Abstract
NF-κB is activated in many types of cancer. Phosphorylation of p65 at serine 276 is required for the expression of a subset of NF-κB regulated genes, including vascular cell adhesion molecule-1 (VCAM-1) and interleukin-8 (IL-8). Thus, inhibition of serine 276 phosphorylation may prevent metastasis and angiogenesis in certain tumor types. Using in silico molecular docking, small molecules that are predicted to bind to a structural pocket near serine 276 were identified. One compound, NSC-127102, hinders serine 276 phosphorylation and the expression of IL-8 and VCAM-1. Small molecules such as NSC-127102 may be optimized for the future treatment of cancer.
Keywords: NF-κB, p65, TNF-α, IL-8, in silico molecular docking, serine 276
1. Introduction
The nuclear factor kappa B (NF-κB) transcription complexes exist as a variety of homo- and heterodimers formed by the subunits p50, p52, c-Rel, Rel A (p65), and Rel B. These factors regulate the expression of hundreds of genes involved in many diverse physiological responses, including cell proliferation, cell survival, inflammation, innate immunity, and the cellular stress response. Mounting evidence suggests a role for NF-κB in oncogenesis. Many studies suggest that aberrant activation of NF-κB is responsible for the initiation of tumorigenesis including evasion of apoptosis, malignant transformation, sustained cell proliferation, metastasis, and angiogenesis [1,2]. Furthermore, deregulated activation of NF-κB has been observed in a number of human cancers, including breast cancer, leukemia, lung cancer, melanoma, colon cancer, and several virally induced tumors [3-9].
It is well established that the phosphorylation of the p65 subunit of NF-κB is important for the transcriptional activity of NF-κB. Phosphorylation of serine 276 has been studied in detail. Phosphorylation of this site by the PKA catalytic subunit or MSK-1 dramatically enhances NF-κB transcriptional activity by recruiting the histone acetyltransferase CBP to p65:DNA complexes [10,11]. Phosphorylation of serine 276 is necessary for the transcription of several genes, including IL-6, IL-8, and VCAM-1 [11-14]. Phosphorylation of serine 276, however, is not required for the transcription of all NF-κB activated genes. For instance, the regulation of MnSOD and MHC class I is not affected by mutation of serine 276 [12].
VCAM-1 plays a critical role in metastasis, while IL-8 has been reported to promote angiogenesis and metastasis. Specific inhibition of serine 276 phosphorylation, therefore, may be useful to prevent metastasis and angiogenesis, and thus reduce the metastatic potential of various cancers. Although over 750 NF-κB inhibitors have been identified [15], most inhibit the ability of NF-κB to target all NF-κB regulated genes and are likely to have unwanted side effects. It is therefore more desirable to specifically target serine 276 phosphorylation and only inhibit the transcription of a subset of NF-κB-dependent genes.
In silico molecular docking was utilized to identify specific inhibitors of serine 276 phosphorylation. This technique has become a primary screening method for the discovery of ligands, and recent analysis of this approach indicates that successful hit rates are significantly higher using the molecular docking approach compared to high-throughput screening [16]. To utilize this technique, the crystal structure of the protein complex containing the p65-p50 heterodimer bound to the Ig/HIV-κB DNA element (RCSB Protein Data Bank code: 1LE9) was examined. During this analysis, a structural pocket adjacent to serine 276 was identified. It was hypothesized that a small molecule that specifically binds to this cleft would inhibit serine 276 phosphorylation. After screening 220,000 compounds available from the National Cancer Institute/Developmental Therapeutics Program (NCI/DTP), the top 10 compounds, as ranked by predicted energy scores (composed of predicted electrostatic interactions and van der Waals’ forces), were tested for inhibition of serine 276 phosphorylation. One compound, NSC-127102, strongly inhibited serine 276 phosphorylation and NF-κB mediated regulation of IL-8 and VCAM-1 gene expression. Thus, it is possible that this compound or its derivatives could be developed as inhibitors of metastasis and angiogenesis in the future.
2. Materials and methods
2.1. In silico molecular docking
The protein crystal structure used for docking site identification and in silico screening was a p65-p50 heterodimer bound to the Ig/HIV-κB DNA element (RCSB Protein Data Bank code: 1LE9). A molecular surface of 1LE9 was prepared using the MSROLL program, which was then used as input for the sphere generating program SPHGEN. A cluster of spheres within the pocket of interest was selected and edited manually to leave a cluster of 21 spheres. The SHOWBOX program was used to construct a 3-dimensional rectangle, 4 Angstroms in any direction from the sphere cluster. The program SYBL 7.0 was used to convert the PDB file of 1LE9 into the appropriate mol2 format. The box file generated was then used as input for the GRID program, which calculates and saves the information concerning the steric and electrostatic environment within the box area of the 1LE9 mol2 file. DOCK was then used to screen the entire National Cancer Institute/Developmental Therapeutics Program (NCI/DTP) database of small molecules (which consisted of approximately 220,000 small molecules at the time of docking) within the 1LE9 grid, using the selected spheres as theoretical binding sites. The small molecule output was ranked based on predicted energy scores (composed of predicted electrostatic interactions and van der Waals’ forces). The top 10 compounds were obtained from the NCI/DTP for cell culture analysis. All of the computer programs listed for this procedure are part of the DOCK5.0 suite developed at UCSF.
2.2. Cell culture and transient transfection
HepG2 human hepatoma cells were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) fetal bovine serum. Transient transfections were performed using FuGENE (Roche Applied Science) according to the manufacturer's instructions. Cells were incubated for 48 h posttransfection, and cell extracts were prepared and analyzed by luciferase assays. Background readings were subtracted from the luciferase assay values. Luciferase activity was normalized to the protein concentration in the cell lysates as determined using the BioRad protein assay reagent. The constructs containing −166/+44 IL-8/Luc (IL-8/Luc) and −166/+44 IL-8/Luc with a mutated κB element located between −80 and −70 bp relative to the transcriptional start site (mκB IL-8/Luc) were kindly provided by Dr. James J. Pestka, Michigan State University.
