Abstract
A major goal of the life sciences is to understand how molecular processes control phenotypes. Because understanding biological systems relies on the work of multiple laboratories, biologists implicitly assume that organisms with the same genotype will display similar phenotypes when grown in comparable conditions. We investigated to what extent this holds true for leaf growth variables and metabolite and transcriptome profiles of three Arabidopsis (Arabidopsis thaliana) genotypes grown in 10 laboratories using a standardized and detailed protocol. A core group of four laboratories generated similar leaf growth phenotypes, demonstrating that standardization is possible. But some laboratories presented significant differences in some leaf growth variables, sometimes changing the genotype ranking. Metabolite profiles derived from the same leaf displayed a strong genotype × environment (laboratory) component. Genotypes could be separated on the basis of their metabolic signature, but only when the analysis was limited to samples derived from one laboratory. Transcriptome data revealed considerable plant-to-plant variation, but the standardization ensured that interlaboratory variation was not considerably larger than intralaboratory variation. The different impacts of the standardization on phenotypes and molecular profiles could result from differences of temporal scale between processes involved at these organizational levels. Our findings underscore the challenge of describing, monitoring, and precisely controlling environmental conditions but also demonstrate that dedicated efforts can result in reproducible data across multiple laboratories. Finally, our comparative analysis revealed that small variations in growing conditions (light quality principally) and handling of plants can account for significant differences in phenotypes and molecular profiles obtained in independent laboratories.
Growth results from a complex network of processes occurring at multiple interconnected organizational levels (molecules, molecular complexes, cells, organs, and the whole plant) and is characterized by successive stages of cellular differentiation. During leaf emergence, cells first proliferate mitotically (phase I), then expand and enlarge their vacuole (phase II). When leaf growth is completed, the fully expanded cells modify and strengthen their walls (phase III; Donnelly et al., 1999; Beemster et al., 2005). Some of the molecular networks that regulate leaf development have been identified, and their role in the proliferation phase is best understood (Wasteneys and Galway, 2003; Hussey et al., 2006; Inzé and De Veylder, 2006). Gene expression analysis during leaf development of Arabidopsis (Arabidopsis thaliana) showed that the transition from cell proliferation to expansion and differentiation is marked by major transcriptional changes (Beemster et al., 2005).
Complex interactions between the different organizational levels have been revealed through the compensatory mechanisms observed in mutants. For example, certain mutants grow normal-sized leaves in comparison with the wild type while producing fewer cells of larger size (Hemerly et al., 1995; Ferjani et al., 2007). Furthermore, some processes controlled at the organism level, such as leaf production or time to flowering, influence cellular and molecular processes that determine the number and size of leaf cells (Cookson et al., 2007; Tisné et al., 2008). Although leaf development has been studied for many years, the fragmented knowledge about leaf growth remains to be assembled into a global and coherent model. This challenge requires multidisciplinary approaches and collaboration between laboratories with complementary expertise. The AGRON-OMICS European consortium (http://www.agron-omics.eu/) was formed to identify molecular components that control leaf growth and to model how such components coordinate their functions to control quantitative leaf growth phenotypes.
An important bottleneck in systems-oriented approaches is the experimental reproducibility (i.e. the ability to compare results produced by different laboratories or in different experiments; Schilling et al., 2008). The scientific community has launched several initiatives to standardize biological data and to facilitate their exchange. The Gene Ontology (GO) consortium has developed a controlled vocabulary that describes biological processes, molecular functions, and cellular distribution of genes and gene products (Gene Ontology Consortium, 2000). The Plant Ontology consortium has built other vocabularies that define plant structures and developmental stages (Jaiswal et al., 2005). Furthermore, online reference databases and public repositories explicitly document the metadata necessary for accurate interpretation of experimental results, including the origin of the samples, the precise environmental conditions in which they were obtained, and the protocols implemented to produce and analyze the data. Specific protocols have been proposed to record the “minimum set of information” to append to microarray or proteomics data (Brazma et al., 2001; Zimmermann et al., 2006; Taylor et al., 2007). However, few attempts have been made so far to assess whether strict standardization of experimental procedures produces comparable plant macroscopic and molecular phenotypes between different laboratories (Dolezel et al., 1998). Some multisite data exist, but it is often difficult to disentangle genotype × environment interactions. Other data do not take into account the experimental reproducibility of the environmental conditions or, at the molecular level, intersite technical differences of the measurements.
In this study, we investigated whether three Arabidopsis ecotypes can be distinguished unambiguously on the basis of leaf phenotypes and related molecular descriptors when grown in 10 independent laboratories adhering to a standard operating protocol. The data types that were included in this comparative analysis are (1) a set of leaf growth variables collected at three organizational levels (whole plant, leaf, and cells) on fully expanded rosettes and (2) the metabolite and (3) transcript profiles of a fully expanded leaf from the same rosettes. The analysis of this comprehensive data set indicated that standardization is challenging but can produce reproducible results across locations and genotypes and that variability depends on the data types considered.
RESULTS
Rationale for the Reference Experiment
The selected ecotypes compared in this study were Columbia (Col-4), Landsberg erecta (Ler), and Wassilewskija (Ws), the most commonly studied Arabidopsis accessions. To rigorously define their respective leaf growth characteristics, the three genotypes were cultivated in an automated phenotyping platform designed for the strict control of environmental conditions (PHENOPSIS; Granier et al., 2006) in the laboratory hereafter designated L3. In this reference experiment, soil water content, temperature, humidity, light, and nutrition were chosen for optimal leaf growth performance. A large set of rosette and leaf macroscopic and microscopic variables were measured throughout the growth period (from stage 1.0 to stage 6.0; Boyes et al., 2001) to investigate phenotypic differences between the three genotypes.
