Abstract
Streptococcus gordonii is a primary colonizer, involved in the formation of dental plaque. This bacterium expresses several surface proteins. One of them is the adhesin SspB, which is a member of the Antigen I/II family of proteins. SspB is a large multi-domain protein that has interactions with surface molecules on other bacteria and on host cells, and is thus a key factor in the formation of biofilms. Here, we report the crystal structure of a truncated form of the SspB C-terminal domain, solved by single-wavelength anomalous dispersion to 1.5 Å resolution. The structure represents the first of a C-terminal domain from a streptococcal Antigen I/II protein and is comprised of two structurally related β-sandwich domains, C2 and C3, both with a Ca2+ ion bound in equivalent positions. In each of the domains, a covalent isopeptide bond is observed between a lysine and an asparagine, a feature that is believed to be a common stabilization mechanism in gram positive surface proteins. S. gordonii biofilms contain attachment sites for the periodontal pathogen Porphyromonas gingivalis and the SspB C-terminal domain has been shown to have one such recognition motif, the SspB adherence region. The motif protrudes from the protein, and serves as a handle for attachment. The structure suggests several additional putative binding surfaces, and other binding clefts may be created when the full-length protein is folded.
Keywords: Streptococcus gordonii, X-ray crystallography, Porphyromonas gingivalis, surface adhesin, isopeptide bond
Introduction
The oral biofilm, commonly called dental plaque, is a complex mixed-species biofilm that can be assembled from over 700 species of micro-organisms.1 Commensal bacteria, such as oral streptococci and Actinomyces spp., are gram-positive, primary colonizing bacteria that initiate the formation of the oral biofilms by adhering to the tooth surface and thereby provides sites for the adhesion of secondary colonizers, which are usually gram-negative anaerobes. Streptococcal species may constitute over 70% of the bacteria in the early biofilm,2 and interact with salivary proteins, bacteria and host cells. Streptococcus gordonii is an oral commensal, which colonizes multiple sites in the human oral cavity, including saliva-coated tooth surfaces where it provides an attachment substrate for secondary colonization by organisms such as Porphyromonas gingivalis.3 Co-adhesion between S. gordonii and P. gingivalis is believed to play an important role in the development of bacterial communities that are associated with the initiation and progression of severe forms of periodontal disease.4 S. gordonii also has pathogenic potential at non-oral sites, and is suspected to be an etiological agent in infective endocarditis.5
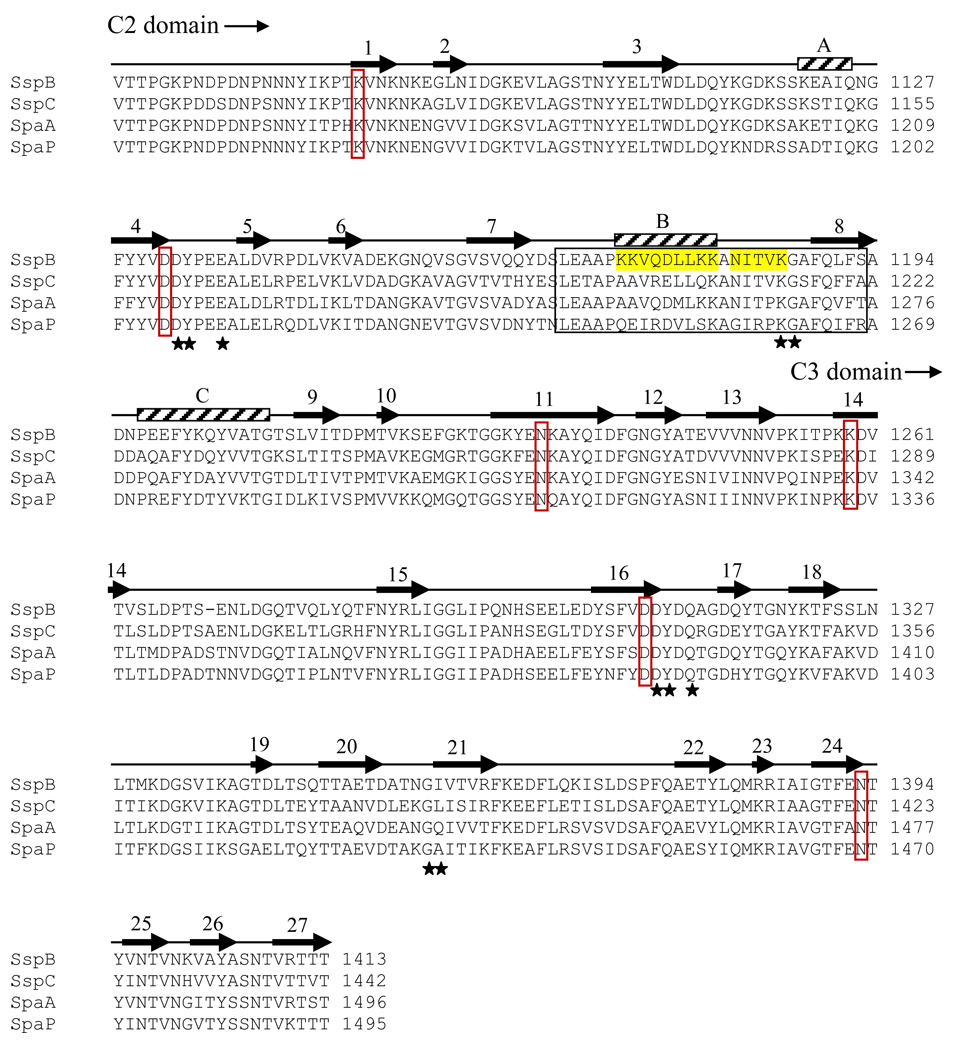
S. gordonii expresses several cell surface adhesins that it uses to interact with cells and proteins in its environment: CshA and CshB which bind fibronectin,6,7 and the serine rich adhesins Hsa (strain Challis (DL1)) and GspB (strain M99), that bind cell surface glycoproteins on leukocytes8 and platelets.9,10 S. gordonii also expresses two cell surface proteins, SspA and SspB, that belong to the antigen I/II (AgI/II) family of proteins, which are expressed by virtually all species of oral streptococci.11 Proteins within the AgI/II family are structurally well conserved. The precursors are between 1310 and 1653 amino acids long and contain seven distinct regions based on primary sequence. These include a signal (leader) peptide, an N-terminal region, an alanine-rich domain, a variable domain, a proline-rich domain, a C-terminal domain and a cell wall-anchored segment.11 Despite this structural similarity, different members of the protein family have different binding specificities. The cell wall-anchored AgI/II family protein SspB of S. gordonii mediates a wide range of interactions with host proteins and other bacteria.12–14 Brooks et al.15 showed that both SspB and SspA bind to the P. gingivalis short fimbrial protein Mfa1, whereas the related AgI/II family polypeptide SpaP from Streptococcus mutans does not, despite the high sequence similarity between the two (Fig. 1a). It has also been shown that the substrate-binding properties of S. gordonii SspA and SspB expressed on the surface of Lactococcus lactis differ in affinity for salivary agglutinin glycoprotein (SAG) and collagen substrates.14,16
Figure 1.
