Abstract
Here, we investigated the physiological role of Arabidopsis (Arabidopsis thaliana) AtNUDX6, the gene encoding ADP-ribose (Rib)/NADH pyrophosphohydrolase, using its overexpressor (Pro35S:AtNUDX6) or disruptant (KO-nudx6). The level of NADH in Pro35S:AtNUDX6 and KO-nudx6 plants was decreased and increased, respectively, compared with that of the control plants, while the level of ADP-Rib was not changed in either plant. The activity of pyrophosphohydrolase toward NADH was enhanced and reduced in the Pro35S:AtNUDX6 and KO-nudx6 plants, respectively. The decrease in the activity of NADH pyrophosphohydrolase and the increase in the level of NADH were observed in the rosette and cauline leaves, but not in the roots, of the KO-nudx6 plants. Notably, the expression level of AtNUDX6 and the activity of NADH pyrophosphohydrolase in the control plants, but not in the KO-nudx6 plants, were increased by the treatment with salicylic acid (SA). The expression of SA-induced genes (PR1, WRKY70, NIMIN1, and NIMIN2) depending on NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), a key component required for pathogen resistance, was significantly suppressed and enhanced in the KO-nudx6 and Pro35S:AtNUDX6 plants, respectively, under the treatment with SA. Induction of thioredoxin h5 (TRX-h5) expression, which catalyzes a SA-induced NPR1 activation, was suppressed and accelerated in the KO-nudx6 and Pro35S:AtNUDX6 plants, respectively. The expression of isochorismate synthase1, required for the regulation of SA synthesis through the NPR1-mediated feedback loop, was decreased and increased in the KO-nudx6 and Pro35S:AtNUDX6 plants, respectively. Judging from seed germination rates, the KO-nudx6 plants had enhanced sensitivity to the toxicity of high-level SA. These results indicated that AtNUDX6 is a modulator of NADH rather than ADP-Rib metabolism and that, through induction of TRX-h5 expression, AtNUDX6 significantly impacts the plant immune response as a positive regulator of NPR1-dependent SA signaling pathways.
Nudix (nucleoside diphosphates linked to some moiety X) hydrolases are a phylogenetically widespread enzyme family and are widely distributed among all classes of organisms, such as bacteria, yeast, algae, nematodes, vertebrates, and plants (Bessman et al., 1996; Xu et al., 2004; Kraszewska, 2008). The enzymes catalyze, with varying degrees of substrate specificity, the hydrolysis of a variety of nucleoside diphosphate derivatives: nucleoside diphosphates and triphosphates and their oxidized forms, dinucleoside polyphosphates, nucleotide sugars, NADH, CoA, and the mRNA caps (McLennan, 2006; Kraszewska, 2008; Gunawardana et al., 2009). Since these compounds are often toxic to cells, Nudix hydrolases seem to play protective, regulatory, and signaling roles in metabolism by hydrolytically removing such compounds (Bessman et al., 1996; Xu et al., 2004).
We reported the molecular and enzymatic characteristics of Nudix hydrolases (AtNUDX1–AtNUDX27) in Arabidopsis (Arabidopsis thaliana) plants (Ogawa et al., 2005, 2008). Notably, among 27 types of AtNUDXs, cytosolic AtNUDX2, AtNUDX6, AtNUDX7, and AtNUDX10 had pyrophosphohydrolase activity toward both ADP-Rib and NADH in vitro. Recent studies have shown that the actions of NADH and/or ADP-Rib pyrophosphohydrolases are closely related to defense systems in response to biotic and abiotic stresses in higher plants.
It has been reported that the expression of AtNUDX7 is induced by avirulent pathogenic attacks. Knockout AtNUDX7 mutants (KO-nudx7) showed enhanced resistance against both virulent and avirulent bacterial strains (Bartsch et al., 2006; Jambunathan and Mahalingam, 2006; Adams-Phillips et al., 2008). In addition, it was revealed that AtNUDX7 functions as a negative regulator on ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) signaling required for basal resistance to invasive pathogens (Bartsch et al., 2006); EDS1 regulates accumulation of the phenolic defense molecule, salicylic acid (SA), and other as yet unidentified signal intermediates and controls the defense activation and programmed cell death by collaborating with its interaction partner PHYTOALEXIN-DEFICIENT4 in cells surrounding pathogen infection foci. Furthermore, Ge et al. (2007) reported that AtNUDX7 functions to prevent excessive stimulation of the defense response, which is dependent on and independent of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), a master regulator of SA-induced defense genes (SAIGs), and SA accumulation.
On the other hand, we recently demonstrated the roles of Arabidopsis NADH/ADP-Rib pyrophosphohydrolases (AtNUDX2 and AtNUDX7) in tolerance to oxidative stress using the respective overexpressors (Pro35S:AtNUDX2 and Pro35S:AtNUDX7) or disruptants (KO-nudx7; Ishikawa et al., 2009; Ogawa et al., 2009). Interestingly, overexpression of AtNUDX2 and AtNUDX7 in Arabidopsis plants was responsible for an enhanced tolerance to oxidative stress derived from the treatment with paraquat (an agent producing O2−) and salinity. Taken together, these results revealed that both AtNUDX2 and AtNUDX7 function in accelerating nucleotide recycling from ADP-Rib produced by poly(ADP-Rib) metabolism, leading to suppression of the overconsumption of NAD+ and ATP in Arabidopsis cells under stressful conditions. In addition, AtNUDX7 served to balance between NADH and NAD+ by NADH turnover and to regulate the defense mechanisms against DNA damage by modulation of the poly(ADP-ribosyl)ation (PAR) reaction through NADH metabolism in response to oxidative stress (Ishikawa et al., 2009; Ogawa et al., 2009). These findings clearly indicated that the regulation of NADH and/or ADP-Rib metabolism via Nudix hydrolases is involved in the responses to both biotic and abiotic stresses in higher plants.
The question that we must consider next is whether the other AtNUDXs (AtNUDX6 and AtNUDX10) with pyrophosphohydrolase activities toward ADP-Rib and NADH are involved in the defense systems against oxidative stress and pathogen attack. The expression of AtNUDX6 has been reported to be induced by pathogenic attacks and treatment with the SA analogs 2,6-dichloroisonicotinic acid and acibenzolar-S-methyl benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH; Bartsch et al., 2006; Qiu et al., 2008; Knoth et al., 2009). Furthermore, the expression of AtNUDX6 was strongly dependent on EDS1 (Bartsch et al., 2006). However, the functional significance of AtNUDX6 is still unclear, since a loss-of-function mutant of AtNUDX6 has not yet been found.
In this paper, to assess the physiological function of AtNUDX6, we identified an Arabidopsis mutant in which T-DNA is inserted into AtNUDX6 and subsequently studied the levels of ADP-Rib and NAD(H), PAR activity, expression of genes related to SA signaling, and SA tolerance in the AtNUDX6 overexpressors and disruptants in comparison with the AtNUDX7 disruptants. The results obtained here indicated that AtNUDX6 positively regulates NPR1-dependent SA signaling via modulation of NADH metabolism in the plant immune response.
