila) | SNF8 | SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | GRLF1 | glucocorticoid receptor DNA binding factor 1 |
| NCOA3 | nuclear receptor coactivator 3 | C6orf1 | chromosome 6 open reading frame 1 |
| DAPK1 | death-associated protein kinase 1 | MGMT | O-6-methylguanine-DNA methyltransferase |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats-containing | MAX | MYC associated factor X |
| KIAA0841 | KIAA0841 | ST3GAL2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| CAMK2G | calcium/calmodulin-dependent protein kinase II gamma | PTGER3 | prostaglandin E receptor 3 (subtype EP3) |
| DIP2C | DIP2 disco-interacting protein 2 homolog C (Drosophila) | EXT1 | exostoses (multiple) 1 |
For each meta-signature we have listed the top ten genes, as ranked by meta-q-value (all at q < 0.001). For each gene we have listed the gene symbol an gene name. Complete meta-signature lists for each factor can be found in Supplementary Table 6 http://chibi.ubc.ca/~mmistry/.
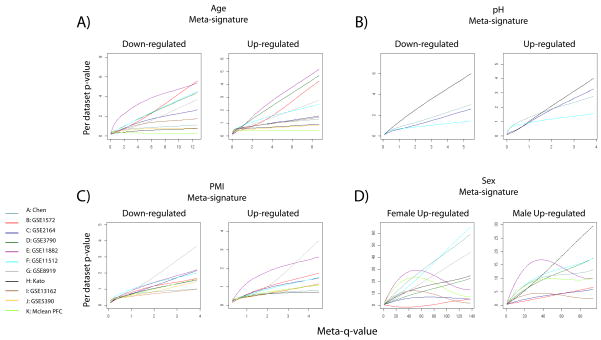
Figure 1. Distribution of dataset p-values across meta-signature q-values.
For each dataset used, gene p-values were plotted against the corresponding meta-q value and a loess fit was computed to generate a smooth curve between points. The fact that most data sets show a rise in p-values correlated with the meta-q-values indicates the contribution of signals of varying strengths to the meta-signatures. An alternative view of the data using heat maps is available in the supplement. The distorted curves for gender are due to the strong effects of a small number of genes with very small meta-q-values (note the difference in scale of D compared to A–C).
Figure 2. Distribution of dataset p-values for individual genes: a magnified view.
For selected genes from each of the meta-signatures we have plotted the log regression p-values from each dataset. Open circles represent the datasets for which the gene was found to be significant after multiple test correction (q < 0.01). Dashed line indicates a per-study p-value significance level of 0.05 for reference.
Figure 3. Top genes down-regulated with age.
The top 50 age down-regulated genes were selected based on meta-analysis q-value ranking. For each gene, the corresponding data from each study was extracted and converted to a heat map. Expression values were normalized across samples within each dataset, and ordered by age. Age is plotted at the top of each heat map. Light values in heat map indicate higher expression. Grey bars indicate missing values. All data sets are at approximately the same horizontal scale except the last, which is compressed to fit on the page.
While we have presented results from an analysis which treated each factor independently (linear regression), we also performed a meta-analysis which models the factors simultaneously in an analysis of covariance, yielding very similar results (available at http://chibi.ubc.ca/~mmistry/). We also attempted to model interactions among factors, but this was difficult due to lack of power.
Meta-profile evaluation
We tested the robustness of our meta-profiles by using a jackknife procedure. This involved sequentially removing a dataset, performing the meta-analysis on the remaining datasets, and then selecting genes at a meta-q < 0.01. This procedure was repeated for each data set in turn, and genes found in all rounds retained as a “core” signature. Each of the core signatures encompass more than half of the genes found in the corresponding meta-profiles, with the pH meta-profiles as an exception. The ”core” signatures can be found in Supplementary Table 6 at http://chibi.ubc.ca/~mmistry/.
