Abstract
Combinatorial readout of multiple covalent histone modifications is poorly understood. We provide insights into how an activating histone mark, in combination with linked repressive marks, is differentially ‘read’ by two related human demethylases, PHF8 and KIAA1718 (also known as JHDM1D). Both enzymes harbor a plant homeodomain (PHD) that binds Lys4-trimethylated histone 3 (H3K4me3) and a jumonji domain that demethylates either H3K9me2 or H3K27me2. The presence of H3K4me3 on the same peptide as H3K9me2 makes the doubly methylated peptide a markedly better substrate of PHF8, whereas the presence of H3K4me3 has the opposite effect, diminishing the H3K9me2 demethylase activity of KIAA1718 without adversely affecting its H3K27me2 activity. The difference in substrate specificity between the two is explained by PHF8 adopting a bent conformation, allowing each of its domains to engage its respective target, whereas KIAA1718 adopts an extended conformation, which prevents its access to H3K9me2 by its jumonji domain when its PHD engages H3K4me3.
The control of gene expression in eukaryotes relies in part on the methylation status of histone proteins. Histone lysine modification is a dynamic process established by specific methyltransferases, including SET-domain proteins1 and a conventional methyltransferase–like protein Dot1 (ref. 2). These methylation marks can be ‘erased’ by protein lysine demethylases that include flavin-dependent monoamine oxidase LSD1 (ref. 3) and α-ketoglutarate-Fe2+–dependent dioxygenases containing jumonji domains4–6. Protein modules, such as PHD finger protein (PHFs), detect the methylation status of histones by recognizing lysine in methylated7–10 and unmethylated states11,12. The jumonji domain often associates with at least one additional recognizable protein domain within the same polypeptide13. For example, JMJD2A contains an N-terminal jumonji domain and C-terminal PHD and tudor domains. The JMJD2A jumonji domain alone is capable of demethylating tri- (H3K9me3) and dimethylated histone H3 Lys9 (H3K9me2) and tri- (H3K36me3) and dimethylated Lys36 (H3K36me2), though it does so with a very low turnover rate (kcat < 1 h−1 and kcat/KM ~ 0.01 h−1 µmol−1) (ref. 14).
Structural studies revealed that the JMJD2A jumonji domain predominantly recognizes the backbone of the histone peptides (unusually for a sequence-specific enzyme), allowing the enzyme to demethylate both H3K9me3 and H3K9me2 as well as H3K36me3 and H3K36me2 (refs. 14–16). Meanwhile, the JMJD2A tudor domain binds two different histone sequences (H3K4me3 and H4K20me3) via radically different approaches17,18. The functional connection between the methyl mark reader and eraser in JMJD2A is not clear. Here, we ask whether regions outside of the catalytic core domain influence the specificity and/or efficiency of the catalytic domain, as in the case of DNA methyltransferase 1 (refs. 19,20), and we explore this possibility in the context of PHF8 and KIAA1718.
PHF8 and KIAA1718 belong to a small family of jumonji proteins with three members in mice and humans (PHF2, PHF8 and KIAA1718) (ref. 13). Each of these proteins harbors two domains in its respective N-terminal half (Fig. 1a): a PHD domain that binds H3K4me3 and a jumonji domain that demethylates H3K9me2, H3K27me2 and H3K36me2 (ref. 21) (see below). The two proteins share 82% identity (87% similarity) between their PHDs and 64% identify (75% similarity) between their jumonji domains (Supplementary Fig. 1). Mutations in the jumonji domain of human PHF8—either deletions or a missense mutation (F279S)—cause inherited X-linked mental retardation21–24. Relative to PHF8, KIAA1718 has a 30-residue alaninerich addition followed by a stretch of 6 prolines in the N terminus, and a 3-residue insertion in the nonconserved linker between PHD and jumonji domains (Supplementary Fig. 1). The sequence and length of the linker of KIAA1718 are highly conserved among the orthologs from different species, whereas the corresponding linker of PHF8 varies in sequence (it contains more glycines) and length (it is often shorter, between 11 and 15 residues) (Supplementary Fig. 2). Here, we focus on residues 1–488 from KIAA1718 and residues 1–447 from PHF8, comparing their demethylase activities on dimethylated lysine at H3K9 (H3K9me2) or H3K27 (H3K27me2) and analyzing the effect of trimethylation of H3K4 (H3K4me3) on demethylation activity.