RESULTS
Characteristics of AtNUDX6-Overexpressed or -Disrupted Arabidopsis Plants
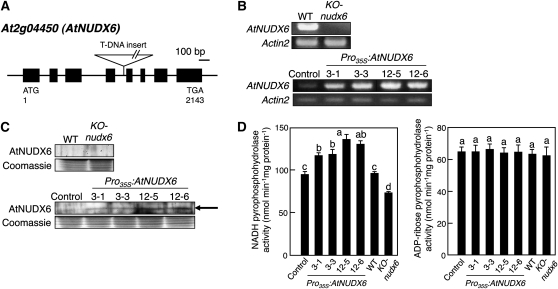
The expression of AtNUDX6 mRNA was detected in the leaves of 2-week-old wild-type Arabidopsis plants (Fig. 1B). However, the protein of AtNUDX6 (deduced molecular mass of 31.9 kD) was not detected in the crude extract (100 μg) prepared from Arabidopsis leaves by protein gel-blot analysis using the anti-recombinant AtNUDX6 antibody (Fig. 1C), although the antibody successfully detected 10 ng or less of the recombinant AtNUDX6 protein (data not shown), indicating that the AtNUDX6 protein is barely accumulated in the leaves under normal conditions. The pyrophosphohydrolase activities toward ADP-Rib and NADH in the crude extract were 63.2 ± 1.7 and 95.6 ± 3.0 nmol min−1 mg−1 protein, respectively (Fig. 1D), consistent with previous reports (Ishikawa et al., 2009; Ogawa et al., 2009).
Figure 1.
Characteristics of AtNUDX6-overexpressed or -disrupted Arabidopsis plants. A, T-DNA insertion site in the Arabidopsis KO-nudx6 plants. Exons and introns are represented by boxes and lines, respectively. The deduced start (ATG) and stop (TGA) codons, and each nucleotide number in the mRNA, are indicated. B, Semiquantitative RT-PCR analysis of the AtNUDX6 mRNA in the wild-type (WT), control (transformed with the empty vector), KO-nudx6, and Pro35S:AtNUDX6 (T3 generations of independent transformed lines) plants. The plants were grown on MS medium for 2 weeks under long-day conditions, and then the leaves were used for the analysis. Equal loading of each amplified cDNA was determined with the control Actin2 PCR product. C, Protein gel-blot analysis of the AtNUDX6 protein in the wild-type, control, KO-nudx6, and Pro35S:AtNUDX6 plants. The AtNUDX6 protein was detected by protein gel blotting using a specific polyclonal antibody raised against the recombinant AtNUDX6 protein. The arrow indicates the band of AtNUDX6 protein (31.9 kD). D, ADP-Rib and NADH pyrophosphohydrolase activities in the leaves of wild-type, control, Pro35S:AtNUDX6, and KO-nudx6 plants. Data are means ± sd for three individual experiments (n = 3) using plants grown independently. Different letters indicate significant differences (P < 0.05).
We obtained an Arabidopsis line containing a T-DNA insert in the fourth intron of AtNUDX6 from the SIGnAL project (signal.salk.edu/tabout.html; Fig. 1A). Semiquantitative reverse transcription (RT)-PCR analysis revealed that the T-DNA insertion in the KO-nudx6 plants resulted in the complete loss of AtNUDX6 expression under normal conditions (Fig. 1B). The activity of NADH pyrophosphohydrolase in the leaves of KO-nudx6 plants was reduced to approximately 75% of that in the wild-type plants (Fig. 1D). On the other hand, the activity of ADP-Rib pyrophosphohydrolase in the KO-nudx6 plants was similar to that in the control plants. The growth and appearance of the KO-nudx6 plants on Murashige and Skoog (MS) medium or soil in an isolated growth chamber were almost the same as those of control plants throughout the growth period under normal conditions (data not shown).
We generated transgenic plants overexpressing AtNUDX6 under the control of the cauliflower mosaic virus 35S promoter (Pro35S:AtNUDX6). In semiquantitative RT-PCR analysis, the levels of the AtNUDX6 mRNA were increased in the T3 generation of Pro35S:AtNUDX6 plants compared with the control plants (transformed with the empty vector; Fig. 1D). The AtNUDX6 protein was detected in the crude extract prepared from the leaves of Pro35S:AtNUDX6 plants but not in that of the control plants (Fig. 1C). The pyrophosphohydrolase activity toward NADH in the Pro35S:AtNUDX6-3-1, Pro35S:AtNUDX6-3-3, Pro35S:AtNUDX6-12-5, and Pro35S:AtNUDX6-12-6 plants was 1.2- to 1.4-fold higher than that in the control plants (Fig. 1D). On the other hand, the pyrophosphohydrolase activity toward ADP-Rib in the Pro35S:AtNUDX6 plants was similar to that in the control plants. The Pro35S:AtNUDX6 plants on MS medium or soil in the isolated growth chamber were indistinguishable from the control plants in growth and appearance throughout the growth period under normal conditions (data not shown).
Intracellular Levels of Pyridine Nucleotides and ADP-Rib, and PAR Activity, in the AtNUDX6 Disruptants and Overexpressors
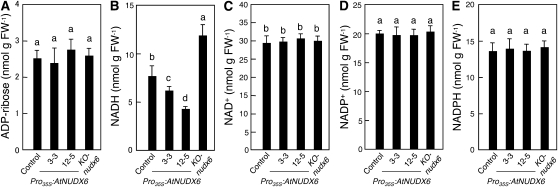
Next, we determined the intracellular levels of pyridine nucleotides and ADP-Rib in the Pro35S:AtNUDX6 and KO-nudx6 plants under normal conditions. The level of NADH was significantly increased in the KO-nudx6 plants compared with the control plants (Fig. 2B). In contrast, the amounts of NADH in the Pro35S:AtNUDX6 plants were markedly reduced. There was no significant difference in the levels of other pyridine nucleotides, NAD, NADP, and NADPH, among the control, Pro35S:AtNUDX6, and KO-nudx6 plants (Fig. 2, C–E). Likewise, the levels of free ADP-Rib in the KO-nudx6 and Pro35S:AtNUDX6 plants were similar to those in the control plants (Fig. 2A). No significant difference was observed in the amount of poly(ADP-Rib), reflecting the degree of the PAR reaction among the control, KO-nudx6, and Pro35S:AtNUDX6 plants (Supplemental Fig. S1).
Figure 2.
Changes in the levels of ADP-Rib and pyridine nucleotides by disruption or overexpression of AtNUDX6. The levels of ADP-Rib (A), NADH (B), NAD+ (C), NADP+ (D), and NADPH (E) in the control, KO-nudx6, and Pro35S:AtNUDX6 plants grown on MS medium for 2 weeks under long-day conditions were determined as described in “Materials and Methods.” Data are means ± sd for at least three individual experiments (n = 3–9) using plants grown independently. Different letters indicate significant differences (P < 0.05). FW, Fresh weight.
Expression of AtNUDX6 and NADH Levels in Different Types of Plant Tissue
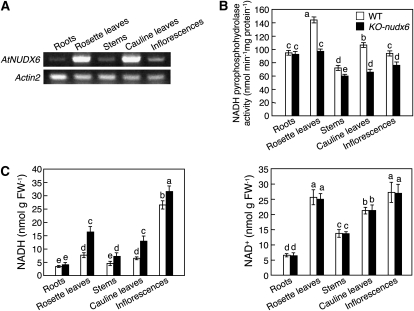
As shown in Figure 3A, the level of AtNUDX6 mRNA was lowest in the roots and highest in the rosette and cauline leaves of 4-week-old control plants grown on soil. Consistent with the transcript levels, the NADH pyrophosphohydrolase activity was highest in the crude extracts prepared from the rosette and cauline leaves of control plants (Fig. 3B). The activity was markedly decreased in the rosette and cauline leaves of the KO-nudx6 plants compared with those of the control plants. Consequently, NADH markedly accumulated in the KO-nudx6 plants (Fig. 3C). There was no significant difference in either the NADH pyrophosphohydrolase activity or the levels of NADH in the roots between the KO-nudx6 and control plants (Fig. 3, B and C). The levels of NAD+ in the respective tissues of the KO-nudx6 plants were similar to those in the control plants.
Figure 3.