The studies we selected for meta-analysis were, in general, not designed to test the effects of age, sex, pH or PMI; in fact attempts may have been made to limit the range of these factors (especially in the case of PMI and pH). However, because human brain samples are difficult to obtain, enough variability is allowed into studies to allow us to perform a meaningful meta-analysis. We still questioned the extent to which our results would agree with more targeted studies. Therefore, for the purpose of validation of our approach, we identified independent gene lists from the literature for age, sex and brain pH (we could not find a comprehensive validation set for PMI). Details on validation sets can be found in Supplementary Table 2. Each validation gene list was then separated into two groups based on direction of change, to correspond with meta-signatures obtained from our meta-analysis. Obviously none of these validation gene lists can be considered true gold standards, but help put our results in the context of previous findings.
To quantify the predictive power of our analysis meta-signatures with respect to the corresponding validation sets, we first preformed a standard receiver operating characteristic (ROC) analysis. The score reported for each profile is the area under the ROC curve (AUC), a value between 0 and 1, where 1.0 is perfect agreement with the external list and 0.5 would reflect a random order. The AUC values for each of the profiles are reported in Table 3. Individual ROC plots can be found in Supplementary Figure 2. We also tested the effect of using a specific statistical threshold for selecting genes from the meta-profiles, by collecting genes at two significance levels (meta-q <0.01, and meta-q < 0.001). The overlap with the validation set was significant (p<0.001, Fisher s exact test; Table 3) for all signatures. We also found a comparable overlap between each of the core signatures and the validation sets.
Table 3.
Comparison of meta-signature profiles against validation gene sets
| No. of profile genes (q < 0.001) | Overlap with validation set (q < 0.001) | No. of profile genes (q < 0.01) | Overlap with validation set (q < 0.01) | AUC score | |
|---|---|---|---|---|---|
| Age Genes | |||||
| Up-regulated | 404 | 40 | 1241 | 69 | 0.74 |
| Down-regulated | 1134 | 113 | 2247 | 136 | 0.76 |
| pH Genes | |||||
| Up-regulated | 25 | 11 | 368 | 131 | 0.91 |
| Down-regulated | 215 | 55 | 1018 | 122 | 0.86 |
| Sex Genes | |||||
| Male | 14 | 7 | 38 | 7 | 0.90 |
| Female | 19 | 1 | 128 | 1 | n/a |
| PMI Genes | |||||
| Up-regulated | 75 | n/a | 691 | n/a | n/a |
| Down- regulated | 4 | n/a | 49 | n/a | n/a |
The brain pH profiles gave high AUC scores of 0.91 and 0.86 (up- and down-regulated genes, respectively), when validated against DLPFC pH-sensitive genes taken from Vawter et al. (2006). Additionally, we obtained reasonably high scores using a smaller independent pH gene list obtained from Mexal et al. (2006), despite a difference in the brain region used between the validation study and the meta-analysis (data not shown). Because pH itself probably covaries across brain regions (Mexal et al., 2006), our results are consistent with the hypothesis that pH-related changes in gene expression are similar across brain regions. The age profiles on the other hand, exhibited slightly lower scores than those obtained for brain pH. Erraji-Benchekroun et al. used samples from Brodmann areas 9 (dorsolateral PFC, BA9) and 47 (orbitofrontal PFC, BA47) from each subject, to evaluate age expression differences, showing comparable changes in both brain regions (Erraji-Benchekroun et al., 2005). As such, our validation set consisted of genes showing age expression changes collectively within both neocortical brain regions BA9 and BA47. While many of these genes appear at the top of our ranked lists, some are also dispersed throughout our ranking. The gender meta-signature from our meta-analysis scored high when validated with a set of genes from Galfalvy et al. (2003), with most validation genes sitting at the top of the ranking.
Overlap with schizophrenia candidate genes
We compared significant genes (meta-q < 0.001) from each of our meta-signatures with genes known to be associated with schizophrenia. We extracted a list of 34 schizophrenia candidate genes provided in a comprehensive literature review (Colantuoni et al., 2008), and searched this list of genes within each of our meta-signatures. We found that 12 of these genes identified with at least one of our meta-signatures, though the majority of overlap was observed amongst the age and pH meta-profiles (Table 4). Overlap of genes with each of the age meta-signatures was significant at p < 0.01.
