Abstract
Introduction
Metagenomics uses gene expression patterns to understand the taxonomy and metabolic activities of microbial communities. Metaproteomics applies the same approach to community proteomes. Previously, we used a novel three-dimensional peptide separation method to identify over 2,000 salivary proteins, including 139 of microbial origin. This study used that data to carry out the first metaproteomic analysis of the human salivary microbiota.
Methods
MEGAN metagenomic software generated a phylogenetic tree, which was checked against the Human Oral Microbiome Database (HOMD). Pathway analyses were done with the Clusters of Orthologous Groups and MetaCyc databases.
Results
37% of the peptides were identifiable only at the level of cellular organisms or bacteria. The rest were distributed among five bacterial phyla (61%), archea (0.5%), and viruses (0.8%). 29% were assignable at the genus level, and most belonged to Streptococcus (17%). 11% of all peptides could be assigned to species. Most taxa were represented in HOMD, and they included well-known species such as periodontal pathogens. However, there also were “exotic” species including aphid endosymbionts, plant, water, and soil bacteria, extremeophiles, and archea. The pathway analysis indicated that peptides were linked to translation (37%), followed by glycolysis (19%), amino acid metabolism (8%), and energy production (8%).
Conclusion
The taxonomic structure of the salivary metaproteome is very diverse, but dominated by streptococci. “Exotic” species may actually represent close relatives that have not yet been sequenced. Salivary microbes appear to be actively engaged in protein synthesis, and the pathway analysis is consistent with metabolism of salivary glycoproteins.
Introduction
Genomic sequencing defines the range of biological activities that can be expressed by any microbial species. Sequence information then can be used as the keystone for genomic and proteomic studies of particular species that are important to dentistry, medicine, animal health, and the environment. Such studies typically focus on one species at a time. However, microbes occur in nature as components of complex, structured, polymicrobial communities. The level of taxonomic diversity in environmental and host-dependent communities is typically very high. Most species have neither been cultured nor sequenced, and have been identified solely on the basis of structural variation in ribosomal RNA. In order to understand how microbes behave in communities, it is necessary to apply a “meta” approach to conventional genomics and proteomics (16, 45).
Metagenomics uses gene expression patterns to understand the taxonomic and metabolic structure of complex microbial communities (16, 45). Recent progress in metagenomics has been greatly aided by improvements in the speed and efficiency of high throughput sequencing of short DNA segments (pyrosequencing) (10, 18, 24, 49, 51). By contrast, progress in the corresponding discipline of metaproteomics has been slower. Relatively few studies have been done. Most of those have focused on simpler environmental communities, such as water treatment systems (28, 33, 39, 40, 60–62). The only host-based community that has been examined was likewise very simple – fecal samples from very young human infants (26). Most of those studies used 2-D or 1-D gels for protein separation and spot/band comparison, often without the use of mass spectrometry for protein identification. Only one group thus far has taken advantage of the high throughput made possible by newer “shotgun” proteomic technologies. They used biofilm populations from extreme mine environments, which again were chosen for simplicity (42).
A question, which has not yet been addressed, is: can useful metaproteomic information be obtained from commensal microbial communities sampled against a high background of host proteins? Our group currently is engaged in a study of the human salivary proteome in oral cancer and health. Whole saliva collected for proteomic studies usually is processed by centrifugation to remove exfoliated epithelial cells, bacteria, and food debris. However, because of the focus on oral cancer, the epithelial cells in the pellet fraction were considered to be potentially quite interesting. Accordingly, the salivary pellet fractions from four oral squamous cell carcinoma patients were pooled for a pilot proteomic study. That study employed a novel “three-dimensional” peptide separation process, which considerably improved the dynamic range of protein discovery. The salivary pellet fraction also contains large numbers of oral bacteria. Thus, it is not surprising that, of over seven thousand peptides identified, 357 were of microbial origin. Those findings have been reported previously (63), but no comprehensive analysis of the bacterial component was done at that time. The purpose of this study was to use bioinformatic software originally developed for metagenomics to test the hypothesis that meaningful metaproteomic information about the taxonomy and metabolic activity of the salivary microbiota could be extracted from this dataset.
Materials And Methods
Proteomic analysis
A detailed account of the methods used for peptide separation and identification has been published previously (63). Briefly, the digest of pooled salivary pellets was separated in a three-stage process. The first step was preparative isoelectric focusing using a free-flow electrophoresis system (FFE). All pI fractions were subject to a preliminary MS/MS analysis to determine which fractions contained the most complex peptide mixtures. Peptides within each of the complex fractions were then further separated according to their absolute number of basic amino acid residues, using strong cation exchange step gradient chromatography (SCX). The contents of each FFE/SCX fraction then were separated according to hydrophobicity, using a microcapillary reverse phase liquid chromatography system coupled to the mass spectrometer. MS/MS was done on an LTQ linear ion trap mass spectrometer.
The primary goal of our previous study was to improve the detection of low abundance human proteins in the salivary pellet, and the majority of peptides in the data set were of human origin. The reader is referred to our previous publication for information on how human proteins were identified (63). To identify microbial proteins, all MS/MS spectra were searched against the Uniprot/Swiss-Prot protein sequence database release 6.0, containing 194,317 entries from over 10,000 different organisms (including over 135,000 protein sequences from bacterial and viral sources). A reversed-sequence version of the database was appended for the purpose of false positive rate estimation. The resulting peptide sequence matches were filtered by comparing predicted peptide pI to the average pI for free flow electrophoresis fractions, and then by a two step filtering process which first determined the lowest PeptideProphet P score needed to maintain the estimated false positive rate at 1%, and then determined the matches at a low stringency P score of 0.2. The results from both steps were combined to obtain a protein catalog with an estimated false positive rate of 1%. Protein identifications corresponding to microbial organisms then were manually extracted. Our previous analysis focused solely on species identification. A species was only considered to be present if peptides matched to two or more proteins derived from that species. If only a single protein from a species was identified, two or more unique peptides from the protein needed to be matched to consider that species as being present. For all species passing the criteria above, at least one of the peptide sequences identified had to be expressed only in that species (i.e. not a redundant peptide sequence found in proteins known to be expressed in multiple taxa). Those criteria resulted in 139 proteins being identified and matched to 34 different microbial species (63). The function of identified proteins was not addressed at that time.
