Abstract
Recent findings have significantly expanded our understanding of the regulation of actin depolymerizing factor (ADF)/cofilin proteins and the profound multifaceted impact that these well-established regulators of actin dynamics have on cell biology. In this review we discuss new aspects of previously documented regulation, such as phosphorylation, but also cover novel recently established modes of regulation and new functions of ADF (also known as destrin)/cofilin. We now understand that their activity responds to a vast array of inputs far greater than previously appreciated and that these proteins not only feed back to the crucially important dynamics of actin, but also to apoptosis cascades, phospholipid metabolism, and gene expression. We argue that this ability to respond to physiological changes by modulating those same changes makes the ADF/cofilin protein family a homeostatic regulator or ‘functional node’ in cell biology.
Introduction
The ADF/cofilin family of actin-binding proteins, which are essential for eukaryotes, has long been known to play a key role in actin filament dynamics in cells and to have highly complex and interesting modes of regulation. However, several recent publications have once again thrust members of this family into the limelight, this time more than ever as central figures in cell biology. The new revelations are in some cases significant expansions or clarifications of previously identified modes of regulating ADF/cofilin activity, but in other cases they are completely novel mechanisms of regulation and function, some not even directly related to actin assembly (Figure 1a). This review explains the implications of these discoveries for a broader understanding of how cofilin impacts cell behavior. Cofilin is emerging as an agent of cellular homeostasis (Figure 1b). This is not too surprising considering the well-established importance of actin dynamics in numerous vital cell processes and cofilin as a key regulator of those dynamics. We will first provide background on the distribution and functions of ADF/cofilin family proteins, then discuss the new modes of regulation, and finally explain how these findings reinforce our suggestion that ADF/cofilin proteins are a functional node in cell biology.
Figure 1. Diverse functions of cofilin including actin dynamics and beyond.
(a) Diagram showing several of the functions of cofilin (cof) within the cell that are described in more detail throughout the text. (b) Many of the factors that regulate cofilin activity are themselves modulated by cofilin and phospho-cofilin. Thus cofilin has a homeostatic role. (c) An example of homeostatic feedback regulation is the oxidative stress cycle. Reactive oxygen species (ROS), made as a result of signaling pathways or from other sources, activate cofilin phosphatases through different mechanisms discussed in the text. The resulting active cofilin can restructure the cortical actin and receptor interactions, modulating signaling. As with all figures in this article, symbols surrounded by a red line are inactive and those surrounded by a green line are active.
Distribution of the ADF/cofilin proteins
The essential ADF/cofilin proteins are expressed in all eukaryotes with three forms in mammals: ADF, cofilin-1, the major form in non-muscle tissue, and cofilin-2, the major form in muscle. ADF and cofilin-1 have similar actin dynamizing activities that differ quantitatively: ADF being somewhat more efficient in monomer sequestering and cofilin being more efficient in nucleation and severing (Box 1). Silencing and rescue experiments of cells in culture show that either protein can rescue most normal behaviors [1]. However, knock-out of the cofilin-1 gene is embryonic lethal in mice [2]; whereas the only gross deficit in ADF knock-out mice is postnatal blindness at about 4 weeks [3]. Although some additional differences in ADF and cofilin have come to light and will be discussed below, we will mostly focus on cofilin-1 and refer to it as ‘cofilin’. We do this because cofilin-1 is the most ubiquitous and has been studied much more widely due to its essential role in development. The ADF/cofilin proteins differ enough that their cellular compartmental localizations and regulation are thought to represent functional specializations, but they are qualitatively similar in many respects.
BOX 1. Summary of the differences between mammalian ADF and cofilin.
Cofilin is a substrate for Src phosphorylation on Tyr68 but ADF is not (Phe68). Src phosphorylation leads to ubiquitin targeting and cofilin degradation [40].
Cofilin has four cysteines that can undergo oxidation to form specific intramolecular or intermolecular disulfide bonds. ADF has seven Cys residues but only three (C39, C80 and C147) that are conserved with those in cofilin. Thus ADF may not be targeted to mitochondria during oxidative stress although this has not been directly tested.
Cofilin is a more potent nucleator of actinADP assembly than is ADF [77].
ADF is a more potent actin depolymerizing protein than cofilin because it has weaker nucleation ability and thus a greater monomer sequestering ability [77].
Expression of ADF, but not cofilin, is down regulated with increasing actin monomer pool [78].
Cofilin null mice are embryonic lethal, but ADF null mice are viable and their most significant abnormality is corneal thickening leading to blindness by 4 weeks postnatal development [2,3].
Other activities that have been directly compared (e.g., PtdIns(4,5)P2 binding/ inhibition, F-actin severing, Ser3 phospho-regulation) are quite similar, but ADF has not been as extensively studied.
Functions of cofilin
Modulation of actin dynamics
Cofilin is best known as a regulator of actin filament non-equilibrium assembly/disassembly (Box 2). Whether cofilin promotes actin assembly or disassembly depends upon the concentration of cofilin relative to actin and the relative concentrations of other actin-binding proteins [4]. In vitro studies have demonstrated that if the ratio of cofilin/actin subunits in a filament is low (<1/100 of the KD), persistent filament severing results [5]. At a high cofilin/actin ratio, cofilin severs rapidly and transiently and then stabilizes filamentous actin (F-actin) in a twisted form [6]. At even higher cofilin/actin ratios in vitro, cofilin can nucleate filaments through severing and through forming an as yet to be elucidated structure with actin monomers [5]. It remains to be determined if and exactly how this concentration dependence contributes to the impressive spatial coordination of actin dynamics that underlies highly motile regions such as lamellipodia and neuronal growth cones [7]. The presence of the many other actin binding proteins may dampen or eliminate such behaviors. One would need to know the instantaneous relative protein concentrations in microregions to predict all behaviors, but these are currently impossible to ascertain.
BOX 2. Background information on cofilin modulation of actin dynamics.