Leaf Growth Kinetics Reveal Phenotypic Differences between Col-4, Ler, and Ws in the Reference Experiment
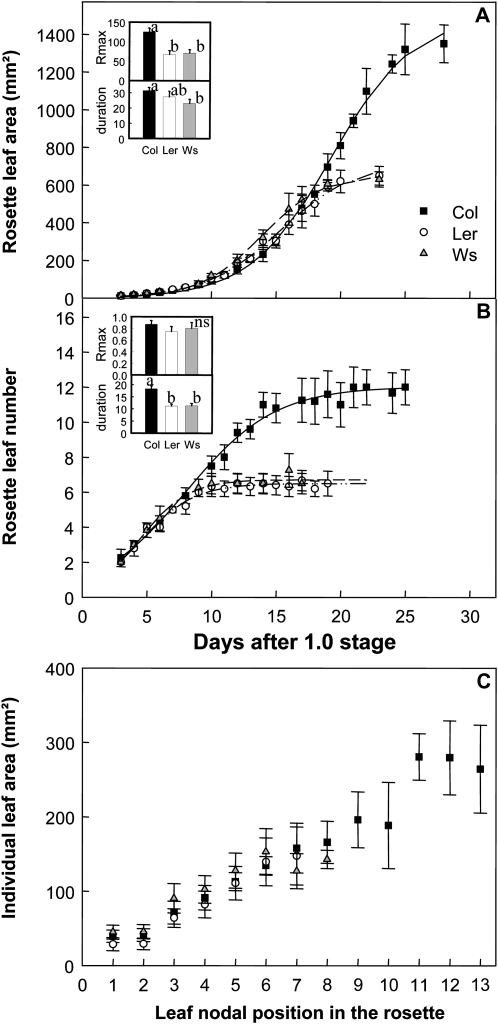
The leaf growth kinetics were followed in the three genotypes from stage 1.0 until the end of growth. The temporal increase in rosette leaf area (RA) of the three genotypes was best fitted with a sigmoid curve (Fig. 1A). During phase I, the increase in RA was small and almost identical for all genotypes. Then, the rosette leaf expansion rate peaked and finally declined until full expansion was complete (Fig. 1A). In Col-4, the maximal rate was higher compared with the other two genotypes and the duration of rosette expansion was longer than for Ws (Fig. 1A, inset). Thus, final RA was significantly larger for Col-4 (Col-4, 1,574 mm2; Ler, 621 mm2; Ws, 661 mm2; Fig. 1A). Col-4 formed more rosette leaves (leaf number [LN]; Col-4, 12; Ler and Ws, six; Fig. 1B), but the final area of leaves at the same nodal position did not differ significantly between genotypes (Fig. 1C). The higher number of leaves observed for Col-4 was due to a longer duration of leaf production, while the rate of leaf production did not differ significantly between genotypes (Fig. 1B, inset).
Figure 1.
Macroscopic kinematic leaf growth phenotypes. A, Changes with time of RA. B, Changes with time of rosette LN. C, Individual final leaf area according to leaf nodal position in the rosette. The increase of RA and rosette LN is described by y = A/[1 + e (−(X−X0)/B)]. The inset in A shows mean and sd values of maximal rate and duration of rosette leaf expansion (Rmax in mm2 d−1 and duration in d), and the inset in B shows mean and sd values of maximal rate and duration of leaf production (Rmax in leaf no. d−1 and duration in d). Lowercase letters indicate significant differences (P < 0.05). ns, No significant difference.
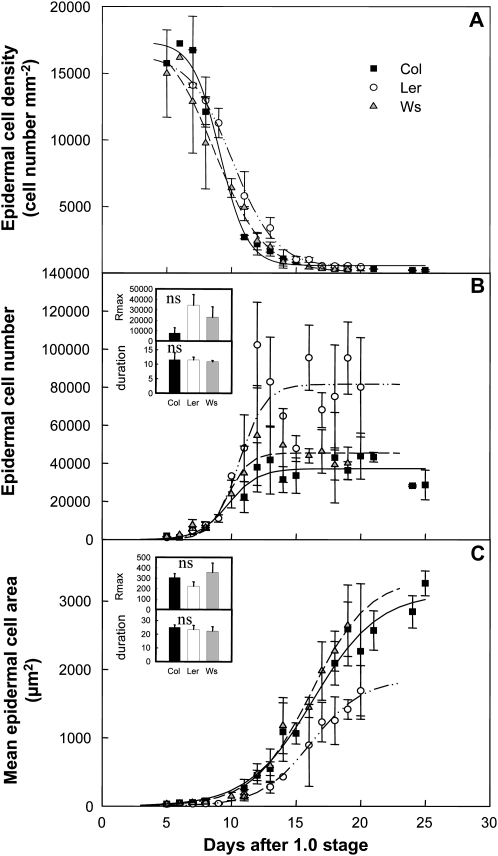
At the cellular level, changes in epidermal cell density (CD) followed a similar curve over time for the three genotypes (Fig. 2A). In young leaves, CD was high during the initial phase coinciding with active cell proliferation and then declined until the end of leaf expansion (Fig. 2A). The epidermal cell number (CN) and cell area (CA) kinetics were not synchronous: in all genotypes, the rapid increase in CN (Fig. 2B) occurred earlier than the increase in CA (Fig. 2C). The maximal cell production rate tended to be higher in Ler than in Ws and Col-4, whereas the duration of cell production did not differ between genotypes (Fig. 2B, inset). The final CN did not differ significantly between Col-4 and Ws but was twice as high in Ler (Fig. 2B). The maximal cell expansion rate tended to be lower in Ler leaves than in Col-4 and Ws, but the duration of cell expansion did not differ between the genotypes (Fig. 2C, inset). The final CA in Ler was half that of the other two genotypes (Fig. 2C). Thus, Ws and Col-4 had very similar leaf cellular phenotypes, whereas Ler differed significantly with a higher number of cells of smaller size.