Changes in the expression levels of AtNUDX6 and the levels of NADH in different plant tissues. A, Semiquantitative RT-PCR analysis of the AtNUDX6 mRNA in the roots, rosette leaves, stems, cauline leaves, and inflorescences of wild-type Arabidopsis plants grown on soil for 4 weeks under long-day conditions. Equal loading of each amplified cDNA was determined with the control Actin2 PCR product. B, NADH pyrophosphohydrolase activities in respective tissues of the 4-week-old wild-type (WT) and KO-nudx6 plants. C, The levels of NADH and NAD+ in respective tissues of the 4-week-old wild-type and KO-nudx6 plants. The NADH and NAD+ levels were determined as described in “Materials and Methods.” Data are means ± sd for at least three individual experiments (n = 3–6) using plants grown independently. Different letters indicate significant differences (P < 0.05). FW, Fresh weight.
Effect of SA Treatment on the AtNUDX6 Expression
We have demonstrated that the expression of AtNUDX2 and AtNUDX7 was regulated in response to various stressful conditions (Ishikawa et al., 2009; Ogawa et al., 2009). However, the level of AtNUDX6 mRNA was not changed significantly in response to various stressful conditions, such as high-light illumination, drought, salinity, and treatment with paraquat (an agent producing O2−) under high-light illumination (data not shown), suggesting that AtNUDX6 is irrelevant to the response to oxidative stress.
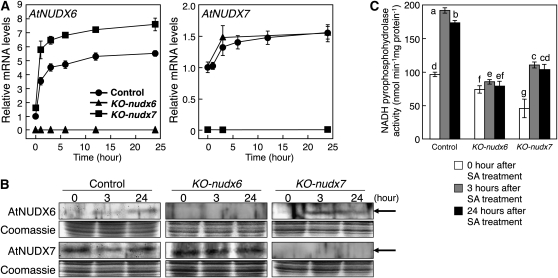
On the other hand, following treatment with 0.5 mm SA, the transcript level of AtNUDX6 was rapidly increased within 1 h in the leaves of the control plants (Fig. 4A), which are associated with expression of AtNUDX6 in the Genevestigator Arabidopsis microarray database (Zimmermann et al., 2004). In addition, according to the database, AtNUDX6 was highly expressed in response to the treatment with SA or an analog molecule, BTH. The AtNUDX6 protein was detected after 3 h of the SA treatment, and its level was increased after 24 h (Fig. 4B). The manipulation for transferring plants in the treatments had no effect on the AtNUDX6 expression (data not shown). No obvious difference was found in the mRNA and protein levels of AtNUDX7 by the treatment with SA. The levels of AtNUDX6 mRNA and protein were increased in KO-nudx7 compared with the control plants under normal conditions and by treatment with SA, although the levels of AtNUDX7 mRNA and protein in the KO-nudx6 plants were similar to those in the control plants (Fig. 4A). The activity of NADH pyrophosphohydrolase in the crude extract prepared from the leaves of control and KO-nudx7 plants was increased by the SA treatment, although the basal activity in the KO-nudx7 plants was significantly lower than that in the control plants (Fig. 4C). On the other hand, the increase in the activity under the treatment was markedly suppressed in the KO-nudx6 plants.
Figure 4.
Changes in the expression levels of AtNUDX6 and AtNUDX7 in response to SA treatment. A, Quantitative RT-PCR analysis of AtNUDX6 and AtNUDX7 expression in the leaves of control, KO-nudx6, and KO-nudx7 plants under the SA treatment. SA treatment was imposed by growing 2-week-old plants on MS medium containing 0.5 mm SA for 0 to 24 h. The relative amounts were normalized to Actin2 mRNA. B, Protein gel-blot analysis of the AtNUDX6 and AtNUDX7 proteins in the leaves of control, KO-nudx6, and KO-nudx7 plants under the SA treatment. Arrows indicate the bands of AtNUDX6 (31.9 kD) and AtNUDX7 (31.8 kD) proteins. C, NADH pyrophosphohydrolase activities in the leaves of control, KO-nudx6, and KO-nudx7 plants under the SA treatment. Data are means ± sd for three individual experiments (n = 3) using plants grown independently. Different letters indicate significant differences (P < 0.05).
Intracellular Levels of NADH, NAD+, and Antioxidants in the AtNUDX6 Disruptants and Overexpressors under SA Treatment
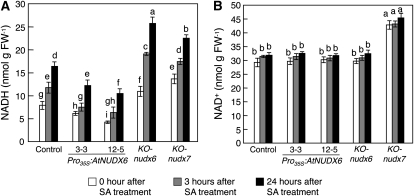
Next, we analyzed the effects of SA treatment on the levels of NADH and NAD+ in the control, Pro35S:AtNUDX6, KO-nudx6, and KO-nudx7 plants. The levels of NADH in the control plants were increased by the SA treatment (Fig. 5). The levels in the Pro35S:AtNUDX6 plants under the SA treatment were significantly low compared with those in the control plants (Fig. 5A). On the other hand, the levels of NADH in both KO-nudx6 and KO-nudx7 were markedly high compared with those in the control plants. Importantly, the degree of increase in NADH level in the KO-nudx6 and Pro35S:AtNUDX6 plants under the treatment was higher and lower, respectively, than that in the control and KO-nudx7 plants. No significant difference was observed in the levels of NAD+ between the control, KO-nudx6, and Pro35S:AtNUDX6 plants, although the levels were constitutively high in the KO-nudx7 plants (Fig. 5B), in agreement with the findings reported previously (Ishikawa et al., 2009). These results clearly indicated that AtNUDX6 regulates NADH levels in response to SA accumulation. There was no difference in the PAR activity regulated by AtNUDX7 between the Pro35S:AtNUDX6 and KO-nudx6 plants and the control plants (Supplemental Fig. S1).
Figure 5.
Changes in the levels of NADH and NAD+ by disruption or overexpression of AtNUDX6 under SA treatment. Experimental conditions are the same as in Figure 4. The levels of NADH (A) and NAD+ (B) in the control, KO-nudx6, Pro35S:AtNUDX6, and KO-nudx7 plants under SA treatment were determined as described in “Materials and Methods.” Data are means ± sd for at least three individual experiments (n = 3–6) using plants grown independently. Different letters indicate significant differences (P < 0.05). FW, Fresh weight.
The levels of reduced glutathione (GSH), a major thiol-disulfide redox buffer in plant cells, and its oxidized form (GSSG) in the Pro35S:AtNUDX6 and KO-nudx6 and KO-nudx7 plants before and after the SA treatment were similar to those in the control plants (Supplemental Fig. S2, A and B). Similarly, there was no difference in the levels of ascorbate (reduced form: AsA) and oxidized AsA (dehydro-AsA; Supplemental Fig. S2, C and D). These findings suggest that changes in NADH level in the cytosol caused by AtNUDX6 are not involved in the perturbation of cellular redox status, probably due to their subcellular distributions; a large amount of AsA and GSH in green tissues are present in the chloroplasts, whereas 45% to 55% of total NADH is in the cytosol (Wigge et al., 1993; Noctor and Foyer, 1998; Meyer and Hell, 2005; Mullineaux and Rausch, 2005; Ishikawa and Shigeoka, 2008). Similar results have been reported in the AtNUDX7 disruptants and overexpressors (Ishikawa et al., 2009).