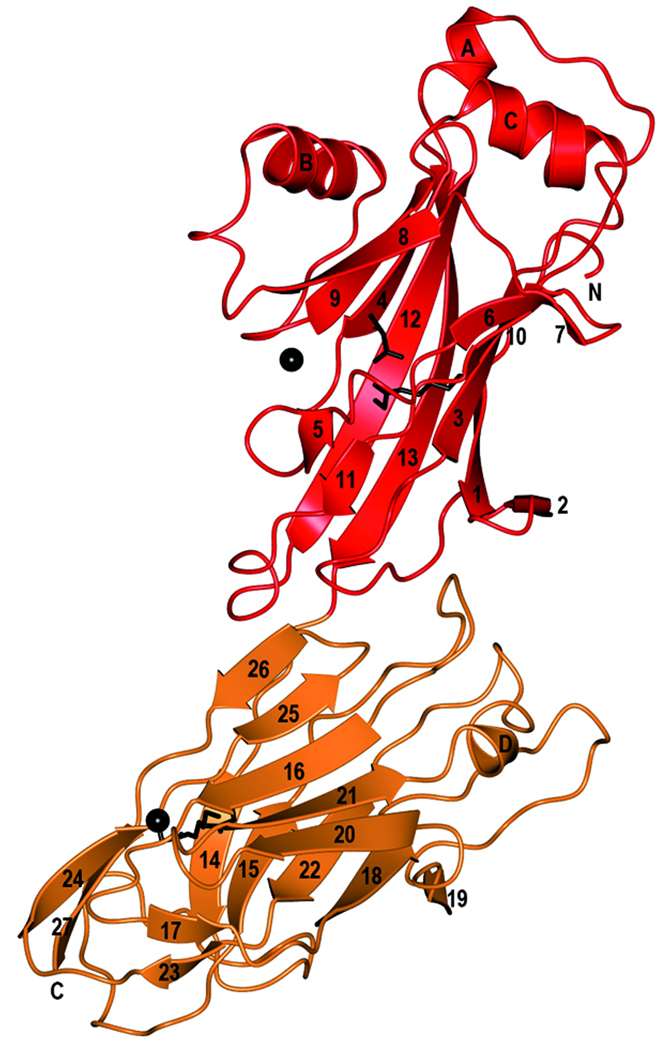
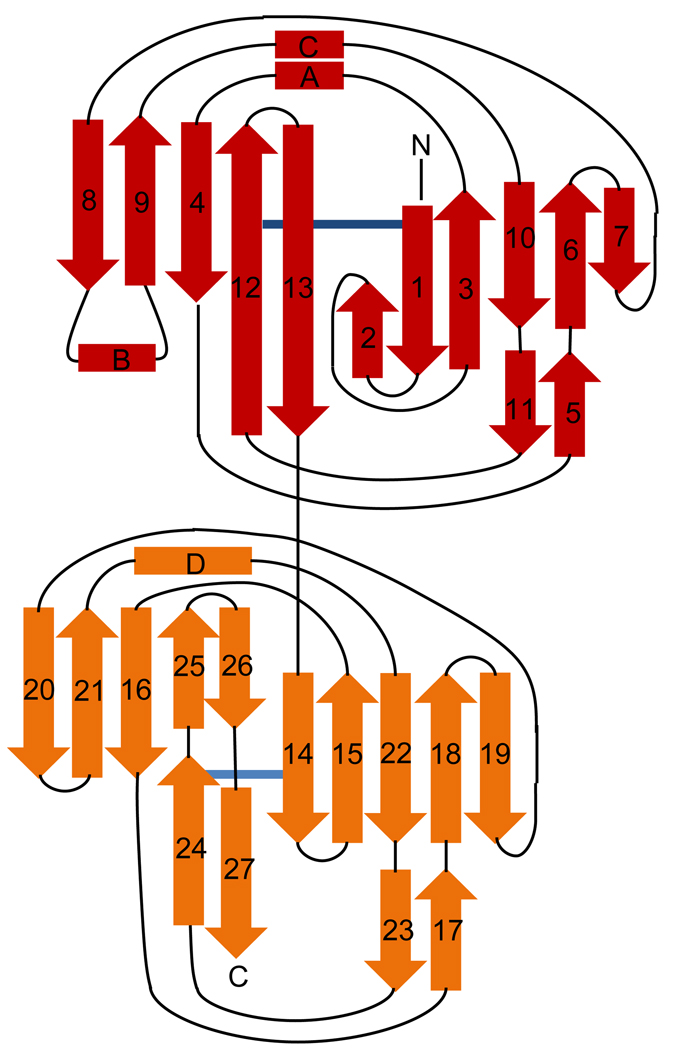
Sequence alignment and overall structure of SspB-C1061–1413. a) Multiple sequence alignment of SspB-C1061–1413 and related AgI/II C-terminal domains. Sequences from the following streptococcal strains were compared; S. gordonii M5 (SspB), S. sanguinis SK36 (SspC), S. sobrinus MUCOB 263 (SpaA) and S. mutans UA159 (SpaP). Secondary structure elements of S.gordonii SspB-C1061–1413 are indicated above the sequence. The metal coordinating residues are marked with filled stars and residues involved in the isopeptide bonds are indicated by red boxes. The BAR region is boxed and the KKVQDLLKK and NITVK motifs are highlighted in yellow. b) Ribbon representation of SspB-C1061–1413. The C2 domain (residues1061-1253) is depicted in red and the C3 domain (residues 1254–1413) in orange. Ca2+ ions are depicted as black spheres. The isopeptide bonds between Lys 1082 and Asn1232 and between Lys1259 and Asn1393 are represented as stick models in black. c) Topology diagram of SspB-C1061–1413. β-strands are represented as arrows, helices as rectangles and loops as lines. Colouring as in (b).
The SspB region that mediates adhesion to P. gingivalis comprises residues 1167–1193 (designated BAR for SspB Adherence Region) within the C-terminal domain of the protein.17,18 Mapping of the BAR region and its interaction with P. gingivalis using synthetic peptides has identified two structural motifs (NITVK and KKVQDLLKK) within BAR that are important for adherence of P. gingivalis to the oralis group of streptococci.15,17 The region spanning the two structural motifs found within BAR resembles that of the eukaryotic nuclear receptor box protein-protein interaction domain, suggesting that a charge-clamp interaction may be utilized for BAR-Mfa1 interaction.19
Dental plaque biofilms can exist in the oral cavity without causing any evident disease. However, populational shifts resulting in increased numbers of gram-negative anaerobes contribute to the onset of periodontal disease. P. gingivalis is considered a primary pathogen in adult periodontitis and is found mostly in the anaerobic environment of the subgingival crevice. Initial colonization of the oral cavity by P. gingivalis likely occurs in supragingival areas where established biofilm communities provide reduced oxygen tension and physiologic support.20,21 The SspB-Mfa1 interaction is essential for the development of P. gingivalis communities on streptococcal substrates.18 This initial colonization mechanism provides an attractive target for developing therapeutic agents that may interfere with its adherence to S. gordonii and other bacteria of the oralis group of streptococci and subsequently may impede P. gingivalis colonization of subgingival locations.
The three dimensional structure of the C-terminal domain of SspB provides valuable information regarding the Mfa1-interacting epitopes of the SspB protein and is a first step towards the design of potential agents to target P. gingivalis adherence and biofilm formation. In addition, structural information will also prove useful in the identification of potential epitopes that are important for interactions of SspB with other molecules.
Since there were no structures available for any AgI/II C-terminal domains, we undertook to determine the crystal structure of the S. gordonii SspB C-domain. Here, we report the 1.5 Å resolution crystal structure of a proteolytically stable truncated form of the C-terminal domain of S. gordonii SspB, which comprises two domains. Each domain contains a central β-sandwich that is stabilized by a Ca2+ ion. In addition, a stabilizing intramolecular isopeptide bond between a lysine and an asparagine is found within each domain. Furthermore, the crystal structure reveals that the BAR motif protrudes from the core of the protein like a handle, making it accessible for recognition by the Mfa1 fimbriae.
Results and Discussion
Structure determination
Two different forms of the protein were crystallized, SspB-C1061–1413 and SspB-C1083–1413, the latter of which yielded more reproducible crystals. The structure of SspB-C1083–1413 was solved by single anomalous dispersion (SAD) to a resolution of 1.5 Å using a single crystal of Selenomethionine (SeMet)-substituted protein. The asymmetric unit contains one molecule of SspB-C. The final model is well ordered with an overall B-factor of 17 Å2 and a crystallographic R factor of 0.18 (Rfree= 0.21). The high resolution SeMet structure consists of residues 1083–1413 as well as one structural Ca2+ per domain. 372 water molecules have also been included in the model as well as two additional metal ions involved in crystal packing. The native protein structures of SspB-C1083–1413 and SspB-C1061–1413 were solved to 1.7 and 2.1 Å respectively. In the long form, residues 1076–1413 are included in the model. The structures are very similar to the SeMet model except for some variations in the conformations of the looped regions in the upper part of domain C2. The amino acid numbering is based on the full-length SspB protein.