Figure 2.
Microscopic kinematic leaf growth phenotypes. A, Epidermal CD in leaf 6. B, Epidermal CN in leaf 6. C, Mean epidermal CA in leaf 6. All data were collected from the sixth leaf of the rosette. The decline of epidermal CD is described by y = D + [(A − D)/(1 + 10(X−log(C))B)], and the increases of epidermal CN and CA are shown by y = A/[1 + e (−(X−X0)/B)]. The inset in B shows mean and sd values of maximal rate and duration of cell production (Rmax in cell no. d−1 and duration in d), and the inset in C shows mean and sd values of maximal rate and duration of cell expansion (Rmax in μm2 d−1 and duration in d). ns, No difference.
Because leaf growth variables at rosette, leaf, and cell levels at the final stage of rosette development were sufficient to discriminate the three genotypes, the phenotypic comparison between laboratories in the comparative experiment was based exclusively on end point data sets, without considering growth dynamics.
Rationale and Conditions for the Comparative Experiment
Based on the growth conditions defined in the reference experiment, a very detailed protocol was established and shared with the other nine laboratories (Supplemental Protocol S1 with the aim to reproduce the reference growth conditions as closely as possible within the constraints of each facility. All growth chambers involved were maintained by expert plant biologists and have been operating routinely for years. To test the reproducibility of growth variables and molecular profiles, multiple plants of each genotype were grown by each laboratory using identical seed batches (see “Materials and Methods”). Sample preparation and data collection were organized to minimize technical sources of variability. Pots, soil substrate, and nutrients were distributed by L3 to the other nine laboratories and therefore were exactly the same as in the reference experiment. In each laboratory, samples (frozen fifth leaf) and images of the plants (scans and imprints) were collected at the same stage of rosette leaf development when all leaves were fully expanded (stage 6.0; Boyes et al., 2001) and at the same time of the day (middle of the light period). At the end of the experiments, scans and imprints were sent to L3 for the measurements of macroscopic and microscopic leaf growth variables obtained in each laboratory. Harvested frozen plant material was sent to L4 for grinding and preparing aliquots. L4 and L6 profiled the metabolites and the transcripts, respectively.
A Consensus Phenotype Was Reached by a Core Group of Four Laboratories in the Comparative Experiment
The actual growth chamber conditions did not always match exactly those defined in the reference protocol and were sometimes not fully documented (Table I). However, this variability was small considering such a large-scale analysis in 10 different laboratories. Furthermore, such variations offer a fair representation of those that are typically encountered when reproducing published protocols.
Table I. Growth conditions reported by each laboratory.
| Laboratory | Watering Regime | Daylength | Air Temperature Day/Night | Light Quality: Type of Lamps (Manufacturer) | Light Intensity | Air Humidity | Distance between Lamps and Rosettes | No. of Repetitions for Col-4/Ler/Ws | Environmental Sensors for Light/Temperature and Humidity |
| h | °C | μmol m−2 s−1 | % | cm | |||||
| L1 | Daily adjustment to 40% | 16 | 20/21.5 | Fluo tubes (Osram Fluora –77, Osram Lumlux 31–830) | 120–130 | 70–75 | 85 | 10/3/7 | Sensors that equip the phytotron VB1014 from Vötsch Industrietechnik |
| L2 | Subirrigation to maximum capacity | 16 | 20/21.5 | Tubes (Osram Lumlux 36W/865) | 100 | 70–75 | 120 | 9/10/10 | Licor LI-185B (Licor)/Excel 500 (Honeywell) |
| L3 | Daily adjustment to 40% | 16 | 20/21.5 | Mix of cool-white fluorescent tubes (Sylvania Grolux 36 W) and HQI lamp (Philips, HPI T+ 400 W E40) | 150 | 70–75 | 140 | 10/10/9 | SKP215 (Skye Instruments)/HMP45C (Campbell Scientific) |
| L4 | Daily adjustment to 40% | 16 | 20/21.5 | Mix of sodium (Philips SON-T-AGRO 400 W) and HQI lamps (Philips HPI-T+ 400 W) | 130–170 | 70–75 | 200 | 10/4/10 | No information |
| L5 | Daily adjustment to 40% | 16 | 20/21.5 | Cool and warm white (Sylvania Luxline plus, six lamps 36 W) | No information | 70–75 | 29 | 9/7/9 | LI-210SA (Licor)/Pt-100 (Rotronic)/Hygro C-94 (Rotronic) |
| L6 | 5 mL added daily | 16 | 20/22 | Fluo tubes (Sylvania, F96T12-215W, F72T12-160W, F48T12-115W) and normal light (Sylvania 100 W) | 180 | 70–75 | 200 | 10/10/9 | LI-190SA (Licor)/HMD40Y (Vaisala) |
| L7 | Daily adjustment to 40% | 16 | 20/18 | Tungsten bulbs and HQI lamp (household 60 W and Osram T 400 W) | 140 | 70–75 | Not recorded | 9/10/6 | SKP 215 (Skye Instruments)/1–200 series (Rotronic) |
| L8 | Daily adjustment to 40% | 16 | 20/21.5 | Cool-white fluorescent tubes (Philips, 830HF 32 W) | 150 | 70–75 | Not recorded | 8/8/6 | SKP 200 (Skye Instruments)/H290 (Rotronic) |
| L9 | Daily adjustment to 40% | 16 | 19.3/21.1 | Sodium lamps (Osram, L58W/535) | 150 | 70–75 | 100 | 10/6/3 | SKP 215 (Skye Instruments)/Pen type thermo-hygrometer (Huger Electronics) |
The experiment underscored that precise control of environmental conditions is challenging and pointed out how laboratories can best achieve the standardization of a growth protocol in practice. Nine of the 10 laboratories successfully cultivated the plants, although not all succeeded in harvesting 10 individuals per genotype by the end of the experiment. The main problem encountered by the researchers in charge of the experiment was related to early stages of plant development. Seeds germinated but plants died before emergence of the first pair of leaves (stage 1.02; Boyes et al., 2001), mostly as a result of soil drying at the surface of the pots. Data from the laboratory that was apparently unable to get proper germination and/or early growth were excluded from the subsequent analyses.