Effect of Disruption or Overexpression of AtNUDX6 on the Expression of NPR1-Dependent SA-Induced Genes under SA Treatment
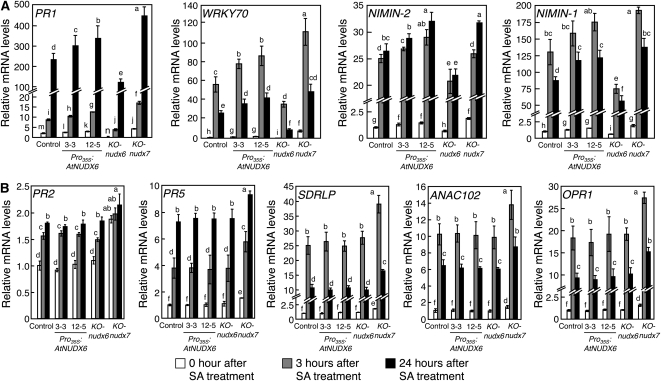
To clarify the roles of AtNUDX6 in the SA signaling pathways, we analyzed the expression of SA-induced genes in the Pro35S:AtNUDX6, KO-nudx6, and KO-nudx7 plants treated with SA (Fig. 6). The transcript levels of NPR1-dependent SAIGs (PATHOGENESIS-RELATED GENE1 [PR1], NIM1-INTERACTING1 [NIMIN1], NIMIN2, and WRKY DNA-BINDING PROTEIN70 [WRKY70]) were significantly decreased and increased in the KO-nudx6 and Pro35S:AtNUDX6 plants, respectively, compared with those in the control plants under normal conditions and SA treatment (Fig. 6A). On the other hand, the transcript levels of NPR1-independent SAIGs (PR2, PR5, ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN102 [ANAC102], 12-OXOPHYTODIENOATE REDUCTASE1 [OPR1], and SDRLP [for short-chain dehydrogenase/reductase family protein]; Blanco et al., 2009; Zhang and Mou, 2009) in the KO-nudx6 and Pro35S:AtNUDX6 plants were mostly similar to those in the control plants (Fig. 6B). The expression levels of both NPR1-dependent and -independent SAIGs were significantly increased in KO-nudx7 compared with those in the control plants under normal conditions and SA treatment.
Figure 6.
Changes in the expression of NPR1-dependent and -independent SA-induced genes by disruption or overexpression of AtNUDX6 under SA treatment. Experimental conditions are the same as in Figure 4. Quantitative PCR analysis was performed to determine the expression levels of NPR1-dependent SA-induced genes (PR1, WRKY70, NIMIN1, and NIMIN2 [A]) and NPR1-independent SA-induced genes (PR2, PR5, SDRLP, ANAC102, and OPR1 [B]) in the control, KO-nudx6, Pro35S:AtNUDX6, and KO-nudx7 plants. The relative amounts were normalized to Actin2 mRNA. Basal levels in the control plants under normal conditions were set at 1.0, and results are expressed as fold increase over the control levels. Data are means ± sd for three individual experiments (n = 3) using plants grown independently. Different letters indicate significant differences (P < 0.05).
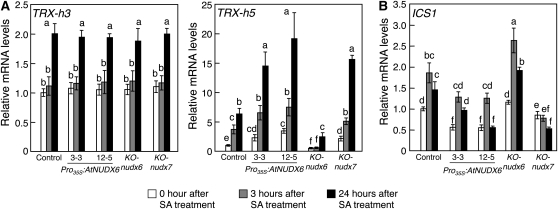
Next, we analyzed the expression levels of genes encoding thioredoxins, TRX-h3 and TRX-h5, which catalyze an SA-induced NPR1 oligomer-to-monomer reaction. The expression level of TRX-h3 and TRX-h5 was markedly increased by treatment with 0.5 mm SA in the control plants (Fig. 7A). The induction of TRX-h5 expression was suppressed in the KO-nudx6 plants, while the induction was accelerated in the Pro35S:AtNUDX6 and KO-nudx7 plants. There was no difference in the levels of TRX-h3 between the control and transgenic plants.
Figure 7.
Changes in the expression of TRXs and ICS1 by disruption or overexpression of AtNUDX6 under SA treatment. Experimental conditions are the same as in Figure 4. Quantitative PCR analysis was performed to determine the expression levels of genes encoding TRXs (TRX-h3 and TRX-h5 [A]) and ICS1 (B) in the control, KO-nudx6, Pro35S:AtNUDX6, and KO-nudx7 plants. The relative amounts were normalized to Actin2 mRNA. Basal levels in the control plants under normal conditions were set at 1.0, and results are expressed as fold increase over the control levels. Data are means ± sd for three individual experiments (n = 3) using plants grown independently. Different letters indicate significant differences (P < 0.05).
Recently, it has been reported that nuclear NPR1 suppresses the expression of isochorismate synthase1 (ICS1), which is required to synthesize SA, through a feedback loop, thus preventing the already elevated SA content from continuing to escalate (Zhang et al., 2009). Under normal conditions and SA treatment, the transcript levels of ICS1 were significantly increased in the KO-nudx6 compared with the control plants, while the levels in Pro35S:AtNUDX6 were decreased similar to the KO-nudx7 plants (Fig. 7B).
Effect of Disruption or Overexpression of AtNUDX6 on SA Tolerance
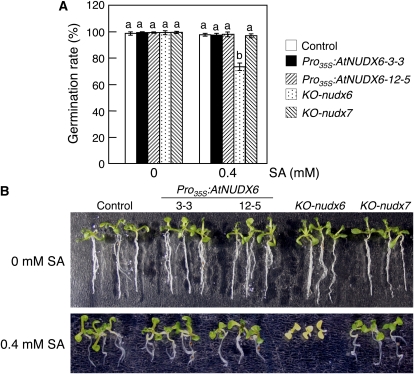
Seeds of control, KO-nudx6, Pro35S:AtNUDX6, and KO-nudx7 plants were plated on MS medium containing 0.4 mm SA, and their germination rates were determined (Fig. 8). In the absence of SA, there was no significant difference in the germination rates among these plants. At 0.4 mm SA, the germination rates of KO-nudx6 plants were significantly lower than those of the control, Pro35S:AtNUDX6, and KO-nudx7 plants, although the rates of Pro35S:AtNUDX6 and KO-nudx7 plants were not altered (Fig. 8A). The KO-nudx6 seedlings developed chlorotic cotyledons and failed to develop beyond the cotyledon stage, while the seedlings of control, Pro35S:AtNUDX6, and KO-nudx7 developed green cotyledons (Fig. 8B).
Figure 8.
Enhancement of SA tolerance by disruption of AtNUDX6 seed germination rates (A) and growth (B) of the control, KO-nudx6, KO-nudx7, and Pro35S:AtNUDX6 plants under SA treatment. A, Seeds plated on MS medium with or without 0.4 mm SA were incubated at 4°C for 3 d and then grown under long-day conditions for 3 d. Germination rates were calculated from results of three replicates using 100 to 150 seeds. Data are means ± sd for at least three individual experiments (n = 3) using plants grown independently. Different letters indicate significant differences (P < 0.05). B, Growth of the control, KO-nudx6, KO-nudx7, and Pro35S:AtNUDX6 seedlings under SA treatment. Seeds plated on MS medium with or without 0.4 mm SA were grown for 7 d.
DISCUSSION
AtNUDX6 Functions as an NADH Pyrophosphohydrolase in Vivo
Nudix hydrolases act in a variety of cellular processes, including maintenance of cellular homeostasis and transduction of accurate signaling for both biotic and abiotic stress (Bartsch et al., 2006; Jambunathan and Mahalingam, 2006; Ge et al., 2007; Adams-Phillips et al., 2008, Ishikawa et al., 2009; Ogawa et al., 2009). First, we studied the physiological properties of an ADP-Rib/NADH pyrophosphohydrolase, AtNUDX6, in the leaves of 2-week-old wild-type (control), KO-nudx6, and Pro35S:AtNUDX6 plants.
