Abstract
We report here genome sequences and comparative analyses of three closely related parasitoid wasps: Nasonia vitripennis, N. giraulti, and N. longicornis. Parasitoids are important regulators of arthropod populations, including major agricultural pests and disease vectors, and Nasonia is an emerging genetic model, particularly for evolutionary and developmental genetics. Key findings include the identification of a functional DNA methylation tool kit; hymenopteran-specific genes including diverse venoms; lateral gene transfers among Pox viruses, Wolbachia, and Nasonia; and the rapid evolution of genes involved in nuclear-mitochondrial interactions that are implicated in speciation. Newly developed genome resources advance Nasonia for genetic research, accelerate mapping and cloning of quantitative trait loci, and will ultimately provide tools and knowledge for further increasing the utility of parasitoids as pest insect-control agents.
Parasitoid wasps are insects whose larvae parasitize various life stages of other arthropods (for example, insects, ticks, and mites). Female wasps sting, inject venom, and lay eggs on or in the host, where the developing offspring consume and eventually kill it. Parasitoids are widely used in the biological control of insect pests, and they are very diverse, with estimates of over 600,000 species (1, 2). Nasonia is the second genus of Hymenoptera to have whole-genome sequencing, after Apis mellifera (Fig. 1), and Nasonia comprises four closely related parasitoid species: N. vitripennis, N. giraulti, N. longicornis, and N. oneida (3, 4). Nasonia are genetically tractable organisms with short generation time (~2 weeks), large family size, ease of laboratory rearing, and cross-fertile species. Like other hymenopterans, haploid males develop from unfertilized eggs, and diploid females develop from fertilized eggs. Cross-fertile species facilitate the mapping and cloning of genes that are involved in species differences. Haploid genetics assist efficient genotyping, mutational screening (5), and evaluation of gene interactions (epistasis) without the added complexity of genetic dominance. As a result, Nasonia are now emerging as genetic model organisms, particularly for complex trait analysis, developmental genetics, and evolutionary genetics (4).
Fig. 1.
Phylogenetic relationships of Nasonia and the DNA methylation tool kit. (A) Nasonia relationships to other sequenced genomes (6). Right: DNA methyltransferase subfamilies (Dnmt1, Dnmt2, Dnmt3) in these taxa. (B) Relationships among the three sequenced Nasonia genomes. (C) Crossing scheme used for mapping scaffolds on the Nasonia chromosomes and for studies of nuclear-cytoplasmic incompatibility.
We sequenced, assembled, annotated, and analyzed the genome of N. vitripennis from sixfold Sanger sequence genome coverage by using a highly inbred line of N. vitripennis (6). The draft genome assembly comprises 26,605 contigs [total length of 239.8 Mb, with half of the bases residing in contigs larger than 18.5 kb (N50), 40.6% guanine plus cytosine content (GC)]. Contigs were placed with mate-pair information into 6,181 scaffolds (total size 295 Mb, N50 =709 kb). We assessed the N. vitripennis assembly for completeness and accuracy by comparing it with 19 finished bacterial artificial chromosome (BAC) sequences and 18,000 expressed sequence tags (ESTs). The genome assembly contained 98% of the BAC and 97% of the EST sequences, with an error rate of 5.9×10−4. Thus the assembly is a high-quality representation of both genomic and transcribed N. vitripennis sequences.
Highly inbred lines of the two sibling species N. giraulti and N. longicornis (Fig. 1B) were sequenced with onefold Sanger and 12-fold, 45–base pair (bp) Illumina genome coverage. Assembled by alignment to the N. vitripennis reference using stringent criteria (6), these reads cover 62% and 62.6% of the N. vitripennis assembly, and 84.7% and 86.3% of protein coding regions, respectively. These were used for genome comparisons and provided resources [for example, single nucleotide polymorphisms (SNPs) and microsatellites] for scaffold, gene, and quantitative trait loci (QTL) mapping. Sequence error rates for the N. giraulti alignment are estimated to be 3.8×10−3 for the entire alignment and 1.47×10−4 for coding sequences on the basis of comparison to three finished N. giraulti BACs (6). Sequences of 25 coding genes in both species perfectly matched their respective aligned sequences.
