Abstract
Objective
DNA promoter methylation is an epigenetic phenomenon for long-term gene silencing during tumorigenesis. The purpose of this study is to identify novel hypermethylated loci associated with clinicopathologic variables in endometrioid endometrial carcinomas.
Methods
To find hypermethylated promoter loci, we used differential methylation hybridization coupling with microarray and further validated by combined bisulfite restriction analysis and MassARRAY assay. Methylation levels of candidate loci were corrected with clinicopathologic factors of endometrial carcinomas.
Results
Increased promoter methylation of CIDE, HAAO and RXFP3 was detected in endometrial carcinomas compared with adjacent normal tissues, and was associated with decreased gene expression of all three genes. In a clinical cohort, promoter hypermethylation on CIDEA, HAAO and RXFP3 was detected in 85, 63 and 71% of endometrial carcinomas, respectively (n=118, P<0.001) compared with uninvolved normal endometrium. Methylation status of CIDEA, HAAO and RXFP3 had significant association with microsatellite instability in tumors (P<0.001). Furthermore, methylation levels of HAAO were further found to relate to disease-free survivals (P=0.034).
Conclusions
Hypermethylation of CIDEA, HAAO and RXFP3 promoter regions appears to be a frequent event in endometrial carcinomas. Hypermethylation at these loci is strongly associated with microsatellite instability status. Moreover, HAAO methylation predicts disease-free survival in this cohort of patients with endometrioid endometrial cancer.
Keywords: Endometrial carcinoma, Hypermethylation, CIDEA, HAAO, RXFP3
Introduction
The 2009 estimated new cases and death for uterine corpus (endometrium) cancer are 42,160 and 7,780, respectively [1]. Most uterine cancers are endometrial adenocarcinomas, among which endometrioid endometrial carcinoma is the most common histological subtype. Tumorigenesis is a multistep process in which genetic defects are progressively accumulated [2]. These DNA changes, including mutations, loss of heterozygosity and gene amplification, can lead to inactivation of tumor suppressor genes and activation of oncogenes, all of which contribute to uncontrolled growth of transformed cells. More recently, epigenetic defects have been found to be equally important in cancer development. These molecular alterations do not involve changes in primary DNA sequences, but are frequent events observed in tumors [3, 4], including endometrial cancer.
Although tumor suppressor genes can be inactivated by deletions and/or mutations in cancer cells, epigenetic mechanisms including aberrant methylation of CpG sites within the promoter region also contribute to gene silencing [3, 4]. In the past, candidate gene approaches were used to identify potential biomarkers for endometrial cancer. Promoter hypermethylation of hormone-receptor genes, ERα or PR, is usually associated with loss of their expression in more advanced stages of this disease [5, 6]. This hypermethylation event may also occur early in endometrial tumorigenesis [7]. Recently, hypermethylation of more tumor suppressor genes has been reported in endometrial tumors. Aberrant methylation is frequently correlated with clinicopathologic features of endometrial cancer patients. For example, hypermethylation of RAR-β2 was found in 75% of endometrial hyperplasia samples and 92% of carcinomas [8]. Functional analysis has implicated that this methylation-mediated silencing may contribute to high proliferative activities of endometrial hyperplasia without differentiation [8]. Hypermethylation of RASSF1A is frequently associated with tumors of advanced stage disease (FIGO stage III and IV), lymph node involvement, and high grade [9, 10]. Reduced expression of GSTP1 as a result of hypermethylation of its promoter is found to be associated with myometrial invasion potential of endometrial carcinoma [11]. Taken together, these previous studies have demonstrated that hypermethylated CpG islands are potential biomarkers for early detection and disease recurrence of endometrial cancer.
Promoter hypermethylation of MLH1, one of DNA mismatch repair genes, contributes frequently to microsatellite instability (MSI) in sporadic endometrial carcinomas [12, 13]. MSI is a phenomenon of the accumulation of insertions and/or deletions at short DNA repeats, caused by the loss of DNA mismatch repair. The impact of MSI on outcomes in patients with endometrial cancer is still inconclusive [14–18]. The contradictory findings could be explained by the heterogeneity of patient population and by the methodologies to assess DNA mismatch repair [15]. We previously showed that MSI+ tumors without MLH1 methylation were associated with younger age but the combined MSI/MLH1 methylation status did not predict overall survival (OS) or disease-free survival (DFS) [15].
Herein, we report that the expression of CIDEA, HAAO and RXFP3 was lost and their promoters were hypermethylated in endometrial cancer when compared with adjacent normal tissues. Endometrial cancer cells exposed to inhibitors of DNA methylation and/or of histone deacetylation, reactivated CIDEA, HAAO and RXFP3 gene expression. We further show that CpG methylation of all three genes was associated with microsatellite instability. Particularly, hypermethylation of HAAO is related to disease-free survival. This study provides novel hypermethylated loci corrected with MSI+ phenotype in endometrial cancer.