Metaproteomic analysis
In retrospect, we considered that the decision to exclude non-redundant peptides might likewise have excluded a considerable amount of useful information about the taxonomy and metabolic activity of the salivary microbiota. The current analysis therefore used all peptides that matched to any microbe, regardless of whether they had been included in the initial analysis. That set of 357 peptides was submitted to protein BLAST for sequence matching using the NR database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The resulting BLASTP file then was parsed using MEGAN metagenomic software (19). MEGAN uses a homology-matching algorithm to generate a phylogenetic tree based on the NCBI taxonomy database. Because the number of reads in this proteomic dataset was considerably smaller than the thousands usual in a metagenomic dataset, the number of reads required for a taxon assignment was set to one. The resulting phylogenetic tree was checked manually against the 16S–rRNA-based Human Oral Microbiome Database (HOMD) (11), to determine if any taxa not previously reported in the mouth were represented in this peptide dataset. Software for pathway analysis of microbes is much less well developed than what is available for the human genome. Functional analysis of the peptide dataset was done by manually matching protein names against gene function groups defined within the Clusters of Orthologous Groups (COGs) prokaryotic database (54). This was supplemented by manually searching the MetaCyc database for matching prokaryotic pathways (5). Those manually curated databases do not include all currently sequenced microbial species, so COGs group/MetaCyc pathway matches sometimes had to be based on data from closely related taxa.
Results
Taxonomic assignments
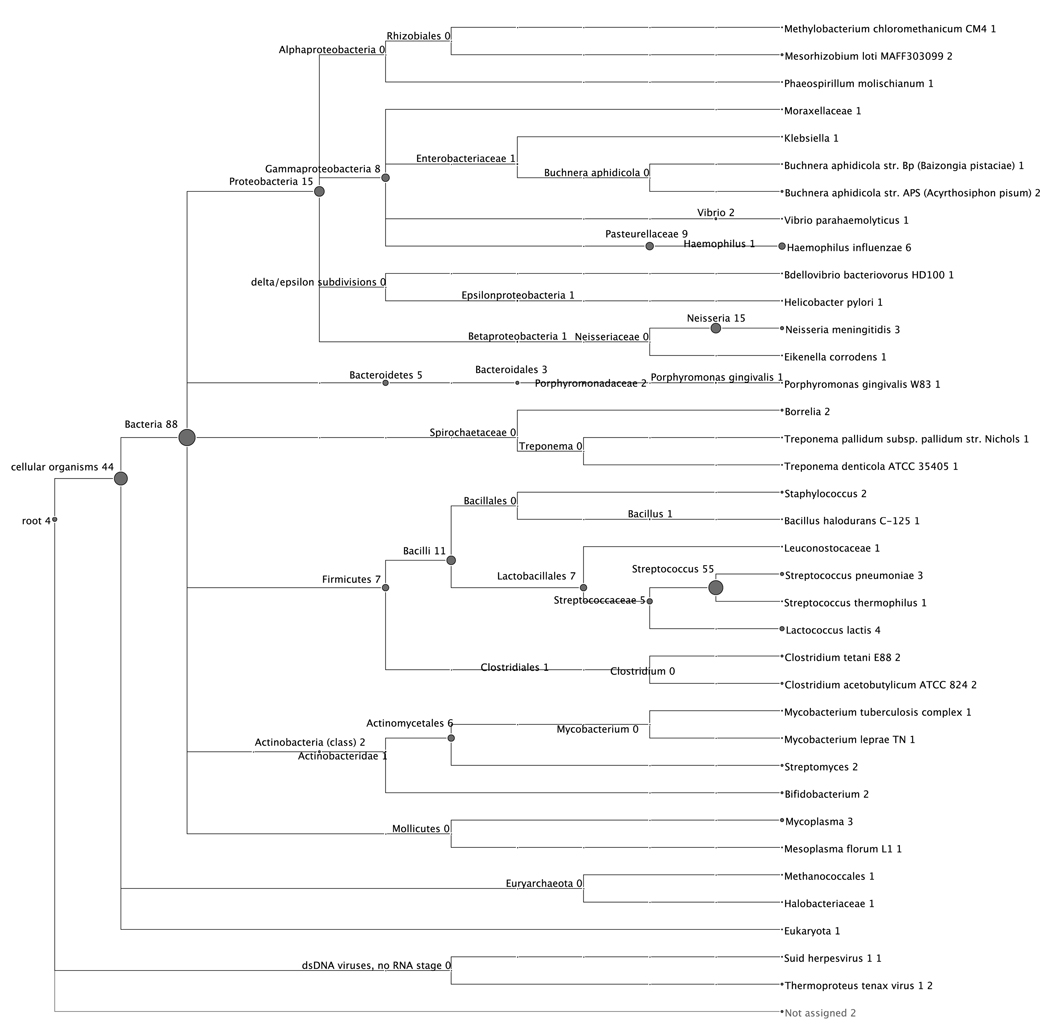
The phylogenetic tree generated by MEGAN is shown in Fig. 1. The number by each node indicates the number of peptides assigned to that taxonomic level. This is indicated graphically by the size of the node. The peptides in the dataset were broadly distributed among five major bacterial phyla, but a few were assigned to Archaea and viruses. Only 11% of all peptides could be assigned at the species level. Those remaining were assignable only to higher taxonomic levels. More peptides were assigned to the genus Streptococcus than to any other bacterial taxon (17%). However, it is important to note that 25% of peptides were assignable only at the kingdom level, to Bacteria.
Fig. 1.
Phylogenetic tree generated by MEGAN, incorporating all taxa to which peptides were assigned. The size of each circular node is proportional to the number of assignments at that particular taxonomic level. That number also is indicated by the number after the name of each taxon. It is important to note that those numbers are not summed across the lower taxonomic levels for any given taxon. For example, the 88 following Bacteria indicates that 88 peptides could not be assigned below the kingdom level.