Subunit release is a function of both nucleotide bound to actin and the structural state of the filament following assembly [80–82]. For mature filaments in vitro at steady state, actinATP adds to the growing barbed end and actinADP is released from the pointed end in a process called ‘treadmilling’ [4] during which ATP is hydrolyzed and the γ phosphate (Pi) is released. However, immediately after assembly, recent in vitro work shows that, the filaments are found in a less stable twisted form from which subunits dissociate rapidly; in this case they dissociate from the barbed end [81]. The filaments undergo spontaneous transitions between the twisted and untwisted forms before assuming the more stable untwisted form ~2 hours after assembling [83]. The binding of cofilin to these mature filaments has been suggested to destabilize them by inducing or stabilizing a twist [84]. Whether cofilin binds the filaments depends on the release of Pi after actinATP hydrolysis since Pi binds antagonistically with cofilin [85]. In addition release of Pi is increased ~10-fold by cofilin binding [86]. Cofilin’s ability to depolymerize faster at higher pH is thought to result from the pH dependence of Pi release [87] because the binding of Pi is stronger at low pH (6.5) than high pH (8.0) [85].
In some cells, when filaments are severed, conditions favor assembly and the generation of barbed ends for polymerization [88]. A high concentration of pointed-end capping proteins favors assembly after severing while barbed-end capping proteins favor disassembly. The roles of some other actin-binding proteins in modulating cofilin’s effects on actin dynamics in vivo are discussed later.
If the concentration of cofilin/actin complex exceeds some critical threshold, yet another mode of regulation sets in. This occurs when cofilin is overexpressed or when almost all cells types are energetically or oxidatively stressed, the exception being cells expressing low levels of cofilin (Figure 1c). Excessive levels of active cofilin drive the formation of cofilin–actin bundles or ‘rods’ [8] that sequester a large fraction of the total cofilin, thus rendering cofilin incapable of promoting disassembly or severing. This additional autoregulatory mode will be explained in more detail below.
Modulation of actin filament branching
Cofilin is now understood to have a role that underlies the rapid assembly and disassembly of long unbranched filaments in the lamellipodium of motile cells. This role derives from the recently discovered ability of cofilin to compete directly with Arp2/3 complex and to reduce dramatically the affinity of Arp2/3 complex for filaments [6]. The Arp2/3 complex, consisting of two actin related proteins and five others, nucleates the growth of new filaments at a distinctive 70° angle relative to pre-existing filaments, thus generating a dendritic network typical of the lamellipodium [9]. Newly reported results from in vitro studies show that cofilin enhances the removal of these branches (Figure 1a) [6]. Arp2/3 complex is involved in the forward movement of cells, which is critical for processes as fundamental as embryonic development. The cyclical appearance and disappearance of Arp2/3- branched filaments in vivo may result in part from the spatial and temporal coordination of cofilin activation and cofilin interaction with Arp2/3, i.e., the generation of active cofilin at a time and place in the cell that renders it effective in modulating Arp2/3-induced actin branch growth. Regulation of cofilin within the leading edge of migrating cells will be discussed more fully below.
Chaperoning of actin to nucleus
Several studies have now pointed to functions of cofilin surprisingly unrelated to actin-assembly regulation. The first is that of chaperoning actin to the nucleus (Figure 1a). Actin itself has no nuclear localization sequence but its binding partner cofilin does. Because actin is recognized to have important functions in chromatin remodeling, the formation of heterogeneous nuclear ribonucleoprotein complexes, and gene expression [10,11], we consider the ability to enable actin nuclear functions to be one of the crucial cellular roles of cofilin. Moreover, plant ADF may have a specific effect on gene regulation per se, in addition to its ability to chaperone actin [12].
Release of cytochrome c
The second function of cofilin that is not related to the regulation of actin assembly comes from a proteomic study in which cofilin was found to translocate to mitochondria after the staurosporine induction of apoptosis in a neuroblastoma cell (Figure 1a) [13]. In neutrophils cofilin oxidation and its mitochondrial translocation was found also to induce apoptosis [14]. Cofilin translocation appears necessary for the opening of the mitochondrial permeability transition pore and subsequent release of cytochrome c, an early step in apoptosis. Actin is not carried with cofilin to mitochondria. Neither the translocation to mitochondria nor the release of cytochrome c requires actin binding although apoptosis is blocked by mutating the actin binding domain [13]. Apoptotic death in tissues is preferable to necrosis in that it reduces damage to surrounding cells. Curiously CAP1, an actin-cofilin binding protein (see below) also moves to mitochondria after apoptosis induction, and, while overexpression of wild-type CAP1 alone does not induce apoptosis, it does potentiate cofilin-induced apoptosis [15]. These effects require both the actin-binding domain of CAP1 and its mitochondrial targeting domain. CAP1-knockdown in HeLa cells makes them resistant to mitochondrial-dependent apoptosis. The mitochondrial targeting and functioning of cofilin and CAP1 are not unique among actin-binding proteins. Gelsolin, an actin severing and capping protein, with very different regulation from cofilin, also translocates to mitochondria, but, unlike cofilin, gelsolin is thought to inhibit cytochrome c release by closing the voltage-dependent anion channel [16]. These properties of gelsolin, CAP1, and cofilin tie together the cytoskeleton and apoptosis, as does actin turnover itself. A reduction in actin dynamics is thought to cause a decrease in mitochondrial membrane potential and increased ROS levels [17].
Activation of phospholipase D1
An unexpected and important new cofilin function is also the first assigned to phospho-cofilin, previously described only as an inactive form. It is novel in that it involves neither the direct regulation of actin assembly nor the binding of an actin regulatory protein. This function of phospho-cofilin is the direct activation of phospholipase D1 (PLD1; Figure 2a) [18], an enzyme essential for chemotaxis [19], the movement of cells along chemical gradients that figures centrally in processes, such as neutrophil tracking of pathogens and cell migration toward growth factors (cancer cells, neuronal development). The key role of cofilin in these processes has previously focused on the two-stage activation of cofilin through release from phosphatidylinositol (4,5) bisphosphate (PtdIns(4,5)P2)-inhibitory binding and dephosphorylation [4]. The activation of PLD1 increases the levels of phosphatidic acid (PtdOH). This activation was demonstrated to be downstream of a G-protein-coupled receptor, the muscarinic cholinergic receptor [18]. Rac activation at the leading edge, a process essential for neutrophil chemotaxis, requires the precise localization of an atypical guanine exchange factor, DOCK2, which depends upon both PtdIns(3,4,5) P3 and PtdOH [19]. Interestingly, this now suggests that cofilin has the dual job of getting Rac properly positioned on the membrane by assuring a supply of PtdOH and then controlling actin dynamics. What could be more efficient than having a single component in the cell with such a diversity of functions? It secures the certainty of the two activities needed being coincidentally present since they both reside in one molecule!