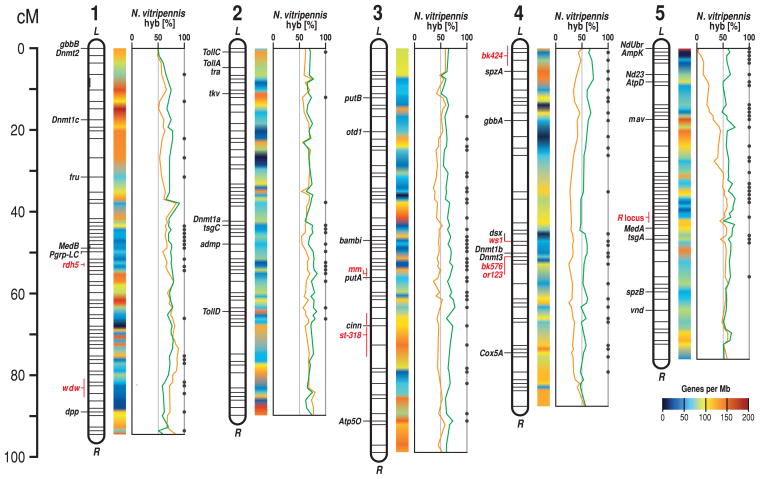
Normally, the intracellular bacteria Wolbachia prevent the formation of interspecies hybrids; however, antibiotically cured strains are cross-fertile (7). Hybrid crosses (Fig. 1C) (6) were used to map scaffolds and visible mutations onto the five chromosomes of Nasonia (Fig. 2). Several interspecies QTL have already been mapped using genetic/genomic resources, including wing size (8, 9), host preference (10), female mate preference (11), and in this study, sex-ratio control and male courtship (6). Linkage analysis has revealed that the genome-wide recombination rate in Nasonia is 1.4 to 1.5 centimorgans (cM)/Mb, which is lower than that of honeybees (12, 13), and shows a 100-fold difference in rate between high- and low-recombination regions of the genome (Fig. 2) (6).
Fig. 2.
A high-resolution recombination map of the five Nasonia chromosomes is shown (6), with estimated gene density and locations of visible markers, landmark genes, and QTL. The hybridization percentage to N. vitripennis alleles is shown among surviving adult N. vitripennis × N. giraulti F2 hybrid males with either N. vitripennis (green curve) or N. giraulti (orange curve) mitochondria. Dots specify genome regions with significant differences in the hybridization ratio between the reciprocal crosses (P < 0.01).
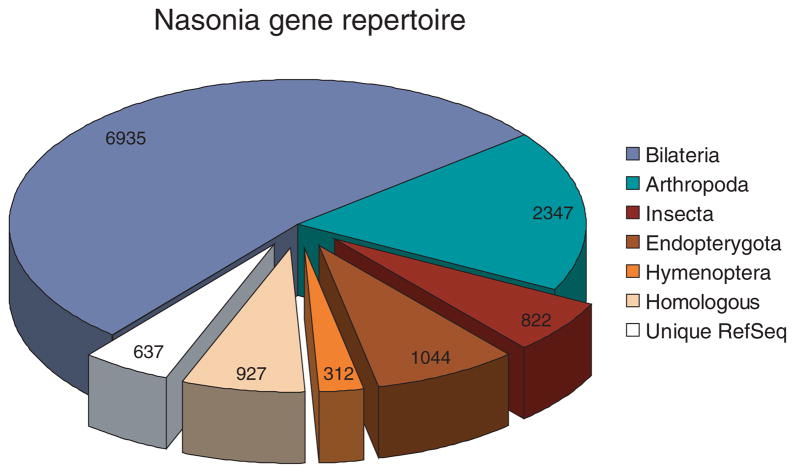
An official gene set (OGS v1.1) was generated from comparisons to A. mellifera, Tribolium castaneum, Drosophila melanogaster, Pediculus humanus, Daphnia pulex, and Homo sapiens [details are given in (6)]. Overall, Nasonia encodes a typical insect gene repertoire (Fig. 3) (6), of which 60% of genes have a human ortholog, 18% are arthropod-specific, and 2.4% appear to be hymenoptera-specific, showing high conservation between Nasonia and Apis and low conservation or absence in other taxa. An additional 12% are either Nasonia-specific or without clear orthology. Many (63%) single-copy orthologs shared between Nasonia and Apis occur in microsynteny blocks, which is similar to the amount of microsynteny blocks in Aedes aegypti/Anopheles gambiae and H. sapiens/Gallus gallus (14). Four hundred and forty-five orthologs between Nasonia and humans lack a candidate homolog in D. melanogaster (table S1), including the human transcription factors E2F7 and E2F8, which are involved in cell-cycle regulation. Further refinement of the gene set resulted in OGS v1.2 (15), which totals 17,279 genes, of which 74% have tiling micro-array or EST support (6).
Fig. 3.
Distribution of recognizable Nasonia orthologs and Nasonia-specific genes among gene models with expression sequencing support (6).