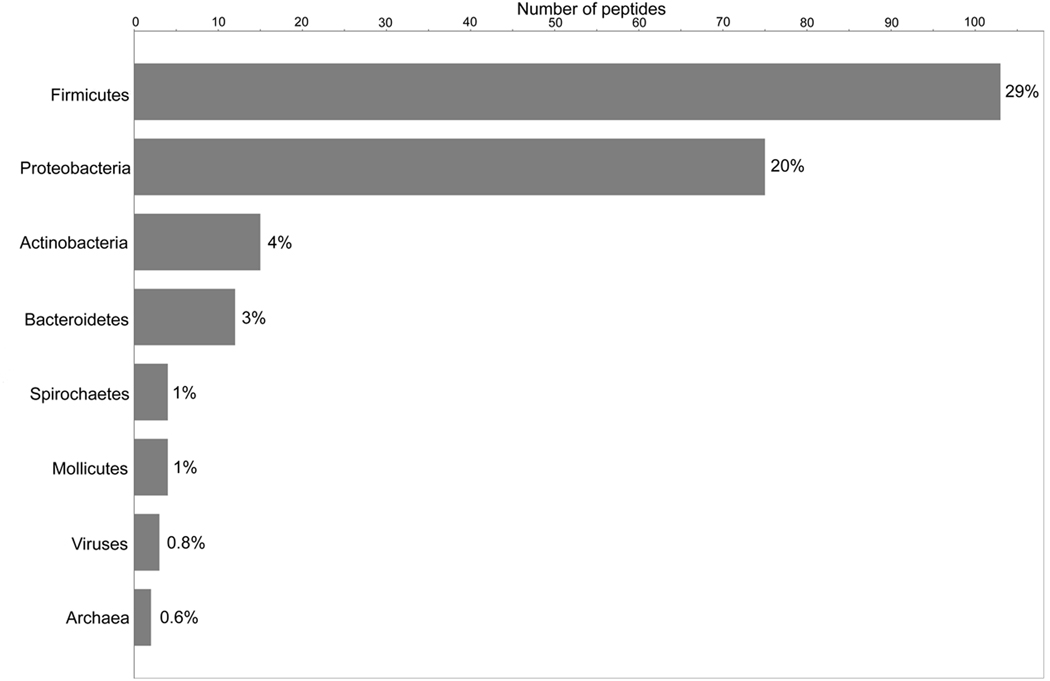
The distribution of peptides at the phylum level is depicted graphically in Fig. 2. The largest numbers of peptides were assigned to Firmicutes (29%), and approximately half of those were assigned to Streptococcus. The second largest group was Proteobacteria, at 20%. Peptides assignable to Actinobacteria, Bacteroidetes, Spirochaetes, and Mollicutes were present in much smaller numbers (4%, 3%, 1%, and 1% respectively). Finally, 0.8% of peptides were assigned to double-stranded DNA viruses, and 0.6% to Euryarcheota.
Fig. 2.
Bar chart showing the distribution of peptides among five bacterial phyla, Mollicutes, Archaea, and viruses. The x-axis indicates the number of peptides represented by each bar; that number is expressed as a percentage of 357 total peptides, at the end of each bar. This chart does not include the peptides that were assignable to bacteria only at the kingdom level, so the percentages do not sum to 100.
Thirty-four species assignments had been made using our previous criteria (63). Only 16 of those were concordant with the MEGAN phylogenetic tree. The non-concordant assignments all were referred to higher taxonomic levels within their respective lineages (not shown). However, MEGAN did assign peptides to four species that did not meet the previous criteria. Of the peptides assignable by MEGAN at the species level, 43% were assigned to species that are considered pathogenic (Table 1). Haemophilus influenzae had the largest number hits (2% of all peptides), followed by Neisseria meningitidis (1%), and Streptococcus pneumoniae (1%). All three of those species are associated primarily with the nasopharynx (55), but also can be constituents of the oral flora (11). Peptides also were attributed to putative periodontal disease pathogens, including Eikenella corrodens, Porphyromonas gingivalis, and Treponema denticola (50). Peptides likewise were associated with agents of serious diseases, including Treponema palidum (syphilis), Helicobacter pylori (peptic ulcers), Mycobacterium tuberculosis (tuberculosis), and Mycobacterium leprae (leprosy) (55). All of those species have been detected in the mouth by 16S rRNA sequencing, as reported in HOMD (11). However, peptides also were assigned to two pathogens not yet included in HOMD. These were Clostridium tetani, the agent of tetanus, found in soil and feces (55), and Vibrio parahaemolyticus, a marine bacteria which can cause diarrheal illness when consumed in raw seafood (13).
Table 1.
Pathogenic species to which peptides were assigned
| Species | Percent of peptides | In HOMD? |
|---|---|---|
| Haemophilus influenzae | 2% | Yes |
| Neisseria meningitidis | 1% | Yes |
| Streptococcus pneumoniae | 1% | Yes |
| Eikenella corrodens | 0.2% | Yes |
| Porphyromonas gingivalis | 0.2% | Yes |
| Treponema denticola | 0.2% | Yes |
| Treponema palidum | 0.2% | Yes |
| Helicobacter pylori | 0.2% | Yes |
| Mycobacterium tuberculosis | 0.2% | Yes |
| Mycobacterium leprae | 0.2% | Yes |
| Clostridium tetani | 0.5% | No |
| Vibrio parahaemolyticus | 0.2% | No |
Six percent of all peptides were assigned to taxa that can be considered “exotic”, in the sense that they have not been previously reported in the mouth, and would be considered unlikely to occur there (Table 2). Perhaps the most unusual of those is Buchnera aphidicola, an obligate aphid endosymbiont with a truncated genome. Peptides were assigned to two strains of B. aphidicola, and each strain is considered to be specific to a different species of aphid (14). Another taxon with an insect host was Borrelia, which includes the agent of Lyme disease (55). Taxa typically found on plants included Mesorhizobium loti (legume root nodules) (21), Leuconostocaceae (3), and Mesoplasma florum (a plant mycoplasma) (35). There also were assignments to environmental microbes, including Streptomyces (found in soil) (22), and Phaeospirillum molischianum, a phototrophic freshwater purple spiral bacterium (20). Bacteria associated with industrial activities included Clostridium acetobutylicum, which is used industrially for making acetone (25), and Methylobacterium chloromethanicum, a chloromethane-utilizing methylotroph originally recovered from polluted soil (36). Extremophiles included akaliphilic Bacillus halodurans (34), archeal halophiles (Halobacteriaceae) (38), and archeal methanogens (Methanococcales) (59). Viruses were represented by peptides attributed to Suid herpesvirus 1, the agent of pseudorabies in swine (41), and Thermoproteus tenax virus 1, a phage parasitic on Archaea (12). None of the above taxa previously have been reported in HOMD (11).