Figure 1.
PHF8 PHD domain binding of H3K4me3 enhances its jumonji domain– mediated demethylation of H3K9me2. (a) Schematic representation of PHF8. (b) Effect of H3K4me3 on the demethylation of H3K9me2 by PHF8. Top panels show progression of demethylation as a function of reaction time. Supplementary Figure 11a shows representative mass spectra at various time points. Bottom panels show kinetics of PHF8 on two peptide substrates, with calculated kinetic parameters. (c) ITC measurement of binding of PHF8 to doubly methylated H31–24K4me3-K9me2 peptides, carried out under the conditions of 11 µM protein concentration and 0.2 mM peptide concentration in 100 mM NaCl and 50 mM HEPES, pH 7.0. (d) The inhibitory effect of adding an equimolar ratio of H31–12K4me3 (top) or H31–21K4me3 peptides (bottom) on the demethylation of H31–24K9me2 by PHF8. (e) The PHD (blue) and jumonji (green) collaborate in binding the H3 peptide (magenta) containing H3K4me3 and H3K9me2. Omit electron densities, Fo – Fc (black mesh), contoured at 4σ above the mean, are shown for the trimethlyated H3K4me3 and dimethlyated H3K9me2, respectively. (f) The surface representation of PHF8, colored with blue (PHD), green (jumonji) and magenta (H3 peptide). (g) H3K4me3 binding in the cage, surrounded on four sides by Tyr14, Met20 and Trp29 of PHD (blue) and Ser354 of jumonji (green). The carbonyl oxygen of Ser354 is in van der Waals contact with one of the methyl groups. Tyr7 (in thin lines) covers the top of the cage. (h) H3K9me2 binds in the active site.
RESULTS
H3K4me3 binding by the PHF8 PHD enhances demethylation
PHF8, containing both the PHD and jumonji domains (Fig. 1a), has slow activity on H31–24 K9me2, with t1/2 (the time required for 50% demethylation under the assay conditions) of ~1 h (Fig. 1b, top left). We probed the demethylation activity of PHF8 on a histone H3 peptide modified with both H3K4me3 and H3K9me2. The presence of H3K4me3 enhanced PHF8 activity on H3K9me2 by 12-fold, with t1/2 ~ 5 min (Fig. 1b, top right). PHF8 has lower KM by a factor of 16 and higher kcat by a factor of 4 on the doubly methylated peptide than that of H3K9me2 peptide (Fig. 1b, lower panels). In addition, PHF8 binds the doubly methylated peptide with a KD of approximately 1 µM (Fig. 1c), whereas it has negligible binding on H31–24K9me2 under the conditions tested (data not shown). In contrast, when present in trans, the H3K4me3 modification either in a short H31–12 or a long H31–21 peptide inhibits the PHF8 activity on H31–24K9me2 (Fig. 1d).