Nasonia is abundant in transposable elements (TEs) and other repetitive DNA (table S2 and fig. S1). This contrasts with a paucity of TEs in A. mellifera (16). TE diversity in Nasonia is 30% higher (2.9 TE types/Mb) than the next most diverse insect (Bombyx mori, 2.1 TE types/Mb), and is 10-fold higher than the average dipteran (6, 17). Nasonia also contains an unusual abundance of nuclear-mitochondrial insertions and a higher density of microsatellites (10.9 kb/Mb) than most other arthropod species (18, 19), suggesting that the accumulation of repetitive DNA is a feature of these insects.
The Nasonia genome encodes a full DNA methylation tool kit, including all three DNA cytosine-5-methyltransferase (Dnmt) types (Fig. 1A). In vertebrates, Dnmt3 establishes DNA methylation patterns, Dnmt1 maintains these patterns, and Dnmt2 is involved in tRNA methylation (20). The Nasonia genome encodes three Dnmt1 genes, one Dnmt2, and one Dnmt3, in contrast with D. melanogaster, which has only Dnmt2. The presence of all three subfamilies in both Nasonia and Apis (Fig. 1) raises the question of whether methylation has similar regulatory functions in Hymenoptera as it does in vertebrates. DNA methylation is important in Apis caste development (21) and is suggested for Nasonia sex determination (22). Coding exons of both Nasonia and Apis show bimodal distributions in observed/expected CpG (fig. S2) (6, 23), which is consistent with mutational biases due to DNA methylation of hyper- and hypomethylated genes. We confirmed methylated CpG dinucleotides in five examined N. vitripennis genes by bisulfite sequencing (fig. S3). These results suggest that epigenetic modifications by DNA methylation may be important in Hymenoptera. Nasonia also has the largest number of ankyrin (ANK) repeat–containing proteins (over 200) so far found in any insect (table S3) (6), suggesting a regulatory importance through protein-protein interactions (24).
Systemic RNA interference (RNAi) in Nasonia allows for gene expression knockdowns (4, 25). The Nasonia genome encodes homologs for the majority of genes implicated in small RNA processes (table S4). However, as in Tribolium and Apis, Nasonia lacks an RNA-dependent RNA polymerase (RdRp) ortholog, indicating a different systemic RNAi mechanism than in Caenorhadbitis. Using various computational approaches (6), we identified 52 putative micro RNAs (miRNAs) with homologies to known miRNAs (26), nine that were previously unknown, and 11 additional Hymenoptera-specific miRNAs (table S5). Small-RNA library sequencing confirmed 39 predicted and identified 59 additional miRNAs (table S6).
Nasonia shares a long germ-band mode of embryonic development with Drosophila, but exhibits significant differences in the genetic mechanisms involved (5, 27, 28) (see fig. S4). All major components of the dorso-ventral patterning system are present, with many Nasonia-specific gene duplications in the Toll pathway. Orthologs of vertebrate genes absent from Drosophila include the transforming growth factor–β (TGFβ) ligands ADMP and myostatin, and the bone morphogenesis protein (BMP) inhibitors BAMBI and DAN, but their functions in Nasonia are not yet known. A. mellifera shows an expansion of the yellow/major royal jelly (yellow/MRJP) genes that are linked to caste formation and sociality (29). Nasonia has the largest number of yellow/MRJP genes so far found in any insect, including an independent amplification of MRJP-like proteins (fig. S5) (6, 29). Although their function in Nasonia is unknown, these genes are expressed broadly in different tissues and life stages (table S7). The insect sex peptide/receptor system, which causes female re-mating refractoriness (30), is highly conserved in insects but is absent in Nasonia and Apis (table S8) (6). Instead, Nasonia males inhibit female re-mating behaviorally with a special “post-copulatory display” (31). Additional features analyzed (6) include those related to sex determination (fig. S6), pathogens and immunity (fig. S7), neuro-peptides (tables S9 and S10), cuticular proteins (table S11), xenobiotics (fig. S8), and diapause (table S12).