Table 2.
“Exotic” non-oral taxa to which peptides were assigned
| Taxon | Attributes | In HOMD? |
|---|---|---|
| Buchnera aphidicola str. APS | Obligate aphid endosymbiont | No |
| Buchnera aphidicola str. Bp | Obligate aphid endosymbiont | No |
| Methylobacterium chloromethanicum | Chloromethane-utilizing | No |
| Mesorhizobium loti | Legume root nodules | No |
| Phaeospirillum molischianum | Phototrophic, fresh-water | No |
| Borrelia | Includes Lyme disease agent | No |
| Bacillus halodurans | Akaliphilic extremophile | No |
| Leuconostocaceae | Typically isolated from food | No |
| Clostridium acetobutylicum | Used for making solvents | No |
| Mesoplasma florum | Plant mycoplasma | No |
| Streptomyces | Predominantly in soil | No |
| Methanococcales | Methanogenic, Archaea | No |
| Halobacteriaceae | Archaea, extreme halophiles | No |
| Suid herpesvirus 1 | Herpesvirus from swine | No |
| Thermoproteus tenax virus 1 | Phage parasitic on archaea | No |
Protein pathways
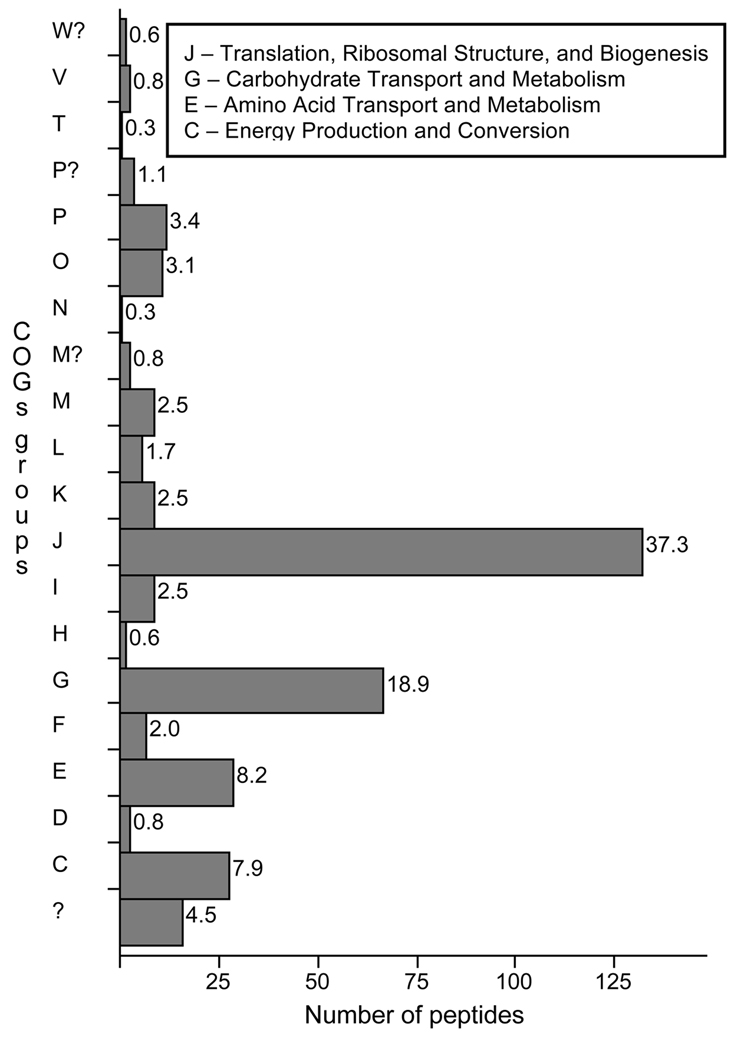
The overall distribution of peptides among COGs groups is shown in Fig. 3. Four groups accounted for 72% of the peptides. The largest single group was J – Translation, Ribosomal Structure, and Biogenesis (37%). Peptides were assigned to tRNA synthetases, elongation factors, and ribosomal proteins, with elongation factor Tu, elongation factor G, and 50S ribosomal proteins L7/L12 and L5 being the most prevalent (Table 3). The second largest group was G – Carbohydrate Transport and Metabolism (19%). The majority of peptides in Group G were assigned to the glycolysis pathway, with enolase and phosphoglycerate kinase being the most predominant proteins (Table 3). The third most common group was E – Amino Acid Transport and Metabolism (8.2%). This encompassed various amino acid biosynthesis/degradation pathways, with more peptides being assigned to methionine biosynthesis (homocysteine transmethylase), valine/leucine/isoleucine biosynthesis (ketol-acid reductoisomerase), arginine biosynthesis/degradation (ornithine transcarbamylase), and glutamate degradation (NAD-specific glutamate dehydrogenase) (Table 3). The final common group was C – Energy Production and Conversion (7.9%). Gluconeogenesis (phosphoenolpyruvate carboxykinase) was the predominant pathway, followed by mixed acid fermentation (pyruvate oxidase and formate dehydrogenase) (Table 3).
Fig. 3.
Bar chart depicting the assignment of all 357 peptides to COGs groups, denoted by capital letters (a ? indicates an uncertain assignment). The x-axis indicates the peptide count for each bar, whereas the number at the end of each bar indicates the percent of peptides assigned to that group. The functions represented by the four COGs groups that were most prevalent in the dataset are defined in the box. A full list of the functional categories represented by each COGs group letter is available at http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi
Table 3.