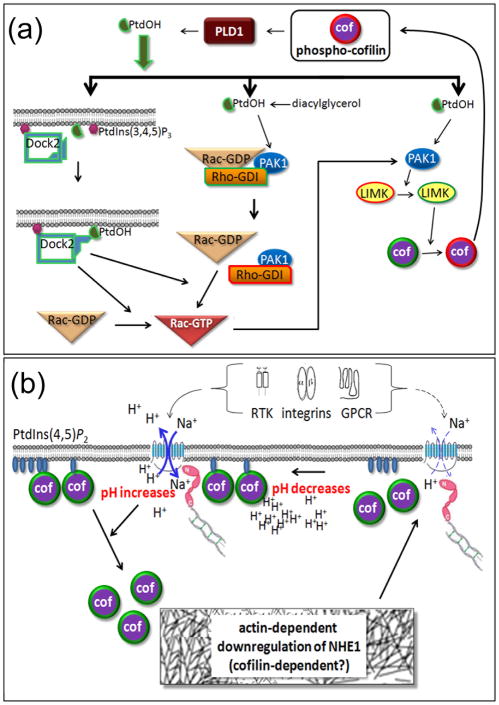
Figure 2. Recently discovered roles for cofilin in lipid metabolism and signaling and in pH-regulated actin remodeling.
(a) Phospho-cofilin is an activator of phospholipase D1 (PLD1) and thus a regulator of phosphatidic acid (PtdOH) production. PtdOH plays multiple roles in the activation of Rac1, a Rho family GTPase, as well as directly activating PAK1, a kinase that is also downstream of active Rac1. PAK1 is an activator of the cofilin kinase LIMK1. Thus a phospho-cofilin/PtdOH positive feedback cycle exists. (b) Many receptors along with intracellular acidity activate the Na+/H+ exchanger (NHE1), linked via molecules in the ezrin/moesin/radixin family (shown in pink) to the actin cytoskeleton. The influx of Na+ and efflux of H+ locally elevates pH that enhances the release of cofilin from its inhibitory membrane binding to PtdIns(3,4)P2. Released cofilin may then contribute to actin remodeling that can decrease the flux of the NHE1 and lower pH. Thus cofilin is a likely regulator of pH homeostasis.
The activation of PLD1 by phospho-cofilin has implications more far-reaching than chemotaxis. The generation of PtdOH through phosphorylation of diacylglycerol (DAG) was recently shown to release Rac-GDP from the guanine nucleotide exchange inhibitor Rho-GDI [21]. Such release, if also true for PLD1-generated PtdOH, could be part of a positive-feedback cycle from Rac-GTP to p21 activated kinase 1 (PAK1) to LIM kinase (LIMK), the predominant enzyme that phosphorylates cofilin (Figure 2a). Active LIMK generates more phospho-cofilin which activates PLD1 and produces more PtdOH. The likelihood of a more direct cycle is quite high as PtdOH is known to stimulate PAK1 whose mediation of increased phospho-cofilin drives PtdOH generation [22]. Thus all the nuances of the phospho-regulation of cofilin now interweave with cellular phospholipid metabolism and put cofilin in the center of a growing functional cycle that even extends to the synthesis of reactive oxygen species (ROS). Phospho-cofilin can negatively feed forward via PtdOH, Rac-GTP, and NADPH oxidase to induce ROS. ROS is now known to decrease phospho-cofilin levels as described below.
Regulation of cofilin
Phosphorylation of Ser3
Phosphorylation of cofilin on Ser3 inhibits its binding to G-actin (monomeric actin) and F-actin (Box 3). Research on the this regulation has focused on the signaling pathways involved [23,24]. This work has exploded and now fills complex schematics of review articles with multiple, cross-talking pathways of two kinase families, LIM kinases and Tes kinases [4]. LIMK 1 is a target of Rho GTPases via PAK1, PAK2, PAK4, and MRCKα (myotonic dystrophy kinase-related Cdc42-binding protein kinase [25]). LIM kinase 2 (LIMK2) is a target of Rho GTPases via ROCK1 and ROCK2 and MRCKα TES kinases can be regulated through cell-adhesion [26] and other upstream factors [27] but have been much less studied. Cofilin is activated by two phosphatase enzymes: chronophin, highly specific for cofilin, and slingshot 1L (SSH1L), which also recognizes LIMK1 and coronin 1B as substrates [28]. Each of these phosphatases is regulated by different upstream signaling pathways [4,23].
BOX 3. Front and side views of human cofilin structure.
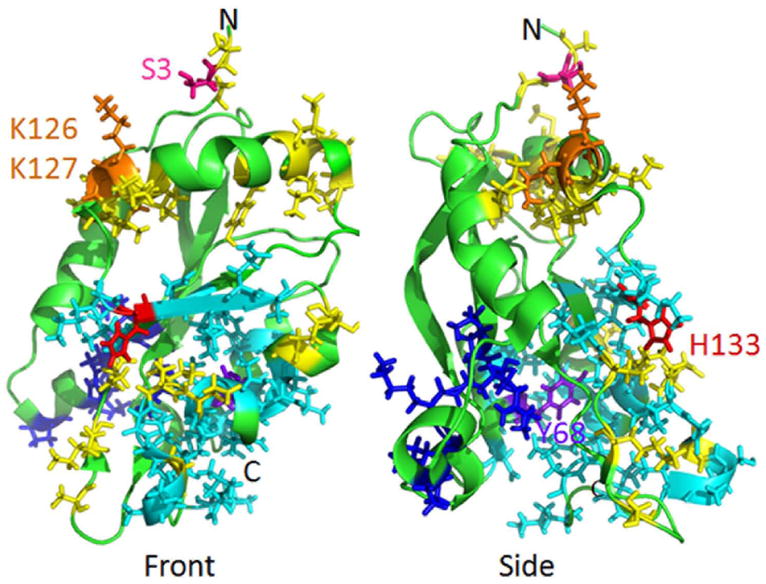
The residues shown in Figure I in yellow are ones that are in the actin contact regions (right side of the molecule in the side view), those in cyan are in the PtdIns(3,4)P2-binding region [79], and those in dark blue are in the nuclear localization sequence. Also shown is the Ser3 regulatory phosphorylation site, the two Lys residues (K126 and K127) to which Ser3 binds when it is phosphorylated, His133 that confers pH-sensitivity to the PtdIns(3,4)P2-binding, and Tyr68 that serves as a Src substrate for phosphorylation prior to ubiquitination and degradation of cofilin.