To understand how H3K4me3 binding enhances demethylase activity, we determined the cocrystal structure of PHF81–447 with an H31–24 peptide containing H3K4me3 and H3K9me2 (Fig. 1e) in the presence of Fe2+ and N-oxalylglycine (the cofactor analog) to form a catalytically inert complex (Table 1). The structure, determined at a resolution of 2.2 Å, shows that the PHD and jumonji domains act in concert in substrate recognition. The first 11 residues of the H3 peptide were nearly buried in a deep cleft between the PHD and the jumonji domains with a disordered linker (residues 66–78) (Fig. 1e). The H3 N terminus was buried in the PHD-jumonji interface, and Gly12-Gly13-Lys14 emerged from an opening on the surface of the jumonji domain (Fig. 1f); the remainder of the peptide (residues 15–24) was unstructured.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Data collection | PHF8 (residues 1–447) | KIAA1718 (residues 1–488) | KIAA1718 (residues 92–488) | |||
|---|---|---|---|---|---|---|
| Cofactors | Zn2+, Fe2+, N-oxalylglycine |
Zn2+, Fe2+, N-oxalylglycine |
Zn2+, Fe2+, α-ketoglutarate |
Fe2+ | Fe2+, α-ketoglutarate |
Fe2+, N-oxalylglycine |
| Peptide | H3(1–24)K4me3-K9me2 | |||||
| Space group | P43212 | P212121 | P6122 | |||
| Cell dimensions | (α= β= γ= 90°) | (α= β= γ= 90°) | (α= β= 90°, γ= 120°) | |||
| a, b, c (Å) | 73.5, 73.5, 210.8 | 62.7, 125.6, 206.1 | 63.4, 125.2, 206.0 | 77.9, 77.9, 289.7 | 78.1, 78.1, 290.8 | 78.3, 78.3, 289.1 |
| Resolution (Å) | 34.72–2.19 (2.27–2.19)a |
34.82–2.39 (2.48–2.39) |
34.54–2.89 (2.99–2.89) |
34.32–2.29 (2.37–2.29) |
34.40–2.79 (2.89–2.79) |
33.67–2.69 (2.79–2.69) |
| Rmerge | 0.035 (0.647) | 0.055 (0.637) | 0.119 (0.557) | 0.089 (0.793) | 0.088 (0.381) | 0.085 (0.394) |
| I/σI | 7.2 (2.3) | 9.7 (1.7) | 16.5 (4.0) | 21.9 (4.2) | 15.5 (4.1) | 13.3 (2.6) |
| Completeness (%) | 100.0 (99.9) | 94.8 (81.3) | 97.6 (95.6) | 99.8 (100) | 99.5 (99.3) | 98.3 (86.3) |
| Redundancy | 6.2 (5.7) | 8.0 (4.2) | 7.8 (7.4) | 11.1 (11.9) | 7.0 (7.3) | 14.4 (8.0) |
| Refinement | ||||||
| Resolution (Å) | 2.19 | 2.39 | 2.89 | 2.29 | 2.79 | 2.69 |
| No. reflections | 28,629 | 57,397 | 35,289 | 23,158 | 12,680 | 14,477 |
| Rwork/Rfree | 0.217/0.255 | 0.216/0.245 | 0.215/0.253 | 0.222/0.258 | 0.203/0.269 | 0.216/0.267 |
| No. atoms | ||||||
| Protein | 3,564 | 7,084 | 7,145 | 2,922 | 2,925 | 2,931 |
| Heterogen | 56 | 17 | 30 | 4 | 14 | 14 |
| Water | 206 | 329 | 166 | 111 | 50 | 38 |
| B-factors (Å2) | ||||||
| Protein | 25.7 | 41.0 | 32.7 | 42.9 | 47.3 | 53.9 |
| H3 peptide | 22.1 | |||||
| Cofactor | 32.0 | 75.5 | 82.2 | 57.0 | 69.3 | 92.6 |
| Water | 29.8 | 37.5 | 22.6 | 47.1 | 40.7 | 41.0 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.008 | 0.009 | 0.007 | 0.006 |
| Bond angles (°) | 1.2 | 1.3 | 1.3 | 1.4 | 1.3 | 1.3 |
Values in parentheses are for highest-resolution shell.
The limited solvent exposure of bound histone peptide implies that the substrate did not diffuse into a preformed binding site. The surface area buried at the PHF8-peptide interface is approximately 1,550 Å2 (with ~30% contribution from the PHD and ~70% from the jumonji domain). The H3 peptide shows an extended conformation from Ala1 to Lys9, then makes a sharp, nearly 90° turn at Lys9 (Fig. 1e and Supplementary Fig. 3a). Besides main chain interactions, all 11 side chains of H3 (Ala1–Thr11) are involved in interactions, either with both domains (Ala1, Arg2, Lys4me3 (K4me3), Thr6; Supplementary Fig. 3b–e), PHD alone (Thr3) or jumonji alone (Gln5 and Ala7–Thr11; Supplementary Fig. 3f–i). Intramolecular interactions occur between Thr3 and Gln5 (Supplementary Fig. 3f) and Arg8 and Ala7 (Supplementary Fig. 3g). This is in sharp contrast to the structures of JMJD2A jumonji domains in complex with substrate peptides that have little side chain contact14–16.