Predominant pathway/protein assignments within major COGs groups
| COGs group/Pathway/Protein | Peptide counta | COGs %b |
|---|---|---|
| J – Translation, Ribosomal Structure, and Biogenesis/ | 132 | |
|
|
||
| Elongation factor Tu | 30 | 23% |
| Elongation factor G | 14 | 10% |
| 50S ribosomal protein L7/L12 | 20 | 15% |
| 50S ribosomal protein L5 | 10 | 8% |
| G – Carbohydrate Transport and Metabolism/ | 67 | |
| Glycolysis/ | 51 | 76% |
| Enolase | 18 | 27% |
| Phosphoglycerate kinase | 11 | 16% |
| E – Amino Acid Transport and Metabolism/ | 29 | |
| Met biosynthesis/Homocysteine transmethylase | 6 | 21% |
| Val/Leu/Ile biosynthesis/Ketol-acid reductoisomerase | 5 | 17% |
| Arg synthesis-degradation/Ornithine transcarbamylase | 6 | 21% |
| Glu degradation/Glutamate dehydrogenase | 4 | 14% |
| C – Energy Production and Conversion | 28 | |
|
|
||
| Gluconeogenesis/phosphoenolpyruvate carboxykinase | 8 | 28% |
| Mixed acid fermentation/ | 8 | 28% |
| Pyruvate oxidase | 4 | 14% |
| Formate dehydrogenase | 4 | 14% |
Number of peptides assigned to each COGs group, pathway, or protein.
Expressed as a percent of the total number of peptides in the parent COGs group. Only prevalent pathways/proteins are listed, so percentages do not sum to 100.
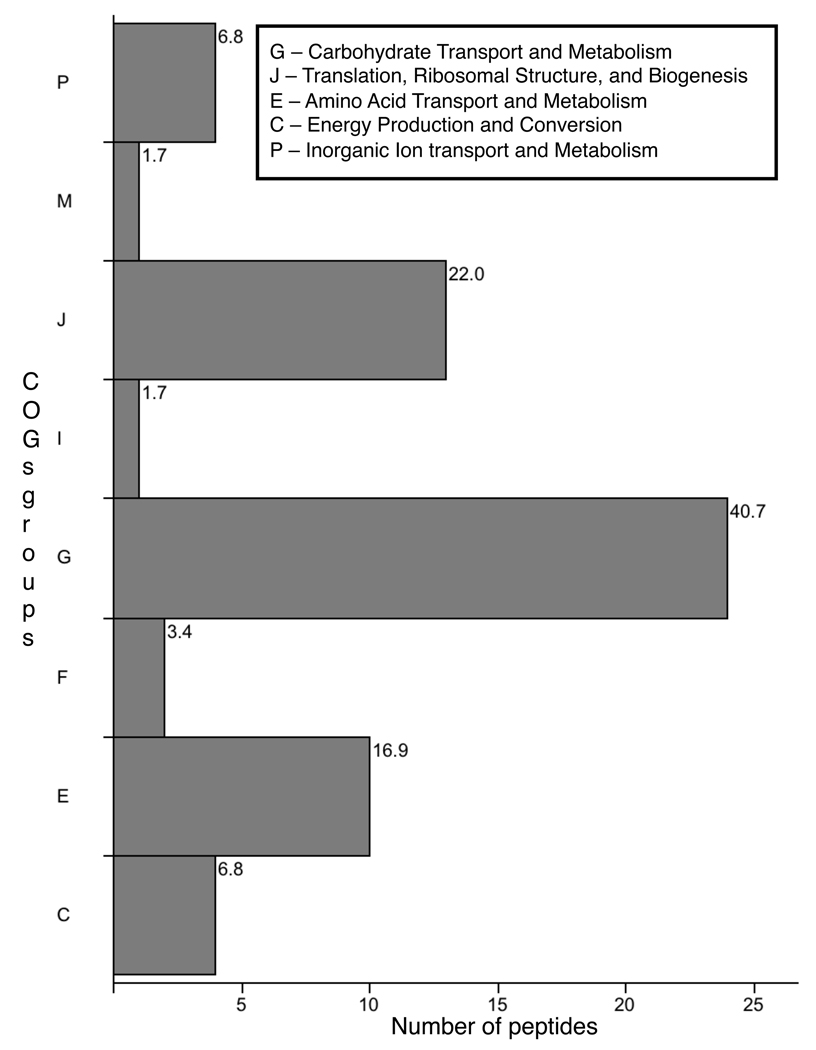
The COGs group distribution for Streptococcus, the most prevalent taxon, was similar to the distribution for the entire dataset (Fig. 4). However, Group G was more prevalent than Group J. Peptides in Group G incorporated almost the entire range of steps involved in sugar transport, and glycolysis, including components of sugar phosphotransferase systems, glucose-6-phosphate isomerase, fructose-1,6-bisphosphate aldolase, triosephosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate mutase, and enolase. Group C was represented entirely by pyruvate oxidase, the subsequent step in aerobic fermentation of sugars to acetate. Protein distributions for Groups J and E were broadly similar to the corresponding groups in the full dataset (not shown). Group P – Inorganic Ion Transport and Metabolism was represented solely by peptides assigned to superoxide dismutase.
Fig. 4.
Bar chart indicating the distribution of the 59 peptides assigned to Streptococcus (at the genus or species level) among COGs groups. The x-axis indicates the peptide count for each bar, whereas the number at the end of each bar indicates the percent of peptides assigned to that group. The functions represented by the four COGs groups that were most prevalent among streptococci are defined in the box. A full list of the functional categories represented by each COGs group letter is available at http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi
Discussion
To our knowledge, this study is the first metaproteomic analysis of the human salivary microbiota. Several types of potential bias have to be considered when interpreting the data. A metaproteomic approach is unlikely to give as complete a picture of community taxonomic structure as 16S rRNA, because it cannot be targeted to exclude conserved regions of proteins. The peptides that could not be assigned below the kingdom level of Bacteria were heavily weighted towards proteins involved in translation. Such proteins are probably fundamental to microbial life (4), and thus likely to include highly conserved amino acid sequences that may interact with highly conserved regions of rRNA. It is possible that such peptides will always be relatively abundant in metaproteomic samples, simply because ribosomes are relatively abundant in active bacteria, and conserved peptides will be the most abundant of all.