We investigated genome microevolution, including rapidly evolving genes that are potentially involved in species differences and speciation, by using the genomes of the three closely related Nasonia species. Synonymous divergence between N. vitripennis and its sibling species N. giraulti and N. longicornis is 0.031 ± 0.0002 SE and 0.030 ± 0.0002 SE, respectively, and between N. giraulti and N. longicornis is 0.014 ± 0.0001 SE (6), which is comparable to those among Drosophila sibling species (32). We compared the ratio of synonymous-to-nonsynonymous substitutions (dN/dS) between Nasonia species pairs with respect to gene ontology (GO) term categories, using genes with high-quality alignments and 1:1 orthologs between Nasonia and Drosophila. Nuclear genes that interact with mitochondria revealed significantly elevated dN/dS [by comparison of dN/dS distributions for each GO term to resampled distributions, see (6) and table S13], specifically those encoding mitochondrial ribosomes (P < 0.003 for all species pairs) and oxidative phosphorylation complex I (P < 0.03 for N. vitripennis/N. giraulti and N. vitripennis/N. longicornis) and complex V (P < 0.04 for all species pairs). This finding is consistent with the rapid evolutionary rate of Nasonia mitochondria (33) and studies implicating nuclear-mitochondrial incompatibilities in F2 hybrid breakdown (7, 31). For example, reciprocal crosses between N. giraulti × N. vitripennis have identical F1 nuclear genotypes, but their mitochondrial haplotypes differ. Yet, micro-array hybridization (Fig. 2) (6) of DNA from pooled surviving adult F2 haploid males shows distortion in the recovery of particular regions of the genome, which is dependent upon their mitochondrial haplotype (giraulti versus vitripennis). Because hybrid mortality is post-embryonic (7) and embryo ratios are Mendelian (33), these distortions reflect larval to adult mortality. In particular, F2 males with N. vitripennis alleles on the left arm of chromosome 5 and N. giraulti mitochondria suffer nearly 100% mortality (Fig. 2). This region contains three genes encoding mitochondrial interacting proteins, atpD, ampK, and nadh-ubiquinone oxireductase (Fig. 2). Coevolution of nuclear and mitochondrial genomes can accelerate evolution (34, 35), and these findings indicate that such interactions contribute to reproductive incompatibility and speciation in Nasonia.
Sequences of 25 gene regions from multiple strains for the three Nasonia species (6) show low levels of intraspecific variation (table S14) with synonymous site variation ranging from 0.0005 in N. giraulti to 0.0026 in N. vitripennis, which are much lower than in Drosophila species and more akin to levels observed in humans (36). This low nuclear variation could be explained by founder events, purging of deleterious mutations in haploid males, or inbreeding.
Recent lateral gene transfers from the bacterial endosymbiont Wolbachia into the genomes of Nasonia and other arthropods have been identified (37). Detecting ancient lateral transfers is more problematic. By examining protein domain arrangements in Nasonia relative to other organisms, we uncovered an ancient lateral gene transfer involving Pox viruses, Wolbachia, and Nasonia. Thirteen ANK repeat–bearing proteins encoded in the N. vitripennis genome also contain C-terminal PRANC (Pox proteins repeats of ankyrin–C terminal) domains. This domain was previously only described in Pox viruses, where it is associated with ANK repeats and inhibits the nuclear factor κB (NF-κB) pathway in mammalian hosts (38). A computational screen revealed ANK-PRANC–bearing genes in some Wolbachia and a related Rickettsiales (Fig. 4). Screening additional Wolbachia confirmed the presence of ANK-PRANC genes in diverse Wolbachia. The Nasonia PRANC genes are clearly integrated in the genome (6) and are expressed in different life stages (table S15). Phylogenetic analysis of the PRANC-domain sequences suggests that the Nasonia lineage acquired one or more of these proteins from Wolbachia, with subsequent amplification and divergence (Fig. 4). Such lateral gene transfers between bacteria and animals could be an important source of evolutionary innovation (37).
Fig. 4.
PRANC domain proteins in Nasonia, Pox viruses, and Wolbachia. (A) Maximum-likelihood tree of PRANC-domain sequences found in Pox viruses, rickettsia (Wolbachia and Orientia), and parasitoids (N. vitripennis and Cotesia congregata). The tree was estimated using RaxML with 1000 bootstrap replicates and model settings estimated by ProtTest [see (6); alignment deposited in Tree-base with ID SN4709]. Bootstrap values above 50% are shown by the corresponding nodes. The phylogenetic relationships suggest lateral transfer from Wolbachia to the Nasonia lineage. (B) Representative domain arrangements for ANK-PRANC proteins.
Nasonia is a carnivore, feeding on an amino acid–rich diet both as larva and adult (4). Mapping of Nasonia genes onto metabolic pathways (39) revealed loss or rearrangement in some amino acid metabolic pathways, including tryptophan and aminosugar metabolism (fig. S9) (6). The changes may reflect its specialized carnivorous diet and can inform efforts to produce artificial diets for more economical parasitoid rearing.
The venom of parasitoids, injected into a host before oviposition, serves to condition the host for successful development of wasp progeny (1, 2). Unlike the defensive Apis venom that inflicts pain and damage, parasitoid venoms have diverse physiological effects on hosts, including developmental arrest; alteration in growth and physiology; suppression of immune responses; induction of paralysis, oncosis, or apoptosis; and alteration of host behavior (40). The identification of Nasonia genes with venom features and proteomic analyses of venom reservoir tissues have uncovered a rich assemblage of 79 candidate venom proteins (table S16) (41). Some Nasonia venom reservoir proteins belong to previously known insect venom families such as serine proteases; however, nearly half were not related to any known insect venoms. As expected, many of these venom candidates show highly elevated expression in the female reproductive tract, which includes the venom glands and reservoirs. Venom genes also showed significantly higher dN/dS ratios between N. vitripennis and N. giraulti than nonvenom genes did (Mann-Whitney U test, P < 2 × 10−6), suggesting that changes in host use between the species may be accompanied by rapid evolution of venom proteins. The large venom protein set found in Nasonia with diverse physiological effects (40) and abundance of parasitoid species (1, 2) suggests that parasitoids may contain a rich venom pharmacopeia of potential new drugs.