The best diffracting crystals were obtained by adding 1% (w/w) α-chymotrypsin to the protein immediately before crystallization. Mass determination of the purified protein and the crystal contents by MALDI-TOF-MS indicates that 3.3–3.4 kDa is cleaved off by α-chymotrypsin (Table S1 and Fig. S1). In the SspB-C1083–1413 models, electron density for the two linker residues preceding the SspB construct is visible.
Overall structure of the truncated SspB C-terminal domain
The proteolytically stable SspB-C is elongated with a length of 90 Å and a width of 30 Å. The structure is organized into two domains that we have named C2 and C3 (Fig. 1b and c). C1 is assigned to the protease sensitive domain representing the first ~200 residues of the SspB C-terminal domain. Each domain consists of a central β-sandwich composed by two anti-parallel sheets. In domain C2 (here modelled by residues 1076–1253), the β-sandwich is made up of two sheets, S1 and S2, consisting of six and five strands, respectively. β3 and β4 are connected by a short helix, αA, and a coiled region. An additional helix, αC, is located parallel to the coiled region and links β9 and β10. The helix region is located on top of the C2 domain. Linking β8 with β9 is a region containing a helix (αB) packed against the concave face of S2.
The C3 domain encompasses residues 1254–1413 and consists of a β-sandwich formed by two β-sheets (S3 and S4), each containing five strands. Most strands are connected by long loops on the side of the β-sandwich that is facing the C2 domain. A long loop region connects β15 with β16 (14 aa), β18 and β20 are connected by a long coiled region (16 aa) containing a short β-strand, β19, and β21 and β22 are connected by a coiled region comprising a short helix, αD. Only β25 and β26 are linked by a turn. On the opposite side of the sandwich, the connecting loops are short, except for a long coiled region (18 aa) that connects β14 with β15. Two strands, β24 and β27 form an elongation of the S4 sheet.
There is one Ca2+ ion bound in each domain. Each domain is also stabilized by a covalent isopeptide bond that is formed between Lys1082 (β1) and Asn1232 (β12) in the C2 domain and between Lys1259 (β14) and Asn1393 (β24) in the C3 domain.
Comparison between the SspB C2 and C3 domains
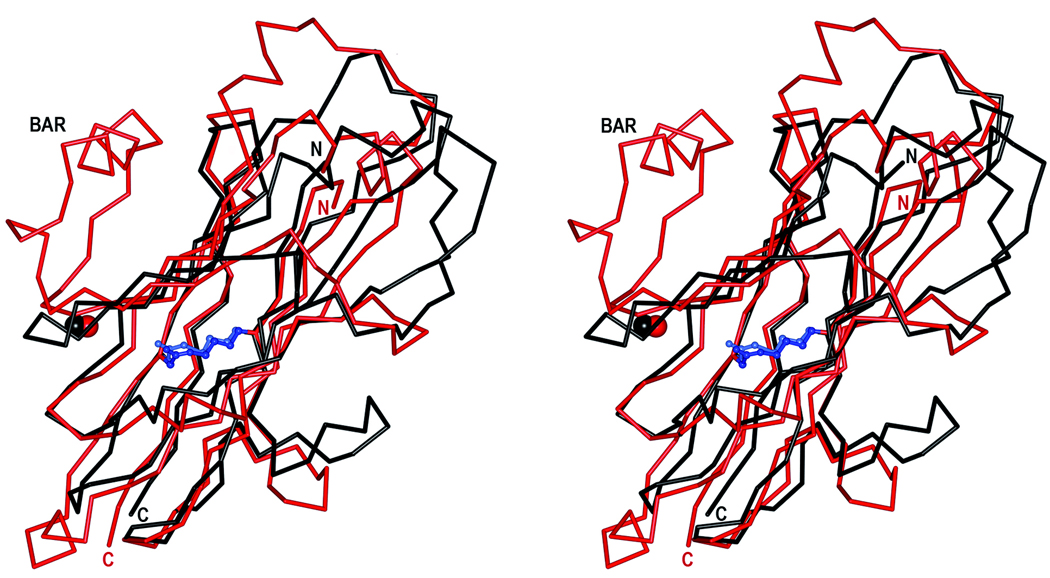
A pairwise DALI comparison22 was used to compare the C2 and C3 domains of SspB. The two domains were superimposed with a root-mean-square deviation (r.m.s.d.) of 2.1 Å for 141 common Cα atoms despite sharing only 18% sequence identity in the aligned regions (Fig. 2a). Despite their similar topology, there are significant differences between the two domains. In the C2 domain, the β8 and β9 strands are connected by a stretch of 21 residues that includes a helix located perpendicular to the S2 sheet, whereas the corresponding strands in C3 are connected by a short turn. In C2, strands β1 and β3 are connected by a short strand (β2) and a turn, whereas the comparable strands in C3 are connected by a long coiled region. Also, strands β6 and β7 in C2 are connected by a short turn, but in C3 the equivalent strands (β18 and β19) are connected by a long coil. Furthermore, the connections between strands β3 and β4, and β9 and β10 in C2 are made by helices, whereas all of the connections in the C3 domain are made by loops and coils except for one very short helical segment.
Figure 2.
Domain structure and stabilization. a) Superposition of the C2 and C3 domains. The C2 domain is coloured in red and the C3 domain in black. The isopeptide bonds are represented as ball-and-stick models in blue and light blue. The N-, and C-termini and the BAR motif are labelled. The picture is shown in stereo. b) Superposition of the metal binding sites in the C2 and C3 domains. The C2 residues are coloured in green and the C3 residues in yellow. The Ca2+ ions are depicted as brown spheres. The bonds involved the metal coordination are represented by dashed lines. c) The isopeptide bond in the C2 domain. The Lys-Asn isopeptide bond is represented as a stick model in a simulated annealing omit Fo-Fc map, contoured at 4σ. Hydrogen bonds to the aspartate residue are shown as dashed lines. Hydrophobic residues surrounding the isopeptide are shown in coral. The picture is shown in stereo. d) The isopeptide bond in the C3 domain. The figure is prepared as in c).