Materials and methods
Endometrial specimens and cell lines
Tissue specimens (118 tumors and 22 uninvolved controls) were obtained as part of our ongoing work and were described in a previous report [19]. All participants consented to both molecular analyses and follow-up studies, and the protocols were approved by the Human Studies Committee at Washington University and the Ohio State University. Tumor specimens and adjacent normal tissues were collected from primary endometrioid endometrial carcinomas at the time of hysterectomy. Normal controls were procured from women (pre-menopausal, age<50, except one case), also undergoing hysterectomy. All specimens were evaluated by at least one pathologist and confirmed the diagnoses from hematoxylin and eosin-stained tissue sections. Tumor specimens had high neoplastic cellularity (mean 74%, median 80%) while normal tissues did not contain any malignant portion by direct microscopic visualization. The presence of MSI and MLH1 methylation status was determined and reported previously [15, 20]. Tumor characteristics were summarized in Supplementary Table S1, including patient age, tumor grade and stage, and menopausal status. Standard methods were used to extract DNA and RNA from tumors, corresponding non-neoplastic tissues and normal controls.
Human endometrial cancer cell lines, AN3CA, ECC-1, HEC1A, Ishikawa, KLE, RL95-2 and SK-UT-1B, were routinely maintained in our laboratory [19]. For epigenetic studies, these cells were treated with 5-aza-2′-deoxycytidine (DAC, 5 μM, Sigma) for 48 h and/or trichostatin A (TSA, 0.5 μM, Sigma) for 24 h. DNA and RNA from treated and untreated cells were isolated as described previously [19].
Differential methylation hybridization (DMH) analysis
DMH was done to profile global methylation of two pools of DNA from endometrial cancer samples (10 samples/pool) and a reference control pooled from two normal endometrial DNA samples. A CpG island microarray (Agilent) contains probes spanning all the 27,800 islands annotated to the human reference sequence (NCBI Build 36.1, UCSC hg18). Tumor and control amplicons were labeled with Cy5 and Cy3, respectively, and cohybridized onto microarray slides using our established protocols [20]. Normalized Cy5/Cy3 hybridization intensities were calculated, and candidate loci with ratios >1.5 were identified as hypermethylated in tumors compared with normal endometrial tissues according to previously published methods [20].
Reverse transcription and quantitative PCR (RT-qPCR)
Total RNA (1 μg) was reverse transcribed with the Superscript III reverse transcriptase (Invitrogen). PCR was performed as described previously [19]. Specific primers for amplification are available upon request. The relative expression of a coding gene in cells was determined by comparing the threshold cycle (Ct) of the gene against the Ct of GAPDH.
Combined bisulfite restriction analysis (COBRA)
Genomic DNA (500 ng) was treated with sodium bisulfite using the EZ DNA Methylation kit (Zymo Research) following manufacturers’ recommended protocols. COBRA was used to evaluate methylation of CIDEA, HAAO and RXFP3. Target sequences were amplified by PCR, and the products were digested with AciI, (New England Biolabs) to identify methylated sequences. Primer sequences and PCR conditions are available upon request. Digested and non-digested PCR products were resolved on 2% agarose gels stained with ethidium bromide. DNA fragments digested by AciI were scored as “methylated” in a given sample.
MassARRAY analysis
To quantify methylation levels of the CpG islands of CIDEA, HAAO and RXFP3 in clinical samples, the high-throughput MassARRAY platform (Sequenom) was carried out as described previously [19]. Briefly, bisulfite-treated DNA was amplified with primers and the PCR products were spotted on a 384-pad SpectroCHIP (Sequenom) and followed by spectral acquisition on a MassARRAY analyzer. Methylation data of individual units (1–4 CpG sites per unit) were generated by the EpiTyper software (Sequenom).
Statistical and survival analyses
The mRNA expression and methylation levels of the paired tumors and adjacent normal endometrial tissues were compared by using paired two-sample t-test. A significance was assigned as * if P≤0.05. The marginal relationship between continuous methylation levels of CIDEA, HAAO and RXFP3 and a relevant categorical clinicopathologic covariate including MSI was examined using the t-test for binary variables or ANOVA for categorical variables. Linear model was performed to examine the effect of a clinicopathologic covariate on methylation levels after adjusting for others. Overall survival (OS) and disease-free survival (DFS) were defined in a previous report [19]. The Kaplan-Meier analysis, and univariate and multivariate Cox proportional hazard models were used to evaluate the effect of continuous or dichotomized methylation levels on the survival outcomes as described previously [19]. All tests were two-sided and all analyses were performed using R 2.8.1.