Substrate specificity is contributed by both domains, as exemplified by the interactions of H3K4me3 with PHD (through the hydrophobic cage) and jumonji (through a van der Waals contact with the main chain carbonyl oxygen of Ser354) (Fig. 1g). The target K9me2 lies in the active site right next to the Fe2+ and N-oxalylglycine (Fig. 1h). One of its terminal N-CH3 groups projects toward the aromatic ring of Tyr234, and the other methyl group points toward Asp249 and Asn333, forming two hydrogen bonds of C-H–O type (Fig. 1h)—a type of hydrogen bond that occurs in the active sites of both SET and jumonji domains14,25. The dimethylated terminal nitrogen atom carrying the lone pair of electrons forms a hydrogen bond with one of the oxygen atoms of N-oxalylglycine (Fig. 1h). The active site cannot accommodate a trimethylated lysine because the third methyl group would cause repulsive tension with N-oxalylglycine. Phe279, substitution of which (to serine) causes inherited X-linked metal retardation22–24, makes van der Waals contacts with Ile248 and Ile318 (Supplementary Fig. 3j), forming a hydrophobic core supporting the backbone of Fe2+-coordinating residues His247, Asp249, and His319.
H3K4me3 binding by KIAA1718 PHD inhibits its jumonji activity
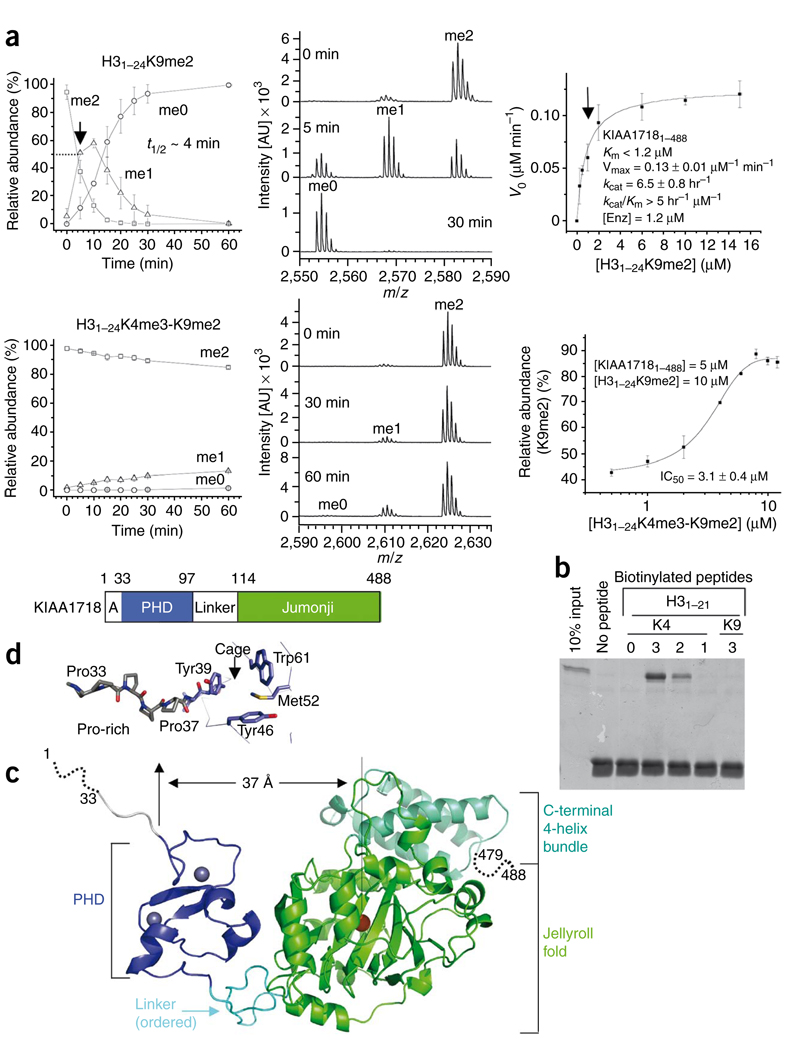
Unlike PHF8, KIAA1718 (Fig. 2) is highly active on H31–24K9me2, with t1/2 ~ 4 min and kcat = 6.5 h−1 (Fig. 2a, top panels). Unexpectedly, the presence of both H3K4me3 and H3K9me2 on the same peptide, H31–24K4me3-K9me2, has exactly the opposite effect on KIAA1718 compared to PHF8: it nearly abolishes the demethylation activity of KIAA1718 on K9me2 (Fig. 2a, lower left). Apparently, binding of the PHD to H3K4me3 (Fig. 2b) prevents demethylation of K9me2 by the linked jumonji domain, which on its own is active on H3K9me2 with kcat = 6 h−1 and kcat/KM = 0.5 h−1 µmol−1 (Supplementary Fig. 4e). The doubly methylated peptide inhibits KIAA1718 activity on H31–24K9me2 with a half-maximal inhibitory concentration (IC50) of 3.1 µM (Fig. 2a, lower right) under MS assay conditions (Supplementary Fig. 5).