Table 4.
Schizophrenia candidate gene analysis
| Schizophrenia genes identified in meta-profiles | ||
|---|---|---|
| Age | Down | Opcml, Pldn, Nrg1, Rgs4, Bdnf, Dlg4, Gad67 |
| Up | Ntrk2, Ppp1r1b, Erbb3 | |
| pH | Down | Ntrk2, Slc1a2 |
| Up | Rgs4, Gad67 | |
| PMI | Up | Nrg1 |
Affected Biological Pathways
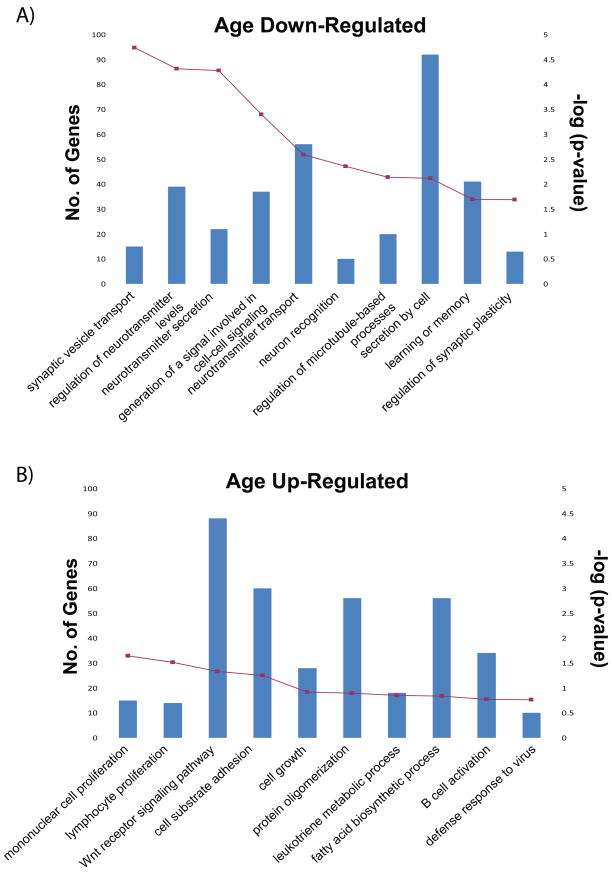
To derive a high-level biological interpretation of our meta-signatures we performed a Gene Ontology (GO) (Ashburner et al., 2000) enrichment analysis using ErmineJ (Lee et al., 2005). We extracted the ‘top’ GO categories for each of the profiles and a comparison amongst them revealed various ‘biological processes’ unique to each meta-signature, in addition to a number of shared categories. In Figure 4, we have displayed the top ten categories for each age meta-signature depicting the number of genes represented in each GO category and the associated p-value (corrected for multiple testing). The ORA data generated for all eight meta-signatures can be found in Supplementary Table 5 at http://chibi.ubc.ca/~mmistry/.
Figure 4. Gene Ontology Enrichment Analysis.
For the age meta-signatures, we have displayed the top 10 GO terms identified using a GO over-representation analysis. The primary y-axis displays the number of meta-signature genes that fall in the given ‘biological process’ category, while the secondary axis displays the associated p-value. GO terms were collapsed to parent term if parent and child both appeared in the top ten.