2.3. cDNA synthesis and reverse transcriptase PCR
Total RNA was isolated from HepG2 cells with Trizol Reagent (Invitrogen) using the instructions provided by the manufacturer. RNA was reverse transcribed to first-strand cDNA under the following conditions: 25°C for 10 min, 42°C for 30 min, and 95°C for 5 min. PCR was subsequently performed using IL-8, VCAM-1, and Cyclophilin primers in a total volume of 50 μl. The primers used to amplify IL-8 are as follows: 5’-GGCAGCCTTCCTGATTTCTG-3’ and 5’-CTCTCAATCACTCTCAGTTC-3’. The primers used to amplify VCAM-1 are as follows: 5’-GATGGTCGTGATCCTTGGAG-3’ and 5’-CAAGTCAATGAGACGGAGTC-3’. The primers used to amplify Cyclophilin are as follows: 5’-ACCATGTTCTTCGACGTTGC-3’ and 5-ACGTTTTCTGCTGTCTTTGG-3’. The reactions were performed under the following conditions: 96°C for 5 min followed by 35 cycles of 96°C for 1 min, 59°C for 1 min, and 72°C for 1 min. All reaction products were visualized on a 2% agarose gel.
2.4. Immunoblot analysis
HepG2 hepatoma or human mammary MDA-MB-231 carcinoma cells were grown in DMEM supplemented with 10% fetal bovine serum. Cells at 60% confluence were infected with adenoviruses and incubated for 24 h. Cells were subsequently treated with the compounds obtained from the NCI/DTP for 24h. Cell extracts were prepared with a detergent lysis buffer (20 mM Tris-HCl, pH 7.6, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 20 mM NaP2O7, 0.2% Triton X-100, 10 nM microcystin, 0.1% β-mercaptoethanol, 0.1 M PMSF, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin). Lysates were dissolved in an equal volume of 2X Laemmli sample buffer, boiled for 3 min and proteins (20 μg per lane) were resolved by SDS-PAGE. The proteins were transferred to nitrocellulose membrane and probed with specific antibodies for 2 h (p65, sc-109 from Santa Cruz Biotechnology, Inc.; phospho-serine 276 p65, #3037 from Cell Signaling Technology; Actin, sc-1616-R from Santa Cruz Biotechnology, Inc.; CREB, #9192 from Cell Signaling Technology; and phospho-serine 133 CREB, #9196 from Cell Signaling Technology) followed by incubation with an anti-rabbit or anti-mouse IgG conjugated to alkaline phosphatase (Santa Cruz Biotechnology, Inc).
2.5. In vitro kinase assays
MSK-1 kinase assays were performed in MSK-1 Kinase Assay Buffer (40 mM MOPS, pH 7.0, 1 mM EDTA) including 10 mM magnesium acetate and 100 μM ATP. F-p65 (purified from HepG2 cells) or CREB (Biomol) served as a substrate. Seventy-five nanograms of MSK-1 (Millipore) was added to the reaction and incubated at 30°C for 30 min. Kinase reactions were stopped by the addition of 4X Laemmli sample buffer and boiled for 5 min. Proteins were resolved by SDS-PAGE and immunoblot analysis was performed for phospho-serine 276 p65 or phospho-serine 133 CREB. PKA assays were performed in Kinase Assay Buffer (50 mM Hepes, pH 7.5, 10 mM MgCl2, 2.5 EGTA, 1 mM DTT, 100 μM NaF, 100 μM Na3V04) including 125 μM ATP. One hundred fifty nanograms of PKA (Sigma) was added to the reaction and incubated at 30°C for 30 min. Kinase reactions were stopped by the addition of 4X Laemmli sample buffer and boiled for 5 min. Immunoblot analysis was performed as described for the MSK-1 assays.
3. Results
3.1. In silico molecular docking analysis of p65
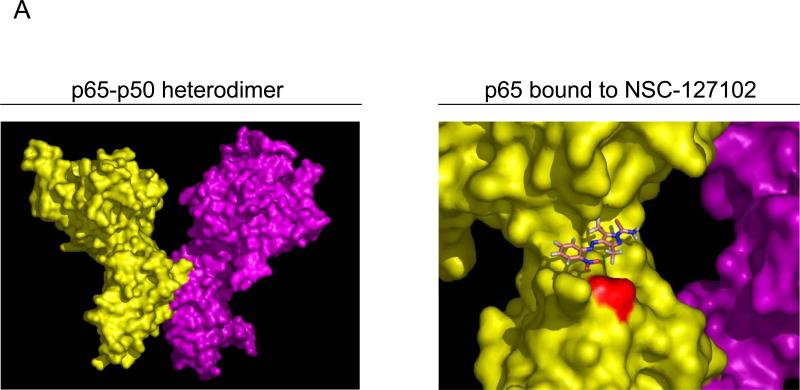
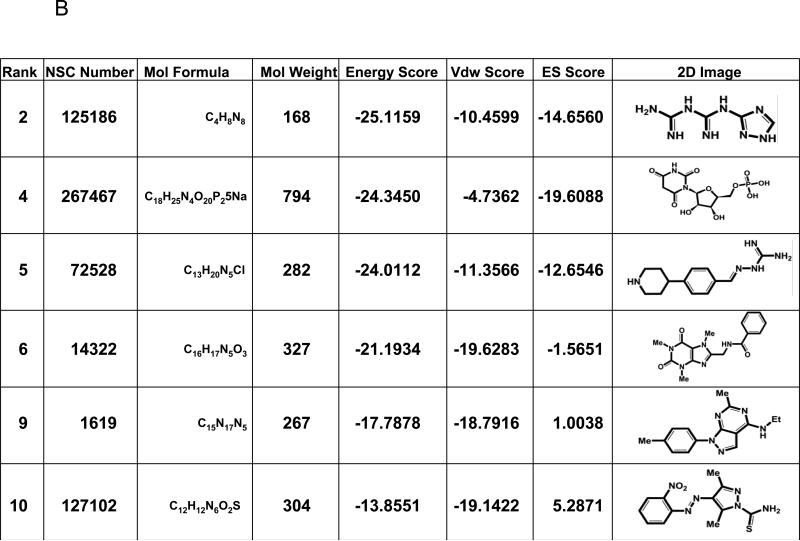
The crystal structure of the protein:DNA complex containing the p65-p50 heterodimer bound to the Ig/HIV-κB DNA element (RCSB Protein Data Bank code: 1LE9) was used in in silico molecular docking (Fig. 1A). This κB element is present in the promoters of both the Ig κ light chain gene as well as the long terminal repeat of HIV-1. A structural pocket adjacent to serine 276 (shown in red) was identified. We hypothesized that a small molecule that specifically binds in this cleft would prevent the phosphorylation of serine 276. In silico molecular docking experiments were performed to identify compounds that bind to this p65 structural pocket. Approximately 220,000 compounds available from the NCI/DTP database were screened. Each small molecule was positioned in the selected site in 1000 different orientations and the best orientations and their corresponding van der Waals (vdw), electrostatic (ES) and energy scores were calculated. The top ten compounds, ranked according to their energy scores, were obtained from the NCI/DTP. Of these, six were soluble in DMSO (Fig. 1B). These six compounds were tested for inhibition of constitutive serine 276 phosphorylation in HepG2 hepatoma cells and MDA-MB-231 breast carcinoma cells, as described below.