Metal binding sites
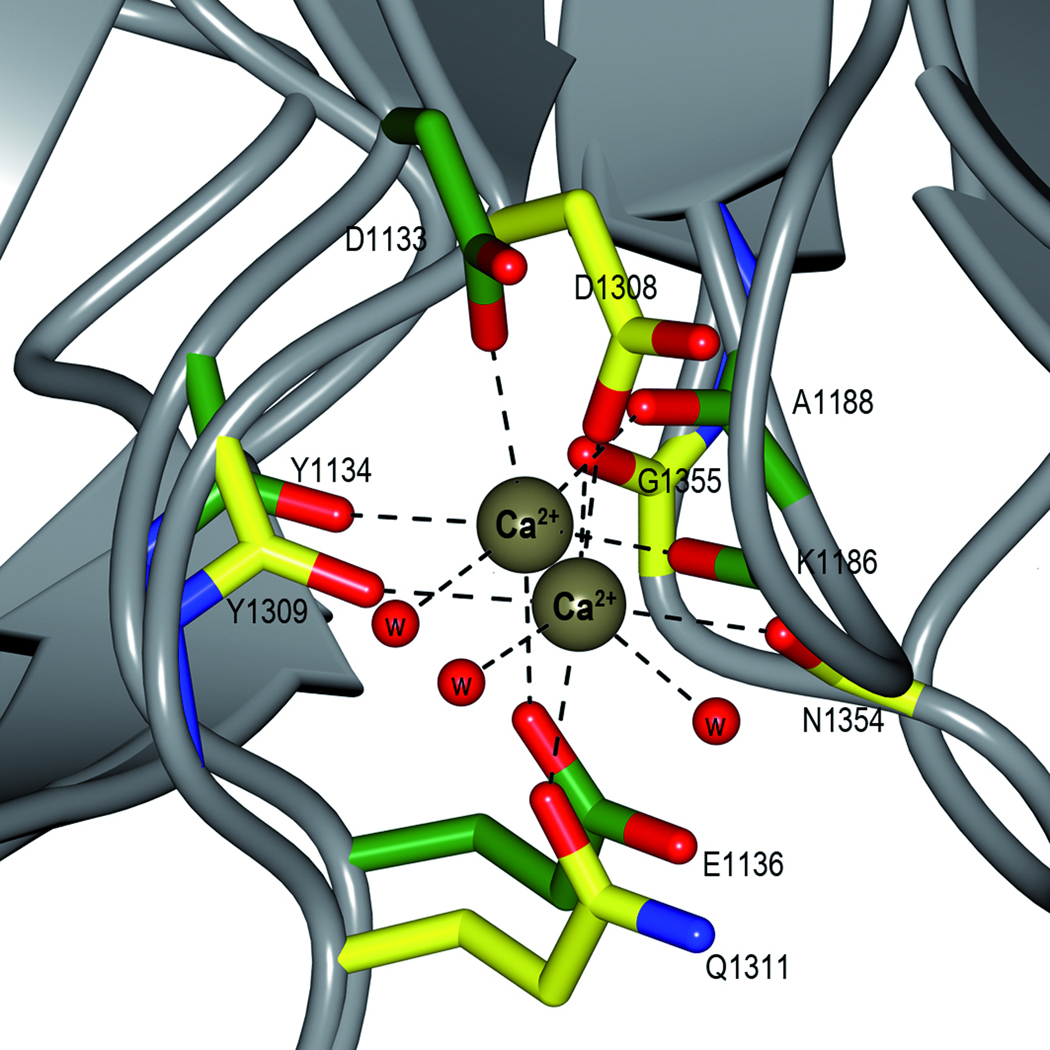
The SeMet protein was crystallized in the presence of calcium and each domain has a tightly bound Ca2+ ion located at equivalent positions (Fig. 2a). In the C2 domain, the Ca2+ ion is coordinated by Lys1186 (O) and Ala1188 (O) from the loop between β4 and the β5, and Asp1133 (OD1), Tyr1134 (O) and Glu1136 (OE1) from the loop before β9 (Fig. 2b). The metal is also coordinated by one water molecule.
In the C3 domain, the Ca2+ ion is coordinated by Asp1308 (OD1), Tyr1309 (O) and Gln1311 (OE1) from the loop between β16 and β17, and Asn1354 (O) and Gly1355 (O) from the loop bridging β20 and β21, adding to the stability of the structure. The ion is further coordinated by two water molecules. The SeMet structure also has two more Ca2+ ions, which are more loosely coordinated and are located between symmetry related molecules.
The native SspB-C1083–1413 has structural metal ions bound in the same locations as the SeMet model. Though the crystals were grown in 0.2 M MgCl2, the metal ions were modelled as Ca2+ since the metal-protein distances refine to an average of 2.36 Å which is more consistent with Ca2+ (2.36–2.39 Å) than with Mg2+ (2.07–2.26 Å).23 In addition, the structure has one Mg2+ ion involved in crystal packing.
The native SspB-C1061–1413 has one Ca2+ atom involved in crystal packing in addition to the structural Ca2+ atoms.
The presence of Ca2+ in the structure supports the observation that Ca2+ is needed for aggregation to occur between salivary gp340 and L. lactis expressing AgI/II proteins on their surfaces24 and for the adherence of SspA and SspB proteins to β1-integrin.25
The isopeptide bonds
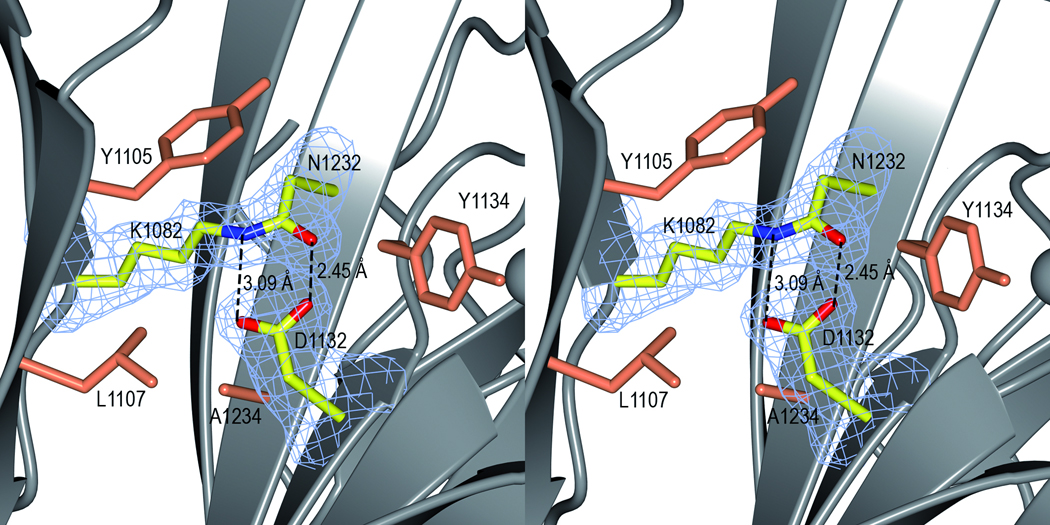
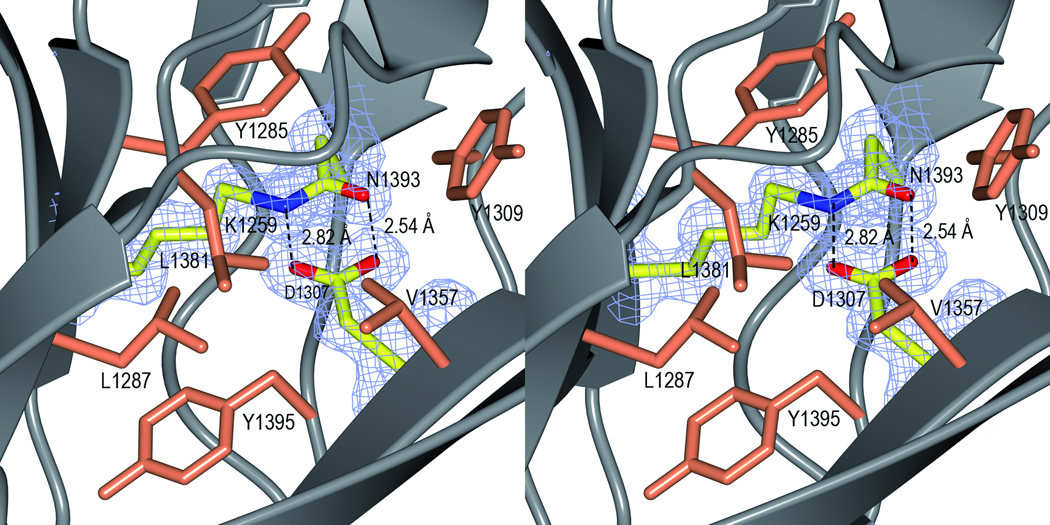
The structure is further stabilized by one isopeptide bond in each domain. In the C2 domain, the S1 and S2 sheets are linked by an isopeptide bond between Lys1082 (β1) and Asn1232 (β12). Similarly, in the C3 domain a covalent bond is formed between Lys1259 from β14 (S3) and Asn1393 from β24 (the extension of S4). Both isopeptide bonds are formed between the NZ atom of the lysine and the CG of the asparagine (Fig. 2c and d). An aspartate residue, Asp1132 and Asp1307 in C2 and C3 respectively, completes the interaction by hydrogen bonding to the isopeptide C=O and NH groups. The crosslink is surrounded by aromatic and hydrophobic residues, which results in an increased pKa of the Asp and a decreased pKa of the lysine amino group. This environment facilitates the nucleophilic attack on the Asn CG carbon by the unprotonated Lys amino group, resulting in a covalent bond and the release of ammonium.26 The native protein (SspB-C1061–1413 and SspB-C1083–1413) as well as the mutants, D1132A, N1232A, D1307A and N1393A were analysed by ESI-TOF MS to examine if the isopeptide bonds were present also in solution. The mutants were measured to 42471 Da which equals the loss of one NH3 group (17 Da) from the calculated mass of 42487 Da, thus the mutants form one isopeptide bond as expected. Similarly, the SspB-C1083–1413 form was measured to 40108 Da, which is 16 Da less than the expected 40124 Da. Surprisingly, the presence of isopeptide bonds in the native SspB-C1061–1413 protein could not be detected. The mass of the native protein was found to be 42533 Da, which is approximately the same as the calculated mass (42531 Da) (Table S2 and Fig. S2). Evidently, the isopeptide bonds are not necessarily formed immediately when the protein is folded. We speculate that formation of isopeptide bonds is facilitated by the folding of the full-length protein, and also by crystal packing. The reason why the remaining isopeptide bond in SspB-C1083–1413 and in the mutants is more prone to form in solution is unclear and needs to be studied further.