Results
CIDEA, HAAO and RXFP3: three novel methylation markers in endometrial carcinoma
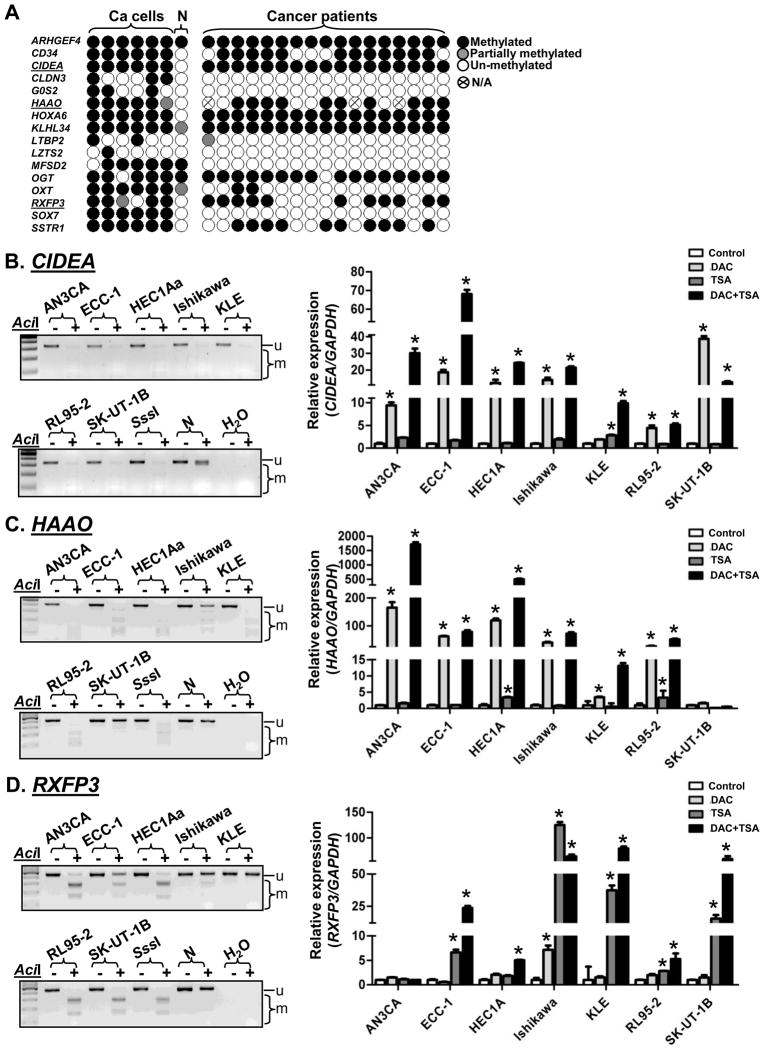
In initial microarray data analysis, we identified 16 loci with increased levels of promoter methylation in tumors compared with normal endometrial tissues (normalized Cy5/Cy3>1.5) (Fig. 1A, Table S2). Among these loci, the expression of five (CD34, LTBP2, LZTS2, OGT and SSTR1) have been reported with cancer development [21–25]. The expression of six other (CIDEA, CLDN3, G0S2, HAAO, HOXA6 and SOX7) have previously been shown to be regulated by DNA methylation [26–31]. All 16 loci were evaluated by COBRA in six endometrial cancer cell lines, one pooled sample derived from two noncancerous endometrial samples, as well as seventeen primary endometrial tumors (Fig. 1A). Promoter hypermethylation of CIDEA was detected in all six endometrial cancer cell lines, ECC-1, HEC1A, Ishikawa, KLE, RL95-2, and SK-UT-1B (Fig. 1B, left panel). Hypermethylation of HAAO and RXFP3 was also found in five out of six cancer cell lines (Fig. 1C, 1D, left panels). Based on COBRA (see Fig. 1A), CIDEA, HAAO and RXFP3 were selected for further analysis because these three genes were predominantly hypermethylated in cancer cell lines and primary tumors, but not in normal specimens. In addition, our unpublished MassARRAY analysis indicated that HOXA6 was methylated in both normal and cancer specimens (data not shown). Optimized methylation assays could not be developed for CD34 and SSTR1, due to poor PCR amplification or high GC-content of their CpG islands. These three loci, therefore, were not analyzed in the present study.
Fig. 1.
CIDEA, HAAO and RXFP3 are novel hypermethylated markers in endometrial cancer. (A) Summary of the methylation status of sixteen loci by COBRA in endometrial cancer cell lines as well as in primary tissues. (B–D, left panels) COBRA in endometrial cancer cell lines. u, unmethylated band; m, methylated bands; SssI, 100% methylated control; N, one pooled sample derived from two noncancerous endometrial tissues as negative control; +, AciI restriction enzyme added; −, without AciI. (B–D, right panels) Relative expression levels of CIDEA, HAAO and RXFP3 mRNA in endometrial cancer cell lines treated with DAC and/or TSA in relation to untreated controls was determined by RT-qPCR analysis. GAPDH was used as internal control gene. Error bar, SD; *, P<0.05 compared with untreated control of the same cell type.
Since the hypermethylation of CIDEA, HAAO and RXFP3 was observed in endometrial cancer cells (Fig. 1A) and the 5′-end of these loci has a canonical CpG island (Fig. S1), we determined whether these mRNAs can be reactivated in endometrial cancer cell lines. When these cancer cells were treated with a demethylating agent, DAC (5 μmol/L), a histone deacetylase inhibitor, TSA (0.5 μmol/L), or their combination, reactivation of CIDEA, HAAO and RXFP3 was observed in these cells (Fig. 1B–1D, right panels). These results suggest that the silencing of CIDEA, HAAO and RXFP3 is mediated through promoter hypermethylation in endometrial cancer.