For genes increasing in expression with the progression of age, top GO categories included those involved in cell growth and proliferation, and cell-cell interaction, consistent with previous studies (Erraji-Benchekroun et al., 2005, Hong et al., 2008). Other processes included the insulin receptor signaling pathway, encompassing a number of genes involved in longevity and aging (Bartke, 2008). The age down-regulated genes presented an enrichment in synaptic and/or receptor activity with GO categories such as neuron recognition, neurotransmitter transport and secretion, and regulation of neurotransmitter levels. This finding is concordant with existing aging studies in mouse and human (Erraji-Benchekroun et al., 2005, Oh et al., 2009). Similarly, we found an enrichment of genes involved in neurotransmitter secretion in the pH up-regulated meta-signature, in addition to genes implicated in metabolism and a different array of pathways (i.e. G-protein signaling). The female and male meta-signatures identified a similar enrichment of terms including relevant processes such as sex determination. However, the genes associated with these shared terms were quite different between the two meta-signatures. Functional enrichment analysis of our meta-signatures, although far from providing hard cellular evidence, still provided a useful indication of the biological processes altered by each factor and greater insight at the molecular level.
Correlation between factors and meta-profiles
Because we analyzed each factor independently, we wished to check whether factors were correlated with each other across the 415 samples (Table 5). Age and PMI displayed the highest correlation of 0.35, consistent with a positive correlation reported in (Galfalvy et al., 2003). Age and brain pH displayed a slight negative correlation of −0.2. Further investigation of these two factors revealed that categorizing the values of age into ‘young’ (< 50 years of age) and ‘old’ (≥ 50 years of age) groups resulted in a lower mean pH in the ‘old’ group versus the ‘young’. This was the general trend within each dataset (Supplementary Figure 3).
Table 5.
Rank correlations between factors
Spearman rank correlations were computed using sample characteristic information for individual subjects.
indicates a p-value of ≤ 0.05
indicates a p-value of ≤ 0.01
indicates a p-value of ≪ 0.001
Due to these correlations, we expected that individual genes in some meta-profiles might overlap with other meta-profiles (Table 6). Accordingly, the two factors displaying the highest number of overlapping genes were those [up or down] in age and those [down or up] with brain pH, respectively. However, these effects were weak and were even weaker in the ‘core’ signatures (Supplementary Table 4). We also found that a number of genes up-regulated with PMI were also identified amongst the profiles for brain pH and age in both directions, but were unable to extract any definite patterns.
Table 6.
Evaluating gene overlap between meta-signatures
| No. of Profile genes | Age | pH | PMI | Sex | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Female | Male | |||
| Age | Up | 404 | ||||||||
| Down | 1134 | 2 | ||||||||
| pH | Up | 25 | 0 | 18 | ||||||
| Down | 215 | 75 | 1 | 0 | ||||||
| PMI | Up | 75 | 2 | 32 | 0 | 2 | ||||
| Down | 4 | 1 | 2 | 0 | 0 | 0 | ||||
| Sex | Female | 19 | 4 | 1 | 0 | 1 | 0 | 0 | ||
| Male | 14 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
Using genes for each meta-profile at q < 0.001, we compared them against one another to evaluate the overlap and potential relationships between the factors. The observation of two genes in the age meta-signature that change in both directions is a consequence of multiple probes of different specificity for the same gene, which show different directions of change.
Discussion
We have conducted a meta-analysis of gene expression in human cortex, examining gender, age, postmortem interval and brain pH, as well as expression level. This meta-analysis was made possible by the fact that many gene expression analyses have useful data for each of these factors, even though they were usually considered potential “confounds” to be controlled. Our results have at least two potential uses for future studies. First, the results of our meta-analysis provide new information on the effects of each of the factors on gene expression and can be studied further independently or used to bolster support for other studies. Second, the identification of robust signatures associated with these factors will provide a ‘watch list’ of genes which might be viewed cautiously if they are found to be implicated in neuropsychiatric disease by expression studies. To facilitate the use of these lists in future studies, they are provided in Supplementary Table 6 at http://chibi.ubc.ca/~mmistry/, with the top ten from each list displayed in Table 7.