Fig. 1.
Identification of a structural pocket near serine 276 of p65 and interacting molecules identified by high-throughput in silico screening. (A) The crystal structure of p65 from a p65-p50 heterodimer bound to the Ig/HIV-κB DNA element (RCSB Protein Data Bank code: 1LE9) is displayed with serine 276 highlighted in red. NSC-127102 docked into the structural pocket near serine 276 is shown. (B) The structures of six of the top lead compounds identified by in silico docking are ranked according to their energy scores. Van der Waals (Vdw) and electrostatic (ES) scores are also displayed along with the molecular formulas and molecular weights of these compounds.
3.2. NSC-127102 inhibits p65 serine 276 phosphorylation
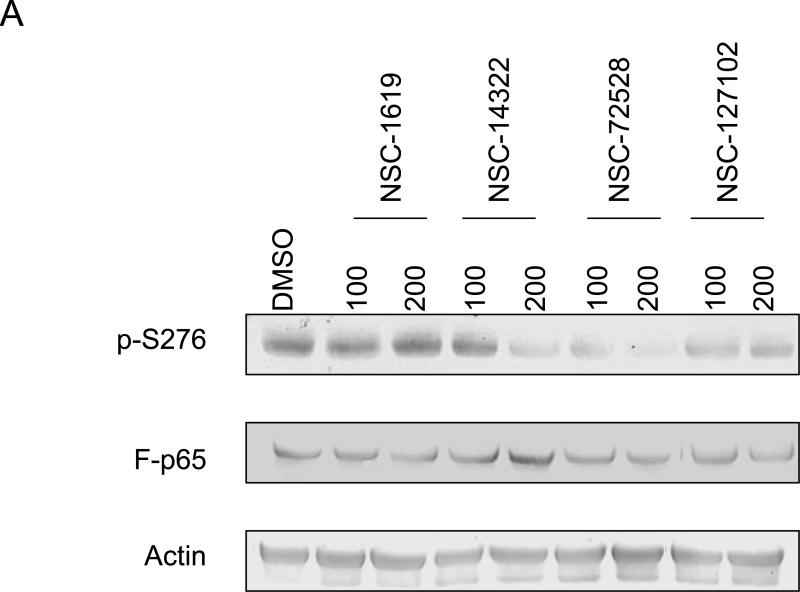
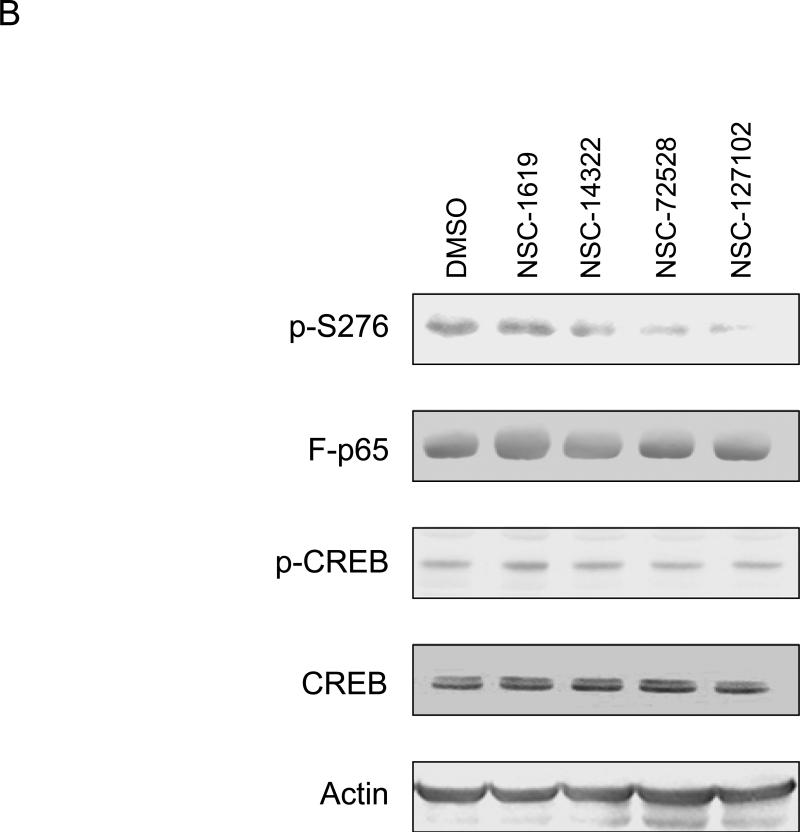
The effect of six compounds on serine 276 phosphorylation in HepG2 cells was tested. Immunoblot analysis showed that compounds NSC-14322, NSC-72528, and NSC-127102 inhibit serine 276 phosphorylation, while the other compounds were without effect (Fig. 2A). These experiments were repeated to determine if NSC-14322, NSC-72528, or NSC-127102 affect the constitutive phosphorylation of another transcription factor, CREB. Both the serine 276 phosphorylation site of p65 and the phosphorylation site of CREB at serine 133 are consensus sites recognized by PKA [17,18]. HepG2 cells were treated with 100 μM NSC-14322, NSC-72528, or NSC-127102. While NSC-14322, NSC-72528 and NSC-127102 inhibited serine 276 phosphorylation, constitutive CREB phosphorylation was not affected (Fig. 2B). NSC-127102 was consistently the most effective inhibitor of serine 276 phosphorylation, so we focused our experiments on this compound, the structure of which is shown bound to p65 near serine 276 in Fig. 1A. The MDA-MB-231 human breast carcinoma cell line was treated with NSC-127102 to determine if this compound affected serine 276 phosphorylation in another cell line. NSC-127102 also inhibited serine 276 phosphorylation in MDA-MB-231 cells, demonstrating that this effect occurs in other cell types (Fig. 2C).