N. vitripennis is a generalist parasitoid with a wide host utilization of many fly species, whereas the other Nasonia species are specialists (4, 10). Using genomic tools, a major host preference locus has been mapped to a region of ~2 cM (10). Other genes in the Nasonia genome that are potentially involved in host finding include odorant binding proteins (table S17) and chemoreceptors (42), which show expansions, contractions, and pseudogenization, indicative of rapid turnover.
A suite of genetic tools and resources is available or under development for the Nasonia system (4, 6, 11, 28), and the genome resources presented here can be used for fine-scale mapping (6, 9–11) and positional cloning (8) of QTLs. By combining haploid genetics, ease of rearing, short generation time, systemic RNAi, interfertile species, and new genome resources for three species, Nasonia shows promise as a genetic model system for evolutionary and developmental genetics. Genome resources described here and our resulting enhanced understanding of parasitoid biology will also open avenues for improving parasitoid utility in biological control of pests of agricultural and medical importance.
Supplementary Material
Author List: The Nasonia Genome Working Group
John H. Werren,1*† Stephen Richards,2*† Christopher A. Desjardins,1 Oliver Niehuis,3‡ Jürgen Gadau,3 John K. Colbourne,4 Leo W. Beukeboom,5 Claude Desplan,6 Christine G. Elsik,7 Cornelis J. P. Grimmelikhuijzen,8 Paul Kitts,9 Jeremy A. Lynch,10 Terence Murphy,9 Deodoro C. S. G. Oliveira,1§ Christopher D. Smith,11,12 Louis van de Zande,5 Kim C. Worley,2 Evgeny M. Zdobnov,13,14,15 Maarten Aerts,16 Stefan Albert,17 Victor H. Anaya,18 Juan M. Anzola,19 Angel R. Barchuk,20 Susanta K. Behura,21 Agata N. Bera,22 May R. Berenbaum,23 Rinaldo C. Bertossa,24 Márcia M. G. Bitondi,25 Seth R. Bordenstein,26,27 Peer Bork,28 Erich Bornberg-Bauer,29 Marleen Brunain,30 Giuseppe Cazzamali,8 Lesley Chaboub,2 Joseph Chacko,2 Dean Chavez,2 Christopher P. Childers,7 Jeong-Hyeon Choi,4 Michael E. Clark,1 Charles Claudianos,31 Rochelle A. Clinton,32 Andrew G. Cree,2 Alexandre S. Cristino,31,33 Phat M. Dang,34 Alistair C. Darby,35 Dirk C. de Graaf,30 Bart Devreese,16 Huyen H. Dinh,2 Rachel Edwards,1 Navin Elango,36 Eran Elhaik,37 Olga Ermolaeva,9 Jay D. Evans,38 Sylvain Foret,39 Gerald R. Fowler,2 Daniel Gerlach,13,14 Joshua D. Gibson,3 Donald G. Gilbert,40 Dan Graur,37 Stefan Gründer,41 Darren E. Hagen,7 Yi Han,2 Frank Hauser,8 Da Hultmark,42 Henry C. Hunter IV,11 Gregory D. D. Hurst,35 Shalini N. Jhangian,2 Huaiyang Jiang,2 Reed M. Johnson,43 Andrew K. Jones,22 Thomas Junier,13 Tatsuhiko Kadowaki,44 Albert Kamping,5 Yuri Kapustin,9 Bobak Kechavarzi,45 Jaebum Kim,46 Jay Kim,11 Boris Kiryutin,9 Tosca Koevoets,5 Christie L. Kovar,2 Evgenia V. Kriventseva,47 Robert Kucharski,48 Heewook Lee,45 Sandra L. Lee,2 Kristin Lees,22 Lora R. Lewis,2 David W. Loehlin,1 John M. Logsdon Jr.,49 Jacqueline A. Lopez,4 Ryan J. Lozado,2 Donna Maglott,9 Ryszard Maleszka,48 Anoop Mayampurath,45 Danielle J. Mazur,49 Marcella A. McClure,32 Andrew D. Moore,29 Margaret B. Morgan,2 Jean Muller,28 Monica C. Munoz-Torres,7,50 Donna M. Muzny,2 Lynne V. Nazareth,2 Susanne Neupert,51 Ngoc B. Nguyen,2 Francis M. F. Nunes,25,52 John G. Oakeshott,53 Geoffrey O. Okwuonu,2 Bart A. Pannebakker,5,54 Vikas R. Pejaver,45 Zuogang Peng,36 Stephen C. Pratt,3 Reinhard Predel,51 Ling-Ling Pu,2 Hilary Ranson,55 Rhitoban Raychoudhury,1 Andreas Rechtsteiner,4,56 Justin T. Reese,7,57 Jeffrey G. Reid,2 Megan Riddle,58|| Hugh M. Robertson,23 Jeanne Romero-Severson,59 Miriam Rosenberg,6 Timothy B. Sackton,60 David B. Sattelle,22 Helge Schlüns,61 Thomas Schmitt,62 Martina Schneider,8 Andreas Schüler,29 Andrew M. Schurko,49 David M. Shuker,63 Zilá L. P. Simões,25 Saurabh Sinha,46 Zachary Smith,4 Victor Solovyev,64 Alexandre Souvorov,9 Andreas Springauf,41 Elisabeth Stafflinger,8 Deborah E. Stage,1 Mario Stanke,65 Yoshiaki Tanaka,66 Arndt Telschow,29 Carol Trent,58 Selina Vattathil,2¶ Eveline C. Verhulst,5 Lumi Viljakainen,67 Kevin W. Wanner,68 Robert M. Waterhouse,15 James B. Whitfield,23 Timothy E. Wilkes,35 Michael Williamson,8 Judith H. Willis,69 Florian Wolschin,70,3 Stefan Wyder,13 Takuji Yamada,28 Soojin V. Yi,36 Courtney N. Zecher,27 Lan Zhang,2 Richard A. Gibbs2