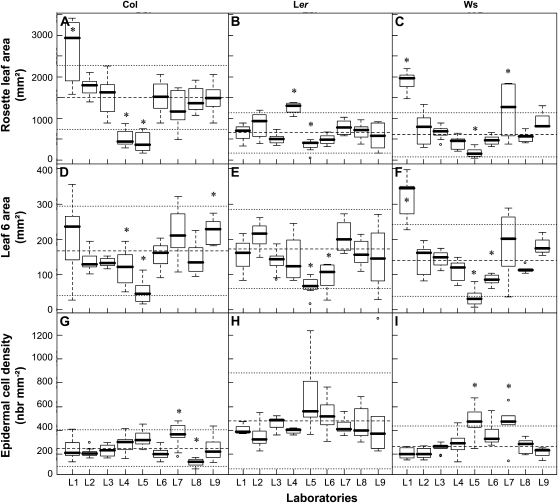
The variability of the leaf growth phenotype was assessed through the comparison of three growth variables at three organizational levels: RA, area of leaf 6 (AL6), and epidermal CD of leaf 6 (Fig. 3). Rosette fresh weight was also analyzed but showed the same trends as RA (Supplemental Fig. S1A). All data were collected at stage 6.0, when the sixth leaf is mature and fully expanded. Overall, the results revealed a large variability both within and between laboratories (Fig. 3). RA values varied 20-, 28-, and 58-fold for Col-4, Ler, and Ws, respectively, AL6 values varied 21-, 17-, and 56-fold, and CD values varied 6-, 6-, and 5-fold. In some cases, the variability within a laboratory was as large as the variability between laboratories (e.g. AL6 for Col-4 in L1 and for Ler in L7). In general, variability and ratios between maximum and minimum values were smaller for CD than for AL6 and RA. Ler had the least variability across all variables.
Figure 3.
Leaf growth phenotypes of three genotypes across nine independent laboratories. Data are represented as box plots with a representation of the first, median, and third quartiles in the box. A to C, RA. D to F, Sixth leaf area. G to I, Epidermal CD in the sixth leaf. Laboratories (L1–L9) with significantly different phenotypes are noted with asterisks for each genotype and each variable (ANOVA and Tukey's posthoc test results). Mean and sd values of homogenous groups are represented on each plot by dashed and dotted lines, respectively. A core group of four laboratories with similar results for all genotypes and at the different organizational levels was identified: L2, L3, L8, and L9.
Noticeably, the laboratories belonging to the largest group with a consensus phenotype for one variable sometimes differed for other variables. For example, L7 and L8 significantly differed from the largest group with the same CD in Col-4 but not for the other two variables in the same genotype. L1 had higher RA in Col-4 compared with the other laboratories but similar AL6 and CD. L5 differed from the others with a clear “laboratory effect,” since it had lower RA and AL6 than the consensus group for all genotypes and a higher CD in Ws.
Considering the complete phenotypic data set, L2 and L3 always belonged to a consensus group, while L8 and L9 differed from it in only one case (CD and AL6 in Col-4, respectively). Therefore L2, L3, L8, and L9 were grouped as the core laboratories.
Genotype Rank Based on Growth Phenotypes Was Not Always Conserved across Laboratories
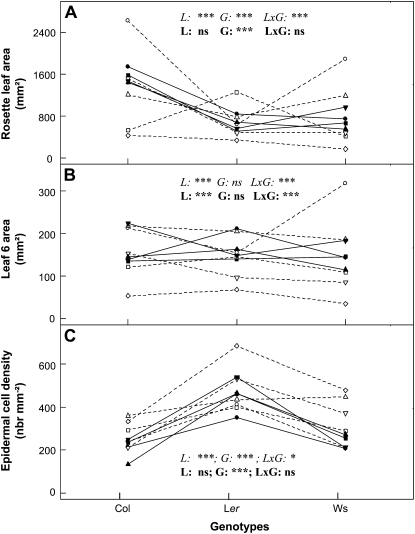
The mean value of each leaf growth variable was compared per genotype for all laboratories (Fig. 4; Supplemental Fig. S1B). ANOVA revealed significant differences between genotypes (G effect) for both RA and CD but not for AL6 (Fig. 4). This observation is consistent with the results of the reference experiment (Figs. 1 and 2). ANOVA also revealed significant differences between laboratories (L effect) and different genotype ranking between laboratories (L × G effect).
Figure 4.