Fig. 2.
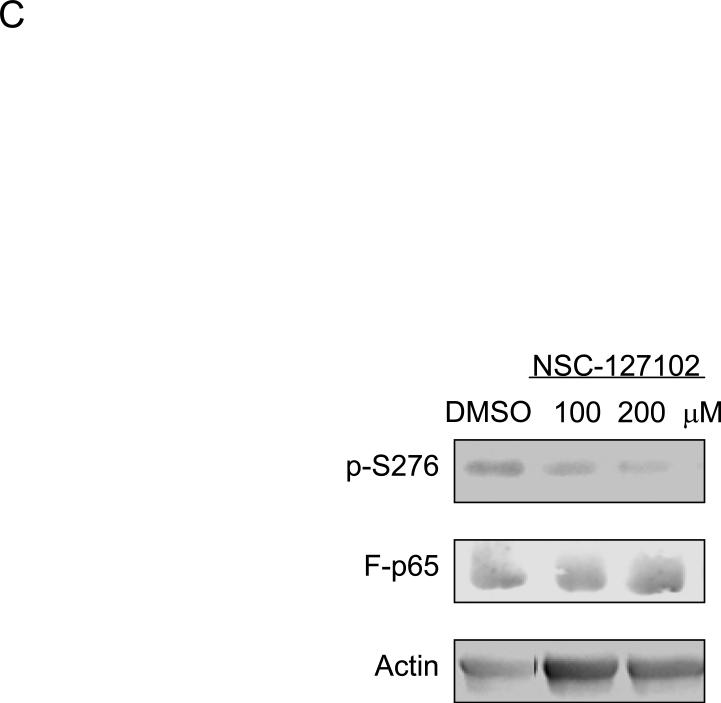
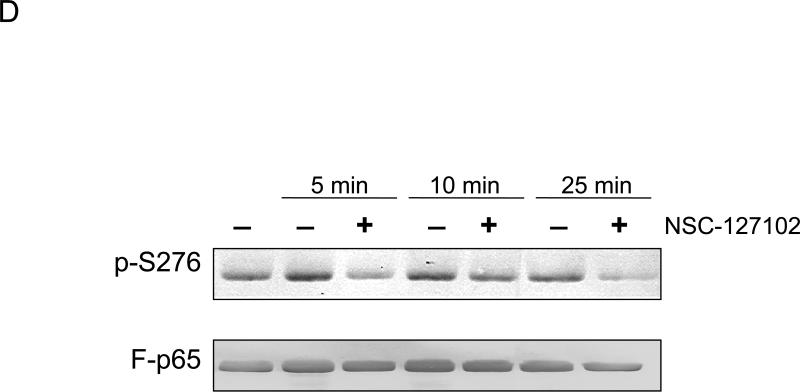
NSC-127102 inhibits serine 276 phosphorylation of p65. (A) HepG2 hepatoma cells were infected with an adenovirus expressing flag epitope tagged p65 (F-p65) for 24 h. Cells were subsequently treated for another 24 h with 100 or 200 μM of the compounds obtained from the NCI/DTP. DMSO served as vehicle control. F-p65 was immunoprecipitated using anti-Flag-agarose and immunoprecipitates were analyzed by immunoblot using antibodies that specifically recognize p65 or p65 phosphorylated on serine 276 (p-S276). Prior to flag-agarose immunoprecipitation, whole cell lysates were prepared and analyzed by immunoblot analysis for β-actin to confirm that all lanes were equally loaded. (B) HepG2 hepatoma cells were infected with an adenovirus expressing F-p65, as described in panel A and subsequently treated with 100 μM NSC-1619, NSC-14322, NSC-72528, or NSC-127102 for 24h. Whole cell lysates prior to immunoprecipitation were also prepared. Immunoblot analysis for p65, p-S276, CREB, CREB phosphorylated on serine 133, and β-actin, as a loading control, was performed. (C) Experiments performed in HepG2, as described in panel A, were repeated in MDA-MB-231 cells. MDA-MB-231 cells were treated with 100 or 200 μM NSC-127102 for 24 h, and immunoblot analysis for p65, p-S276 and β-actin was performed. (D) HepG2 cells were pre-treated with DMSO or 100 μM NSC-127102 for 24 h. Cells were subsequently treated with 40 ng/ml TNF-α for 5, 10, and 25 min. F-p65 was immunoprecipitated using flag-agarose and immunoprecipitates were analyzed by immunoblot using antibodies that specifically recognize p65 or p65 phosphorylated on serine 276 (p-S276).
To determine if NSC-127102 inhibits TNF-α stimulated phosphorylation of serine 276, HepG2 cells were treated with TNF-α in the presence or absence of NSC-127102. As shown by others, TNF-α stimulates serine 276 phosphorylation in a time-dependent manner [13,19] (Fig. 2D). NSC-127102 inhibited the phosphorylation of serine 276 induced by TNF-α, showing that NSC-127102 inhibits both constitutive and stimulated phosphorylation of this site.