There are some limitations of our study. We used a relatively simple meta-analysis method, and other techniques may have even higher sensitivity. We combined datasets generated using differing platforms, which may contribute noise and potentially reduce the power of our meta-analysis. The MAQC (MicroArray Quality Control) project recently initiated a number of studies to specifically address these concerns, and in general, reported a high agreement between platforms (Canales et al., 2006, Shi et al., 2006, Shippy et al., 2006). A number of other studies have been conducted to this end, showing agreement between platforms (Dobbin et al., 2005, Irizarry et al., 2005, Petersen et al., 2005), and a high concordance between the top functions identified by each platform (Pedotti et al., 2008, Li et al., 2009). While we acknowledge that there still remain small differences between studies, we are only focussed on the consistencies and combining them to extract more robust expression changes than can be derived from single dataset studies. To maximize total sample size in our study, we used a coarsely defined neocortical brain region. The majority of our datasets utilized samples from the frontal cortex, but we also included a number of samples from the temporal and parietal cortices. Various groups have studied regional patterns of gene expression in the postmortem human brain, revealing cortical regions to generally cluster together indicating a shared global expression profile (Khaitovich et al., 2004, Roth et al., 2006, Ernst et al., 2007). Finally, we only considered linear model fits to age, pH and PMI. Future studies can address some of these issues, and also include more data as studies become available.
Comparison of our results to the validation sets strongly supported the relevance of the meta-signatures. However, the overlap in ‘top genes’ between meta-profiles and validation sets, while statistically significant, was only a subset of the genes in the validation lists. There are several possible explanations for this effect. One is that most of the studies we used treated these factors as nuisances to be eliminated, which may have reduced our power to find real changes. For example, the majority of the datasets have no subjects under the age of 30 at the time of death, and most are over 40. In particular the Kato and Chen data sets, which use samples from the Stanley Foundation, have a particularly well-controlled (narrow) age range. In contrast the age validation set used a broader age range (13 to 79 years of age) (Erraji-Benchekroun et al., 2005). Additionally, we expect biological variation among sample groups (and therefore studies). Strong signals in any given data set, including the validation sets, may be specific to that study. That is, none of the validation sets are truly gold standards. Further examination of genes on the validation lists within each individual dataset supports this notion. The agreement between the meta-analysis and the validation lists is better than the agreement between the validation and any single dataset, with only a few to none of the validation genes being identified in each dataset. In summary, the limited overlap of the meta-signatures with the validation sets may simply be contingent on the data we used, and does not call into question the validation sets or the meta-analysis.
Using a jackknife analysis, we obtained core signatures for each of the factors. We found a large proportion of meta-profile genes overlap with the ‘core’ signatures, illustrating the ability of our meta-analysis to extract gene profiles robust to influences from individual datasets. The exception was brain pH, for which the ‘core’ signatures consisted of very few genes (Supplementary Table 4). This was due to a large pH effect in the Kato dataset. However, examination of the other data sets revealed that many genes showing large effects in the Kato data set also show trends with pH (Supplementary Figure 4). Thus, even though the pH meta-profiles are arguably biased by strong effects from the Kato dataset, these genes appear to show weaker changes in many other data sets.
The inverse relationship we observed between our age and brain pH meta-profiles is in agreement with a previous study (Harrison et al., 1995). A review of the literature also reveals that results from independent gene expression studies examining changes with age are strikingly similar to results derived from brain pH profiling studies, (Li et al., 2004, Erraji-Benchekroun et al., 2005, Berchtold et al., 2008, Hong et al., 2008, de Magalhaes et al., 2009), further supporting this relationship. It has been suggested that the relationship between age and pH is likely a result of slower modes of death experienced by elderly subjects (Harrison et al., 1995), but this has not yet been fully explored. Previous studies however, have found brain pH to be a proxy for agonal stress (Li et al., 2007). Subjects experiencing a longer terminal phase of death results in lower brain pH levels than would be observed in subjects experiencing a sudden death. We were unable to obtain cause of death information for the majority of our datasets, and thus were unable to incorporate it into the meta-analysis. This raises the question of whether the reasoning behind the inverse relationship is as hypothesized by Harrison et al. (1995), or if the process of aging actually results in a general decline of brain pH.