In C3, the isopeptide stacks with the aromatic ring of Tyr1285, and Asp1307 is surrounded by Tyr1395, Leu1287, Leu1381, Val1357 and Tyr1309. In C2, the isopeptide bond stacks with Tyr1105 and Asp1132 is surrounded by Ala1234, Leu1107 and Tyr1134. The equivalents of Val1357 and Leu1381 in C2 are exchanged for Asp1125 and Gln1190 in the C2 domain creating an environment less hydrophobic. Kang et al.26 reported similar self-generated intramolecular isopeptide bonds that stabilize the structure of the Streptococcus pyogenes pili Spy0128. They hypothesize that gram-positive bacteria may use such intramolecular crosslinks as a mode of stabilization of cell surface structures that are exposed to physical and chemical stress. The same means of stabilization has been reported in each of the three domains of the Bacillus cereus pilus27 and most recently, isopeptide bonds were reported for the Corynebacterium diphtheriae pilin SpaA.28 As SspB also is a cell surface protein from a gram-positive bacterium, its structure supports the hypothesis that intramolecular isopeptide bonds are used for stabilization of cell surface adhesive proteins. Oral bacteria are dependent on extreme stability of their surface adhesins. They will be removed from surfaces by the physical shear forces of salivary fluid flow and tongue movement, and eventually lost from the oral cavity by swallowing or expectoration unless they are firmly attached to a surface.
Streptococci are predominant in the early biofilm and constitute the framework for numerous of layers of bacteria,29 providing sites of attachment even after the first layers of bacteria are no longer viable. Moreover, the architecture of the biofilm must resist extreme changes in environment such as pH and temperature fluctuations due to food intake or the presence of acid generating bacteria. The biofilm is also exposed to mechanical stress induced by chewing or the flow of saliva. In addition, the building blocks of the biofilm must also withstand the release of proteases from the salivary glands and oral bacteria.30 Treating the SspB-C1061–1413 protein and the isopeptide mutants with trypsin showed that they were equally stable and were barely affected by the protease (Fig. S3). This experiment indicates that the C-terminal domain is intrinsically stable, and that the isopeptide bonds are not crucial for resisting proteases. Therefore we hypothesize that the isopeptide bonds have evolved for other purposes, such as to withstand mechanical stress.
It is important to remember that the C-terminal domain, except for being the putative binding partner to several other molecules, constitutes only the stalk of the 1500 residue Antigen I/II protein. Its main purpose may be to expose the variable and N-terminal domains to facilitate biofilm formation; and therefore its stability is of extreme importance.
A homology search
A homology search on the entire SspB-C1061–1413 structure using the DALI server22 identified several distant relatives, all of which are surface proteins. Separate searches for the individual C2 and C3 domains resulted in similar hits. The closest relative found for both the C2 and C3 domains was the middle domain of the shaft pilin SpaA from C. diphtheriae28 (PDB code 3HTL , Z-score 12.4 with an r.m.s.d. of 2.8 Å on 140 aligned Cα residues for C2 and a Z-score 12.6, r.m.s.d. of 2.4 Å on 134 aligned Ca residues for C3). The β-sheets of these structures superimpose well and share the same topology with some variation in the connecting loops and helices. Notably the three structures contain isopeptide bonds located in identical positions. In addition, AgI/II proteins have been reported to interact with collagen.31 Both domains show structural similarities, including the same topology as the collagen binding domain (N2 domain) of CnaA from Staphylococcus aureus32 (PDB code 2F6A) and the collagen binding subdomain from Enterococcus faecalis33 (PDB code 2OKM). Their collagen binding surfaces are located on the concave sides of their β-sandwiches, and superpose well with pronounced symmetric grooves formed by the concave sides of the SspB S1 β-sheet in C2 and the S3 β-sheet in C3. Whereas the collagen binding surface is rich in aromatic residues that can stack with the proline residues of collagen, the corresponding surfaces of SspB-C contain no such residues. While these comparisons support a concept of a binding surface of SspB, interaction with collagen at this particular surface is unlikely. The collagen binding proteins discussed above have isopeptide triads (Lys-Asn-Asp) in identical positions as SspB, and even though such covalent bonds were not discussed,32,33 it has been shown that the presence of isopeptide bonds are likely also in these proteins.26 An additional close relative found for the C3 domain is the exo-alpha-sialidase protein-protein interaction module NanJX82 from Clostridium perfringens34 (PDB code 2VO8, Z-score 10.8, with and r.m.s.d. of 2.7 Å on 119 aligned Cα residues). The superposed parts of the proteins were the β-sandwiches, but the regions connecting the strands differ in length and secondary structure, especially on the sides of the sandwiches that face the C2 domain.
The protease sensitive domain, C1, is predicted to have the same fold as C2 and C3. A MetaServer35 3D-PSSM search returned the collagen binding domain from the S. aureus adhesin (pdb code 1AMX)36 as the closest structural relative.