3.3. NSC-127102 inhibits p65, but not CREB phosphorylation, in vitro
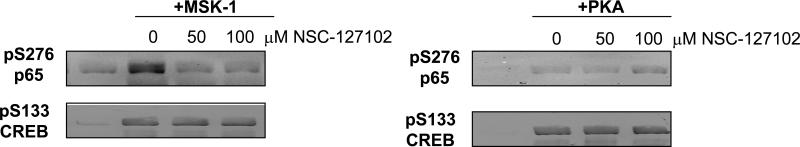
Both serine 276 of p65 and serine 133 of CREB are known PKA substrates [17,18]. Furthermore, MSK-1 phosphorylates serine 276 of p65 both in vivo and in vitro [11,20]. To show that NSC-127102 selectively inhibits p65 phosphorylation, in vitro kinase assays were performed using either p65 or CREB as a substrate. As shown in Fig. 3, both PKA and MSK-1 phosphorylate p65 at serine 276 and CREB at serine 133. While NSC-127102 did not inhibit phosphorylation of CREB by either PKA or MSK-1, this compound strongly inhibited phosphorylation of p65 by MSK-1. Interestingly, NSC-127102 did not affect phosphorylation of p65 at serine 276 by PKA, even at higher concentrations (data now shown). One possible explanation for this difference is that NSC-127102 interacts with p65 in such a way to sterically inhibit access of MSK-1 to serine 276, but not access of PKA to serine 276. These data also show that NSC-127102 does not directly inhibit PKA or MSK-1 kinase activity, but likely binds to p65 near serine 276 to inhibit phosphorylation.
Fig. 3.
NSC-127102 inhibits serine 276 phosphorylation in vitro. CREB or p65 was used as a substrate for phosphorylation of CREB at serine 133 or phosphorylation of p65 at serine 276 by MSK-1 or PKA. CREB and p65 were pre-incubated with 50 or 100 μM NSC-127102 for five minutes prior to the addition of MSK-1 or PKA. Immunoblot analysis for p-S133 of CREB and p-S276 of p65 was performed. Results are representative of three separate experiments.
3.4. NSC-127102 inhibits transcription mediated by p65 and CBP
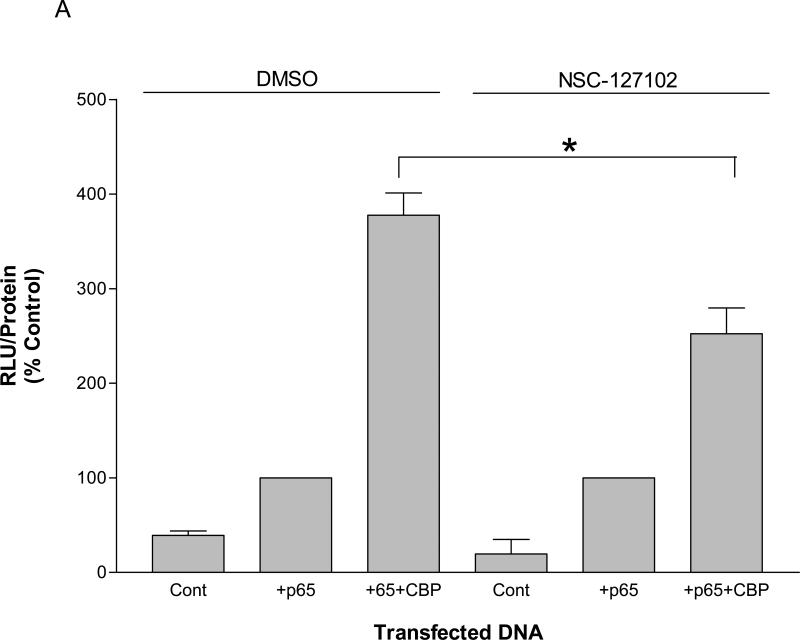
The phosphorylation of p65 at serine 276 is required for the interaction of p65 with CBP. To determine if NSC-127102 affects transcription regulated by p65 and CBP, HepG2 cells were transfected with a construct that contains five κB elements upstream of a luciferase reporter gene in the presence or absence of co-transfected p65 and CBP. As shown in Fig. 4A, p65 enhanced luciferase activity approximately three-fold over control. The presence of CBP further enhanced p65-mediated activity by an additional three-fold over control levels. NSC-127102, however, inhibited the enhancement of activity by CBP, showing that NSC-127102 regulates the effects of CBP on p65-mediated activity.
Fig. 4.
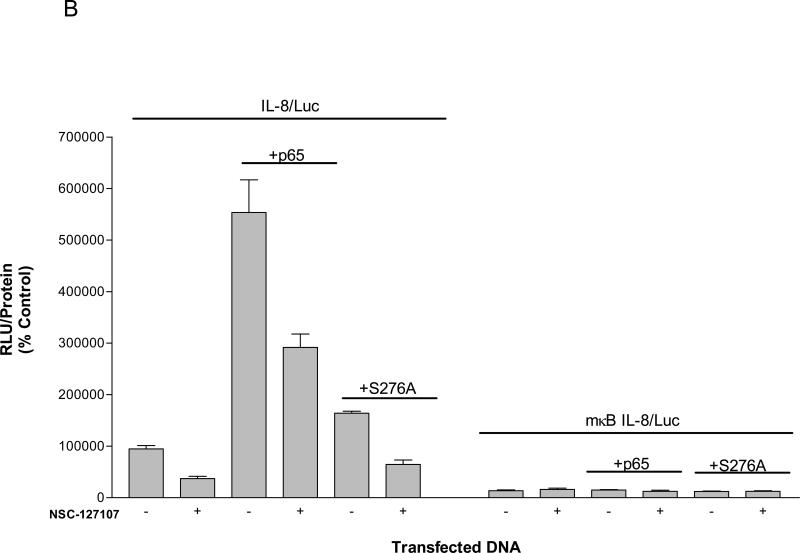
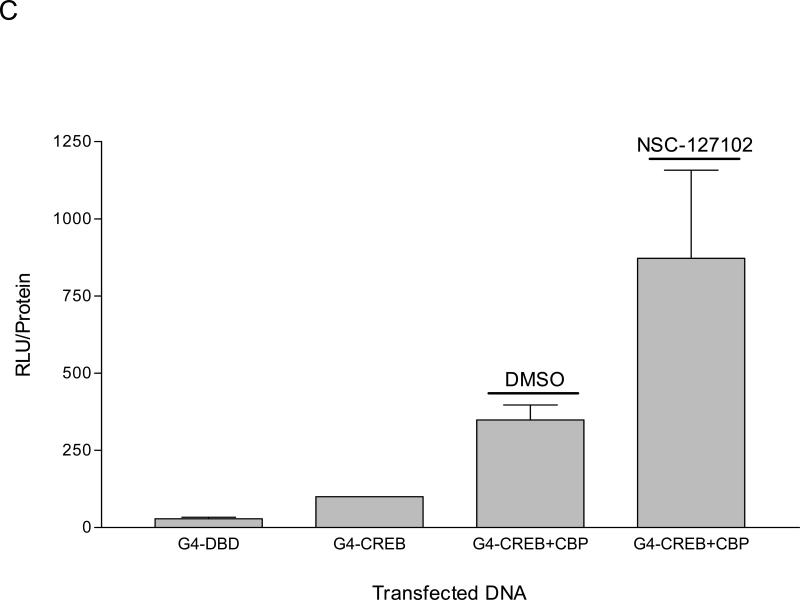
NSC-127102 inhibits activity mediated by p65 and CBP. (A) HepG2 hepatoma cells were transfected with a construct that contains five κB elements upstream of a luciferase reporter gene in the presence or absence of co-transfected p65 and CBP. Twenty-four hours later, cells were treated with 200 μM NSC-127102 for an additional 24 h. Cell lysates were prepared and luciferase activity was measured. Luciferase activity was normalized to protein concentration. The data represent the average of triplicate determinations ± S.E. (*p<0.05, Student's t test), and is representative of four independent experiments. (B) HepG2 hepatoma cells were transfected with IL-8/Luc or mκB IL-8/Luc the presence or absence of co-transfected p65 or S276A. Twenty-four hours later, cells were treated with 200 μM NSC-127102 for an additional 24 h. Cell lysates were prepared and luciferase activity was measured. Luciferase activity was normalized to protein concentration. The data are representative on three independent experiments. (C) HepG2 cells were co-transfected with a construct that has CREB fused to the Gal4 DNA binding domain (G4-CREB) and a construct that has five Gal 4 DNA binding sites upstream of the E1B TATA box and luciferase reporter gene ((GAL4)5E1bLuc). After 24 h, cells were treated with 200 μM NSC-127102 for an additional 24 h. Cells were subsequently harvested, as described in panel A, and luciferase activity was measured and normalized to protein concentration.