To this point we have focused on evaluating the meta-analysis in light of other data sets, but clearly one of the reasons to do a meta-analysis is to integrate information on weak patterns. Indeed, our meta-analysis has confirmed previous finding and also added to them. By assembling the significant genes (q<0.01) from individual datasets, we were able to generate a ‘union’ signature, for each of the factors (Table 2). A comparison of the union signatures against the corresponding meta-signatures revealed greater than 50% of the genes in each of our meta-signatures to be novel (not found in any of the individual studies, and only revealed by the meta-analysis). These novel genes span a broad range of cellular functions, implicating various gene families with each of the different factors. An example is alterations of the gamma-aminobutyric acid (GABA)-related transcriptome in age. We found two GABA receptors (Gabbr and Gabrg2) and two glutamic acid decarboxylases (Gad67 and Gad65) to be down-regulated with age. Animal studies have demonstrated that GABA receptors are markedly decreased with age, and there is evidence to suggest this plays a role in age-related cognitive changes (Jiang et al., 2001, El Idrissi, 2008). Evidence of reduced inhibitory neurotransmission in the human brain is supported by evidence from a recent study (Loerch et al., 2008) using the Lu et al. (2004) dataset, and is also observed in the results of our meta-analysis. Also consistently altered in our age meta-signature are members of the regulator of G-protein signalling (RGS) family. RGS family members, expressed in the brain and periphery, plays a critical role in signal transduction by negatively regulating G-protein-coupled receptors (GPCR) by means of their GTPase accelerating activity. These proteins have been implicated in normal behavioural processes and various brain disorders. In the brain, they function to modulate neurotransmission resulting from the activation of metabotropic GPCRs. Rgs4, a member of this family has been previously shown to be down-regulated with age (Colantuoni et al., 2008). In our study, we confirm this finding and additionally report four other members of the family, (Rgs6, Rgs7, Rgs12 and Rgs17) that display an age-related decline in expression. Alterations in expression of RGS genes presents a possible molecular mechanism that could affect neuronal functioning during aging.
One motivation of our study was to identify gene expression changes which need to be accounted for when studying potential expression changes in neuropsychiatric disorders such as schizophrenia. This is important because changes in expression due to the factors we studied can be large, compared to the reported effects of psychiatric disease (Mirnics et al., 2006). Thus even a mild bias in age might cause a change in expression which is as large, or larger than, the effect of disease. Therefore a gene which is known to change expression with age (for example) must be analyzed very carefully if it is to be considered a candidate marker for disease, because it is difficult to control perfectly for age. We searched our meta-signatures for a list of schizophrenia associated genes and found that 12 of these genes identified with at least one of our meta-signatures (Table 4). One such example is Rgs4, a gene that has been extensively characterized in schizophrenia studies (Levitt et al., 2006) and which we find to be down-regulated with age. Our results confirm previous work showing that Rgs4 is down-regulated with age (Colantuoni et al., 2008). We also identified the receptor-ligand pair ErbB3 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 3) and Nrg1 (neuregulin 1) in our age up- and down-regulated meta-signatures, respectively. Nrg1 and its receptor ErbB3 are implicated in key neurodevelopmental processes in the nervous system (Falls, 2003), and have also implicated in schizophrenia (Corfas et al., 2004). Evidence of a role for Nrg1-ErbB in schizophrenia includes reduction in the level of ErbB3 expression in human postmortem prefrontal cortex samples and genetic association evidence linking Nrg1 to schizophrenia (Corfas et al., 2004). Another notable candidate is Gad67, a gene that is down-regulated across our meta-signatures for age (consistent with some reports in the literature (Cashion et al., 2004, Siegmund et al., 2007)), and up-regulated with pH. The reduction of Gad67 expression in schizophrenia is arguably one of the best established changes for this disorder (Mirnics et al., 2006). Together these findings of expression alterations of genes implicated in schizophrenia with respect to these factors contribute an added complexity to their pre-existing relationships with the disorder.