Putative interaction surfaces
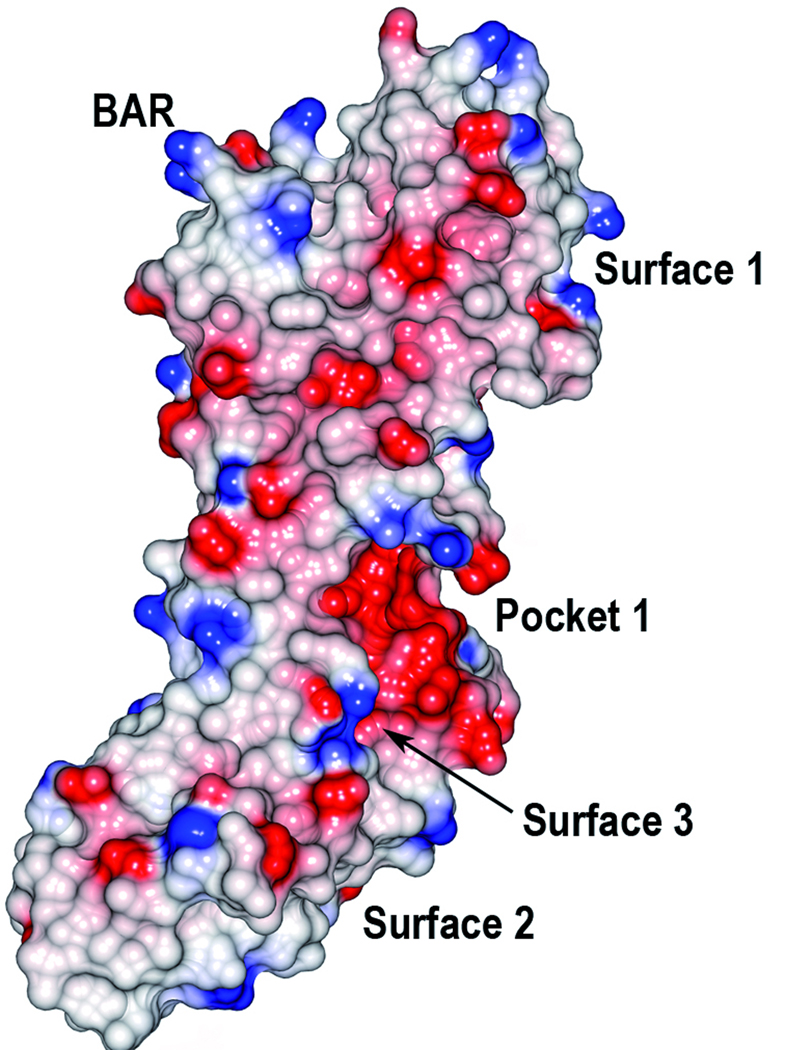
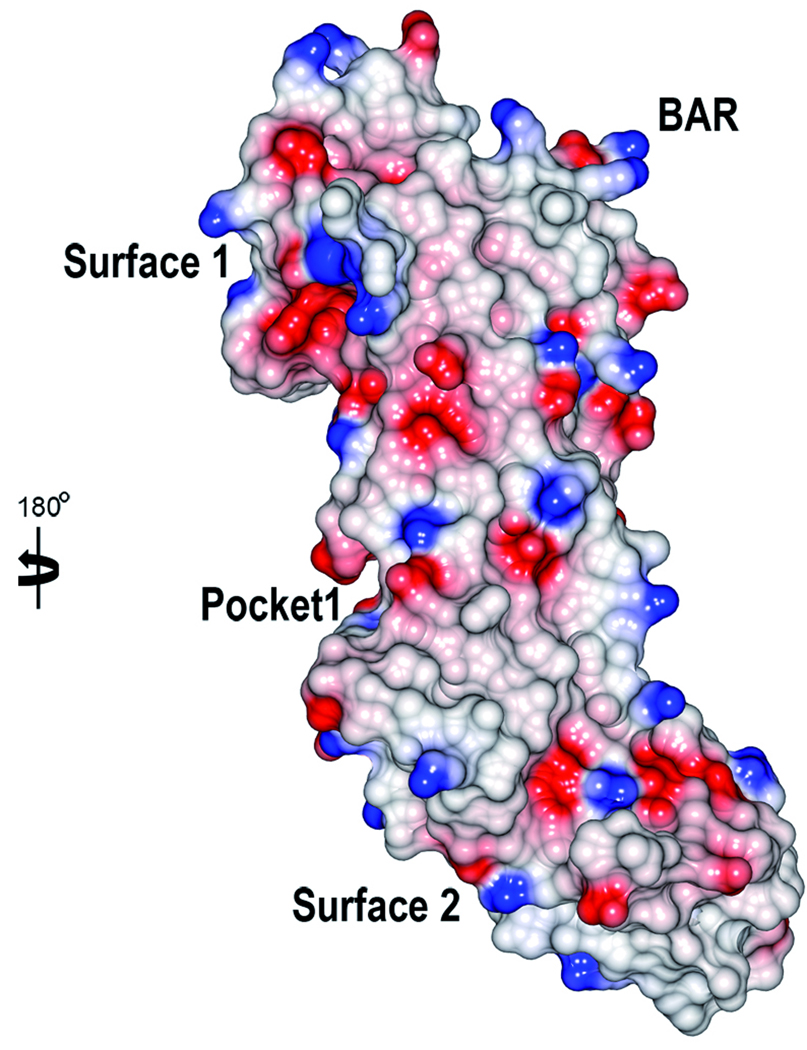
Analysis using the CASTp server37 identified only small putative binding pockets (Fig. 3). The most pronounced is a cleft that is located on interface between the C2 and C3 domains (Pocket 1, 167 Å3). By visually analyzing the structure and the surface representation of SspB, a few putative binding surfaces were found. The S1 and S3 β-sheets each form a bent surface (Surface 1 and 2 respectively) that superpose well with the collagen binding surfaces of the collagen adhesins discussed above. In the C3 domain, a region above the S4 β-sheet and close to αD (Surface 3) superposes well with the dockerin interaction surface of NanJX82. It should be noted that that the full-length SspB protein is composed of several domains and that other binding surfaces may be generated when the full-length protein is folded.
Figure 3.
Electrostatic surface of SspB-C1061–1413. The molecular surfaces are colored in red and blue according to positive and negative electrostatic potential, respectively. The two views are rotated 180° with respect to each other. The BAR motif, the putative binding pocket and surfaces are indicated.
The P. gingivalis recognition region
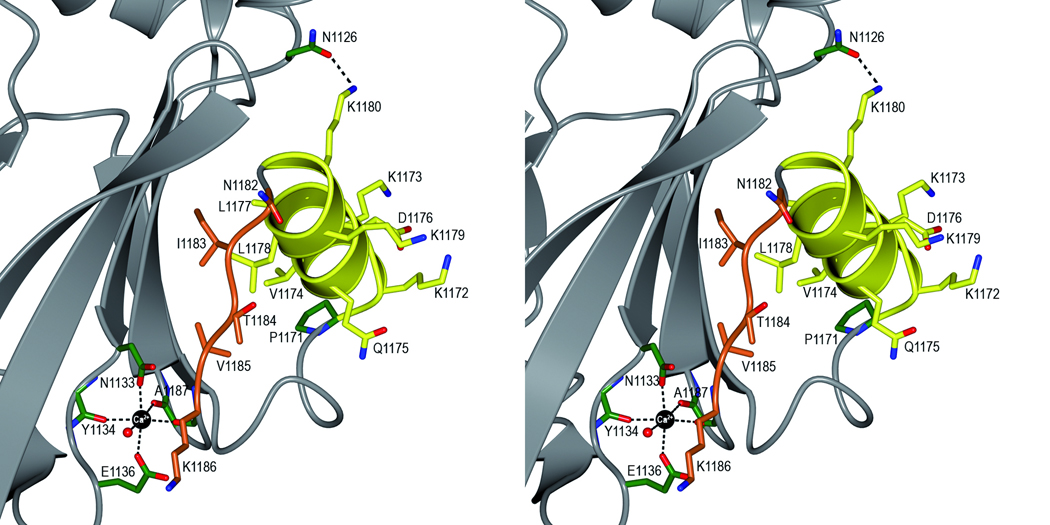
It was previously shown that the interaction between SspB and the minor fimbrial subunit protein Mfa1 of P. gingivalis is needed for colonization of P. gingivalis on a streptococcal substrate.38 The specific interaction of SspB with Mfa1 has been mapped to a region within the C-terminal domain of SspB, designated the SspB Adherence Region (BAR), comprising residues 1167–1193.15,18 In the crystal structure, the BAR region, with sequence LEAAPKKVQDLLKKANITVKGAFQLFS is located in domain C2 (Fig. 1a and b). BAR comprises a short 310 helix, a ten residue α-helix (αB) followed by an extended conformation and a part of β9 (Fig. 4). Within the BAR region, two structural motifs (KKVQDLLKK and NITVK) have been identified that are important for adherence between SspB and the Mfa1 fimbrial protein.17 The BAR motif-region resides between β8 and β9 and protrudes from the core of domain C2, like a handle for facilitating recognition by the Mfa1 fimbriae. The position of the BAR handle is stabilized by the Ca2+ ion coordinated by residues from β4 and β9.
Figure 4.
Stereoview of the BAR handle. The BAR handle is important for recognition by the Porphyromonas gingivalis fimbriae Mfa1. The handle is constituted by two motifs; a helix (KKVQDLLKK, in yellow) followed by an extended region (NITVK, in orange). The side chains are represented as stick objects. A hydrogen bond between K1180 and N1126 in the β-sheet (grey) is marked with a dashed line. The position of the BAR handle is stabilized by a Ca2+ ion (black), coordinated by three main chain and two side chain oxygen atoms and a water molecule.