Phenotype ranking per genotype across laboratories. Mean values of RA (A), sixth leaf area (B), and epidermal CD in the sixth leaf (C) calculated per genotype and per laboratory are shown. L1, White circles; L2, black circles; L3, black squares; L4, white squares; L5, white diamonds; L6, white lower triangles; L7, white upper triangles; L8, black upper triangles; L9 black lower triangles. The core laboratories (L2, L3, L8, and L9) are represented by black symbols and straight lines and the others with white symbols and dashed lines. For each variable, ANOVA was performed either on the data including all nine laboratories (italics) or the core laboratories (boldface) to evaluate laboratory (L), genotype (G), and interaction (L×G) effects. Asterisks indicate significant differences (*** P < 0.001, * P < 0.05), and ns indicates the absence of a significant difference.
When only considering the core laboratories, ANOVA revealed that RA and CD mean values were significantly different between genotypes (G effect) but did not show laboratory (L) or interaction (L × G) effects (Fig. 4, A and C). Nevertheless, AL6 still displayed a laboratory effect, probably emphasized because the difference between genotypes was not significant for this variable (Fig. 4B). Rosette fresh weight presented very similar results to RA (Supplemental Fig. S1B) due to significant correlations between RA and fresh weight for all laboratories (Supplemental Fig. S1C). However, the slopes of the correlations between RA and fresh weight were tested by analysis of covariance, and the test assumed that differences in the slopes were highly significant between laboratories.
Data obtained by the four core laboratories (L2, L3, L8, and L9) were in agreement with those of the reference experiment (L3). RA was significantly higher for Col-4 due to a larger number of rosette leaves, whereas individual leaf area did not differ when comparing leaves at the same nodal position (Supplemental Fig. S2, A–C). CD was significantly higher in Ler due to a smaller CA and a higher CN (Supplemental Fig. S2, D–F).
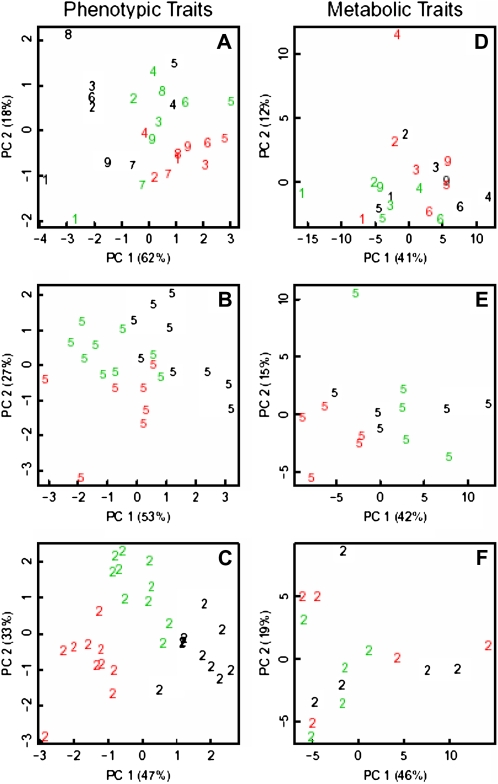
A principal component analysis (PCA) combining five leaf growth variables (LN, RA, AL6, CD, and CA) from all laboratories indicated a good separation between genotypes (Fig. 5A). Considering ranking differences among genotypes between laboratories, genotype discrimination was also tested separately for each laboratory. In all laboratories, within the core group (e.g. L2; Fig. 5C) or outside (e.g. L5; Fig. 5B), PCA clearly discriminated the genotypes.
Figure 5.
PCA of leaf growth variables (rosette LN, RA, AL6, epidermal CD in leaf 6, and mean epidermal CA in leaf 6; A–C) and metabolic profiles (D–F). Graphs represent data averaged from all laboratories (A and D) or individual replicates from L5 (B and E) and L2 (C and F). Colors indicate the genotype: Col, black; Ler, red; Ws, green. Numbers identify laboratories.
Genotypes Were Not Well Separated by Metabolite Profiles
Leaf growth variables are complex readouts integrating numerous cellular and biochemical processes. To determine to what extent phenotypic variation between laboratories and genotypes is reflected at the molecular level, leaf samples were also analyzed for metabolite content with gas chromatography-mass spectrometry (GC-MS)-based profiling techniques (Lisec et al., 2008). Metabolic profiles are very sensitive phenotypes that can differentiate between genotypes, environments, and developmental stages (Fernie et al., 2004). To this end, five biological replicates of the complete fifth leaf (stage 6.0) of each genotype from seven of the 10 laboratories were analyzed, and more than 80 annotated metabolites were detected in every sample.
PCAs integrating metabolite profiles or leaf growth variables (Fig. 5) were compared to assess the discriminative power of the two data types. Contrary to growth variables, the metabolic traits in this experiment did not strongly discriminate genotypes, as illustrated by their interspersed distribution in the graph representing the two first components (Fig. 5, compare D with A).
Assuming that metabolite profiles are more sensitive to small environmental changes, the discrimination of genotypes was also tested separately for each laboratory. The five metabolite profiles (one per leaf) recorded for each laboratory and genotype were treated as separate data points in PCA, thus diminishing the potential impact of different growth conditions between sites. In this case, the influence of the genotype was noticeable by visual inspection in four of the seven laboratories for which metabolic data were available (L1, L3, L4, and L5). As illustrative examples of these two scenarios, at least two genotypes are clearly separated in L5 data but no such separation can be observed in L2 (Fig. 5, E and F). Taken together, these data suggest that the metabolite profiles are very sensitive even to small environmental changes within the same growth chamber and therefore may yield readouts suitable for measuring genotype × environment interaction.