We previously reported that AtNUDX7 accounts for 53% and 23% of the total pyrophosphohydrolase activities toward NADH and ADP-Rib, respectively, in the leaves of wild-type plants and thus acts as the predominant ADP-Rib/NADH pyrophosphohydrolase in leaves under normal conditions (Ishikawa et al., 2009). The differences in the amounts of ADP-Rib and NADH, and the activities of pyrophosphohydrolase toward both molecules in the leaves of control, KO-nudx6, and Pro35S:AtNUDX6 plants (Figs. 1 and 2), suggest that AtNUDX6, accounting for 25% of the total NADH pyrophosphohydrolase activity, prefers NADH to ADP-Rib as a physiological substrate but barely contributes to the activity toward ADP-Rib in the leaves under normal conditions. It is well known that pyridine nucleotides, including NADH, mediate hundreds of redox reactions and thus impact on virtually every metabolic pathway (Berger et al., 2004; Hunt et al., 2004; Ying, 2006); therefore, the intracellular NADH level or its redox state are important not only for maintaining a balance between anabolic and catabolic pathways but also for signaling processes in higher organisms (Moller, 2001; Apel and Hirt, 2004; Hunt et al., 2004; Mittler et al., 2004; Foyer and Noctor, 2005; Noctor et al., 2006). By the action of AtNUDX6 as an NADH pyrophosphohydrolase, NADH is hydrolyzed to reduced nicotinamide mononucleotide and AMP (Ogawa et al., 2005). In addition to de novo and salvage pathways, NAD+ is thought to be produced via a novel salvage pathway, by which NADH can be directly generated from nicotinamide mononucleotide and ATP by the action of nicotinamide mononucleotide adenylyl transferase (Berger et al., 2005). Therefore, like AtNUDX7 (Ishikawa et al., 2009), AtNUDX6 may play a regulatory role in the maintenance of pool size and/or redox state of NADH without perturbations of the other pyridine nucleotide levels.
To assess the physiological importance of AtNUDX6 in respective tissues, the expression of AtNUDX6 was analyzed in the roots, rosette leaves, stems, cauline leaves, and inflorescences of 4-week-old Arabidopsis plants. We demonstrated that the transcript levels of AtNUDX6 in the leaves were higher than those in the roots and stems (Ogawa et al., 2005). In agreement with the previous reports, the transcript level and the NADH pyrophosphohydrolase activity of AtNUDX6 were lowest in the roots and highest in the rosette and cauline leaves (Fig. 3). Consistent with the tissue-specific activities, the decrease in the activity of NADH pyrophosphohydrolase and the marked increase in the level of NADH were observed in the rosette and cauline leaves, but not in the roots, of the KO-nudx6 plants. These results suggest that AtNUDX6 distributes primarily in the leaves of Arabidopsis plants and is associated with NADH metabolism.
AtNUDX6 Acts Primarily in NADH Metabolism in Response to SA Signaling
The innate immune responses involve the production of signaling molecules, including reactive oxygen intermediates, nitric oxide, and SA (Malamy et al., 1992; Jabs et al., 1997; Dangl, 1998; Delledonne et al., 2001). SA accumulation has been shown to be necessary for the onset of systemic acquired resistance (SAR), which protects plant cells against further pathogen infection, since the treatment of plant tissues with SA or its functional analogs is sufficient to trigger a defense reaction resembling SAR (Cao et al., 1994; Lawton et al., 1996). The transcript level of AtNUDX6 was increased by not only the infection of virulent and avirulent pathogens but also the application of the SA analogs 2,6-dichloroisonicotinic acid and BTH (Bartsch et al., 2006; Ge et al., 2007; Qiu et al., 2008; Knoth et al., 2009). In addition, AtNUDX7, having enzymatic properties similar to those of AtNUDX6 in vivo, is known to act as a negative regulator of immune responses (Bartsch et al., 2006; Ge et al., 2007). Therefore, we explored the physiological function of AtNUDX6 in SA signaling. Increases in the AtNUDX6 transcript and subsequent accumulation of the protein were observed in the leaves of control plants within 24 h after treatment with 0.5 mm SA, whereas changes in the expression of AtNUDX7 were barely detected under the treatment (Fig. 4). Notably, the SA treatment caused a marked increase in NADH pyrophosphohydrolase activity in the control and KO-nudx7 plants but not in the KO-nudx6 plants. The levels of NADH under normal conditions and the treatment with SA were negatively correlated with the expression levels of AtNUDX6 (Figs. 4 and 5).
The findings presented here indicated that the physiological role of AtNUDX6 considerably differs from that of AtNUDX7. Under normal and stressful conditions, AtNUDX7 accounts for the majority of the total pyrophosphohydrolase activity toward NADH and ADP-Rib, thereby controlling energy and redox homeostasis and modulating defense mechanisms via activation of the PAR reaction against stress (Ishikawa et al., 2009). Importantly, the treatment with SA had no effect on the AtNUDX7 expression (Fig. 4). Since AtNUDX7 was rapidly expressed by various types of oxidative stresses (Jambunathan and Mahalingam, 2006; Ishikawa et al., 2009), the induction may be dependent on the accumulation of ROS or subsequent perturbation of the cellular redox state. On the other hand, AtNUDX6 accounts for the majority of the NADH pyrophosphohydrolase activity in response to the accumulation of SA, probably due to pathogen attacks, and therefore may play a regulatory role in the immune responses through NADH metabolism or subsequent changes in pool size and/or the redox state of the molecule. The increased expression of AtNUDX6 in KO-nudx7 under normal conditions and by the SA treatment in the KO-nudx7 plants (Fig. 4A) may be caused by an accumulation of SA through excess activation of immune responses by depletion of the AtNUDX7 expression (Bartsch et al., 2006; Ge et al., 2007).
AtNUDX6 Serves as a Positive Regulator of the NPR1-Dependent SA Signaling Pathway
NPR1, a positive regulator of SAR, functions in multiple nodes of the SA signaling network (Tada et al., 2008). Innate immune responses are associated with changes in cellular redox states sensed by NPR1 (Cao et al., 1994; Mou et al., 2003; Tada et al., 2008). In uninfected plants, NPR1 resided in the cytoplasm as an oligomer maintained through redox-sensitive intermolecular disulfide bonds (Mou et al. 2003). Either following pathogenic infection or in response to SA treatment, the NPR1 oligomer becomes a monomer by reduction of the disulfide bonds and then moves into the nucleus to activate the transcription of NPR1-dependent SAIGs. However, little is known about the signaling mechanisms from the cellular redox changes to the activation of NPR1. Recent data revealed that the actions of AtNUDX7 in the immune defense response are both dependent on, and independent of, NPR1 and SA (Ge et al., 2007). Therefore, we analyzed the expression of NPR1-dependent SAIGs (PR1, WRKY70, NIMIN1, and NIMIN2) and NPR1-independent SAIGs (PR2, PR5, SDRLP, ANAC102, and OPR1) in the control, Pro35S:AtNUDX6, and KO-nudx6 plants treated with SA (Fig. 6). Notably, the expression of NPR1-dependent SAIGs paralleled that of AtNUDX6 under normal conditions and by the SA treatment, while that of NPR1-independent SAIGs did not change together with expression of AtNUDX6. Depletion of AtNUDX7 expression caused the increased expression of NPR1-dependent and -independent SAIGs under normal conditions and SA treatment (Fig. 6), in agreement with previous reports (Ge et al., 2007). These findings suggest that AtNUDX6 positively regulates the expression of NPR1-dependent SAIGs in the course of SAR against pathogen attacks.