Figure 2.
KIAA1718 PHD binding of H3K4me3 inhibits its jumonji domain activity targeting H3K9me2. (a) Effect of H3K4me3 on the demethylation of H3K9me2 by KIAA1718. Left panels show progression of demethylation as a function of reaction time. Middle panels show representative mass spectra at various time points. Right top panel shows kinetics of KIAA1718 on substrate H31–24K9me2. KM is estimated to be less than 1.2 µM (indicated by an arrow), which is the amount of enzyme used to generate sufficient fluorescence signal. Right bottom shows the IC50 value of H31–24K4me3-K9me2 peptide [I] on H31–24K9me2 [S] demethylase activity of KIAA1718 [E]. The relative abundance of the substrate [S] was measured after eight minutes incubation at 37 °C by adding varying amounts of [I] to the reaction mixture. No demethylation of the doubly methylated peptide occurred. (b) Peptide pulldown assays with peptides H31–21 that were unmodified or mono-, di- or trimethylated (1, 2 or 3) at H3K4 or H3K9 using a GST-tagged PHD domain of KIAA1718. (c) KIAA1718 contains four segments: a disordered alanine-rich sequence followed by a stretch of prolines, a PHD domain (blue) containing two zinc metals (gray balls), a rigid linker (cyan) and a jumonji domain (green) followed by a four-helix bundle. (d) The KIAA1718 PHD domain contains a surface hydrophobic cage, a presumptive site for binding of H3K4me3. In the crystal lattice, the cage is blocked by the N-terminal prolines of a crystallographic symmetry-related molecule (Supplementary Fig. 6c).
The crystal structure of the apo-form KIAA1718, determined at the resolution of 2.4 Å in the presence of N-oxalylglycine (Table 1), suggests a mechanism for this inhibition. Residues 32–479 of KIAA1718 (Fig. 2c) have continuous electron density through the main chain. The disordered residues include the C-terminal tail (residues 480–488) and the N-terminal tail (residues 1–31), the latter of which is absent in the KIAA1718 orthologs of zebrafish and Xenopus laevis (Supplementary Fig. 2) and whose deletion has no impact on activity (Supplementary Fig. 6). The six-proline stretch (Pro32–Pro37) displays an extended conformation (Fig. 2d). An identical surface hydrophobic cage to that of PHF8 is formed by two tyrosines, one tryptophan and one methionine (Fig. 2d). The cage is likely where H3K4me3 would bind, as the PHD alone binds tri- and dimethylated H3K4 peptide (Fig. 2b). Unlike PHF8, the linker between PHD and jumonji (residues 98–113) seems ordered, as indicated by the continuity of electron density and similarity of crystallographic thermal factors for all three regions (25 Å2 for PHD, 30 Å2 for the linker and 35 Å2 for jumonji).
The distance between the PHD cage (the H3K4me3 binding site) and the jumonji Fe2+-binding site, immediately adjacent to the presumed target methyl-lysine binding site, is approximately 37 Å (Fig. 2c). This is more than twice the linear distance of ~16 Å between H3K4 and H3K9 in an extended form of the substrate. Isothermal titration calorimetric (ITC) analysis shows that KIAA1718 binds the doubly methylated peptide, H31–24K4me3-K9me2, with KD ~ 0.3 µM; removing the three methyl groups at H3K4 increased KD to ~60 µM, a reduction by a factor of 200 in binding affinity (Fig. 3a). Therefore, the dominating interaction between KIAA1718 and the doubly methylated peptide is the binding of H3K4me3 by the PHD. This interaction would place H3K9me2 too far from the active site of the linked jumonji domain in this extended conformation.