However, although this study cannot equal the level of microbial diversity seen in a recent pyrosequencing study of 16S rRNA variable regions amplified from saliva of healthy subjects (24), it is striking that the relative abundance of bacteria at the phylum level was similar in both cases. The pyrosequencing study identified over 70,000 distinct 16S rRNA sequences, but 99% of them were in seven different phyla. Firmicutes, in particular streptococci, were by far the most abundant group. That was also the case in this study. Proteobacteria were highly prevalent, and they were dominated by Haemophilus and Neisseria. That likewise was the case in this study. The only major difference was for Bacteroidetes, which encompassed 27% of 16S rRNA sequences, but only 3% of the peptides in this study. That may be a reflection of sampling variation, since the pyrosequencing study included more subjects.
Identifications at the species level are more problematic, for several reasons. One problem is that different classification algorithms may yield different results. That was the case here, since the homology-based approach used by MEGAN was generally more conservative than the manual classification criteria employed in our previous study (63). This metaproteomic dataset did include many peptides assigned by MEGAN to species previously reported in the mouth, including periodontal pathogens. That provides some confidence in the data, although the likelihood of error will be greater for taxa that are represented by only a single peptide. One must also consider the presence of a number of species that have never been reported previously in the mouth. It is possible that some of those were simply false positives. However, the extent of diversity in the oral microbiota is still being discovered, and some of these “exotic species” may literally be present. Others seem more improbable. Buchnera aphidicola is a case in point, since it is considered to be an obligate endosymbiont of aphids. Different strains are specific to different aphid species, and the genome of B. aphidicola has become truncated to the point that it is not able to survive outside of its host (14). How then, does it come to be detected in the mouth? The answer may reflect an intrinsic bias in the sequence databases, in that the pace of genomic sequencing lags far behind that of 16S rRNA sequencing. The majority of bacteria in the human and environmental microbiomes have not yet been sequenced. Thus, the extent of protein sequence conservation among bacterial species is unknown. It is likely that peptides attributed to B. aphidicola and at least some other “exotic” species actually represent oral relatives of those species, which have not yet been sequenced. The periodontal pathogen Tannerella forsythia provides a precedent, since it has been found by 16S rRNA sequencing to have uncultured relatives in termite guts (53).
The same issue applies to species that would be expected in the mouth. For example, the single peptide assigned by MEGAN to Treponema denticola originated from an adenine phosphoribosyltransferase. Purine salvage is a fundamental biological process, and it is entirely possible that the same sequence may exist in some of the many species of oral treponemes that have not yet been sequenced (6).
Despite these concerns, it seems clear that the metaproteomic approach can be used to obtain meaningful information about the community structure of the salivary pellet microbiota. Moreover, pathway analysis of the metaproteome also provides meaningful information about the activities of the pellet community at the time of saliva collection. Bacteria in the salivary pellet originate from two distinct environmental sources, biofilms on tooth surfaces, and oral mucous membranes. As oral biofilms mature, bacteria near the periphery may detach, and become planktonic in saliva (23, 29, 37, 52). That transition is likely to be accompanied by major changes in patterns of gene expression (44, 64), which presumably would be accompanied by a corresponding increase in mRNA translation. Less is known about bacteria associated with mucosal cells, but it has become clear that many oral epithelial cells harbor polymicrobial intracellular communities (46–48). Since whole saliva contains exfoliated epithelial cells, it is likely that some of those cells also contain bacteria. Intracellular bacteria in oral epithelial cells can readily be detected by in situ hybridization, using probes directed against 16S rRNA (46–48). This indicates that they contain large numbers of ribosomes (2), which presumably are engaged in translation. Thus, the relative abundance of peptides from proteins involved in translation is quite consistent with what would be predicted from other lines of evidence.
The pathway analysis further suggests that pellet microbial communities are metabolically active, with glycolysis being the predominant activity. Subjects were instructed to refrain from eating, drinking (except water), chewing gum, or using mints etc. for at least 1.5 hours prior to saliva collection. Thus, it is unlikely that host diets were the source of the sugar substrates for this glycolytic activity. However, it has been demonstrated that some oral microbes are capable of growing on media consisting solely of sterilized human saliva (7–9). The substrate for growth on saliva was presumed to be salivary glycoproteins. The salivary proteome is notable for a high abundance of high molecular weight proteins with complex oligosaccharide side chains. These include the salivary mucins MUC5B and MUC7, the agglutinin GP340, and proline-rich glycoprotein (17), as well as epithelial cell membrane mucins MUC1 and MUC4 (31). In vitro incubation of mucins with oral streptococci results in degradation of the oligosaccharide chains, and different streptococcal species can act synergistically to promote that process (56–58). Although specific bacterial enzymes involved in oligosaccharide breakdown (e.g. sialidase) were not detected in the streptococcal metaproteome, several proteins involved in the streptococcal phosphotransferase sugar transport system were identified. In combination with the relative abundance of proteins involved in glycolysis and acid fermentation, this pattern suggests that the salivary microbiota is actively engaged in metabolizing salivary glycoproteins. The presence of streptococcal pyruvate oxidase and superoxide dismutase suggests that this is occurring under aerobic conditions (1), as would be expected in saliva.
This first metaproteomic analysis of the salivary microbiota serves as a proof of concept. The three-dimensional peptide separation technique developed by our group improves the detection of low abundance proteins (63). That in turn, facilitates metaproteomic analysis of the community and structure of the salivary microbiota against a high background of host proteins. The saliva samples used were obtained from oral squamous cell carcinoma patients, and that raises the question of whether different results might have been obtained from healthy subjects. The salivary microbiota of healthy people and oral cancer patients have previously been compared for 40 species, using checkerboard DNA-DNA hybridization. Samples from cancer patients showed a greater relative abundance of three common commensal species, Prevotella melaninogenica, Capnocytophaga gingivalis, and Streptococcus mitis (32). The first two are Bacteroidetes, which were not common in this dataset. However, the high abundance of peptides from streptococci might be consistent with their results for S. mitis. A 16S rDNA cloning study detected a high prevalence of H. influenzae in oral cancer lesions (30), and that may be consistent with our finding that H. influenzae was represented by more peptides than any other single species. It is intriguing that bacterial production of acetaldehyde has been suggested as a possible risk factor for oral cancer (15). However, the present dataset did not include peptides assigned to enzymes capable of converting acetate to acetaldehyde. It is likewise intriguing that hydrogen peroxide, a potential carcinogen, is a byproduct of streptococcal pyruvate oxidase activity (27). The salivary glands secrete an endogenous peroxidase. However, salivary peroxidase is inhibited by cigarette smoke, and this has been suggested to play a role in oral carcinogenesis (43).