Since AtNUDX7, but not AtNUDX6, acts as a pyrophosphohydrolase toward not only NADH but also ADP-Rib in vivo (Ishikawa et al., 2009), metabolism of ADP-Rib by AtNUDX7 may cause the differences in the physiological actions between AtNUDX6 and AtNUDX7. Notably, it has been suggested that metabolism of ADP-Rib is important for the defense mechanisms against pathogenic attacks, since both the PAR reaction and its degradation reaction catalyzed by poly(ADP-Rib) glycohydrolases (PARG), which hydrolyzed protein-bound poly(ADP-Rib) due to the PAR reaction to free ADP-Rib, were accelerated in response to the attacks (Adams-Phillips et al., 2008, 2010). In addition, the knockout mutation of AtPARG caused accelerated onset of disease symptoms when infected with Botrytis cinerea (Adams-Phillips et al., 2010). These findings strongly support the importance of ADP-Rib metabolism by AtNUDX7 in immune responses.
In addition, accumulation of SA caused by depletion of AtNUDX7 expression (Bartsch et al., 2006; Ge et al., 2007) may at least partially cause the different gene responses between the disruptants of AtNUDX6 and AtNUDX7. Furthermore, it remains a possibility that the actions of AtNUDX6 and/or AtNUDX7 in immune responses are independent of the activity of hydrolysis; these enzymes may regulate the activity of other protein(s) through protein-protein interaction. It has been reported that a Nudix hydrolase in the cauliflower (Brassica oleracea) acts as an inhibitor of Gln synthetase and nitrate reductase (Moorhead et al., 2003).
It has been demonstrated that the SA-induced NPR1 oligomer-to-monomer reaction is regulated by thioredoxin through reduction or oxidation of its intermolecular disulfide bonds (Tada et al., 2008). However, it remains unclear how the expression of thioredoxin is regulated in response to SA accumulation. Among the eight cytosolic TRX-h genes in Arabidopsis, the expression of TRX-h5 was substantially up-regulated upon infection with pathogens, whereas TRX-h3 was the most highly and constitutively expressed TRX-h (Laloi et al., 2004; Tada et al., 2008). The induction of TRX-h5 expression under SA treatment was significantly accelerated and suppressed in the KO-nudx6 and Pro35S:AtNUDX6 plants, respectively, compared with that of the control plants (Fig. 7). On the other hand, the induction of TRX-h5 expression was significantly accelerated in the KO-nudx7 plants, probably due to the accumulation of SA in the mutants reported previously (Bartsch et al., 2006; Ge et al., 2007). These results suggest that AtNUDX6 enhances the expression of TRX-h5, probably via NADH metabolism, in response to SA accumulation and thereby activates subsequent SA signal transduction.
Furthermore, an inverse correlation was observed between the levels of ICS1 and AtNUDX6 under both normal conditions and SA treatment (Fig. 7). An Arabidopsis plant expressing a npr1 with a dysfunctional nuclear localization signal had enhanced sensitivity to the toxicity of high-level SA and an overaccumulation of SA in response to pathogen infection caused by an increase in expression of ICS1, whose product acts on SA synthesis from chorismate, suggested that accurate activation of NPR1 is required to maintain an adequate level of SA (Zhang et al., 2009). Like the npr1 mutants, the KO-nudx6 plants showed enhanced sensitivity to SA compared with the control, Pro35S:AtNUDX6, and KO-nudx7 plants (Fig. 8). These results strongly suggest that AtNUDX6 contributes to controlling the SA level in the cells through modulation of NPR1 activity.
There is fairly general agreement that fine-tuning by both positive and negative regulation of the expression of defense-related genes is important for precise immune responses; excess activation of the responses often causes deleterious effects on normal cell functioning. In conclusion, it was indicated that, through induction of TRX-h5 expression, AtNUDX6 significantly impacts the plant immune response as a positive regulator of NPR1-dependent SA signaling pathways, although it needs further consideration regarding the differences in the regulatory mechanisms of expression and physiological actions between AtNUDX6 and AtNUDX7. A definitive understanding of how NADH metabolism with AtNUDX6 and AtNUDX7 is involved in the regulation of SA signaling will provide important insight into the mechanisms of plant immune responses.
MATERIALS AND METHODS
Materials and Plant Growth Conditions
The vectors for the Gateway cloning system, pDONR201 and pGWB2, were obtained from Dr. T. Nakagawa. Restriction enzymes and modifying enzymes were purchased from TaKaRa. All other chemicals were of analytical grade and used without further purification. Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. Since AtNUDX7 acts as a modulator in immune responses, pathogens and insects that frequently presented in a growth room have been reported to cause delayed growth of KO-nudt7 plants (Ge et al., 2007). Therefore, all experimental data presented here were obtained using ecotype Columbia plants grown on a sterilized MS medium under long-day conditions (16 h of light, 25°C/8 h of dark, 22°C; 100 μE m−2 s−1) in an isolated growth chamber, unless otherwise specifically indicated in the text.
Generation of Transgenic Plants
Total RNA was isolated from the leaves of 4-week-old Arabidopsis plants (1.0 g fresh weight) as described previously (Yoshimura et al., 1999). First-strand cDNA was synthesized using ReverTra Ace reverse transcriptase (Toyobo) with an oligo(dT) primer. The vector for generation of the AtNUDX6-overexpressed plants was constructed using Gateway cloning technology (Invitrogen). The cDNA encoding the open reading frame of AtNUDX6 was cloned into the donor vector, pDONR201, and then recloned into the destination vector, pGWB2. The specific primers with attB1 and attB2 sequences were as follows: attB1-AtNUDX6 (5′-AAAAAGCAGGCTATGGACAATGAAGATCAG-3′) and attB2-AtNUDX6 (5′-AGAAAGCTGGGTCACGTTCTGAAGAAGGGT-3′). PCR and in vitro BP and LR recombination reactions were performed according to the manufacturer's instructions (Invitrogen).
Agrobacterium tumefaciens, which was transformed with the obtained constructs by electroporation, was used to infect Arabidopsis via the vacuum infiltration method. T1 seedlings were selected on basic MS medium in petri dishes containing 3% Suc, 20 mg L−1 hygromycin, and 20 mg L−1 kanamycin for 2 weeks and then transferred to soil. Homozygous T3 seeds were harvested and used for the experiments. The knockout Arabidopsis line (the KO-nudx6 plant; obtained through the SIGnAL project [http://signal.salk.edu/]) containing a T-DNA insert in the AtNUDX6 gene (At2g04450) was outcrossed and selfed to check for segregation and to obtain a purely homozygous line. The knockout Arabidopsis line (the KO-nudx plant) containing a T-DNA insert in the AtNUDX7 gene (At4g12720) was reported as described previously (Ishikawa et al., 2009).
Determination of mRNA Levels
Semiquantitative RT-PCR analysis was performed as described previously (Ogawa et al., 2008, 2009). Total RNA extracted from Arabidopsis leaves was converted into first-strand cDNA using ReverTra Ace (Toyobo) with the oligo(dT)20 primer. The specific primers used for amplification of the cDNAs encoding AtNUDX6 and Actin2 were as follows: AtNUDX6-F (5′-TGGACAATGAAGATCAGGAG-3′), AtNUDX6-R (5′-CAACCAGAGGTGGAGGCTAG-3′), Actin2-F (5′-GAGATCCACATCTGCTGG-3′), and Actin2-R (5′-GCTGAGAGATTCAGGTGCCC-3′). PCR amplification was performed for 20 to 30 cycles of 95°C for 60 s, 55°C for 60 s, and 72°C for 60 s, followed by 72°C for 10 min. Equal loading of each amplified cDNA was determined using the control Actin2 PCR product. The transcript levels were estimated from densitometric readings of three independent experiments and expressed as relative expression ratios.