Loss expression and hypermethylation of CIDEA, HAAO and RXFP in endometrial primary tumors
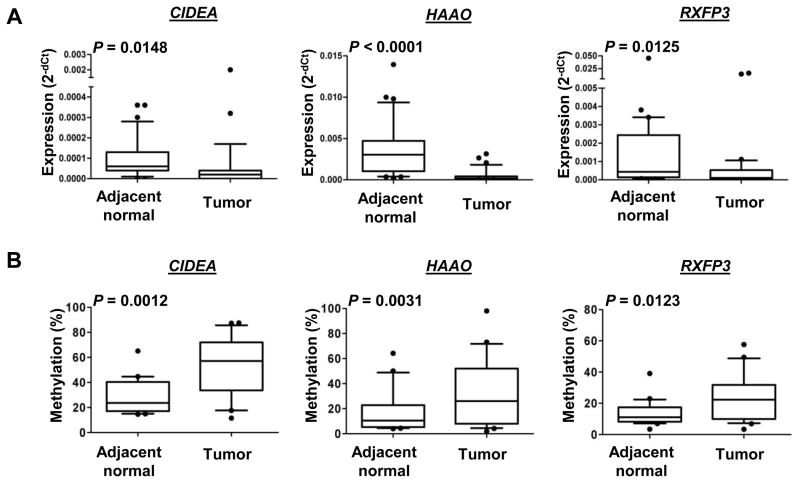
Since the expression of these genes has not been reported in endometrial cancer, we first examined their mRNA expression in 31 paired specimens. RT-qPCR results demonstrated that mRNA levels of all three genes were lower in tumors compared with the adjacent normal counterparts (P<0.05; Fig. 2A). We confirmed an inverse relationship between mRNA expression and DNA methylation in 21-paired samples randomly chosen from the aforementioned specimens (n=31) (Fig. 2). The detailed methylation level of each CpG unit (1–4 CpG sites per unit) was showed in Supplementary Fig. S1. These results suggest that CIDEA, HAAO and RXFP3 loss their expression in endometrial carcinoma due to promoter hypermethylation.
Fig. 2.
CIDEA, HAAO and RXFP3 are loss of mRNA expression and are hypermethylated in endometrial tumors. (A) Box plots indicating the relative expression levels of CIDEA, HAAO and RXFP3 mRNA in 31 paired endometrial tissues. GADPH served as internal controls. P value estimated from paired two-sample t-test. (B) Box