In the past year, this work has taken a few new interesting twists and turns [29–31], providing a more detailed understanding of the upstream regulators of the kinases and phosphatases that turn off and on cofilin binding to actin (Figure 1c and Figure 2a): (i) cysteine residues in the scaffolding protein 14-3-3ζ are targets of oxidative stress, thus releasing SSH1L from its inhibiting complex [29]; (ii) alternatively, oxidative stress also can inhibit SSH1L by activating protein kinase D1 (PKD1) phosphorylation of SSH1L [30,32], which creates the phosphorylated sites in SSH1L to which 14-3-3ζ binds [33]; (iii) energetic stress, i.e., ATP depletion, frees chronophin from an inhibitory complex with HSP90 [31]; (iv) fluctuations in intracellular calcium have the capacity to activate or inactivate cofilin through SSH1L and LIMK [4]. In neurons the calcium-sensitive upstream factors are the following: calcineurin, an activator of SSH1L; CaMKII, a modulator of a synaptic GTPase activating protein, synGAP; and RasGRF1, a RAS guanine exchange factor [34]. (v) cAMP/protein kinase A can activate LIMK1 directly in mouse embryonic fibroblasts [35]; however elevated cAMP may also lead to cofilin dephosphorylation indirectly [36,37]; (vi) type I and II coronins have different localizations, but depletion of either one results in increased phospho-cofilin; phospho-coronin 1B, like cofilin, is activated by SSH1L [38]. Coronin 1B is found in the leading edge; whereas coronin 2A is predominantly a stress fiber and focal adhesion protein [39].
Tyrosine phosphorylation and ubiquitination
Tyrosine 68 (Y68) is a residue common to mammalian, chicken, and Xenopus cofilin but not mammalian or chicken ADF (Box 1). Phosphorylation of Y68 was shown this year to be another mode of cofilin regulation [40]. It does not affect actin dynamizing activity, but, when 293T cells are transfected with v-Src, Y68 phosphorylation does increase ubiquitination and proteosome degradation enough to reduce cofilin levels and cell spreading. Surprisingly F-actin levels also decline, showing the difficulty of predicting the state of actin assembly based only on the levels of cofilin. The relationship between cofilin level and cell migration is biphasic [41]. A moderate increase in cofilin accelerates cell migration, clearly a factor in metastasis, but greater increases reverse this effect.
pH and phosphatidylinositol (4,5) bisphosphate
A greater understanding of other modes of cofilin regulation has also been achieved. It has long been recognized that pH in vitro [42,43] and in vivo [44,45] modulates mammalian but not yeast cofilin activity. The in vivo mechanism was recently clarified. Frantz et al demonstrated that the ability of cofilin to act as a cellular pH sensor, with higher activity at higher pH, involves the well-established inhibition of cofilin activity by binding to PtdIns(4,5)P2 (Figure 2b) [46]. They used a fibroblast lacking H+ efflux and thus incapable of generating the transient barbed end increase needed for membrane protrusion in response to growth factors, integrin engagement, and wounding. They demonstrated that the cofilin H133A mutant, which is pH unresponsive with limited PtdIns(4,5)P2 binding but still capable of actin severing in vitro, rescues the growth factor response of fibroblasts lacking H+ efflux. In contrast, the S3A cofilin mutant, which cannot be inactivated by phosphorylation but can be inactivated by protonation of H133 and so has strong PtdIns(4,5)P2 binding at the lower pH of resting cells, cannot rescue the growth factor response in fibroblasts lacking H+ efflux. Thus they confirmed suggestions of cofilin as a coincidence detector and brought together two modes of cofilin regulation previously considered to be separate: pH and PtdIns(4,5)P2 binding. If a “sticky” mutant of cofilin (D122K) with increased PtdIns(4,5)P2 affinity is overexpressed in migrating fibroblasts, chemotaxis is disrupted, confirming the importance of cofilin release [47].
In a series of molecular dynamic simulations using NMR structure [48], the structural basis for the independence of pH and phosphorylation regulation was deduced [46]. Increased pH has little effect on cofilin’s overall structure but, as shown, deprotonates His133 which reduces PtdIns(4,5)P2 binding to that of the H133A mutant. The inhibition of phospho-cofilin binding to actin was also explained: the phosphorylation of Ser3 enables salt bridges to form between itself and two residues, Lys126 and Lys127 that are involved in actin binding of dephospho-cofiilin (Box 3) [48].
Oxidation
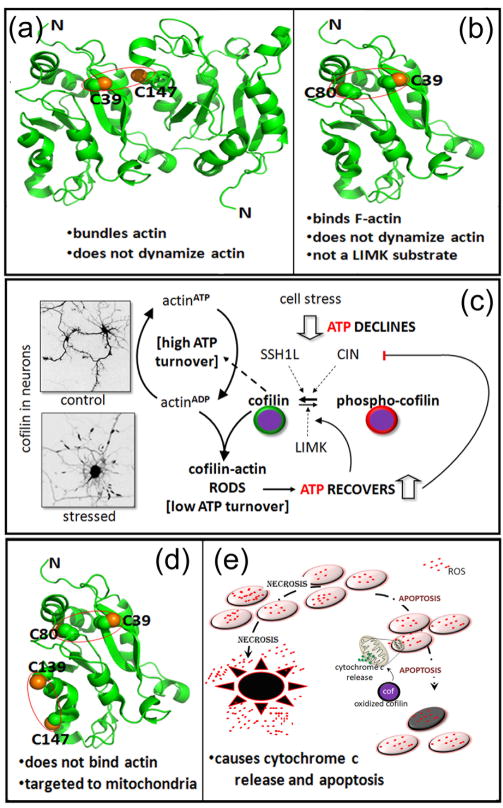
Several years ago, it was found that the major intracellular reducing agent, glutathione, when oxidized, can oxidize and dimerize cofilin in vitro through intermolecular disulfide bonding (Figure 3a). Moreover, the dimerized cofilin no longer has actin depolymerizing or severing activity [49]. Klemke et al have extended oxidation as a regulatory mechanism for cofilin in vivo; however, in their studies, intra- rather than inter-molecular disulfide bonds form [50]. In T cells the generation of a C39–C80 intra-molecular bond, delivered by the oxidative burst of granulocytes, does not eliminate actin binding, but increased F-actin results, suggesting a new mode of cofilin-actin inhibition: oxidation (Figure 3b). Further studies on the activity of oxidized cofilin were performed with taurine chloramine, the primary oxidant generated by activated neutrophils [14]. Both C39–C80 and C139–C147 intra-molecular disulfide bonds form in taurine-chloramine treated lymphoma cells in which cofilin is a main target of oxidation (Figure 3d) [14]. Neither disulfide bond alone inhibits cofilin-actin binding in vitro, but actin binding is eliminated when both internal disulfide bonds form. Furthermore, mutation of any one of these cysteines or ser3 phosphorylation blocks cofilin’s induction of cytochrome c release from mitochondria and the taurine-chloramine induced apoptosis (Figure 3e). Because antioxidants inhibit staurosporine-induced apoptosis in neurons [51], cofilin oxidation may also be the mechanism for cytochrome c release induced by staurosporine in neuroblastoma cells [13].