Looking specifically at findings from microarray studies of schizophrenia (Middleton et al., 2002, Vawter et al., 2002, Iwamoto et al., 2005, Arion et al., 2007), we find additional overlap with the results from our meta-analysis. Synaptic machinery transcripts such as Syn2 and Synj1, reported as down-regulated in subjects with schizophrenia, are also down-regulated with age in our meta-signatures. We see similar patterns between our data and other genes reported to be down-regulated in schizophrenia such as Mapk1, Kcnk1, and Crym. Careful analysis of such genes will allow us to explore the potential of interrelationships between these factors and schizophrenia, and reveal the underlying factors driving the changes in gene expression.
In summary, our results show that meta-analysis of human brain gene expression data is both feasible and informative. We hope our study will encourage future meta-analyses on the effects of neuropsychiatric disease on human brain gene expression.
Supplementary Material
Acknowledgments
MM was partly supported by the Canadian Institute for Health Research (CIHR) bioinformatics training program. PP is supported by a career award from the Michael Smith Foundation for Health Research, a CIHR New Investigator award, the Canadian Foundation for Innovation, and a Human Brain Project grant from the National Institutes of Health (GM076990). We thank Kelsey Hamer for technical support. We thank Clare Beasley for critical reading of the manuscript. We also thank the groups and institutions who made their data publicly available, including the SMRI and the Harvard Brain bank. We thank Nicole Berchtold (University of California, Irvine), Mehmet Somel (Max Planck Institute for Evolutionary Anthropology), Alice Chen-Plotkin (Center for Neurodegenerative Disease Research) for providing additional information on their data sets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7:285–290. doi: 10.1111/j.1474-9726.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borozan I, Chen L, Paeper B, Heathcote JE, Edwards AM, Katze M, Zhang Z, McGilvray ID. MAID: an effect size based model for microarray data integration across laboratories and platforms. BMC Bioinformatics. 2008;9:305. doi: 10.1186/1471-2105-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Rovegno F, Mooney D, Newman JC, St Laurent G, 3rd, McCaffrey TA. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007;401:12–18. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- Cashion AB, Smith MJ, Wise PM. Glutamic acid decarboxylase 67 (GAD67) gene expression in discrete regions of the rostral preoptic area change during the oestrous cycle and with age. J Neuroendocrinol. 2004;16:711–716. doi: 10.1111/j.1365-2826.2004.01225.x. [DOI] [PubMed] [Google Scholar]
- Choi KH, Elashoff M, Higgs BW, Song J, Kim S, Sabunciyan S, Diglisic S, Yolken RH, Knable MB, Torrey EF, Webster MJ. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry. 2008;8:87. doi: 10.1186/1471-244X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Hyde TM, Mitkus S, Joseph A, Sartorius L, Aguirre C, Creswell J, Johnson E, Deep-Soboslay A, Herman MM, Lipska BK, Weinberger DR, Kleinman JE. Age-related changes in the expression of schizophrenia susceptibility genes in the human prefrontal cortex. Brain Struct Funct. 2008 doi: 10.1007/s00429-008-0181-5. [DOI] [PubMed] [Google Scholar]
- Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7:575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin KK, Beer DG, Meyerson M, Yeatman TJ, Gerald WL, Jacobson JW, Conley B, Buetow KH, Heiskanen M, Simon RM, Minna JD, Girard L, Misek DE, Taylor JM, Hanash S, Naoki K, Hayes DN, Ladd-Acosta C, Enkemann SA, Viale A, Giordano TJ. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- El Idrissi A. Taurine improves learning and retention in aged mice. Neurosci Lett. 2008;436:19–22. doi: 10.1016/j.neulet.2008.02.070. [DOI] [PubMed] [Google Scholar]
- Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ, Barci BM, Torrey EF. Meta-analysis of 12 genomic studies in bipolar disorder. J Mol Neurosci. 2007;31:221–243. doi: 10.1385/jmn:31:03:221. [DOI] [PubMed] [Google Scholar]