Our ongoing studies will provide more definitive answers to these questions. We presently are engaged in using three-dimensional separation and iTRAQ labeling for quantitative comparisons of the salivary proteomes from large numbers of oral cancer patients, oral dysplasia patients, and healthy controls. The primary focus of those studies is the discovery of salivary protein biomarkers of oral cancer. However, present findings suggest that a reasonably large number of bacterial proteins will be present in those datasets. Metaproteomic analysis then should allow us to determine whether those populations differ in the community structure and activities of their salivary microbiota.
Acknowledgments
Portions of this work were supported by NIH grant 5R01DE17734, American Cancer Society Institutional Research Grant IRG 58-001-46, and Eli Lilly and Company. We thank Dr. Daniel Huson for his advice on adapting MEGAN for use with metaproteomic data.
References
- 1.Abbe K, Carlsson J, Takahashi-Abbe S, Yamada T. Oxygen and the sugar metabolism in oral streptococci. Proc Finn Dent Soc. 1991;87:477–487. [PubMed] [Google Scholar]
- 2.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björkroth J, Holzapfel W. Genera Leuconostoc, Oenococcus and Weissella. In: Dworkin M, editor. The Prokaryotes. New York, NY: Springer; 2006. pp. 267–319. [Google Scholar]
- 4.Callister SJ, McCue LA, Turse JE, et al. Comparative Bacterial Proteomics: Analysis of the Core Genome Concept. PLoS ONE. 2008;3:e1542. doi: 10.1371/journal.pone.0001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspi R, Foerster H, Fulcher CA, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucl Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jong MH, Van Der Hoeven JS, Van Os JH, Olijve JH. Growth of oral Streptococcus species and Actinomyces viscosus in human saliva. Appl Environ Microbiol. 1984;47:901–904. doi: 10.1128/aem.47.5.901-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jong MH, Van Der Hoeven JS, Van Os JH. Growth of micro-organisms from supragingival dental plaque on saliva agar. J Dent Res. 1986;65:85–88. doi: 10.1177/00220345860650021601. [DOI] [PubMed] [Google Scholar]
- 9.De Jong MH, Van der Hoeven JS. The growth of oral bacteria on saliva. J Dent Res. 1987;66:498–505. doi: 10.1177/00220345870660021901. [DOI] [PubMed] [Google Scholar]
- 10.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewhirst FE, Izard J, Paster BJ, et al. The Human Oral Microbiome Database. 2008 http://www.HOMD.org.
- 12.Fauquet C. Virus Taxonomy: Classification and Nomenclature of Viruses : Eighth Report of the International Committee on the Taxonomy of Viruses. 2 edn. Academic Press; 2005. [Google Scholar]
- 13.FDA. Bacterial Analytical Manual Online: http://www.cfsan.fda.gov/∽ebam/bam-9.html.
- 14.Gil R, Sabater-Munoz B, Latorre A, Silva FJ, Moya A. Extreme genome reduction in Buchnera spp.: Toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci. 2002;99:4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillison ML. Current topics in the epidemiology of oral cavity and oropharyngeal cancers. Head & Neck. 2007;29:779–792. doi: 10.1002/hed.20573. [DOI] [PubMed] [Google Scholar]
- 16.Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86:680–693. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 18.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imhoff JF, Petri R, Suling J. Reclassification of species of the spiral-shaped phototrophic purple non-sulfur bacteria of the {alpha}-Proteobacteria: description of the new genera Phaeospirillum gen. nov., Rhodovibrio gen. nov., Rhodothalassium gen. nov. and Roseospira gen. nov. as well as transfer of Rhodospirillum fulvum to Phaeospirillum fulvum comb. nov., of Rhodospirillum molischianum to Phaeospirillum molischianum comb. nov., of Rhodospirillum salinarum to Rhodovibrio salinarum comb, nov., of Rhodospirillum sodomense to Rhodovibrio sodomensis comb. nov., of Rhodospirillum salexigens to Rhodothalassium salexigens comb. nov. and of Rhodospirillum mediosalinum to Roseospira mediosalina comb. nov. Int J Syst Bacteriol. 1998;48:793–798. doi: 10.1099/00207713-48-3-793. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis BDW, Van Berkum P, Chen WX, et al. Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int J Syst Bacteriol. 1997;47:895–898. [Google Scholar]
- 22.Kämpfer P. The Family Streptomycetaceae, Part I: Taxonomy. In: Dworkin M, editor. The Prokaryotes. New York, NY: Springer; 2006. pp. 538–604. [Google Scholar]
- 23.Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J Bacteriol. 2003;185:1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 25.Keis S, Bennett CF, Ward VK, Jones DT. Taxonomy and phylogeny of industrial solvent-producing Clostridia. Int J Syst Bacteriol. 1995;45:693–705. doi: 10.1099/00207713-45-4-693. [DOI] [PubMed] [Google Scholar]
- 26.Klaassens ES, de Vos WM, Vaughan EE. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl Environ Microbiol. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacerda CMR, Choe LH, Reardon KF. Metaproteomic analysis of a bacterial community response to cadmium exposure. J Proteome Res. 2007;6:1145–1152. doi: 10.1021/pr060477v. [DOI] [PubMed] [Google Scholar]
- 29.Lee SF, Li YH, Bowden GH. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect Immun. 1996;64:1035–1038. doi: 10.1128/iai.64.3.1035-1038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leshem O, Klein EA, Mager DL, Deschler DG, Spiegel JH, Dewhirst FE. The diversity of bacteria on oral squamous cell carcinomas. J Dent Res. 2005;84:2611. Special Issue A: Abstract. [Google Scholar]
- 31.Liu B, Lague JR, Nunes DP, et al. Expression of membrane-associated mucins MUC1 and MUC4 in major human salivary glands. J Histochem Cytochem. 2002;50:811–820. doi: 10.1177/002215540205000607. [DOI] [PubMed] [Google Scholar]