Figure 3. Cofilin as a mediator of oxidative stress.
(a) Hypothetical model of a cofilin dimer linked by a disulfide bridge between C39 of one cofilin and C147 of another. This dimer promotes actin bundling in vitro, has been formed by oxidized glutathione [49] but was not detected in vivo following cell exposure to taurine chloramine, the major oxidant of neutrophils [14]. Instead, taurine chloramine induces one or two intramolecular disulfide bonds from the four cofilin cysteine residues. (b) The intramolecular disulfide bond between C39 and C80. The two residues are not close enough to form a disulfide bond without some alteration in the protein structure, which causes a loss of actin dynamizing activity without eliminating F-actin binding. (c) Cofilin-actin rods form when abnormally high levels of active cofilin and ADP-actin are reached, as in response to oxidative stress and ATP decline. The inverted images show cofilin immunostained neurons under control and stressed conditions. Rods can sequester a large percentage of the active cofilin, thus slowing the decline in ATP brought about by cofilin-stimulated actin turnover and its associated ATP hydrolysis. That turnover consumes considerable ATP. The ability to maintain some critical level of ATP is necessary for the cell to recover as stress subsides. Thus cofilin contributes to energy homeostasis. (d) The fully oxidized cofilin, with disulfide bonds between C39–C80 and C139–C147, does not bind to F-actin but is targeted to mitochondrial outer membrane. Preliminary modeling of the structure showed a disruption in the alpha-5 helix, which includes C147. Two F-actin-binding residues (Box 3) reside nearby, and the alteration in structure probably accounts for the loss of F-actin binding. (e) Fully oxidized cofilin (d) is targeted to the mitochondrial outer membrane where it causes leakage of cytochrome c and activation of an apoptotic cascade. Apoptosis causes much less tissue damage than necrosis, a process responsible for inflammation, increased ROS, and extensive cell death.
Proteins modulating cofilin–actin interactions
The number and nuances of the proteins modulating cofilin-actin interactions continues to grow. These proteins include tropomyosins, cortactin, CAP1/Srv2p, coronins, and Aip1 (actin-interacting protein; reviewed in [52]). The plethora of tropomyosins encoded by four genes and alternative splicing extends to over 40 isoforms. The better-known longer forms are competitors of cofilin for F-actin and stabilize filaments while at least one of the shorter forms is a cofilin collaborator in enhancing actin dynamics ([53]; reviewed in [54]). In contrast, cortactin, an actin-binding protein of the cell cortex, competes with cofilin for binding to actin subunits. However, cortactin binds preferentially to ATP and ADP·Pi subunits of actin; whereas cofilin binds preferentially to ADP-actin [4]. In addition, in the highly specialized invadopodium compartment of metastatic cells, cofilin binds cortactin in a complex with Arp2/3, the Arp2/3 activator N-WASp, and the N-WASp activator Nck1. Cofilin is released for the essential function of severing after phosphorylation of a cortactin tyrosine [55].
A third type of interaction with cofilin is seen with CAP1/Srv2p, found in all eukaryotes, and Aip1. Like Aip1, CAP1/Srv2p alone has no effect on actin stability but does potentiate cofilin-stimulated filament turnover [56]. CAP1/Srv2p was first identified as part of the yeast adenylyl cyclase complex [57]. It displaces cofilin from actinADP [58]. This displacement enhances nucleotide exchange since cofilin binding to G-actin retards exchange. Such in vitro studies of actin with two proteins can give us useful information, but we are still far from a clear understanding of how the combination of these proteins in a particular cellular compartment might interact to effect actin regulation.
Another three-protein study, this a recent one on yeast coronin (Crn1), illustrates well how the complexity of actin regulation, involving nucleotide states, combined with a third protein (Crn1), can amplify the polarizing effects of cofilin on actin turnover [59]. The report also helps to resolve a number of apparently conflicting reports on coronin’s effect on actin. Crn1 has three domains: N-terminal β propeller domain that binds F-actin, a middle domain, and a C-terminal coiled-coil (CC) domain that modulates the Arp2/3 complex. The β propeller and CC domains allow Crn1 to have opposing effects on cofilin/actin interaction. Which effect prevails depends upon the nucleotide bound to actin. The CC domain competitively binds to ATP/ADP·Pi- actin, thus reducing the already low binding of cofilin to this actin. In ADP-rich actin regions, the β propeller domain synergizes cofilin severing. When the dominant inhibitory effects of the CC domain are negated by phosphorylation, Crn1 can switch from its filament protective role to its cofilin- synergizing role via the β propeller domain. Additional insight into the likely behavior of some of these proteins in intact cells can be gained in vitro when still more proteins are included.