The KKVQDLLKK motif constitutes an amphipathic α-helix (αB), which is in agreement with secondary structure predictions.17 The charged and polar residues in the helix, K1172, K1173, Q1175, D1176, K1179 and K1180 face the solvent, and are exposed to binding partners. They are also important for stabilization of the helix via ionic and hydrogen bonding interactions. K1172(NZ) forms a salt bridge with D1176(OD2) within the helix. Q1175(NE2) is located within hydrogen bonding distance to three main chain carbonyls in the turn that precedes the helix (L1167, E1168 and A1170). K1180(NZ) forms a hydrogen bond with an asparagine residue (N1126 (OD1)) in the S2 β-sheet. The hydrophobic residues V1174, L1177 and L1178 face the hydrophobic contact surface between the BAR helix and the upper part of the S2 sheet. The helix is sterically constrained by the proline (P1171) on the N-terminal end and by the K1180-N1126 hydrogen bond on its C-terminal end.
The NITVK motif, important for specific binding by Mfa1, is found within the extended region following the helix and preceding β9. This motif also contains polar residues that face the solvent and the hydrophobic residues packed against the β-sheet. The single mutations N1182G and V1185P create a protein more like the S. mutans SpaP and result in loss of P. gingivalis adherence.18 The specificity of P. gingivalis adherence to SspB opens the possibility of future drug design based on the BAR handle structure.
Material and Methods
Cloning and site directed mutagenesis
A protocol for cloning, purification and crystallization of a truncated form of SspB-C was previously described.39 When screening for reproducible crystal forms two constructs were generated, SspB-C1083–1413 and SspB-C1061–1413. The cloning procedure for SspB-C1083–1413 was identical to the one described for SspB-C1061–141339 except that the forward primer 5’-TTTTTCCATGGTAACTACTCCTGGTAAA-3’ was used (NcoI site underlined). In both constructs, the cloned fragment is preceded by a hexa-histidine and a cloning vector linker (PMSDYDIPTTENLYFQGAM). The hexa-histidine is preceded by two residues, MK. Mutation of isopeptide residues were performed using the QuickChange site directed mutagenesis kit (Strategene) as descibed by the manufacturer. The following primers were used: D1132A, 5'-GGCTTCTACTACGTAGCTGATTATCCAGAGG-3', 5'-CCTCTGGATAATCAGCTACGTAGTAGAAGCC-3'; N1232A, 5'- 5'- CAGGTGGTAAGT ATGAAGCCAAGGCTTATCAAATTGATTTC-3', 5'- GAAATCAATTTGATAAGCCTTGGCTTCATACTTACCACCTG-3'; D1307A, 5'- GATTACAGCTTTGTGGCTGATTATGACCAAGC-3', 5'- GCTTGGTCATAATCAGCCACAAAGCTGTAATC-3'; N1393A, 5'- GCTATTGGAACATTTGAAGCTACTTATGTAAATACTGTTAA-3' 5'- TTAACAGTATTTACATAAGTAGCTTCAAATGTTCCAATAGC-3'. The underlines show the locations of the mutated codons. The mutations were verified by DNA sequencing.
Protein expression and purification
To express SeMet-substituted SspB-C1083–1413, cells were grown in M9 media supplemented with glucose at 37°C. At an optical density of ~0.6 at 600 nm, lysine, threonine, phenylalanine at 100 mg/l, leucine, isoleucine, valine, proline and SeMet at 50 mg/l were added to downregulate the synthesis of methionine.40 The culture was cooled to 20°C, protein expression induced with 0.4 mM IPTG and the culture grown overnight. Cells were harvested by centrifugation at 5000 rpm and the pellet was frozen at −80°C. Cells were resuspended in 20 mM Tris pH 7.5, 150 mM NaCl and 10 mM imidazole supplemented with EDTA-free protease inhibitor cocktail (Roche). The cells were lysed on ice by sonication and cellular debris was removed by centrifugation at 18000 rpm for 60 min. The supernatant was loaded onto a column packed with Ni-NTA (Qiagen). The protein was eluted in 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 300 mM imidazole, and the buffer was exchanged to 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA and 0.5 mM tris (2-carboxyethyl)-phosphine hydrocholoride (TCEP). The protein was further purified by gel filtration on a Superdex200 16/60 PG column (Amersham Biosciences) and the protein purity was assessed by SDS-PAGE. The protein was concentrated in 20 mM Tris-HCl pH 7.5 and 0.5 mM TCEP using an Amicon Ultra centrifugal filter device (Millipore), frozen in liquid nitrogen and stored at −80°C. The His-tag was not removed prior to crystallization trials.
Expression and purification of the native proteins was performed as described,39 with the exceptions that expression was at 20°C over night and an ion exchange purification was substituted for the gel filtration step using buffers 20 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 0.5 mM TCEP and 20 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 0.5 mM TCEP and 1 M NaCl. The protein was concentrated in 20 mM Tris-HCl pH 7.5, 0.5 mM EDTA and 0.5 mM TCEP and stored as the SeMet-substituted protein. The isopeptide mutants were purified as described for the native protein.
Crystallization, data collection and processing
The crystals were grown by vapour diffusion using the hanging-drop method at room temperature. Droplets of 1 µL of SeMet-substituted protein at 20 mg/ml with 1% (w/w) α-chymotrypsin were mixed with 1 µL reservoir solution containing 0.2 M CaCl2, 0.1 M HEPES pH 7.5 and 20% (w/v) PEG 3350. SeMet-substituted protein crystals grew within two weeks in space group P21 with cell dimensions a=37.7 Å, b=50.3 Å c=94.4 Å, β= 97.4° and one molecule per asymmetric unit. Crystals were equilibrated in a cryoprotectant solution containing reservoir solution with 10% glycerol added, and flash-cooled in liquid-nitrogen. Native SspB-C1083–1413 at 30 mg/ml with 1% (w/w) α-chymotrypsin was mixed with 0.2 M MgCl2, 0.1 M Tris-HCl pH 8.5 and 30% (w/v) PEG 4000. Crystals were obtained in space group P21 with cell dimensions a=37.2 Å, b=61.0 Å, c=74.0 Å, β=100.1°, with one molecule in the asymmetric unit. Similarly, native SspB-C1061–1413 at 15 mg/ml was treated with α-chymotrypsin and crystallized in 0.2 M CaCl2, 0.1 M HEPES pH 7.5 and 20% (w/v) PEG 4000. The crystals grew in space group P212121 with cell dimensions a=48.8 Å b=80.2 Å c=87.4 Å and one molecule per asymmetric unit. The native crystals were equilibrated in reservoir solution complemented with 20% glycerol, and flash-cooled in liquid nitrogen. Single-wavelength anomalous diffraction (SAD) data set of SeMet-substituted SspB-C1083–1413 to 1.5 Å and a native data set to 1.7 Å were collected at beam line ID23-1 at ESRF Grenoble. Native data of SspB-C1061–1413 were collected to 2.1 Å at beam line I911-3 at MAX-lab, Lund. Diffraction images were processed with MOSFLM41 and scaled with SCALA from the CCP4 program suite42. Relevant processing statistics are presented in Table 1.
Table I.