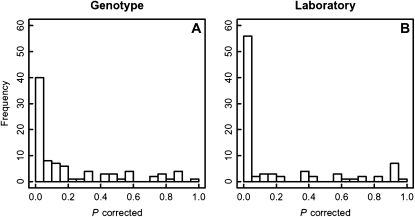
To further investigate the relative contributions of genotype and environment, we performed ANOVA for all 89 metabolites measured in all samples. Genotype significantly influenced only 40 of them (Fig. 6A), and the rank of metabolite relative levels was stable across genotypes (data not shown). In contrast, laboratory (environment) showed a predominant influence on 55 metabolites (Fig. 6B), with 21 metabolites being significantly influenced only by environment at P = 0.05, whereas the 34 others also showed a significant influence of the genotype (although with a smaller statistical power; for a list of all metabolites and their respective P values, see Supplemental Table S2).
Figure 6.
Histograms of P values from an ANOVA of all metabolites with the factors genotype (A) and laboratory (B).
Closer inspection of the 21 metabolites associated mainly with laboratory environment revealed that they were covering all major parts of primary metabolism, including the tricarboxylic acid cycle (malic acid), amino acids (including Leu, Pro, and Tyr), redox state-sensitive metabolites such as ascorbic acid, and sugar and sugar alcohols such as maltose and raffinose.
In contrast, lipids were largely excluded from this group, indicating a smaller environmental influence on lipid composition (at least with respect to the fatty acid composition). Another interesting metabolite that showed in contrast a stronger influence of the genotype is Suc, suggesting a greater buffering of Suc levels.
Genotypes Were Distinguished by Transcript Profiles
To test whether variation in morphological, histological, and metabolic phenotypes is reflected in transcriptional changes, we profiled the transcriptome of 41 samples from the comparative experiment using a tiling microarray representing both strands of the Arabidopsis genome. Each RNA sample was extracted from the fifth leaf (stage 6.0) of a single plant. Signals above background were detected on average for 12,243 genes (minimum, 11,454; maximum, 12,757), demonstrating that at least 40% of all annotated Arabidopsis genes are transcribed in fully expanded rosette leaves.
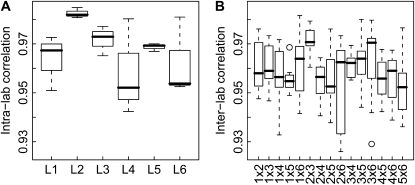
First, we used the 18 Col-4 samples to analyze the correlation of expression values. Intralaboratory correlation was generally good (between 0.943 and 0.985, with a median of 0.970), confirming the concordance between samples from individual laboratories. Correlation between replicates from the same laboratory was generally highest and most uniform for L2, L3, and L5, while it was slightly lower and more variable for L1, L4, and L6 (Fig. 7). Interlaboratory correlation (between 0.927 and 0.982, with a median of 0.959) was only slightly lower than intralaboratory correlation. Correlations between samples from various pairs of laboratories were similar to each other, but correlation was highest for samples from L2 and L3, which belong to the core laboratories identified for leaf growth variables. In addition to global correlation coefficients, we also used gene-wise coefficients of variation (CV; i.e. the ratio of the sd to the mean) to analyze similarity between samples. The CV values for intralaboratory replication were generally small (median values between 3.1% and 4.3%) and lowest for L2 (Supplemental Fig. S3). Interlaboratory CV values were in the range between 4.2% and 5.0%. The similarity of the intralaboratory median CV value (4.0%) and the interlaboratory median CV value (4.7%) suggests that intralaboratory variance is comparable to interlaboratory variance.
Figure 7.
Box plots of genome-wide correlation coefficients for gene expression values in replicate samples. A, Intralaboratory correlation coefficients. B, Interlaboratory correlation coefficients.
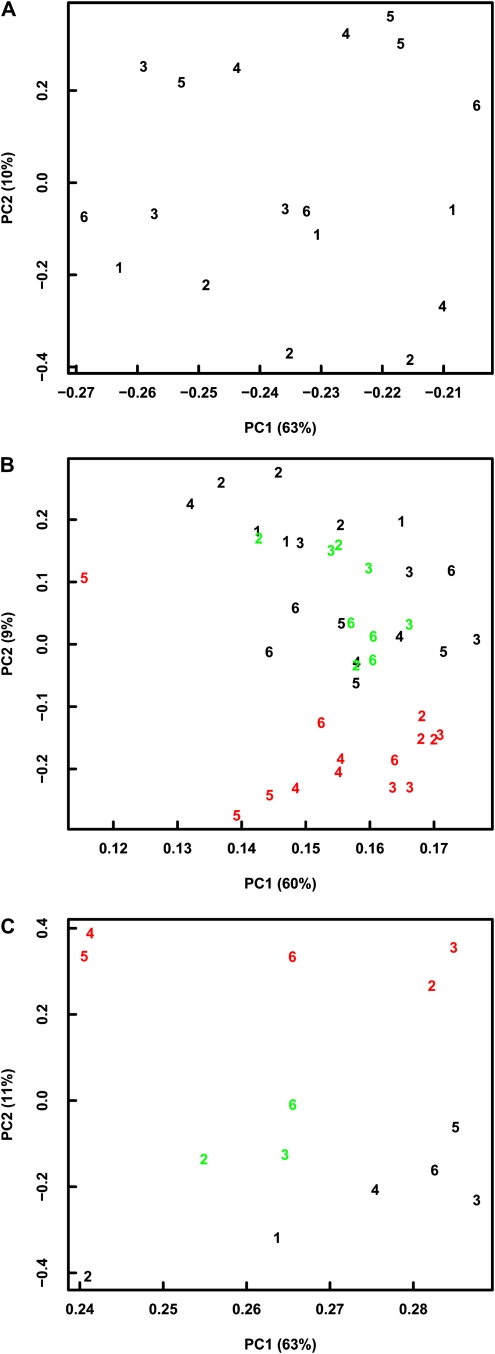
We used hierarchical clustering and PCA to visualize similarities between samples. Based on the 500 genes with maximal relative variance (according to ranked CV values) in the 18 Col-4 samples, hierarchical clustering identified the common origin of samples from L1 as well as L2 but not from L3, L4, L5, and L6 (Supplemental Fig. S4A). In a plot of the first two principal components revealed by PCA, replicates from the same laboratory were often spread along PC1 (compare L1, L5, and L6), which explains more than 60% of the total variance (Fig. 8A). Thus, the major source of variance for the top 500 variable genes is variation between replicate plants within laboratories. The origin of the samples often determined the position along PC2, which explains only about 10% of the total variance. Therefore, variability of gene expression values was to a lesser degree determined by laboratory effects, confirming the successful standardization of growth conditions between laboratories as probed by transcriptional changes. When including the Ler and Ws samples, hierarchical clustering usually identified the common genetic backgrounds of samples, with the exception of one Col-4 and one Ws sample that could not be grouped correctly (Supplemental Fig. S4B). In contrast, the origin of samples was again only poorly recovered. In a plot of the first two principal components revealed by PCA, Ws samples were well separated from Ler and Col-4 samples but Ler and Col-4 samples were not well separated from each other (Fig. 8A). Similarly, samples of identical laboratory origin did not cluster well in the PCA plot. Thus, analysis of correlation, of CV values, and PCA all revealed that variance in the gene expression data set was larger for plant-to-plant differences than for laboratory effects.