plots pointing the quantitative DNA methylation analysis using MassARRAY in 21 paired samples of endometrial tissues. Measured methylation levels of the samples were corrected using a standard curve. P value estimated from paired two-sample t-test.
Methylation status of CIDEA, HAAO and RXFP3 in a large cohort of endometrioid endometrial carcinomas and its association with clinicopathologic variables
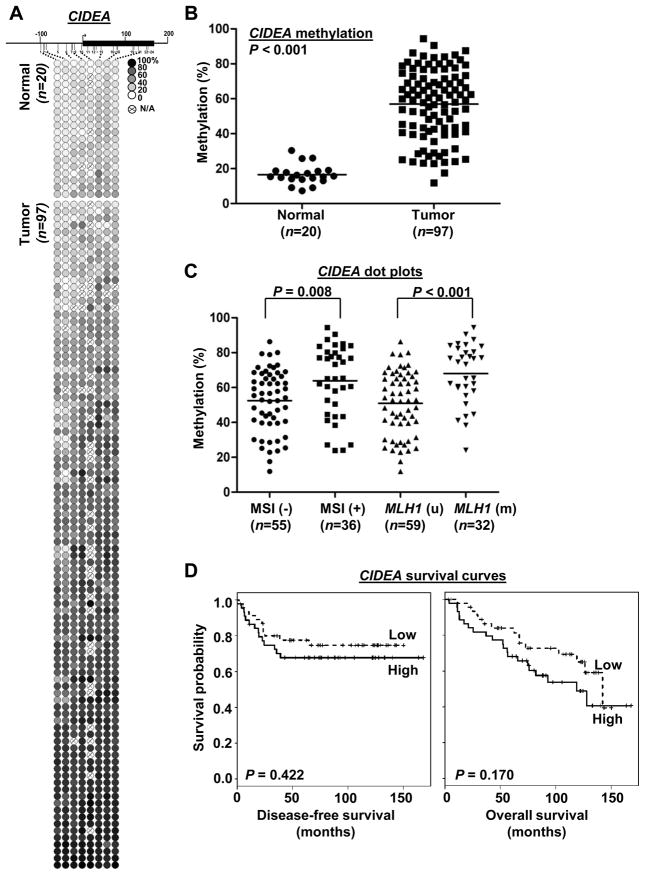
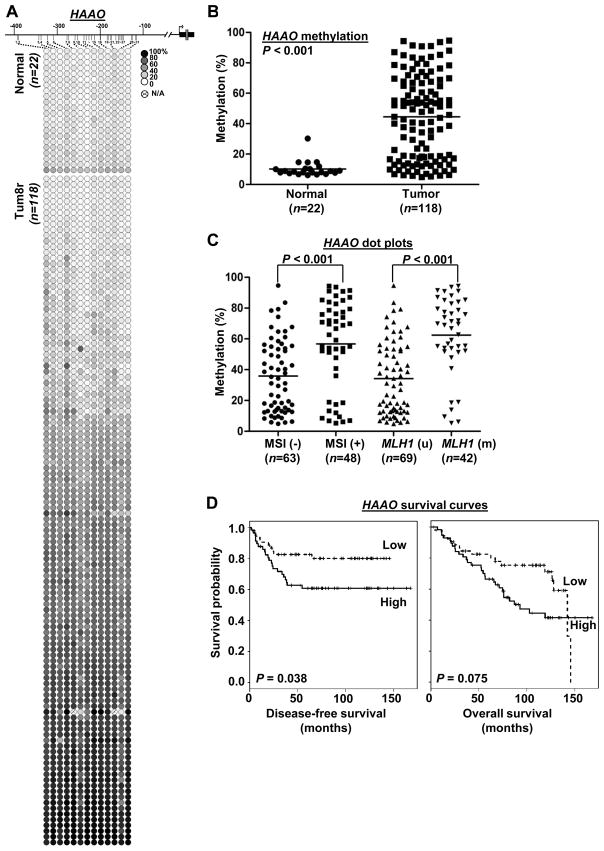
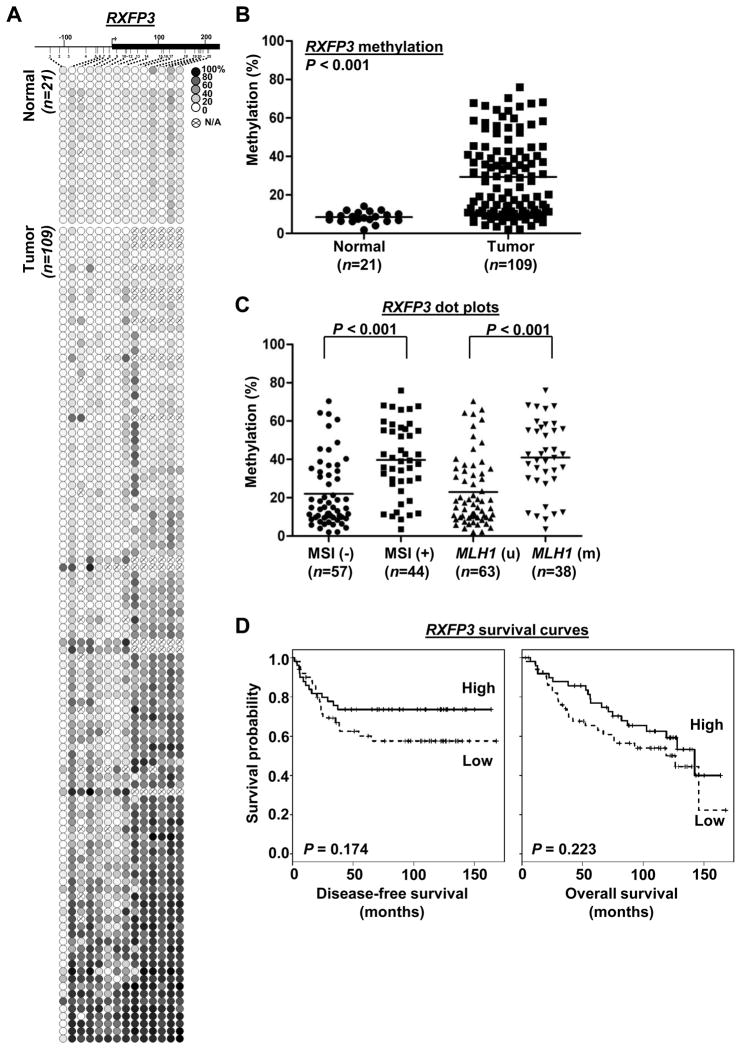
To determine if hypermethylation of CIDEA, HAAO and RXFP3 genes is associated with clinicopathologic variables of endometrioid endometrial carcinomas, we performed MassARRAY in 118 clinical tumor samples and 22 uninvolved controls. Quantitative methylation levels of each CpG unit were shown in Fig. 3A, 4A and 5A. The mean methylation level of each specimen was used to compare the differences between tumor and normal groups. Extensive promoter methylation of CIDEA, HAAO and RXFP3 was found in more than 85, 63 and 71% of the primary tumors relative to those of uninvolved controls, respectively (P<0.001; Fig. 3B, 4B, 5B).
Fig. 3.