The IL-8 gene promoter is regulated by p65 phosphorylated on serine 276 [12-14]. To determine if NSC-127102 also inhibits IL-8 promoter activity, HepG2 cells were transfected with a construct that encodes −162 to +44 bp of the IL-8 promoter upstream of a luciferase reporter construct (IL-8/Luc). As shown in Fig. 4B, the IL-8 promoter has a certain level of activity that is likely regulated by constitutively active p65 phosphorylated at serine 276. This activity if inhibited by NSC-127102. Co-transfection of an expression vector encoding p65 enhanced IL-8 promoter activity approximately six-fold, and this activity was also inhibited by NSC-127102. Co-transfection of an expression vector encoding p65 with a mutation of serine 276 to an alanine (S276A), however, only minimally induced IL-8 promoter activity, underscoring the role of serine 276 phosphorylation for activation of this gene promoter.
As a control, HepG2 cells were also transfected with IL-8/Luc encoding a mutated κB element located between −80 and −70 bp relative to the transcriptional start site (mκB IL-8/Luc) (Fig. 4B). p65 did not enhance the activity of this gene promoter, as expected. Furthermore, NSC-127102 did not inhibit the activity of mκB IL-8/Luc, indicating that NSC-127102 specifically inhibits activity mediated by p65 and does not inhibit the IL-8 basal promoter.
To determine if NSC-127102 affects the activity of a different transcription factor, CREB, HepG2 cells were co-transfected with a construct that has CREB fused to the Gal4 DNA binding domain (G4-CREB) and a construct that has five Gal 4 DNA binding sites upstream of the E1B TATA box and luciferase reporter gene ((GAL4)5E1bLuc). This approach was used since transfection of HepG2 cells with CREB did not affect the transcription of a CRE-luciferase reporter construct (data not shown), likely due to high levels of endogenous CREB in the nucleus. Furthermore, several different transcription factors bind to the CRE, making it difficult to specifically study CREB. Co-transfection of CBP stimulated G4-CREB activity approximately 3-fold (Fig. 4C). Interestingly, NSC-127102 did not inhibit the regulation of G4-CREB by CBP. Together, these experiments show that NSC-127102 suppresses the stimulation of p65 by CBP, but does not inhibit the stimulation of CREB by CBP.
3.5. NSC-127102 inhibits IL-8 and VCAM-1 gene expression
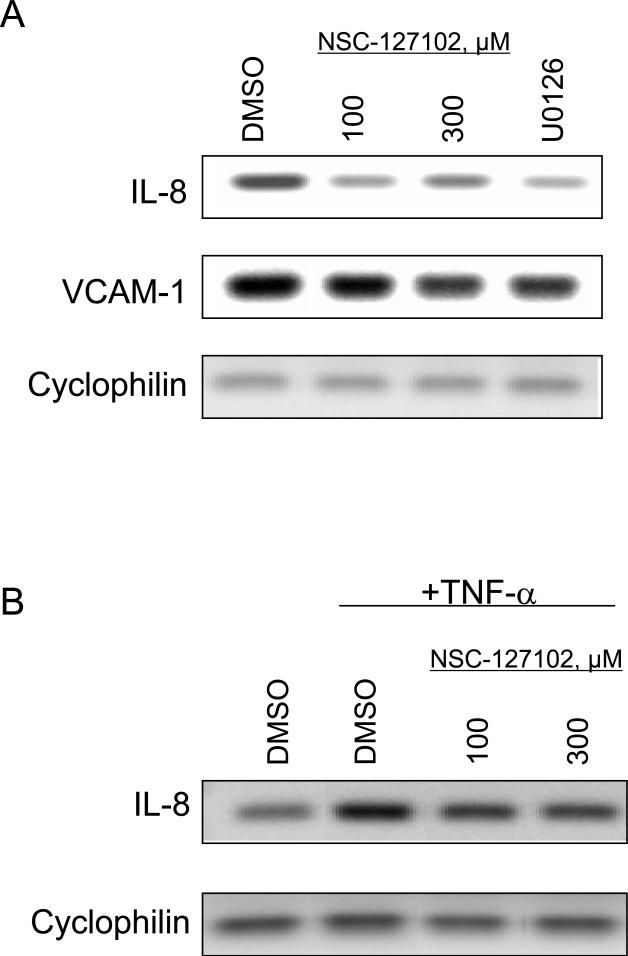
Phosphorylation of p65 at serine 276 is required for IL-8 and VCAM-1 gene expression [12,13]. To assess whether inhibition of serine 276 phosphorylation by NSC-127102 affects the transcription of endogenous genes, HepG2 cells were treated for 24 h with NSC-127102 and IL-8 and VCAM-1 mRNA levels were assessed by RT-PCR. NSC-127102 decreased IL-8 and VCAM-1 gene expression (Fig. 5A). Additionally, the MEK inhibitor U0126, which inhibits serine 276 phosphorylation [20] (data not shown), also decreased IL-8 and VCAM-1 gene expression. HepG2 cells were also treated with TNF-α, which stimulates IL-8 gene expression. As shown in Fig. 5B, TNF-α stimulated IL-8 gene expression, while NSC-127102 inhibited the ability of TNF-α to stimulate IL-8 gene expression.