1Department of Biology, University of Rochester, Rochester, NY 14627, USA. 2Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA. 3School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA. 4The Center for Genomics and Bioinformatics, Indiana University, Bloomington, IN 47405, USA. 5Evolutionary Genetics–Centre for Ecological and Evolutionary Studies, University of Groningen, 9750 AA Haren, Netherlands. 6Department of Biology, New York University, New York, NY 10003, USA. 7Department of Biology, Georgetown University, Washington, DC 20057, USA. 8Center for Comparative and Functional Insect Genomics, Department of Biology, University of Copenhagen, DK-2100 Copenhagen, Denmark. 9National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA. 10Institut für Entwicklungsbiologie, Universität zu Köln, 50923 Köln, Germany. 11Department of Biology, San Francisco State University, San Francisco, CA 94132, USA. 12Drosophila Heterochromatin Genome Project, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA. 13Department of Genetic Medicine and Development, University of Geneva Medical School, CH-1211 Geneva, Switzerland. 14Swiss Institute of Bioinformatics, CH-1211 Geneva, Switzerland. 15Imperial College London, London, SW7 2AZ, UK. 16Laboratory of Protein Biochemistry and Biomolecular Engineering, Ghent University, B-9000 Ghent, Belgium. 17BEEgroup and Institute of Pharmaceutical Biology, University of Würzburg, 97082 Würzburg, Germany. 18Institute for Theoretical Biology, Humboldt University Berlin, 10115 Berlin, Germany. 19Animal Science and Biology, Texas A&M University, College Station, TX 77843, USA. 20Departamento de Ciências Biomédicas, Universidade Federal de Alfenas, Alfenas, Minas Gerais, 37130-000, Brazil. 21Eck Institute for Global Health, Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA. 22Medical Research Council Functional Genomics Unit, Department of Physiology Anatomy and Genetics, University of Oxford, Oxford OX1 3QX, UK. 23Department of Entomology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. 24Chronobiology–Centre for Behavior and Neurosciences, University of Groningen, 9750 AA Haren, Netherlands. 25Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Departamento de Biologia, Universidade de São Paulo, Ribeirão Preto, São Paolo 14040-901, Brazil. 26Department of Biological Sciences, Vanderbilt University, Nashville, TN 37235, USA. 27Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, MA 02536, USA. 28European Molecular Biology Laboratory, 69117 Heidelberg, Germany. 29Institute for Evolution and Biodiversity, University of Münster, 48143 Münster, Germany. 30Laboratory of Zoophysiology, Ghent University, B-9000 Ghent, Belgium. 31The Queensland Brain Institute, The University of Queensland, Brisbane, Queensland 4072, Australia. 32Department of Microbiology and the Center for Computational Biology, Montana State University, Bozeman, MT 59715, USA. 33Instituto de Física de São Carlos, Departamento de Física e Informática, Universidade de São Paulo, São Carlos, São Paolo 13560-970, Brazil. 34Subtropical Insects Research Unit, United States Department of Agriculture–Agricultural Research Service (USDA-ARS), U.S. Horticultural Research Lab, Fort Pierce, FL 34945, USA. 35School of Biological Sciences, University of Liverpool, Liverpool L69 7ZB, UK. 36School of Biology, Georgia Institute of Technology, Atlanta, GA 30332, USA. 37Department of Biology and Biochemistry, University of Houston, Houston, TX 77204, USA. 38Bee Research Lab, USDA-ARS, Beltsville, MD, 20705, USA. 39Australian Research Council Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, Queensland 4811, Australia. 40Department of Biology, Indiana University, Bloomington, IN 47405, USA. 41Institute of Physiology, Rheinisch-Westfaelische Technische Hochschule (RWTH) Aachen University, D-52074 Aachen, Germany. 