- 32.Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maron P-A, Ranjard L, Mougel C, Lemanceau P. Metaproteomics: A new approach for studying functional microbial ecology. Microbial Ecol. 2007;53:486–493. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- 34.Martínez A, Delgado O, Breccia J, Baigorí M, Siñeriz F. Revision of the taxonomic position of the xylanolytic Bacillus sp. MIR32 reidentified as Bacillus halodurans and plasmid-mediated transformation of B. halodurans. Extremophiles. 2002;6:391–395. doi: 10.1007/s00792-002-0269-4. [DOI] [PubMed] [Google Scholar]
- 35.Mccoy RE, Basham HG, Tully JG, Rose DL, Carle P, Bove JM. Acholeplasma florum, a new species isolated from plants. Int J Syst Bacteriol. 1984;34:11–15. [Google Scholar]
- 36.McDonald I, Doronina N, Trotsenko Y, McAnulla C, Murrell J. Hyphomicrobium chloromethanicum sp. nov. and Methylobacterium chloromethanicum sp. nov., chloromethane-utilizing bacteria isolated from a polluted environment. Int J Syst Evol Microbiol. 2001;51:119–122. doi: 10.1099/00207713-51-1-119. [DOI] [PubMed] [Google Scholar]
- 37.Mitrakul K, Loo CY, Hughes CV, Ganeshkumar N. Role of a Streptococcus gordonii copper-transport operon, copYAZ, in biofilm detachment. Oral Microbiol Immunol. 2004;19:395–402. doi: 10.1111/j.1399-302x.2004.00176.x. [DOI] [PubMed] [Google Scholar]
- 38.Oren A. The Order Halobacteriales. In: Dworkin M, editor. The Prokaryotes. New York, NY: Springer; 2006. pp. 113–164. [Google Scholar]
- 39.Park C, Helm RF. Application of metaproteomic analysis for studying extracellular polymeric substances (EPS) in activated sludge flocs and their fate in sludge digestion. Water Sci Tech. 2008;57:2009–2015. doi: 10.2166/wst.2008.620. [DOI] [PubMed] [Google Scholar]
- 40.Pierre-Alain M, Christophe M, Séverine S, Houria A, Philippe L, Lionel R. Protein extraction and fingerprinting optimization of bacterial communities in natural environment. Microbial Ecol. 2007;53:426–434. doi: 10.1007/s00248-006-9121-1. [DOI] [PubMed] [Google Scholar]
- 41.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of Pseudorabies Virus: Impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ram RJ, VerBerkmoes NC, Thelen MP, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- 43.Reznick AZ, Klein I, Eiserich JP, Cross CE, Nagler RM. Inhibition of oral peroxidase activity by cigarette smoke: in vivo and in vitro studies. Free Radic Biol Med. 2003;34:377–384. doi: 10.1016/s0891-5849(02)01297-2. [DOI] [PubMed] [Google Scholar]
- 44.Rollet C, Gal L, Guzzo J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;290:135–142. doi: 10.1111/j.1574-6968.2008.01415.x. [DOI] [PubMed] [Google Scholar]
- 45.Rondon MR, August PR, Bettermann AD, et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudney JD, Chen R, Sedgewick GJ. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect Immun. 2001;69:2700–2707. doi: 10.1128/IAI.69.4.2700-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- 48.Rudney JD, Chen R, Zhang G. Streptococci predominate in a diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- 49.Schluter A, Bekel T, Diaz NN, et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol. 2008;136:77–90. doi: 10.1016/j.jbiotec.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 51.Sundquist A, Bigdeli S, Jalili R, et al. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol. 2007;7:108. doi: 10.1186/1471-2180-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam K, Kinsinger N, Ayala P, Qi F, Shi W, Myung NV. Real-time monitoring of Streptococcus mutans biofilm formation using a quartz crystal microbalance. Caries Res. 2007;41:474–483. doi: 10.1159/000108321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanner ACR, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 54.Tatusov R, Fedorova N, Jackson J, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformat. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Todar K. 2008 Todar's online textbook of bacteriology: http://www.textbookofbacteriology.net.
- 56.Van Der Hoeven JS, Van Den Kieboom CW, Camp PJ. Utilization of mucin by oral Streptococcus species. Antonie Van Leeuwenhoek. 1990;57:165–172. doi: 10.1007/BF00403951. [DOI] [PubMed] [Google Scholar]
- 57.Van Der Hoeven JS, Camp PJM. Synergistic degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures. J Dent Res. 1991;70:1041–1044. doi: 10.1177/00220345910700070401. [DOI] [PubMed] [Google Scholar]
- 58.Van Der Hoeven JS, Camp PJ. The use of lectins in monitoring degradation of oligosaccharide chains in mucin by oral streptococci. Caries Res. 1994;28:257–261. doi: 10.1159/000261979. [DOI] [PubMed] [Google Scholar]
- 59.Whitman W, Bowen T, Boone D. The methanogenic bacteria. In: Dworkin M, editor. The Prokaryotes. New York, NY: Springer; 2006. pp. 165–207. [Google Scholar]
- 60.Wilmes P, Bond PL. Towards exposure of elusive metabolic mixed-culture processes: the application of metaproteomic analyses to activated sludge. Water Sci Tech. 2006;54:217–226. doi: 10.2166/wst.2006.390. [DOI] [PubMed] [Google Scholar]
- 61.Wilmes P, Bond PL. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Wilmes P, Wexler M, Bond PL. Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS ONE. 2008;3:e1778. doi: 10.1371/journal.pone.0001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie H, Onsongo G, Popko J, et al. Proteomic analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteom. 2008;7:486–498. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Ymele-Leki P, Ross JM. Erosion from Staphylococcus aureus biofilms grown under physiologically relevant fluid shear forces yields bacterial cells with reduced avidity to collagen. Appl Environ Microbiol. 2007;73:1834–1841. doi: 10.1128/AEM.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]