Figure 3.
Effect of the linker on the KIAA1718 jumonji activity targeting H3K9me2. (a) ITC measurements of binding of the KIAA1718 to doubly methylated H31–24K4me3-K9me2 peptides (left) and H31–24K9me2 peptides (right). The measurements were carried out under the conditions of 18 µM protein concentration and 0.4 mM peptide concentration in 50 mM NaCl and 50 mM HEPES, pH 7.0. (b) Superimposition of PHF8 (colored) and KIAA1718 (gray) in their respective jumonji domains. (c) The engineered hybrid enzyme of KIAA1718 carrying the PHF8 linker gains substantial activity on H31–24K3me3-K9me2 (right), faster by a factor of more than 100 than that of the wild-type enzyme (see Fig. 2a).
The individual domain structures of KIAA1718 and PHF8 are very similar, with an r.m.s. deviation of less than 0.65 Å when comparing the 58 pairs of PHD Cα atoms and less than 0.89 Å when comparing the 361 pairs of jumonji Cα atoms. Superimposition of the two enzymes by their jumonji domains reveals that the PHF8 PHD adopts a bent conformation toward the jumonji domain in the presence of substrate binding, whereas the PHD and jumonji domains of KIAA1718 adopt an extended conformation in its apo structure with an ordered linker (Fig. 3b). To test the idea that the linkers are critical determinants of demethylase activity, we engineered two hybrid enzymes: KIAA1718 carrying the PHF8 linker and PHF8 carrying the KIAA1718 linker (Fig. 3c and Supplementary Fig. 7a). Notably, the hybrid enzymes lost their parental features and become indistinguishable; both were equally active on doubly methylated H31–24K4me3-K9me2 and had a less than two-fold difference of activity on H31–24K9me2 (Supplementary Fig. 7b). Compared to the wild-type enzymes, the most noteworthy change was in the substantial gain of activity by the KIAA1718-PHF8 linker hybrid on H31–24K4me3-K9me2 with t1/2 = 9 min (Fig. 3c, right graph), in stark contrast to the barely detectable activity of native KIAA1718 on the same peptide (see Fig. 2a, lower left). We thus conclude that the PHF8 linker in the hybrid enzyme enables the PHD and jumonji domains to adopt a closed conformation just like the native PHF8, whereas the native KIAA1718 locks the two domains in an extended conformation.
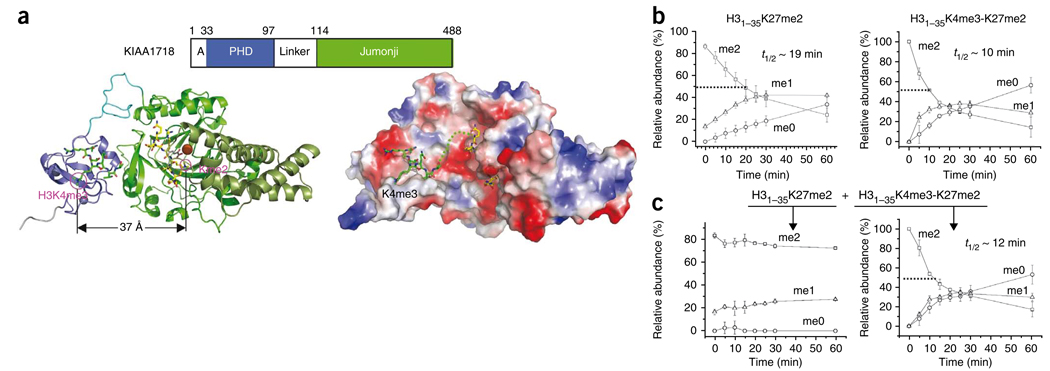
KIAA1718 demethylates H3K27me2 in the presence of H3K4me3
We modeled a H3K4me3 peptide onto KIAA1718-PHD and a H3K9me2 peptide onto KIAA1718-jumonji by superimposing the portions of H3 peptide bound to the PHF8-PHD and PHF8-jumonji domains, respectively (Fig. 4a). The adoption of an extended conformation by KIAA1718 suggests that a peptide carrying H3K4me3 and H3K27me2 (which shares an ARKS tetrapeptide sequence with that of H3K9) could occupy both the PHD and jumonji domains simultaneously and could be demethylated by KIAA1718 in this extended conformation. Indeed, the presence of trimethylated H3K4 enhanced KIAA1718 activity on H3K27me2 by a factor of about 2 (t1/2 = 10 min versus 19 min; Fig. 4b), in contrast to the severe inhibition of H3K9me2 demethylation (see Fig. 2a, lower left). Moreover, in a reaction mixture containing the two substrates in equimolar ratio, KIAA1718 selectively demethylated the peptide containing both H3K4me3 and H3K27me2 (Fig. 4c). This is consistent with the notion that PHD preferentially bound H3K4me3 first and subsequently allowed only H3K27me2 on the same peptide to be demethylated by the jumonji domain.