Fig. 5.
NSC-127102 inhibits IL-8 and VCAM-1 gene expression. (A) HepG2 cells were treated for 24 h with 100 or 300 μM NSC-127102. Cells were also treated with 20 μM U0126 for 24 h. Cells were subsequently harvested and total RNA isolated. The effects of NSC-127102 on IL-8 and VCAM-1 mRNA levels were assessed by RT-PCR. Cyclophilin mRNA served as a loading control. (B) HepG2 cells were treated with 30 ng/ml TNF-α for 4 h to stimulate IL-8 gene expression in the presence or absence of 100 or 300 μM NSC-127102. Total RNA was isolated and IL-8 mRNA levels were assessed by RT-PCR.
4. Discussion
The NF-κB transcription factors control the expression of a wide variety of genes involved in immune and inflammatory responses, cell proliferation, and cell survival. Furthermore, this transcription factor plays a role in the pathogenesis of many diseases, including cancer and autoimmune diseases [21-23]. Inhibitors of NF-κB, therefore, are likely to have benefits against these diseases.
NF-κB subunits are nuclear factors that are responsive to many diverse signaling pathways. The phosphorylation and degradation of IκB is a key step in the activation of NF-κB, as is the nuclear translocation and post-translational modification of this transcription factor. Many NF-κB inhibitors have been identified, including inhibitors of IKK, which are the kinases that phosphorylate IκB; inhibitors of IκB phosphorylation and degradation; and inhibitors of NF-κB nuclear translocation [15]. These inhibitors, however, affect global NF-κB activity, thus making them likely to affect the transcription of most or all NF-κB regulated genes and to have unwanted side effects. We took a unique approach, in silico molecular docking, to identify more selective NF-κB inhibitors.
In silico molecular docking, a primary method for the discovery of ligands, has been used to successfully identify novel ligands for more than 30 targets, including inhibitors of aldose reductase, the retinoic acid receptor, and carbonic anhydrase II [16]. To our knowledge, this is the first application of in silico molecular docking to identify a small molecule inhibitor of protein phosphorylation. A structural pocket is adjacent to serine 276 of p65. We reasoned that a small molecule binding to this site might affect serine 276 phosphorylation. Since the serine 276 phosphorylation affects the regulation of a defined group of genes [12,13,24], inhibition of serine 276 phosphorylation is also expected to have fewer side effects if tested in clinical trials in the future.
The site surrounding serine 276 is a consensus phosphorylation site for PKA, as is the site surrounding serine 133 of CREB. Phosphorylation of these sites is necessary for the interaction of p65 and CREB with CBP [10,25]. Using a different approach, an inhibitor of the CREB:CBP interaction was identified [26]. Using NMR-based screening of a preselected small-molecule library, several compounds that bind to different surfaces on the KIX domain of CBP were identified [26]. One of these compounds, KG-501, at concentrations from 50 – 500 μM, disrupts the CREB:CBP complex and attenuates target gene induction in response to a cAMP agonist. KG-501 does not, however, affect CREB serine 133 phosphorylation. Interestingly, this compound also inhibits p65-mediated transcription [26]. Compound NSC-127102, on the other hand, appears to specifically affect the activity of p65 and not CREB, as neither CREB phosphorylation nor activity is affected by this compound.
Both IL-8 and VCAM-1 promote the growth and metastasis of a wide variety of tumors. Expression of IL-8 by human melanoma cells and human ovarian cancer cells correlates with their metastatic potential [27-29]. Furthermore, IL-8 is a transcriptional target of Ras signaling, with Ras-dependent IL-8 secretion being required for the initiation of tumor-associated inflammation and neovascularization [30]. VCAM-1 plays a role in metastasis and recent studies show that tumor expression of VCAM-1 also plays a role in immune evasion by decreasing the number of tumor-infiltrating CD8+ T cells in the tumors expressing VCAM-1 [31]. Because NSC-127102 decreases IL-8 and VCAM-1 gene expression, this compound may prove beneficial in the treatment of certain types of cancer. It is possible that the concentrations of NSC-127102 (100 – 200 μM) that inhibit serine 276 phosphorylation in cell culture may not be optimal for studies in animals. If this is the case, this compound will be chemically modified to achieve greater potency and efficacy in cell culture. Future experiments include testing this compound and its derivatives in animal models of cancer.