Data collection, refinement and model quality statistics for SspB-C.
| SeMet SAD SspB-C1083–1413 | Native SspB-C1083–1413 | Native SspB-C1061–1413 | |
|---|---|---|---|
| Data processing | |||
| Space group | P21 | P21 | P212121 |
| Cell dimensions a, b, c (Å), β (°) | 37.74, 50.29, 94.37, β=97.4 | 37.20, 61.01, 74.04, β=100.1 | 48.75, 80.15, 87.43 |
| Wavelength (Å) | 0.975 | 1.00 | 0.979 |
| Resolution (Å) | 50.3-1.5 | 46.8-1.7 | 42.6-2.1 |
| Highest resolution shell (Å) | 1.58-1.50 | 1.79-1.70 | 2.19-2.08 |
| Total reflectionsa | 226173 (26754) | 66060 (9664) | 149840 (20799) |
| Unique reflectionsa | 55706 (7546) | 33858 (4906) | 21137 (3030) |
| I/σ (I) a | 15.9 (3.0) | 12.0 (3.0) | 17.8 (6.3) |
| Rmergea,b(%) | 5.9 (39.8) | 5.5 (22.0) | 9.0 (29.6) |
| Completenessa (%) | 98.8 (92.1) | 94.4 (94.6) | 99.7 (99.6) |
| Overall redundancya | 4.1 (3.5) | 2.0 (2.0) | 7.1 (6.9) |
| Refinement | |||
| No. of reflections in working set | 51061 | 31127 | 19713 |
| No. of reflections in test set | 2740 | 1647 | 1073 |
| Rwork/Rfreec (%) | 18.0/20.8 | 18.6/22.8 | 19.2/25.5 |
| Average B-factors (Å2) | |||
| Protein | 15.8 | 15.4 | 24.9 |
| Water | 23.8 | 22.6 | 26.5 |
| Metal ions | 13.0 | 15.2 | 19.4 |
| No of protein atoms | 2636 | 2616 | 2667 |
| No of metal ions | 4 | 3 | 3 |
| No of water molecules | 372 | 294 | 138 |
| RMSD from ideal | |||
| Bond lengths (Å) | 0.011 | 0.011 | 0.012 |
| Bond angles (°) | 1.33 | 1.37 | 1.41 |
| Ramachandran plot (Most favored, allowed, generously allowed, disallowed) (%) |
90.6/9.4/0/0 | 93.0/7.0/0/0 | 90.4/9.6/0/0 |
Values in parentheses indicate statistics for the highest resolution shell
Rmerge = Σhkl Σi |Ii(hkl) - <I(hkl)>|/ Σhkl Σi Ii (hkl), where Ii(hkl) is the intensity of the ith observation of reflection hkl and <I(hkl)> is the average over of all observations of reflection hkl.
Rwork = Σ | |Fobs| - | Fcalc| | / Σ | Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is Rwork calculated using 5% of the data, randomly omitted from refinement.
Structure determination and refinement
The high-resolution SspB-C1083–1413 SeMet structure was solved with SAD-phasing using AutoRickshaw.43 Density modification, molecular averaging and automatic model building using AutoRickshaw and ArpWarp44 resulted in a readily interpretable electron density map.
The derivative data set of 1.5 Å was used for refinement with 5% of the data removed for calculation of the Rfree. The full atomic model was built into the electron density using COOT45 and the model refined using PHENIX.REFINE46. The first refinement step included simulated annealing starting at 5000K and in the last rounds of refinement translational-liberation-screw (TLS) refinement was used, treating each domain as an individual TLS group.47 The native structures were solved by molecular replacement using the MOLREP48 program from the CCP4 suite, with the high-resolution derivative as a starting model. Native data were refined to 1.7 Å for the SspB-C1083–1413 construct and 2.1 Å for the SspB-C1061–1413 construct. The native structures were refined as described for the SeMet structure including simulated annealing and TLS refinement. The quality of the models were analyzed with WHATCHECK.49 Refinement statistics are presented in Table 1. Figures were drawn with CCP4MG.50
Accession Codes
The X-ray coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 2WOY, 2WQS and 2WZA.
Supplementary Material
Acknowledgements
We are grateful for the access to the beam line ID23-1 ESRF, Grenoble, beam line I911-3 MAX-lab, Lund and for the support of beam line scientists. We thank Gunter Stier, EMBL, Germany for cloning vectors. The mass spectrometry analyses were performed by Thomas Kieselbach at Umeå Protein Analysis Facility, Umeå University and by the Protein Analysis Centre at the Karolinska Institute. This work was supported by NIDCR R01 12505, the Swedish Research Council and Magn Bergvall foundation.
Abbreviation
- AgI/II
Antigen I/II
- SspB-C
SspB C-terminal domain
- SAD
single anomalous dispersion
- SeMet
Selenomethionine
- BAR
SspB Adherence Region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 3.Jenkinson HF, Lamont RJ. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 4.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 5.Douglas CW, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 6.McNab R, Holmes AR, Clarke JM, Tannock GW, Jenkinson HF. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 1996;64:4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNab R, Jenkinson HF, Loach DM, Tannock GW. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 1994;14:743–754. doi: 10.1111/j.1365-2958.1994.tb01311.x. [DOI] [PubMed] [Google Scholar]
- 8.Urano-Tashiro Y, Yajima A, Takashima E, Takahashi Y, Konishi K. Binding of the Streptococcus gordonii DL1 surface protein Hsa to the host cell membrane glycoproteins CD11b, CD43, and CD50. Infect. Immun. 2008;76:4686–4691. doi: 10.1128/IAI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 10.Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, Lopez JA, Griffiss JM, Sullam PM. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 2005;58:380–392. doi: 10.1111/j.1365-2958.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 12.Demuth DR, Duan Y, Brooks W, Holmes AR, McNab R, Jenkinson HF. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 13.Egland PG, Du LD, Kolenbrander PE. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 2001;69:7512–7516. doi: 10.1128/IAI.69.12.7512-7516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes AR, Gilbert C, Wells JM, Jenkinson HF. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 1998;66:4633–4639. doi: 10.1128/iai.66.10.4633-4639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks W, Demuth DR, Gil S, Lamont RJ. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakubovics NS, Stromberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 2005;55:1591–1605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- 17.Daep CA, James DM, Lamont RJ, Demuth DR. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect. Immun. 2006;74:5756–5762. doi: 10.1128/IAI.00813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 2001;69:5736–5741. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect. Immun. 2008;76:3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 1998;66:4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuboniwa M, Amano A, Hashino E, Yamamoto Y, Inaba H, Hamada N, Nakayama K, Tribble GD, Lamont RJ, Shizukuishi S. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC. Microbiol. 2009;9:105. doi: 10.1186/1471-2180-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Current Protocols in Bioinformatic. 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5, Unit 5 5. [DOI] [PubMed] [Google Scholar]
- 23.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 24.Jakubovics NS, Kerrigan SW, Nobbs AH, Stromberg N, van Dolleweerd CJ, Cox DM, Kelly CG, Jenkinson HF. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 2005;73:6629–6638. doi: 10.1128/IAI.73.10.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobbs AH, Shearer BH, Drobni M, Jepson MA, Jenkinson HF. Adherence and internalization of Streptococcus gordonii by epithelial cells involves beta1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell. Microbiol. 2007;9:65–83. doi: 10.1111/j.1462-5822.2006.00768.x. [DOI] [PubMed] [Google Scholar]
- 26.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 27.Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc. Natl Acad. Sci. USA. 2008;105:10215–1020. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl Acad. Sci. USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, Haffajee AD, Socransky SS, Oppenheim FG. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 30.Xiuli S, Erdjan S, Frank GO, Eva JH. Activity-based mass spectrometric characterization of proteases and inhibitors in human saliva. PROTEOMICS - CLINICAL APPLICATIONS. 2009;3:810–820. doi: 10.1002/prca.200800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love RM, McMillan MD, Jenkinson HF. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect. Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. A 'Collagen Hug' model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponnuraj K, Narayana SV. Crystal structure of ACE19, the collagen binding subdomain of Enterococus faecalis surface protein ACE. Proteins. 2007;69:199–203. doi: 10.1002/prot.21464. [DOI] [PubMed] [Google Scholar]
- 34.Chitayat S, Gregg K, Adams JJ, Ficko-Blean E, Bayer EA, Boraston AB, Smith SP. Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase. J. Mol. Biol. 2008;375:20–28. doi: 10.1016/j.jmb.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 35.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 36.Symersky J, Patti JM, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas LJ, Hook M, Narayana SV. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 37.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl. Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamont RJ, Hersey SG, Rosan B. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immun. 1992;7:193–197. doi: 10.1111/j.1399-302x.1992.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 39.Forsgren N, Lamont RJ, Persson K. A crystallizable form of the Streptococcus gordonii surface antigen SspB C-domain obtained by limited proteolysis. Acta Crystallogr., Sect F: Struct. Biol. Cryst. Commun. 2009;65:712–714. doi: 10.1107/S1744309109021046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 41.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, N The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr., Sect. D: Biol Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 44.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 48.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 49.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 50.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.