- Ernst C, Sequeira A, Klempan T, Ernst N, Ffrench-Mullen J, Turecki G. Confirmation of region-specific patterns of gene expression in the human brain. Neurogenetics. 2007;8:219–224. doi: 10.1007/s10048-007-0084-2. [DOI] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Combining independent tests of significance. American Statistician. 1948;2:30. [Google Scholar]
- Galfalvy HC, Erraji-Benchekroun L, Smyrniotopoulos P, Pavlidis P, Ellis SP, Mann JJ, Sibille E, Arango V. Sex genes for genomic analysis in human brain: internal controls for comparison of probe level data extraction. BMC Bioinformatics. 2003;4:37. doi: 10.1186/1471-2105-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200:151–154. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- Hess A, Iyer H. Fisher’s combined p-value for detecting differentially expressed genes using Affymetrix expression arrays. BMC Genomics. 2007;8:96. doi: 10.1186/1471-2164-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MG, Myers AJ, Magnusson PK, Prince JA. Transcriptome-wide assessment of human brain and lymphocyte senescence. PLoS ONE. 2008;3:e3024. doi: 10.1371/journal.pone.0003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, Griffin C, Hilmer SC, Hoffman E, Jedlicka AE, Kawasaki E, Martinez-Murillo F, Morsberger L, Lee H, Petersen D, Quackenbush J, Scott A, Wilson M, Yang Y, Ye SQ, Yu W. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci U S A. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, Heissig F, Arendt T, Nieselt-Struwe K, Eichler EE, Paabo S. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: Convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Li JZ, Meng F, Tsavaler L, Evans SJ, Choudary PV, Tomita H, Vawter MP, Walsh D, Shokoohi V, Chung T, Bunney WE, Jones EG, Akil H, Watson SJ, Myers RM. Sample matching by inferred agonal stress in gene expression analyses of the brain. BMC Genomics. 2007;8:336. doi: 10.1186/1471-2164-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney WE, Jr, Myers RM. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- Li Z, Su Z, Wen Z, Shi L, Chen T. Microarray platform consistency is revealed by biologically functional analysis of gene expression profiles. BMC Bioinformatics. 2009;10 (Suppl 11):S12. doi: 10.1186/1471-2105-10-S11-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, Prolla TA, Yankner BA. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Mexal S, Berger R, Adams CE, Ross RG, Freedman R, Leonard S. Brain pH has a significant impact on human postmortem hippocampal gene expression profiles. Brain Res. 2006;1106:1–11. doi: 10.1016/j.brainres.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Oh S, Tseng GC, Sibille E. Reciprocal phylogenetic conservation of molecular aging in mouse and human brain. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedotti P, t Hoen PA, Vreugdenhil E, Schenk GJ, Vossen RH, Ariyurek Y, de Hollander M, Kuiper R, van Ommen GJ, den Dunnen JT, Boer JM, de Menezes RX. Can subtle changes in gene expression be consistently detected with different microarray platforms? BMC Genomics. 2008;9:124. doi: 10.1186/1471-2164-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen D, Chandramouli GV, Geoghegan J, Hilburn J, Paarlberg J, Kim CH, Munroe D, Gangi L, Han J, Puri R, Staudt L, Weinstein J, Barrett JC, Green J, Kawasaki ES. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6:63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Reinius B, Saetre P, Leonard JA, Blekhman R, Merino-Martinez R, Gilad Y, Jazin E. An evolutionarily conserved sexual signature in the primate brain. PLoS Genet. 2008;4:e1000100. doi: 10.1371/journal.pgen.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy R, Fulmer-Smentek S, Jensen RV, Jones WD, Wolber PK, Johnson CD, Pine PS, Boysen C, Guo X, Chudin E, Sun YA, Willey JC, Thierry-Mieg J, Thierry-Mieg D, Setterquist RA, Wilson M, Lucas AB, Novoradovskaya N, Papallo A, Turpaz Y, Baker SC, Warrington JA, Shi L, Herman D. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE., Jr Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, Freed WJ. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, Li J, Lopez JF, Myers R, Cox D, Watson SJ, Akil H, Jones EG, Bunney WE. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.