Methylation of CIDEA CpG island and clinicopathologic covariates analyses in primary endometrial carcinomas. (A) Methylation profiles of 20 normal endometrial tissues and 97 primary tumors created following MassARRAY analysis. Each row represents a sample and each column represents a CpG unit. Color-coding depicts the degree of methyltion with dark black being 100% and white being 0%; N/A, not analyzable. (B) Dot plots pointing that CIDEA hypermethylation was in endometrial tumors. Each dot represents the mean of each specimen on the all CpG sites in Fig. 3A. Horizontal lines, mean values. P value was calculated by two-sample t-test. (C) Dot plots indicating the level of CIDEA promoter methylation was correlated with MSI and moderately associated with MLH1 methylation status. P values were calculated by two-sample t-test. (D) Kaplan-Meier curves for disease-free and overall survivals. Samples were grouped according to the mean level of methylation for the all CpG units of CIDEA while methylation was dichotomized into low/high group based on the median methylation level. Vertical bars represent excluded cases. P values were estimated from Log-rank test.
Fig. 4.
Methylation of HAAO CpG island and clinicopathologic covariates analyses in primary endometrial carcinomas. (A) Methylation profiles of 22 normal endometrial tissues and 118 primary tumors created following MassARRAY analysis. (B) Dot plots pointing that HAAO hypermethylation was found in endometrial tumors. (C) Dot plots indicating the level of HAAO promoter methylation was positively correlated with MSI status. (D) Kaplan-Meier curves indicate HAAO methylation was associated with disease-free survival. (A–D) Assays were described as in Fig. 3.
Fig. 5.
Methylation of RXFP3 CpG island and clinicopathologic covariates analyses in primary endometrial carcinomas. (A) Methylation profiles of 21 normal endometrial tissues and 109 primary tumors created following MassARRAY analysis. (B) Dot plots pointing that RXFP3 hypermethylation was found in primary endometrial tumors. (C) Dot plots indicating the level of RXFP3 promoter methylation was positively correlated with MSI status. (D) Kaplan-Meier curves disease-free and overall survivals. (A–D) Assays were described as in Fig. 3.
Statistical analysis by t-test further revealed that hypermethylation of CIDEA (P=0.0078), HAAO (P=0.0001) and RXFP3 (P<0.0001) was significantly associated with MSI status and MLH1 methylation (Fig. 3C, 4C, 5C). After adjusted for other clinical covariates by linear modeling, the association of the three loci with MSI status was still significant (P<0.001, Table S3). Linear model analysis also indicated that CIDEA methylation was associated with tumor grade whereas RXFP3 methylation was correlated with tumor recurrence, body mass index and tumor grade (Table S3).
We also evaluated the association between DNA methylation and patient survival. On univariate analysis, RXFP3 hypermethylation was significantly correlated with disease-free survival (Cox hazard ratio, 0.11 (0.01~0.93), P=0.042), but not with overall survival (Cox hazard ratio, 0.27 (0.05~1.55) P=0.14) (Table S4).). The Kaplan-Meier survival analysis on methylation levels (dichotomized by median) indicated that patients with HAAO hypermethylation had poor disease-free survival (Fig. 4D left panel; P=0.038, log-rank test). However we did not observe the methylation of CIDEA and RXFP3 correlated with disease-free or overall survival by the Kaplan-Meier survival analysis (Fig. 3D, 5D). Detailed survival analysis of all three genes was shown in Supplementary Table S5. On multivariate Cox analysis, HAAO methylation (dichotomized by median) was also significantly associated with disease-free survival (DFS, P=0.03) and age was also a risk factor for DFS, while stage was not (Table S5).
Discussion
Our previous report identified two hypermethylation markers, SESN3 and TITF1, but their methylation status did not predict overall or disease-free survival within endometrioid endometrial cancers [20]. In this study, we found three additional cancer-specific methylation markers, CIDEA, HAAO and RXFP3, through an evaluation of promoter microarrays containing ~27,800 CpG islands. Both COBRA and MassARRAY assays confirmed that hypermethylation of all three loci was frequent (≥63%) in endometrial carcinomas but was infrequent in normal tissues.
CIDEA is a member of the cell death-inducing DFF45-like effector (CIDE) family. DFF45 is a subunit of the DNA fragmentation factor which is cleaved by active caspase-3 during apoptosis. CIDEA was found to induce apoptosis in mammalian cells, which was inhibited by DFF45 [32]. CIDEA also plays important roles in energy homeostasis. In an animal model, the absence of CIDEA expression may result in lean phenotypes, insulin resistance, and resistance to diet-induced obesity in mice [33]. HAAO, known as 3-Hydroxyanthranilate-3,4-dioxygenase, is an enzyme to catalyze the biosynthetic pathway from tryptophan to quinolonate. While its function in cancer development is still unclear, we demonstrate that HAAO is hypermethylated in ovarian cancers with high sensitivity and specificity as a biomarker [29]. RXFP3, formerly called GPCR135 or SALPR, belongs to the relaxin family peptide receptors and can be activated by relaxin-3, a member of the insulin superfamily [34]. Upon ligand stimulation, RXFP3 activates extracellular signal-regulated kinase signaling via various pathways including protein kinase C [34]. The function of RXFP3 related to cancer is also unknown. Our findings have shown by RT-qPCR that RXFP3 is expressed in normal endometrium and a small subset of endometrial cancers (see Fig. 2C). The loss of RXFP3 expression in tumors is inversely associated with its promoter hypermethylation.