Figure 8.
PCA of gene expression data based on Col samples only (A), based on Col, Ler, and Ws samples (B), and based on Col, Ler, and Ws samples averaged per laboratory (C). Colors indicate the genotype: Col, black; Ler, red; Ws, green. Numbers identify laboratories.
We tested whether reducing the influence of plant-to-plant variances by pooling replicate measurements from the same laboratory would increase the power to differentiate between genetic background and laboratory. When using mean expression values per laboratory and genotype, hierarchical clustering correctly grouped samples according to genotype (Supplemental Fig. S4C). In addition, samples of the same genotype from laboratories L2 and L3 consistently grouped together, illustrating again the high agreement between these two laboratory conditions as observed at the higher organizational level. Similarly, PCA of the averaged data set correctly separated samples according to genotype. Averaging increased median (minimal) correlation coefficients from 0.959 (0.927) to 0.982 (0.971) and decreased median CV values from 0.047 to 0.032. Therefore, averaging or pooling was an effective strategy to reduce sample variance.
Finally, we investigated which genes contributed most to the observed variance. The 18 Col-4 transcript profiles were searched for enriched GO categories within the 500 genes showing highest intralaboratory or interlaboratory variance. The enrichment analysis revealed that intralaboratory variance was often caused by differential expression of stress-related genes, such as those involved in response to jasmonic acid (JA), to fungi, or to hyperosmotic or oxidative stress (Table II). Thus, transcriptome profiles of individual plants react very sensitively to small environmental perturbations that exist even under well-standardized conditions. Interlaboratory variance was affected by similar stress responses, but in this case the largest enrichments were found for genes involved in rhythmic processes and circadian regulation (Table III). This suggests that intralaboratory and interlaboratory standardization were similarly effective. Although we cannot exclude effects of potential variability in harvesting rate, it is more likely that the observed differences reveal environment-specific effects on diurnal processes.
Table II. Enrichment of GO categories for genes with high intralaboratory variance.
| GO Identifier | Fold Enrichment (No. of Genes) | P | Descriptiona |
| GO:0008171 | 10.29 (6) | 2.16E-05 | F, O-methyltransferase activity |
| GO:0009753 | 8.14 (19) | 2.25E-12 | P, response to jasmonic acid stimulus |
| GO:0009828 | 8.07 (5) | 3.55E-04 | P, cell wall loosening (sensu Magnoliophyta) |
| GO:0009861 | 7.73 (21) | 4.41E-13 | P, JA- and ethylene-dependent systemic resistance |
| GO:0050832 | 7.72 (9) | 2.34E-06 | P, defense response to fungi |
| GO:0007155 | 7.62 (5) | 4.66E-04 | P, cell adhesion |
| GO:0009827 | 7.41 (5) | 5.31E-04 | P, cell wall modification (sensu Magnoliophyta) |
| GO:0030508 | 6.86 (10) | 1.95E-06 | F, thiol-disulfide exchange intermediate activity |
| GO:0042538 | 6.86 (4) | 2.62E-03 | P, hyperosmotic salinity response |
| GO:0042542 | 6.86 (4) | 2.62E-03 | P, response to hydrogen peroxide |
| GO:0005618 | 6.86 (30) | 1.19E-16 | C, cell wall |
| GO:0006869 | 6.79 (13) | 6.37E-08 | P, lipid transport |
| GO:0030312 | 6.75 (30) | 1.88E-16 | C, external encapsulating structure |
| GO:0010038 | 6.50 (9) | 9.97E-06 | P, response to metal ion |
| GO:0006972 | 6.45 (4) | 3.28E-03 | P, hyperosmotic response |
| GO:0008289 | 6.43 (17) | 1.42E-09 | F, lipid binding |
| GO:0009814 | 6.31 (26) | 9.73E-14 | P, defense response to pathogen, incompatible interaction |
F, P, and C specify whether a GO term belongs to the main GO categories of molecular function, biological process, or cellular compartment, respectively.