Reconstituted systems are a step toward the complex environment of the cell. Such a system, which contained cofilin, Aip1, and coronin1a, revealed a surprising new mechanism for cofilin depolymerization of actin [60]. Aip1 only potentiates the severing activity of cofilin; it does not sever on its own (reviewed in [52]). In this system of Aip1, coronin1a, and cofilin, designed to study disassembly mechanisms on a single-filament level, the fluorescently labeled actin disassembles in a pattern of successive bursts not previously described and not observed with cofilin alone. Because cofilin has been shown to disrupt subunits between adjacent helical strands of actin in filaments [61] and Aip1 enhances cofilin severing and depolymerizing activity by binding along cofilin-decorated actin [62], one plausible mechanism suggested for the bursting activity is weakening of intra-filament strand interactions and the severing and removal of single-stranded actin subunits in a depolymerizing burst [60]. ‘Cooperative strand separation’ is supported by the fact that cofilin severing by itself does not alter the binding of the barbed-end capping protein CapZ, but cofilin severing in the presence of coronin and Aip1 abrogates CapZ binding. This suggested unwinding of actin filament strands has been seen in electron micrographs [63].
Concluding remarks
Cofilin: a functional node in cell biology
The integration of our current understanding of cofilin regulation and cellular function leads inevitably to the conclusion that cofilin is a functional node in cell biology. The activity of cofilin is modulated by almost any perturbation or fluctuation of normal cell physiology, and in turn it has the potential to correct or dampen those fluctuations (Figure 1b, c; Figure 2). Its actin binding is inhibited by phosphorylation mediated by a wide range of factors, depending upon protein and ion concentrations in micro-domains of compartments that differ from cell to cell: calcium, ROS, cAMP, PtdOH, ATP levels, and RhoGTPases downstream of a plethora of transmembrane receptors (e.g., integrin, receptor tyrosine kinase, G-protein coupled). Its binding to and/or dynamizing of actin is also inhibited directly by oxidation (Figure 3). Its inhibition by PtdIns(4,5)P2 is decreased by elevated pH (Figure 2b). In turn PtdIns(4,5)P2 hydrolysis and subsequent DAG increase and phosphorylation likely drive a feed-forward cycle in which phosphorylated DAG, that is PtdOH, increases the activity of PAK and LIMK, thus generating more phospho-cofilin (Figure 2a). Through PLD1, the increased phospho-cofilin results in perpetuation of the phospholipid cycle, along with its many ramifications. Cofilin feeds back and acts as a functional node by correcting physiological perturbations through its ability to regulate actin dynamics that modulate the following: (i) pH through Na(+)/H(+) exchanger (NHE1) insertion and activity (Figure 2b) [64,65], (ii) ionic fluxes through regulation of transmembrane protein activities [66], (iii) ATP levels through hydrolysis associated with actin turnover; and (iv) receptor delivery to the cell surface [67–71]. Unrelated to actin dynamics, cofilin also feeds back through phospho-cofilin stimulation of PtdOH synthesis [18] and the contribution of cofilin to gene expression through nuclear translocation of actin [10,11]. Thus we suggest that cofilin functions as a node, ‘a centering point of component parts’ because its own activity is controlled by perturbations that can all feed back to mitigate or exacerbate those fluctuations.
A prime example of the ability of cofilin to coordinate cell physiology is the formation of cofilin actin rods (bundles of cofilin-saturated filaments), which occurs when cells are energetically stressed (Figure 3c). The rods appear when abnormally high levels of active cofilin and actinADP arise; this occurs with ATP decline [8]. Abnormal levels of active cofilin follow the release of chronophin from an inhibitory ATP–HSP90–chronophin complex when ATP drops [31]. Rod formation is exacerbated by further depression of ATP levels and rising ROS levels, which then activate the other cofilin phosphatase SSH1L [29] and drive active free cofilin concentration even higher. Stable rods in regions of restricted volume, such as neurites, sequester almost all the immunostainable cofilin present, thus reducing the considerable ATP consumed by actin turnover [72]. Hence rods are neuroprotective, but only transiently because, if they persist, they cause neurite degeneration. Their transient preservation of ATP has the potential of contributing to the maintenance of normal oxidation potential, pH, calcium levels, and phospholipid metabolism (Figure 3).
Cofilin: linkage to disease
A variety of cellular stresses, resulting in ATP decline, causes rod development. These are the same kinds of stress associated with sporadic Alzheimer disease. Moreover, the Aβ1–42, amyloid peptide, infamous in its soluble form for cognitive defects and in its filamentous form for aggregating in Alzheimer disease plaques, also induces rods. The same soluble forms of Aβ 1–42, when used at concentrations that impact cognitive behavior, generate rods in neuronal culture [73]. In addition to being something of an energetic cushion, these rods in neurons also accumulate hyper-phosphorylated tau, perhaps serving as precursors of ‘striated neuropil threads’, a hallmark of Alzheimer disease [74].
If cofilin is a functional node in cell biology, as we are suggesting, it is not surprising that interference with its normal activity is highly likely to have severe repercussions. Repeatedly, the study of unrelated diseases uncovers aberrant regulation of cofilin as the culprit. Klemke and colleagues have suggested that the dysregulation of cofilin interaction with actin underlies the immune deficiencies prevalent in cancer [50]. They found that the production and release of peroxide from macrophages is intense enough to oxidize cofilin in T cells. A single intra-molecular disulfide bond renders it still capable of binding actin but incapable of dynamizing it (Figure 3b). This oxidized cofilin significantly increases F-actin levels, reduces motility, and thus is the likely basis for the suppressed immune response frequently associated with cancer. Kim et al., using experimentally higher levels of peroxide, found no oxidation of cofilin itself but instead oxidation of the scaffolding and SSH1L-inhibiting protein 14-3-3ζ [29]. It is possible that the critical difference between these studies is the fact that Kim et al. used HeLa cells stably transfected with a cofilin protein. The overexpression of cofilin in HeLa cells enormously enhances the generation of cofilin–actin rods when the cells are challenged with peroxide. With a lower level of cofilin–G-actin complex, not favoring rod generation, cofilin might be more susceptible to oxidation that prevents actin depolymerization. Both groups found reduced levels of phospho-cofilin. In T cells, oxidized cofilin was reported to be a poor substrate for LIMK1, but the activation of SSH1L through oxidative release from 14-3-3ζ might also contribute to reduced phospho-cofilin levels as SSH1L activates cofilin directly.