In this study we found that the hypermethylation of CIDEA, HAAO and RXFP3 is associated with MSI+ phenotype. This observation is consistent with previous report that endometrial carcinomas with MSI+ had significantly more epigenetic alterations than MSI- tumors [35]. Promoter hypermethylation of MLH1 contributes frequently to MSI in sporadic endometrial carcinomas [12, 13]. Our previous report showed that MSI+ tumors without MLH1 methylation were associated with younger age while the combined MSI/MLH1 methylation status did not predict OS or DFS [15]. In the current study, the methylation status of HAAO may predict DFS in patients with endometrial cancer. To definitively prove its clinical significance in predicting patient survivals, methylation analysis of a large cohort, such as the GOG-210 trial, is needed to validate this finding. Methylation markers identified in current study and others [5–15, 20] could be specific and informative for endometrial cancer only. Future development of endometrial cancer methylator phenotypes could hold great promise to improve risk assessment, diagnosis, and prognosis.
In conclusion, our findings provide three novel and cancer-specific hypermethylation markers. The methylation levels of all three loci are correlated with MSI status. Although MSI could be either a cause or a consequence of DNA methylation, high-throughput studies can be developed to establish the relationship between MSI status and DNA hypermethylation. This type of omics study may find that loss of DNA mismatch repair together with epigenetically mediated silencing of these genes can be common alterations that contribute to tumorigenesis. As such, the combined epigenetic and genetic alternations can be feasible options for predicting survival rates in cancer patients.
Supplementary Material
Acknowledgments
We thank Jeff Apostolos and Sonya Yamashita for their technical assistance. This study was supported by NIH grants U54 CA113001 (T. Huang), R01 CA071754 and P50 CA134254 (P. Goodfellow) and by funds from The Ohio State University Comprehensive Cancer Center (T. Huang).
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki M, Dharia A, Oh BR, Tanaka Y, Fujimoto S, Dahiya R. Progesterone receptor B gene inactivation and CpG hypermethylation in human uterine endometrial cancer. Cancer Res. 2001;61:97–102. [PubMed] [Google Scholar]
- 6.Sasaki M, Kotcherguina L, Dharia A, Fujimoto S, Dahiya R. Cytosine-phosphoguanine methylation of estrogen receptors in endometrial cancer. Cancer Res. 2001;61:3262–6. [PubMed] [Google Scholar]
- 7.Pijnenborg JMA, Dam-de Veen GC, Kisters N, Delvoux B, van Engeland M, Herman JG, et al. RASSF1A methylation and K-ras and B-raf mutations and recurrent endometrial cancer. Ann Oncol. 2007;18:491–7. doi: 10.1093/annonc/mdl455. [DOI] [PubMed] [Google Scholar]
- 8.Li R, Saito T, Tanaka R, Satohisa S, Adachi K, Horie M, et al. Hypermethylation in promoter region of retinoic acid receptor-beta gene and immunohistochemical findings on retinoic acid receptors in carcinogenesis of endometrium. Cancer Lett. 2005;219:33–40. doi: 10.1016/j.canlet.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Jo H, Kim JW, Kang GH, Park NH, Song YS, Kang SB, et al. Association of promoter hypermethylation of the RASSF1A gene with prognostic parameters in endometrial cancer. Oncol Res. 2006;16:205–9. doi: 10.3727/000000006783981125. [DOI] [PubMed] [Google Scholar]
- 10.Pallares J, Velasco A, Eritja N, Santacana M, Dolcet X, Cuatrecasas M, et al. Promoter hypermethylation and reduced expression of RASSF1A are frequent molecular alterations of endometrial carcinoma. Mod Pathol. 2008;21:691–9. doi: 10.1038/modpathol.2008.38. [DOI] [PubMed] [Google Scholar]
- 11.Chan QK, Khoo US, Chan KY, Ngan HY, Li SS, Chiu PM, et al. Promoter methylation and differential expression of pi-class glutathione S-transferase in endometrial carcinoma. J Mol Diagn. 2005;7:8–16. doi: 10.1016/s1525-1578(10)60003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–7. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 13.Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, et al. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–6. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 14.Cohn DE, Frankel WL, Resnick KE, Zanagnolo VL, Copeland LJ, Hampel H, et al. Improved survival with an intact DNA mismatch repair system in endometrial cancer. Obstet Gynecol. 2006;108:1208–15. doi: 10.1097/01.AOG.0000239097.42987.0c. [DOI] [PubMed] [Google Scholar]
- 15.Zighelboim I, Goodfellow PJ, Gao F, Gibb RK, Powell MA, Rader JS, et al. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients With endometrial carcinomas of the endometrioid Type. J Clin Oncol. 2007;25:2042–8. doi: 10.1200/JCO.2006.08.2107. [DOI] [PubMed] [Google Scholar]