42Department of Molecular Biology, Umeå University, S-901 87 Umeå, Sweden. 43Department of Entomology, University of Nebraska, Lincoln, NE 68583, USA. 44Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya 464-8601, Japan. 45School of Informatics, Indiana University, Bloomington, IN 47405, USA. 46Department of Computer Science, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. 47Department of Structural Biology and Bioinformatics, University of Geneva Medical School, CH-1211 Geneva, Switzerland. 48Research School of Biology, Australian National University, Canberra, Australian Capital Territory 2601, Australia. 49Roy J. Carver Center for Comparative Genomics and Department of Biology, University of Iowa, Iowa City, IA 52242, USA. 50Department of Genetics and Biochemistry, Clemson University, Clemson, SC 29634, USA. 51Institute of General Zoology, University of Jena, D-7743 Jena, Germany. 52Faculdade de Medicina de Ribeirão Preto, Departamento de Genética, Universidade de São Paulo, Ribeirão Preto, São Paolo 14049-900, Brazil. 53Department of Entomology, Commonwealth Scientific and Industrial Research Organisation, Canberra, Australian Capital Territory 2601, Australia. 54Institute of Evolutionary Biology–School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JT, UK. 55Vector Group, Liverpool School of Tropical Medicine, Liverpool L3 5QA, UK. 56Department of Molecular, Cell, and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA 95064, USA. 57Reese Consulting, 157/10 Tambon Ban Deau, Amphur Muang, Nong Khai, 43000, Thailand. 58Department of Biology, Western Washington University, Bellingham, WA 98225, USA. 59Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA. 60Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA. 61School of Marine and Tropical Biology and Centre for Comparative Genomics, James Cook University, Townsville, Queensland 4811, Australia. 62Department of Evolutionary Biology and Animal Ecology, University of Freiburg, 79104 Freiburg, Germany. 63School of Biology, University of St Andrews, St Andrews KY16 9TH, UK. 64Department of Computer Science, Royal Holloway, University of London, Egham, Surrey TW20 0EX, UK. 65Institut für Mikrobiologie und Genetik, Universität Göttingen, 37077 Göttingen, Germany. 66Division of Insect Sciences, National Institute of Agrobiological Science, Tsukuba, Ibaraki 305-8634, Japan. 67Department of Biology and Biocenter Oulu, University of Oulu, 90014 Oulu, Finland. 68Department of Plant Sciences and Plant Pathology, Montana State University, Bozeman, MT 59717, USA. 69Department of Cellular Biology, University of Georgia, Athens, GA 30602, USA. 70Department of Biotechnology, Chemistry, and Food Science, Norwegian University of Life Sciences, N-1432 Ås, Norway.
*These authors contributed equally to this work.
†To whom correspondence should be addressed. E-mail: werr@-mail.rochester.edu (J.H.W.); stephenr@bcm.tmc.edu (S.R.)
‡Current address: Verhaltensbiologie, Universität Osnabrück, 49076 Osnabrück, Germany.
§Current address: Departament de Genètica i de Microbiologia, Universitat Autònoma de Barcelona, 8193 Bellaterra, Spain.
||Current address: Weill Cornell Medical College, New York, NY 10065, USA.
¶Current address: Department of Epidemiology, University of Texas, M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/327/5963/343/DC1
Materials and Methods
SOM Text
References
References and Notes
- 1.Quicke DLJ. Parasitic Wasps. Chapman & Hall; London: 1997. [Google Scholar]
- 2.Heraty J. In: Insect Biodiversity: Science and Society. Tootti R, Alder P, editors. Wiley-Blackwell; Hoboken, NJ: 2009. pp. 445–462. [Google Scholar]
- 3.Raychoudhury R, et al. Heredity. 2010 doi: 10.1038/hdy.2009.147. [DOI] [Google Scholar]
- 4.Werren JH, Loehlin D. Cold Spring Harb Protocols. 2009 doi: 10.1101/pdb.emo134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pultz MA, et al. Genetics. 2000;154:1213. doi: 10.1093/genetics/154.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materials and methods and supplementary text are available as supporting material on Science Online.