Quantitative RT-PCR analysis was performed as described previously (Ogawa et al., 2008, 2009). Total RNA extracted from Arabidopsis leaves was purified with an RNeasy Plant Mini Kit (Qiagen), treated with DNase I to eliminate any DNA contamination (TaKaRa), and converted into first-strand cDNA using ReverTra Ace (Toyobo) with the oligo(dT)20 primer. Primer pairs for the quantitative PCR designed using Primer Express software (Applied Biosystems) were as follows; AtNUDX6-F (5′-TGCCAATGCGTCTCATCGTA-3′), AtNUDX6-R (5′-TGGACCACAAGCACCTCTTTG-3′), AtNUDX7-F (5′-CTTGGGATTCGCCATTGTG-3′), AtNUDX7-R (5′-CATGATCCGCATTGCAGTAGAT-3′), AtPR1-F (5′-CGAAAGCTCAAGATAGCCCACA-3′), AtPR1-R (5′-TTCTGCGTAGCTCCGAGCATAG-3′), AtWRKY70-F (5′-CTCAAAATGCTTCATGTGATAACGA-3′), AtWRKY70-R (5′-CCCTTAACGGGTCCCAATCTT-3′), AtNIMIN1-F (5′-GCACGGAAACGTAGACGAGAAG-3′), AtNIMIN1-R (5′-GACCTTTCTCCGCCGTTAGATT-3′), At NIMIN2-F (5′-CACGAACGGTTGCGAAAGTT-3′), At NIMIN2-R (5′-TTCTGACTCCGTTTCCTCTTCTTAGA-3′), AtPR2-F (5′-CACGGCCAACATCCATCTAGAC-3′), AtPR2-R (5′-AACCGAGTCGAGATTTGCGTC-3′), AtPR5-F (5′-GGCGATGGAGGATTTGAATTG-3′), AtPR5-R (5′-GCGTCAAAGTTGCAGCCTGTA-3′), AtSDRLP-F (5′-CCGCCGTTGAGACAATGG-3′), AtSDRLP-R (5′-ACGCAATTCGCGGTTATCC-3′), AtANAC102-F (5′-TCAATCCATGGGAGCTTCCA-3′), AtANAC102-R (5′-CCGGTCTCTATGCGAGAAGAAGT-3′), AtOPR1-F (5′-GTGATTGAAGCGAGAATGAAAACA-3′), AtOPR1-R (5′-CGCTTTCCTCATCGGCATTA-3′), AtTRX-h3-F (5′-AGATTGGACCGAGAAGCTCAAAG-3′), AtTRX-h3-R (5′-GGCACCATGTTGCAGTGAAGT-3′), AtTRX-h5-F (5′-TGCTTGCCATACCCTCGAA-3′), AtTRX-h5-R (5′-GTCTATCACAATCAGTTTCTTGGATTCA-3′), AtICS1-F (5′-CTAATCTCCGCCGTCTCTGAACT-3′), AtICS1-R (5′-TTGGAACCTGTAACCGAACGA-3′), and Actin2-F (5′-GGCAAGTCATCACGATTGG-3′), Actin2-R (5′-CAGCTTCCATTCCCACAAAC-3′). PCR was performed with an Applied Biosystems 7300 Real Time PCR System using SYBR Premix Ex Taq (TaKaRa). Actin2 mRNA was used as an internal standard in all experiments.
Protein Analysis
A polyclonal mouse antibody raised against the AtNUDX6 protein was prepared using the His-tagged recombinant AtNUDX6 protein, synthesized as described previously (Ogawa et al., 2005). Preparation of a polyclonal mouse antibody raised against the AtNUDX7 protein was reported previously (Ishikawa et al., 2009). Protein gel blot analysis was performed as reported (Yoshimura et al., 2004). The AtNUDX6 and AtNUDX7 proteins were detected using each antibody as the primary antibody and anti-mouse IgG-horseradish peroxidase conjugate (Bio-Rad) as the secondary antibody. Protein bands were detected using the enhanced chemiluminescence detection system (GE Healthcare). The protein concentration was determined by the method of Bradford (1976).
Analysis of ADP-Rib and NADH Pyrophosphohydrolase Activities
The leaves (0.5 g) of Arabidopsis plants were homogenized with 1 mL of 100 mm Tris-HCl (pH 8.0) containing 20% glycerol. After centrifugation (20,000g) for 20 min at 4°C, the supernatant was used for analysis of the enzymatic activity. ADP-Rib and NADH pyrophosphohydrolase activities were assayed by coupling to alkaline phosphatase and measuring colorimetrically the amount of inorganic phosphate formed at 37°C (Ames, 1966; Ribeiro et al., 2001; Ishikawa et al., 2009). The standard assay mixture contained, in a volume of 0.1 mL, 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 0.1 mm substrates, 0.7 units of alkaline phosphatase, 1 mg mL−1 bovine serum albumin, and crude extract (approximately 10.0 μg of protein). The reaction was stopped and color developed by the addition of 1 mL of standard inorganic phosphate reagent (6 volumes of 3.4 mm ammonium molybdate in 0.5 m H2SO4, 1 volume of 570 mm AsA, and 1 volume of 130 mm SDS). Blanks without enzyme and/or substrate were run in parallel. Enzyme activities were linear with time and amount of enzyme.
Determination of NAD+, NADH, NADP+, and NADPH Contents
The leaves (0.5 g) of Arabidopsis plants were homogenized with 1 mL of 0.1 n KOH containing 50% ethanol (for determination of NADPH and NADH) or 5 mL of 0.1 n HCl containing 50% ethanol (for determination of NADP+ and NAD+). After centrifugation (15,000g) for 20 min at 4°C, the oxidized and reduced forms of the nucleotide were determined by enzymatic cycling according to the method reported previously (Maciejewska and Kacperska, 1987; Tamoi et al., 2005). The NADP+ and NADPH contents were determined in a reaction mixture containing 100 mm HEPES-KOH (pH 8.0), 0.5 mm EDTA, 2.5 mm Glc-6-P, 1.66 mm phenazine ethosulfate, 0.42 mm 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide, and an extract or a standard solution corresponding to 0 to 500 pmol of NADP(H) in a final volume of 1 mL. The reaction was started by addition of 0.7 unit of Glc-6-P dehydrogenase. For the determination of NAD+ and NADH, the reaction mixture was the same except that Glc-6-P was replaced by 96% ethanol (0.1 mL). For the determination of NADH, the mixture contained 0.2 mm phenazine ethosulfate. The reaction was started by the addition of 0.7 unit of alcohol dehydrogenase. The rate of thiazolyl blue reduction was recorded by measuring the A570.
Determinations of Antioxidants
The levels of AsA and dehydro-AsA were determined spectrophotometrically using AsA oxidase as described previously (Miyagawa et al., 2000). Levels of GSH and GSSG were determined using a glutathione reductase recycling system coupled to 5,5′-dithiobis(2-nitrobenzoic acid) (Shigeoka et al., 1987).
Immunological Detection of PAR Activity
PAR activity was quantified as described previously (Ogawa et al., 2008; Ishikawa et al., 2009). The protein (7.5 μg) extracted from Arabidopsis plants was spotted on a polyvinylidene difluoride membrane (Bio-Rad). The poly(ADP-ribosyl)ated proteins were detected using anti-PAR antibody (BIOMOL) and anti-mouse IgG-horseradish peroxidase conjugate (Bio-Rad) as a secondary antibody. The quantitative intensity was determined by applying densitometry to video images of the blots (ATTO).
Determination of ADP-Rib
ADP-Rib was quantified by capillary electrophoresis-electrospray-tandem mass spectrometry analyses using a P/ACE MDQ capillary electrophoresis system (Beckman Coulter) coupled to a 4000QTRAP hybrid triple quadrupole linear ion-trap mass spectrometer (Applied Biosystems) followed by semipurification using an ICS-3000 system (Dionex) as described previously (Ogawa et al., 2008; Ishikawa et al., 2009).