- 16.Black D, Soslow RA, Levine DA, Tornos C, Chen SC, Hummer AJ, et al. Clinicopathologic significance of defective DNA mismatch repair in endometrial carcinoma. J Clin Oncol. 2006;24:1745–53. doi: 10.1200/JCO.2005.04.1574. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell GL, Risinger JI, Alvarez AA, Barrett JC, Berchuck A. Favorable survival associated with microsatellite instability in endometrioid endometrial cancers. Obstet Gynecol. 2001;97:417–22. doi: 10.1016/s0029-7844(00)01165-0. [DOI] [PubMed] [Google Scholar]
- 18.Arabi H, Guan H, Kumar S, Cote M, Bandyopadhyay S, Bryant C, et al. Impact of microsatellite instability (MSI) on survival in high grade endometrial carcinoma. Gynecol Oncol. 2009;113:153–8. doi: 10.1016/j.ygyno.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–46. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zighelboim I, Goodfellow PJ, Schmidt AP, Walls KC, Mallon MA, Mutch DG, et al. Differential methylation hybridization array of endometrial cancers reveals two novel cancer-specific methylation markers. Clin Cancer Res. 2007;13:2882–9. doi: 10.1158/1078-0432.CCR-06-2367. [DOI] [PubMed] [Google Scholar]
- 21.Kademani D, Lewis JT, Lamb DH, Rallis DJ, Harrington JR. Angiogenesis and CD34 expression as a predictor of recurrence in oral squamous cell carcinoma. J Oral Maxillofac Surg. 2009;67:1800–5. doi: 10.1016/j.joms.2008.06.081. [DOI] [PubMed] [Google Scholar]
- 22.Vehvilainen P, Hyytiainen M, Keski-Oja J. Latent transforming growth factor-b-binding protein 2 is an adhesion protein for melanoma cells. J Biol Chem. 2003;278:24705–13. doi: 10.1074/jbc.M212953200. [DOI] [PubMed] [Google Scholar]
- 23.Thyssen G, Li TH, Lehmann L, Zhuo M, Sharma M, Sun Z. LZTS2 is a novel β-catenin-interacting protein and regulates the nuclear export of β-catenin. Mol Cell Biol. 2006;26:8857–67. doi: 10.1128/MCB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noach N, Segev Y, Levi I, Segal S, Priel E. Modification of topoisomerase I activity by glucose and by O-GlcNAcylation of the enzyme protein. Glycobiology. 2007;17:1357–64. doi: 10.1093/glycob/cwm105. [DOI] [PubMed] [Google Scholar]
- 25.Min L, Xiaochi W, Wei L, Fei L, Hui Y, Hao W, et al. Somatostatin receptor-1 induces cell cycle arrest and inhibits tumor growth in pancreatic cancer. Cancer Sci. 2008;99:2218–23. doi: 10.1111/j.1349-7006.2008.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Da L, Tang H, Li T, Zhao M. CpG methylation plays a vital role in determining tissue- and cell-specific expression of the human cell-death-inducing DFF45-like effector A gene through the regulation of Sp1/Sp3 binding. Nucleic Acids Res. 2008;36:330–41. doi: 10.1093/nar/gkm1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams L, Roth MJ, Abnet CC, Dawsey SP, Qiao YL, Wang GQ, et al. Promoter methylation in cytology specimens as an early detection marker for esophageal squamous dysplasia and early esophageal squamous cell carcinoma. Cancer Prev Res. 2008;1:357–61. doi: 10.1158/1940-6207.CAPR-08-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusakabe M, Watanabe K, Emoto N, Aki N, Kage H, Nagase T, et al. Impact of DNA demethylation of the G0S2 gene on the transcription of G0S2 in squamous lung cancer cell lines with or without nuclear receptor agonists. Biochem Biophys Res Commun. 2009;390:1283–7. doi: 10.1016/j.bbrc.2009.10.137. [DOI] [PubMed] [Google Scholar]
- 29.Huang YW, Jansen RA, Fabbri E, Potter D, Liyanarachchi S, Chan MW, et al. Identification of candidate epigenetic biomarkers for ovarian cancer detection. Oncol Rep. 2009;22:853–61. [PMC free article] [PubMed] [Google Scholar]
- 30.Strathdee G, Holyoake TL, Sim A, Parker A, Oscier DG, Melo JV, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res. 2007;13:5048–55. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Huang S, Dong W, Li L, Feng Y, Pan L, et al. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009;277:29–37. doi: 10.1016/j.canlet.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–33. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, Toh SY, Chen Z, Guo K, Ng CP, Ponniah S, et al. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 34.van der Westhuizen ET, Werry TD, Sexton PM, Summers RJ. The relaxin family peptide receptor 3 activates extracellular signal-regulated kinase 1/2 through a protein kinase C-dependent mechanism. Mol Pharmacol. 2007;71:1618–29. doi: 10.1124/mol.106.032763. [DOI] [PubMed] [Google Scholar]
- 35.Kang S, Lee JM, Jeon ES, Lee S, Kim H, Kim HS, et al. RASSF1A hypermethylation and its inverse correlation with BRAF and/or KRAS mutations in MSI-associated endometrial carcinoma. Int J Cancer. 2006;119:1316–21. doi: 10.1002/ijc.21991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.