Figure 4.
KIAA1718 selectively demethylates H3K27me2 in the presence of H3K4me3 in cis. (a) A model of KIAA1718 PHD on methylated H3K4 and its linked jumonji active site on a target lysine (left). Surface representation displayed as blue for positive, red for negative and white for neutral (right). The dashed line connects H3K4me3 bound in the aromatic cage and the target lysine in the jumonji domain. (b) The presence of H3K4 methylation in cis enhances KIAA1718 demethylase activities on H3K27me2. (c) When two peptide substrates were mixed in equimolar ratio, H31–35K27me2 (left) and H31–35K4me3-K27me2 (right), KIAA1718 selectively demethylated H31–35 peptides containing both H3K4me3 and H3K27me2 (right).
DISCUSSION
Here we show, enzymatically and structurally, that the PHDs of PHF8 and KIAA1718 bind H3K4me3—a modification associated with transcriptional activation, as their linked jumonji domains remove methyl marks from H3K9 (PHF8) and H3K27 (KIAA1718) that are associated with transcriptional repression. These opposing marks (one for activation and the other for repression) do sometimes coexist, particularly colocalization of H3K27me to genes that are H3K4me modified26–29. We hypothesize that, in vivo, PHF8 and KIAA1718 are recruited to H3K4me3 marks via their PHDs, and that subsequently the jumonji domain amplifies this activating signal by removing any local repressing methyl groups (Supplementary Fig. 8a). In PHF8 the shorter and perhaps more flexible linker between the PHD and jumonji domains enables the active site of jumonji to reach the target H3K9me2 while the PHD binds to H3K4me3. The binding of substrate together by both domains makes the H3K4me3-H3K9me2 a better substrate for PHF8 (higher kcat/KM). In contrast, the corresponding ordered linker in KIAA1718 renders the enzyme inactive on H3K9me2 when H3K4me3 is present, in the process becoming more selective toward H3K27me2 (Supplementary Fig. 8b). We conclude that the structural linkage between the PHD binding to H3K4me3 and the placement of the catalytic jumonji domains relative to this ‘ON’, or active, epigenetic landmark determines which repressive marks are removed in both demethylases.
It remains to be tested whether KIAA1718 also becomes more active or inhibited on H3K36me2 (ref. 21) when H3K4me3 is present on the same peptide (Supplementary Fig. 9). It is also interesting to note that PHF81–447 is nearly inactive on H3K27me2 and H3K36me2 under the conditions tested (Supplementary Fig. 10a). However, a truncated protein with the PHD deleted, PHF862–447, has weak activity on H3K27me2 and H3K36me2 (Supplementary Fig. 10b), suggesting that the linked PHD (probably in the closed conformation) can interfere with substrate access to jumonji domain. Taking these results together, we suggest that the PHF8 and KIAA1718 jumonji domains on their own are promiscuous enzymes; it is the associated PHDs and the linker—a determinant for the relative positioning of the two domains—that are mainly responsible for substrate specificity.