Stress and SA Treatments
Arabidopsis plants were subjected to various forms of stress: treatment with paraquat and salinity, high light, and drought. Paraquat treatment was imposed by growing 14-d-old plants on MS medium containing the agent at 3 μm for 0 to 7 d under normal light (100 μE m−2 s −1) or for 0 to 12 h under high light (1,600 μE m−2 s −1). Salinity stress was imposed by growing the plants on MS medium containing 250 mm NaCl for 0 to 48 h. Drought stress was imposed by subjecting the plants to dehydration on paper towels for 0 to 6 h. SA treatment was imposed by growing the plants on MS medium containing 0.5 mm SA for 0 to 24 h. For mock treatment, the plants were transferred to MS medium without SA for the same period.
Germination Experiments
Germination assays were performed on three replicates of 100 to 150 seeds for independent experiments. Seeds were incubated on the MS medium containing 0 to 0.8 mm SA for 3 d under long-day conditions. The germination rate of seeds was scored when the radicles completely penetrated the seed coat.
Data Analysis
All measurements were repeated at least three times using plants grown independently. Significance of differences between data sets was evaluated by t test. Calculations were performed with Microsoft Excel software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At2g04450 (AtNUDX6), At4g12720 (AtNUDX7), At2g14610 (AtPR1), At3g57260 (AtPR2), At1g75040 (AtPR5), At1g02450 (AtNIMIN1), At3g25882 (AtNIMIN2), At3g56400 (AtWRKY70), At5g63790 (AtANAC102), At1g76680 (AtOPR1), At4g13180 (AtSDRLP), At5g42980 (AtTRX-h3), At1g45145 (AtTRX-h5), and At1g74710 (AtICS1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Changes in the levels of poly(ADP-Rib) by disruption or overexpression of AtNUDX6 under SA treatment.
Supplemental Figure S2. Changes in the levels of AsA and GSH by disruption or overexpression of AtNUDX6 under SA treatment.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tsuyoshi Nakagawa (Shimane University) for his generous donation of the pDONR201 and pGWB2 vectors.
References
- Adams-Phillips L, Briggs AG, Bent AF. (2010) Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis thaliana to biotic stress. Plant Physiol 152: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L, Wan J, Tan X, Dunning FM, Meyers BC, Michelmore RW, Bent AF. (2008) Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact 21: 646–657 [DOI] [PubMed] [Google Scholar]
- Ames BN. (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 1: 115–118 [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M. (2005) Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280: 36334–36341 [DOI] [PubMed] [Google Scholar]
- Berger F, Ramírez-Hernández MH, Ziegler M. (2004) The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci 29: 111–118 [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “Housecleaning” enzymes. J Biol Chem 271: 25059–25062 [DOI] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Bradford M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. (1998) Innate immunity: plants just say NO to pathogens. Nature 394: 525–527 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y. (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145: 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardana D, Likic VA, Gayler KR. (2009) A comprehensive bioinformatics analysis of the Nudix superfamily in Arabidopsis thaliana. Comp Funct Genomics 2009: 820381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Lerner F, Ziegler M. (2004) NAD: new roles in signalling and gene regulation in plants. New Phytol 163: 31–44 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Ogawa T, Hirosue E, Nakayama Y, Harada K, Fukusaki E, Yoshimura K, Shigeoka S. (2009) Modulation of the poly(ADP-ribosyl)ation reaction via the Arabidopsis ADP-ribose/NADH pyrophosphohydrolase, AtNUDX7, is involved in the response to oxidative stress. Plant Physiol 151: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Shigeoka S. (2008) Recent advances in ascorbate biosynthesis and the physiological significance of ascorbate peroxidase in photosynthesizing organisms. Biosci Biotechnol Biochem 72: 1143–1154 [DOI] [PubMed] [Google Scholar]
- Jabs T, Tschope M, Colling C, Hahlbrock K, Scheel D. (1997) Elicitor-stimulated ion fluxes and O2− from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc Natl Acad Sci USA 94: 4800–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N, Mahalingam R. (2006) Analysis of Arabidopsis Growth Factor Gene 1 (GFG1) encoding a Nudix hydrolase during oxidative signaling. Planta 224: 1–11 [DOI] [PubMed] [Google Scholar]
- Knoth C, Salus MS, Girke T, Eulgem T. (2009) The synthetic elicitor 3,5-dichloroanthranilic acid induces NPR1-dependent and NPR1-independent mechanisms of disease resistance in Arabidopsis. Plant Physiol 150: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraszewska E. (2008) The plant Nudix hydrolase family. Acta Biochim Pol 55: 663–671 [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP. (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134: 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Maciejewska U, Kacperska A. (1987) Changes in the level of oxidized and reduced pyridine nucleotides during cold acclimation of winter rape plants. Physiol Plant 69: 687–691 [Google Scholar]
- Malamy J, Hennig J, Klessig DF. (1992) Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG. (2006) The Nudix hydrolase superfamily. Cell Mol Life Sci 63: 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Hell R. (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86: 435–457 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S. (2000) Evaluation of the defense system in chloroplasts to photooxidative stress caused by paraquat using transgenic tobacco plants expressing catalase from Escherichia coli. Plant Cell Physiol 41: 311–320 [DOI] [PubMed] [Google Scholar]
- Moller IM. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Moorhead GB, Meek SE, Douglas P, Bridges D, Smith CS, Morrice N, MacKintosh C. (2003) Purification of a plant nucleotide pyrophosphatase as a protein that interferes with nitrate reductase and glutamine synthetase assays. Eur J Biochem 270: 1356–1362 [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 13: 935–944 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T. (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86: 459–474 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Gakière B. (2006) NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57: 1603–1620 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ishikawa K, Harada K, Fukusaki E, Yoshimura K, Shigeoka S. (2009) Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress on Arabidopsis plants. Plant J 57: 289–301 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ueda Y, Yoshimura K, Shigeoka S. (2005) Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana. J Biol Chem 280: 25277–25283 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, Tanabe N, Shigeoka S. (2008) Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol 148: 1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Carloto A, Costas MJ, Cameselle JC. (2001) Human placenta hydrolases active on free ADP-ribose: an ADP-sugar pyrophosphatase and specific ADP-ribose pyrophosphatase. Biochim Biophys Acta 1526: 86–94 [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Onishi T, Nakano Y, Kitaoka S. (1987) Characterization and physiological function of glutathione reductase in Euglena gracilis Z. Biochem J 242: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. (2005) The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J 42: 504–513 [DOI] [PubMed] [Google Scholar]
- Wigge B, Krömer S, Gardeström P. (1993) The redox levels and subcellular distribution of pyridine nucleotides in illuminated barley leaf protoplasts studied by rapid fractionation. Physiol Plant 88: 10–18 [Google Scholar]
- Xu W, Dunn CA, Jones CR, D'Souza G, Bessman MJ. (2004) The 26 Nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J Biol Chem 279: 24861–24865 [DOI] [PubMed] [Google Scholar]
- Ying W. (2006) NAD+ and NADH in cellular functions and cell death. Front Biosci 11: 3129–3148 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Miyao K, Gaber A, Takeda T, Kanaboshi H, Miyasaka H, Shigeoka S. (2004) Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J 37: 21–33 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Yabuta Y, Tamoi M, Ishikawa T, Shigeoka S. (1999) Alternatively spliced mRNA variants of chloroplast ascorbate peroxidase isoenzymes in spinach leaves. Biochem J 338: 41–48 [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen S, Mou Z. (2009) Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J Plant Physiol 167: 144–148 [DOI] [PubMed] [Google Scholar]
- Zhang X, Mou Z. (2009) Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J 57: 302–312 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.