Acknowledgements
We would like to thank the NCI/DTP Open Chemical Repository (http://dtp.nci.nih.gov) for providing the compounds used in our assays and the makers of the DOCK program Suite (UCSF). This work was supported by the National Institutes of Health grant [R01-CA93651], Florida Department of Health grants [07BB-8] and [09BB-10], and the Komen for the Cure grant [KG080510] to B.L. and the Department of Defense grant [W81XWH-06-1-0633] to M.L. P.C. was supported by NIH/NCI T32 Training Grant in Cancer Biology CA09126.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None declared
References
- 1.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Castranova V. Nuclear factor-κB, an unappreciated tumor suppressor. Cancer Res. 2007;67:11093–11098. doi: 10.1158/0008-5472.CAN-07-1576. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay T, Roth JA, Maxwell SA. Altered expression of the p50 subunit of the NF kappa B transcription factor complex in non-small cell lung carcinoma. Oncogene. 1995;11:999–1003. [PubMed] [Google Scholar]
- 4.Bargou RC, Leng C, Krappmann D, Emmerich R, Mapara MY, Bommert K, Royer HD, Scheidereit C, Dorken B. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood. 1996;87:4340–4347. [PubMed] [Google Scholar]
- 5.Berger C, Brousset P, McQuain C, Knecht H. Deletion variants within the NF-kappaB activation domain of the LMP1 oncogene in acquired immunodeficiency syndrome-related large cell lymphomas, in prelymphomas and atypical lymphoproliferations. Leuk. Lymphoma. 1997;26:239–250. doi: 10.3109/10428199709051773. [DOI] [PubMed] [Google Scholar]
- 6.Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devalaraja MN, Wang DZ, Ballard DW, Richmond A. Elevated constitutive IkappaB kinase activity and IkappaB-alpha phosphorylation in Hs294T melanoma cells lead to increased basal MGSA/GRO-alpha transcription. Cancer Res. 1999;59:1372–1377. [PubMed] [Google Scholar]
- 8.Dejardin E, Deregowski V, Chapelier M, Jacobs N, Gielen J, Merville MP, Bours V. Regulation of NF-kappaB activity by I kappaB-related proteins in adenocarcinoma cells. Oncogene. 1999;18:2567–2577. doi: 10.1038/sj.onc.1202599. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, Lu Y, Zhang Z, Vallyathan V, Ding M, Castranova V, Shi X. Opposite effect of NF-kappa B and c-Jun N-terminal kinase on p53-independent GADD45 induction by aresenite. J. Biol. Chem. 2001;276:11414–11419. doi: 10.1074/jbc.M011682200. [DOI] [PubMed] [Google Scholar]
- 10.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 11.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) EMBO J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anrather J, Racchumi G, Iadecola C. cis-acting, element-specific transcriptional activity of differentially phosphorylated nuclear factor-kappa B. J. Biol. Chem. 2005;280:244–252. doi: 10.1074/jbc.M409344200. [DOI] [PubMed] [Google Scholar]
- 13.Jamaluddin M, Wang S, Boldogh I, Tian B, Brasier AR. TNF-alpha-induced NF-kappaB/RelA Ser(276) phosphorylation and enhanceosome formation is mediated by an ROS-dependent PKAc pathway. Cell Signal. 2007;19:1419–1433. doi: 10.1016/j.cellsig.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase9/cyclin T1 complexes. Mol. Cell. Biol. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore TD, Herscovitch M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene. 2006;25:6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 16.Shoichet BK, McGovern SL, Wei B, Irwin JJ. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 17.Ofir R, Dwarki VJ, Rashid D, Verma IM. CREB represses transcription of fos promoter: role of phosphorylation. Gene Expr. 1991;1:55–60. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong H, SuYang H, Erdjument-Bromage B, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 19.Sethi G, Ahn KS, Sandur SK, Lin X, Chaturvedi MM, Aggarwal BB. Indirubin enhances tumor necrosis factor-induced apoptosis through modulation of nuclear factor-κB signaling pathway. J. Biol. Chem. 2006;281:23425–23435. doi: 10.1074/jbc.M602627200. [DOI] [PubMed] [Google Scholar]
- 20.Joo JH, Jetten AM. NF-kappaB-dependent transcriptional activation in lung carcinoma cells by farnesol involved p65/RelA(Ser276) phosphorylation via the MEK-MSK1 signaling pathway. J. Biol. Chem. 2008;283:16391–16399. doi: 10.1074/jbc.M800945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarov SS. NF-kappaB as a therapeutic target in chronic inflammation: recent advances. Mol. Med. Today. 2000;6:441–446. doi: 10.1016/s1357-4310(00)01814-1. [DOI] [PubMed] [Google Scholar]
- 22.Courtois G, Gilmore TD. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 23.Uwe S. Anti-inflammatory interventions of NF-kappaB signaling: potential applications and risks. Biochem. Pharmacol. 2008;75:1567–1679. doi: 10.1016/j.bcp.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Prasad RC, Wang XL, Law BK, Davis B, Green G, Boone B, Sims L, Law M. Identification of genes, including the gene encoding p27Kip1, regulated by serine 276 phosphorylation of the p65 subunit of NF-κB. Cancer Letters. 2009;275:139–149. doi: 10.1016/j.canlet.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montiminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 26.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Tsaffrir Z, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator-coactivator interaction. Proc. Nat. Acad. Sci. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luca M, Huang S, Gershenwald JE, Singh RK, Reich R, Bar-Eli M. Expression of interleukin-8 by human melanoma cells upregulates MMP-2 activity and increases tumor growth and metastasis. Am. J. Pathol. 1997;151:1105–1113. [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Fidler IJ. Interleukin 8: an autocrine growth factor for human ovarian cancer. Oncol. Res. 2000;12:97–106. doi: 10.3727/096504001108747567. [DOI] [PubMed] [Google Scholar]
- 29.Huang S, Mills L, Mian B, Tellez C, McCarty M, Yang XD, Gudas JM, Bar-Eli M. Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am. J. Pathol. 2002;161:125–134. doi: 10.1016/S0002-9440(10)64164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B Ancrile B, O'Hayer KM, Counter CM. Oncogenic ras-induced expression of cytokines: a new target of anti-cancer therapeutics. Mol. Interv. 2008;8:22–27. doi: 10.1124/mi.8.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin KY, Lu D, Hung CF, Peng S, Huang L, Jie C, Murillo F, Rowley J, Tsai YC, He L, Kim DJ, Jaffee E, Pardoll D, Wu TC. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer Res. 2007;67:1832–1841. doi: 10.1158/0008-5472.CAN-06-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]