Other examples in disease include HIV-1 and inherited cancer. It was recently reported that HIV-1 Nef, directly or indirectly, potently inactivates cofilin through PAK2, presumably via LIMK phosphorylation. The cofilin inactivation in human T lymphocytes impairs their motility and allows evasion of the pathogens by the immune system [75]. Alternatively, cofilin is normally inactive in resting CD4 T cells. In this case activation of cofilin through viral envelope-CXCR4 receptor signaling allows viral nuclear translocation by enhancing the turnover of the otherwise inhibitory cortical actin [76]. Another example is Carney syndrome, the most common cardiac tumor, which is associated with mutation of the adenylate cyclase regulatory protein Prkar1a gene. The downstream effector responsible for abnormal motility in Carney syndrome was found to be cAMP–PKA activation of LIMK1 and cofilin hyper-phosphorylation [35].
Cofilin: future perspectives
Throughout this review we have emphasized how cofilin has, within limits, the capacity to sense and to mitigate a wide variety of physiological perturbations. This assessment is based on pieced together evidence, often from several cell types. It lacks detailed data, particularly protein concentrations and interactions in dynamic processes, that are essential for a complete and clear determination of its value and validity. Technical developments, primarily in imaging, may soon allow the advancement needed for millisecond microdomain analysis of protein interactions and ion concentrations in a single cell. Although the role of cofilin in an ever-expanding number of diseases supports its importance in homeostasis as a functional node, additional information is clearly essential.
Figure I.
Acknowledgments
This work was supported by NIH grant NS40371 and grant 281201 from the Alzheimer Drug Discovery Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Hotulainen P, et al. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurniak CB, et al. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev Biol. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda S, et al. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor) Hum Mol Genet. 2003;12:1029–1036. doi: 10.1093/hmg/ddg112. [DOI] [PubMed] [Google Scholar]
- 4.Van Troys M, et al. Ins and outs of ADF/cofilin activity and regulation. Eur J Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Chan C, et al. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pak CW, et al. Actin-binding proteins take the reins in growth cones. Nat Rev Neurosci. 2008;9:136–147. doi: 10.1038/nrn2236. [DOI] [PubMed] [Google Scholar]
- 8.Minamide LS, et al. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- 9.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 10.Pederson T. As functional nuclear actin comes into view, is it globular, filamentous, or both? J Cell Biol. 2008;180:1061–1064. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng B, et al. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276:2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgos-Rivera B, et al. Actin depolymerizing factor9 controls development and gene expression in Arabidopsis. Plant Mol Biol. 2008;68:619–632. doi: 10.1007/s11103-008-9398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua BT, et al. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol. 2003;5:1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 14.Klamt F, et al. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol. 2009;11:1241–1246. doi: 10.1038/ncb1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, et al. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J Cell Sci. 2008;121:2913–2920. doi: 10.1242/jcs.023911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusano H, et al. Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene. 2000;19:4807–4814. doi: 10.1038/sj.onc.1203868. [DOI] [PubMed] [Google Scholar]
- 17.Gourlay CW, Ayscough KR. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J Cell Sci. 2005;118:2119–2132. doi: 10.1242/jcs.02337. [DOI] [PubMed] [Google Scholar]
- 18.Han L, et al. Direct stimulation of receptor-controlled phospholipase D1 by phospho-cofilin. EMBO J. 2007;26:4189–4202. doi: 10.1038/sj.emboj.7601852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehman N, et al. Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2. Blood. 2006;108:3564–3572. doi: 10.1182/blood-2006-02-005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikimi A, et al. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science. 2009;324:384–387. doi: 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abramovici H, et al. Diacylglycerol kinase ζ regulates actin cytoskeleton reorganization through dissociation of Rac1 from RhoGDI. Mol Biol Cell. 2009;20:2049–2059. doi: 10.1091/mbc.E07-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bokoch GM, et al. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J Biol Chem. 1998;273:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 23.Bamburg JR, Bernstein BW. ADF/cofilin. Curr Biol. 2008;18:R273–R275. doi: 10.1016/j.cub.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Bamburg JR. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 25.Bernard O. Lim kinases, regulators of actin dynamics. Int J Biochem Cell Biol. 2006;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 26.LaLonde DP, et al. Actopaxin interacts with TESK1 to regulate cell spreading on fibronectin. J Biol Chem. 2005;280:21680–21688. doi: 10.1074/jbc.M500752200. [DOI] [PubMed] [Google Scholar]
- 27.Johne C, et al. Spred1 and TESK1--two new interaction partners of the kinase MARKK/TAO1 that link the microtubule and actin cytoskeleton. Mol Biol Cell. 2008;19:1391–1403. doi: 10.1091/mbc.E07-07-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang TY, et al. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Kim JS, et al. Reactive oxygen species (ROS) regulate a slingshot-cofilin activation pathway. Mol Biol Cell. 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eiseler T, et al. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–556. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang TY, et al. Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell. 2008;15:691–703. doi: 10.1016/j.devcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterburs P, et al. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase Slingshot 1 Like. Cancer Res. 2009;69:5634–5638. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 33.Nagata-Ohashi K, et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J Cell Biol. 2004;165:465–471. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlisle HJ, et al. SynGAP regulates steady-state and activity-dependent phosphorylation of cofilin. J Neurosci. 2008;28:13673–13683. doi: 10.1523/JNEUROSCI.4695-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadella KS, et al. Regulation of actin function by protein kinase A-mediated phosphorylation of Limk1. EMBO Rep. 2009;10:599–605. doi: 10.1038/embor.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goeckeler ZM, Wysolmerski RB. Myosin phosphatase and cofilin mediate cAMP/PKA-induced decline in endothelial cell isometric tension and myosin II RLC phosphorylation. J Biol Chem. 2005;280:33083–33095. doi: 10.1074/jbc.M503173200. [DOI] [PubMed] [Google Scholar]