- 7.Breeuwer JAJ, Werren JH. Evolution. 1995;49:705. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 8.Loehlin DW, et al. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loehlin DW, Enders LS, Werren JH. Heredity. 2010 doi: 10.1038/hdy.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desjardins CA, Perfecti F, Bartos JD, Enders LS, Werren JH. Heredity. 2010 doi: 10.1038/hdy.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velthuis BJ, Yang W, van Opijnen T, Werren JH. Anim Behav. 2005;69:1107. [Google Scholar]
- 12.Niehuis O, et al. PLoS ONE. 2010 doi: 10.1371/journal.pone.0008597. [DOI] [Google Scholar]
- 13.Wilfert L, Gadau J, Schmid-Hempel P. Heredity. 2007;98:189. doi: 10.1038/sj.hdy.6800950. [DOI] [PubMed] [Google Scholar]
- 14.Zdobnov EM, Bork P. Trends Genet. 2007;23:16. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 15.The official gene set OGS v1.2. available at http://nasoniabase.org/nasonia_genome_consortium/datasets.html.
- 16.Honeybee Genome Sequencing Consortium. Nature. 2006;443:931. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CD, et al. Gene. 2007;389:1. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viljakainen L, Oliveira DCSG, Werren JH, Behura SK. Insect Mol Biol. 2010;19:27. doi: 10.1111/j.1365-2583.2009.00932.x. [DOI] [PubMed] [Google Scholar]
- 19.Pannebakker BA, Niehuis O, Hedley A, Gadau J, Shuker DM. Insect Mol Biol. 2010;19:91. doi: 10.1111/j.1365-2583.2009.00915.x. [DOI] [PubMed] [Google Scholar]
- 20.Jurkowski TP, et al. RNA. 2008;14:1663. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucharski R, Maleszka J, Foret S, Maleszka R. Science. 2008;319:1827. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 22.Beukeboom LW, Kamping A, van de Zande L. Semin Cell Dev Biol. 2007;18:371. doi: 10.1016/j.semcdb.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Elango N, Hunt BG, Goodisman MAD, Yi SV. Proc Natl Acad Sci USA. 2009;106:11206. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. Protein Sci. 2004;13:1435. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch JA, Desplan C. Nat Protoc. 2006;1:486. doi: 10.1038/nprot.2006.70. [DOI] [PubMed] [Google Scholar]
- 26.Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM. Nucleic Acids Res. 2009;37:D111. doi: 10.1093/nar/gkn707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Nature. 2006;439:728. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- 28.Brent AE, Yucel G, Small S, Desplan C. Science. 2007;315:1841. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- 29.Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Genome Res. 2006;16:1385. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yapici N, Kim YJ, Ribeiro C, Dickson BJ. Nature. 2008;451:33. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- 31.Niehuis O, Judson AK, Gadau J. Genetics. 2008;178:413. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flowers JM, et al. Mol Biol Evol. 2007;24:1347. doi: 10.1093/molbev/msm057. [DOI] [PubMed] [Google Scholar]
- 33.van den Assem J, Visser J. Biol Comport. 1976;1:37. [Google Scholar]
- 34.Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. Mol Biol Evol. 2008;25:2167. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rand DM, Haney RA, Fry AJ. Trends Ecol Evol. 2004;19:645. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Aquadro CF, Dumont VB, Reed FA. Curr Opin Genet Dev. 2001;11:627. doi: 10.1016/s0959-437x(00)00245-8. [DOI] [PubMed] [Google Scholar]
- 37.Dunning-Hotopp JC, et al. Science. 2007;317:1753. [Google Scholar]
- 38.Chang SJ, et al. J Virol. 2009;83:4140. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, et al. Nucleic Acids Res. 2008;36:D480. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivers DB, Yoder YA, Ruggiero L. Trends Entomol. 1999;2:1. [Google Scholar]
- 41.de Graaf DC, et al. Insect Mol Biol. 2010;19:11. doi: 10.1111/j.1365-2583.2009.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson HM, Gadau J, Wanner KW. Insect Mol Biol. 2010;19:121. doi: 10.1111/j.1365-2583.2009.00979.x. [DOI] [PubMed] [Google Scholar]
- 43.Genome sequencing, assembly and annotation were funded by the National Human Genome Research Institute (NHGRI U54 HG003273). The whole-genome shotgun project has been deposited at the DNA Databank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank under accession numbers AAZX00000000 (N. vitripennis), ADAO00000000 (N. giraulti), and ADAP00000000 (N. longicornis). Additional support, acknowledgments, and accession numbers are provided in the supporting online material.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.