The successful generation of hybrid enzymes may lead to a way to engineer enzymes with novel specificities by fusing an epigenetic reader (for highly specific binding of methyl-lysine or acetyl-lysine or other modifications) and a promiscuous enzymatic jumonji domain via a variable linker. Such enzymes could be useful tools in vitro and in vivo for the study of combinations of histone modifications.
The existence of combinatorial readout of multiple covalent histone modifications is an explicit prediction of the ‘histone code hypothesis’ 30–33. Binding of substrate using two or more domains in concert to enhance an enzyme’s activity and its substrate specificity may be a general mechanism for jumonji-containing protein lysine demethylases. For example, JHDM2A-mediated demethylation of histone H3K9me1 or H3K9me2 requires a zinc finger located N-terminally to the jumonji domain for JHDM2A’s enzymatic activity5. JARID jumonji family proteins contain a jumonji domain that demethylates H3K4me3 surrounded by several PHDs, and at least one of them binds H3K9me3 (refs. 34,35) (Supplementary Fig. 8c). Mutation or deletion of this PHD impairs the demethylase activity on H3K4me3 (refs. 34,35). Several histone-methylating enzymes contain components (domains) to both synthesize and bind a specific histone mark, for example mammalian G9a/GLP (for H3K9me1 and H3K9me2) (ref. 36) and Schizosaccharomyces pombe Clr4 (for H3K9me3) (ref. 37). They contain modules within the same polypeptide for both making (via the SET domain) and recognizing (via the ankyrin repeats or chromodomain) a given methyl mark; a mechanism of cross-talk propagates a given methyl mark. PHF8 and KIAA1718 contain modules within the same polypeptide for both recognizing (via the PHD) and removing (via the jumonji domain) two opposing methyl marks—a mechanism of cross-talk removes an ‘OFF’ methyl mark based on an existing ‘ON’ methyl mark. Understanding the function and cross-talk of individual ‘letters’ (one methyl mark, two methyl marks, and so on) will eventually allow researchers to uncover the complex language of the histone code30,38.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Ruiz and R. Gridds for technical assistance, R.M. Blumenthal for critical comments, A.S. Bhagwat and T.W. Roy (Wayne State University) for Escherichia coli strain BH249 overexpressing formaldehyde dehydrogenase and T. Kutateladze (University of Colorado) and A. Mattevi and C. Binda (University of Pavia) for H3K4me3-containing peptides. The Department of Biochemistry at the Emory University School of Medicine supported the use of the SER-CAT synchrotron beamline at the Advanced Photon Source of Argonne National Laboratory, local X-ray facility and MALDI-TOF mass spectrometry. This work was supported by grants GM06860 and DK082678 to X.C. and GM058012 and NCI118487 to Y.S. from the US National Institutes of Health. Y.S. is a cofounder of Constellation Pharmaceutical. X.C. is a Georgia Research Alliance Eminent Scholar.
Footnotes
Accession codes. Protein Data Bank: The coordinates and structure factors have been deposited with accession numbers 3KV4 (for PHF81–447), 3KV5 and 3KV6 (for KIAA17181–488) and 3KV9, 3KVA and 3KVB (for KIAA171892–488), respectively.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
J.R.H. performed crystallographic experiments; A.K.U. performed kinetic experiments; H.H.Q. and Y.S. provided initial expression constructs and the knowledge of specificities of individual PHD and jumonji domains; X.Z. generated hybrid enzymes; X.C. organized and designed the scope of the study and wrote the manuscript, and all others helped in analyzing data and revising the manuscript.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 2.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 5.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 6.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Wysocka J, et al. PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 8.Li H, et al. Molecular basis for site-specific readout of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 14.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 15.Ng SS, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. USA. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 20.Pradhan M, et al. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 21.Loenarz C, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nε-dimethyl lysine demethylase. Hum. Mol. Genet. 2009 October 19; doi: 10.1093/hmg/ddp480. Published online, doi:10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koivisto AM, et al. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 24.Laumonnier F, et al. Mutations in PHF8 are associated with X-linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J. Biol. Chem. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- 26.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 28.Zhao XD, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 32.Turner BM. Defining an epigenetic code. Nat. Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 33.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 34.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Li F, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 38.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.