- 37.Meberg PJ, et al. Actin depolymerizing factor and cofilin phosphorylation dynamics: Response to signals that regulate neurite extension. Cell Motil Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Cai L, et al. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall TW, et al. Coronin 2A regulates a subset of focal-adhesion-turnover events through the cofilin pathway. J Cell Sci. 2009;122:3061–3069. doi: 10.1242/jcs.051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo Y, et al. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene. 2009 Oct 5; doi: 10.1038/onc.2009.319. (in press) [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap CT, et al. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil Cytoskeleton. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- 42.Hawkins M, et al. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 43.Yonezawa N, et al. pH control of actin polymerization by cofilin. J Biol Chem. 1985;260:14410–14412. [PubMed] [Google Scholar]
- 44.Bernstein BW, et al. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil Cytoskeleton. 2000;47:319–336. doi: 10.1002/1097-0169(200012)47:4<319::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 45.Pavlov D, et al. Severing of F-actin by yeast cofilin is pH-independent. Cell Motil Cytoskeleton. 2006;63:533–542. doi: 10.1002/cm.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frantz C, et al. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. 2008;183:865–879. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leyman S, et al. Unbalancing the PI(4,5)P2-cofilin interaction impairs cell steering. Mol Biol Cell. 2009;20:4509–4523. doi: 10.1091/mbc.E09-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pope BJ, et al. Solution structure of human cofilin: actin binding, pH sensitvity, and relationship to actin-depolymerizing factor. J Biol Chem. 2004;279:4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 49.Pfannstiel J, et al. Human cofilin forms oligomers exhibiting actin bundling activity. J Biol Chem. 2001;276:49476–49484. doi: 10.1074/jbc.M104760200. [DOI] [PubMed] [Google Scholar]
- 50.Klemke M, et al. Oxidation of cofilin mediates T cell hyporesponsiveness under oxidative stress conditions. Immunity. 2008;29:404–413. doi: 10.1016/j.immuni.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Maycotte P, et al. Apoptosis and autophagy in rat cerebellar granule neuron death: Role of reactive oxygen species. J Neurosci Res. 2010;88:73–85. doi: 10.1002/jnr.22168. [DOI] [PubMed] [Google Scholar]
- 52.Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: New blades for twisted filament. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- 53.Bryce NS, et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhn TB, Bamburg JR. Tropomyosin and ADF/cofilin as collaborators and competitors. Adv Exp Med Biol. 2008;644:232–249. doi: 10.1007/978-0-387-85766-4_18. [DOI] [PubMed] [Google Scholar]
- 55.Oser M, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- 57.Freeman NL, Field J. Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil Cytoskeleton. 2000;45:106–120. doi: 10.1002/(SICI)1097-0169(200002)45:2<106::AID-CM3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 58.Balcer HI, et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 59.Gandhi M, et al. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kueh HY, et al. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bobkov AA, et al. Cofilin (ADF) affects lateral contacts in F-actin. J Mol Biol. 2004;337:93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 62.Okada K, et al. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J Biol Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- 63.McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J Mol Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- 64.Szaszi K, et al. Regulation of the epithelial Na(+)/H(+) exchanger isoform by the cytoskeleton. Cell Physiol Biochem. 2000;10:265–272. doi: 10.1159/000016358. [DOI] [PubMed] [Google Scholar]
- 65.Ginsberg MH, et al. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 66.Mazzochi C, et al. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am J Physiol Renal Physiol. 2006;291:F1113–F1122. doi: 10.1152/ajprenal.00195.2006. [DOI] [PubMed] [Google Scholar]
- 67.Rosso S, et al. LIMK1 regulates golgi dynamics, traffic of golgi-derived vesicles, and process extension in primary cultured neurons. Mol Biol Cell. 2004;7:3433–3449. doi: 10.1091/mbc.E03-05-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beaulieu V, et al. Modulation of the actin cytoskeleton via gelsolin regulates vacuolar H+ATPase (V-ATPase) recycling. J Biol Chem. 2004;280:2452–2463. doi: 10.1074/jbc.M412750200. [DOI] [PubMed] [Google Scholar]
- 69.Alberts P, et al. Cdc42 and actin control polarized expression of TI-VAMP vesicles to neuronal growth cones and their fusion with the plasma membrane. Mol Biol Cell. 2006;17:1194–1203. doi: 10.1091/mbc.E05-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 71.Noda Y, Sasaki S. The role of actin remodeling in the trafficking of intracellular vesicles, transporters, and channels: focusing on aquaporin-2. Pflugers Arch. 2007;456:737–745. doi: 10.1007/s00424-007-0404-2. [DOI] [PubMed] [Google Scholar]
- 72.Bernstein BW, et al. Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am J Physiol Cell Physiol. 2006:C828–839. doi: 10.1152/ajpcell.00066.2006. [DOI] [PubMed] [Google Scholar]
- 73.Davis RC, et al. Mapping cofilin-actin rods in stressed hippocampal slices and the role of cdc42 in amyloid-beta-induced rods. J Alzheimers Dis. 2009;18:35–50. doi: 10.3233/JAD-2009-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whiteman IT, et al. Activated ADF/cofilin sequesters phosphorylated microtubule-associated protein during the assembly of Alzheimer-like neuritic cytoskeletal striations. J Neurosci. 2009;29:12994–13005. doi: 10.1523/JNEUROSCI.3531-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stolp B, et al. HIV-1 Nef interferes with host cell motility by deregulation of Cofilin. Cell Host Microbe. 2009;6:174–186. doi: 10.1016/j.chom.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Yoder A, et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeoh S, et al. Determining the differences in actin binding by human ADF and cofilin. J Mol Biol. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- 78.Minamide LS, et al. Differential regulation of actin depolymerizing factor and cofilin in response to alterations in the actin monomer pool. J Biol Chem. 1997;272:8303–8309. doi: 10.1074/jbc.272.13.8303. [DOI] [PubMed] [Google Scholar]
- 79.Gorbatyuk VY, et al. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP(2) regulates the cofilin-actin interaction. Mol Cell. 2006;24:511–522. doi: 10.1016/j.molcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 80.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kueh HY, et al. Dynamic stabilization of actin filaments. Proc Natl Acad Sci USA. 2008;105:16531–16536. doi: 10.1073/pnas.0807394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orlova A, et al. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci U S A. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muhlrad A, et al. Inorganic phosphate regulates the binding of cofilin to actin filaments. FEBS J. 2006;273:1488–1496. doi: 10.1111/j.1742-4658.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 86.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 87.Pavlov D, et al. Severing of F-actin by yeast cofilin is pH-independent. Cell Motil Cytoskeleton. 2006;63:533–542. doi: 10.1002/cm.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Condeelis J. How is actin polymerization nucleated in vivo? Trends Cell Biol. 2001;11:288–293. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]