Table III. Enrichment of GO categories for genes with high interlaboratory variance.
| GO Identifier | Fold Enrichment (No. of Genes) | P | Descriptiona |
| GO:0007623 | 18.29 (11) | 8.91E-12 | P, circadian rhythm |
| GO:0048511 | 18.29 (11) | 8.91E-12 | P, rhythmic process |
| GO:0010017 | 16.00 (7) | 1.70E-07 | P, red or far-red light signaling pathway |
| GO:0010114 | 13.30 (8) | 1.07E-07 | P, response to red light |
| GO:0009828 | 11.30 (7) | 2.25E-06 | P, cell wall loosening (sensu Magnoliophyta) |
| GO:0030570 | 10.55 (5) | 9.45E-05 | F, pectate lyase activity |
| GO:0016837 | 10.55 (5) | 9.45E-05 | F, carbon-oxygen lyase activity, acting on polysaccharides |
| GO:0009827 | 10.38 (7) | 4.11E-06 | P, cell wall modification (sensu Magnoliophyta) |
| GO:0010038 | 9.38 (13) | 1.13E-09 | P, response to metal ion |
| GO:0010035 | 9.34 (16) | 1.45E-11 | P, response to inorganic substance |
| GO:0009831 | 8.78 (4) | 1.02E-03 | P, cell wall modification during multidimensional cell growth (sensu Magnoliophyta) |
| GO:0042538 | 8.57 (5) | 2.64E-04 | P, hyperosmotic salinity response |
| GO:0046686 | 8.35 (7) | 1.85E-05 | P, response to cadmium ion |
| GO:0042547 | 8.13 (4) | 1.37E-03 | P, cell wall modification during multidimensional cell growth |
| GO:0006972 | 8.07 (5) | 3.55E-04 | P, hyperosmotic response |
| GO:0009861 | 7.73 (21) | 4.41E-13 | P, JA- and ethylene-dependent systemic resistance |
| GO:0009753 | 7.72 (18) | 2.18E-11 | P, response to JA stimulus |
| GO:0009813 | 7.68 (7) | 3.25E-05 | P, flavonoid biosynthesis |
| GO:0009812 | 6.98 (7) | 6.10E-05 | P, flavonoid metabolism |
| GO:0008171 | 6.86 (4) | 2.62E-03 | F, O-methyltransferase activity |
| GO:0042542 | 6.86 (4) | 2.62E-03 | P, response to hydrogen peroxide |
F, P, and C specify whether a GO term belongs to the main GO categories of molecular function, biological process, or cellular compartment, respectively.
The transcriptomic data revealed that despite standardization, considerable plant-to-plant variation of gene expression profiles remained. This is in accordance with the large plant-to-plant variation in growth variables in most laboratories (Fig. 3). Averaging over replicate individual plants or, by inference, pooling of material from multiple plants greatly reduced the effect of plant-to-plant variation and made genotype or laboratory-of-origin effects easier to recognize. Finally, the standardization between laboratories applied in this study ensured that interlaboratory variation was not considerably larger than intralaboratory variation. At least at the transcriptional level, strict standardization of growth conditions between laboratories led to very similar profiles.
Phenotype, metabolite, and transcriptomics data from this experiment are publicly accessible from https://agronomics.ethz.ch/openbis/.
DISCUSSION
Implementing a Shared Protocol to Enhance Phenotypic Reproducibility
The 10 laboratories involved in this study had a strong experience in growing Arabidopsis plants in specific conditions, mainly in vitro or hydroponics, and a few routinely cultivated plants in soil. To best guide laboratories in their experiment, a reference protocol (Supplemental Protocol S1) was established and described in considerably more detail (micrometeorological conditions, plant and culture management) than is usually the case in textbooks or publications. Nevertheless, the comparative experiment revealed the difficulty of both growing Arabidopsis plants in soil using a standard operating protocol under laboratory conditions and obtaining an identical leaf growth phenotype for plants cultivated in different locations. Indeed, only four out of 10 laboratories obtained similar absolute values for the leaf growth variables at three different levels (rosette, leaf, and cell) for the three tested genotypes, while five groups ranked the genotypes similarly on the basis of these variables. The absolute values for phenotypic variables characterizing the consensus group and the genotype classification obtained by most laboratories were identical to those of the reference experiment that defined the common growth conditions. Therefore, in 40% of the cases in this study, laboratories were able to reproduce a similar quantitative leaf growth phenotype using a shared reference protocol.
At the molecular level, the metabolite analysis did not result in a robust separation of the genotypes across laboratories. In contrast, the comparative analysis of the leaf samples based on transcript profiles suggested that the standardization of growth conditions was successful. Variation in gene expression between plants from different laboratories was not much larger than between plants from the same laboratory. Genotypes could be separated based on transcriptome data, and this separation was most efficient after averaging across replicates per laboratory. But transcript data only poorly discriminated samples according to their laboratory of origin. In brief, in our experimental setup, interlaboratory standardization of growth conditions for Arabidopsis was relatively straightforward for transcriptomics studies but was more challenging for morphological, histological, and metabolic studies.
Potential Sources of Variability
Leaf development depends on environmental conditions, including temperature (Granier et al., 2002; Atkin et al., 2006), intensity and quality of incident light (Shipley, 2000; Cookson and Granier, 2006; Ohashi-Kaneko et al., 2006; Poudel et al., 2008), evaporative demand (Ben-Haj-Salah and Tardieu, 1996), daylength (Cookson et al., 2007; Clerget et al., 2008), size of pots used for the soil-based culture (Ray and Sinclair, 1998), substrate composition (Colla et al., 2007), soil water content (Lecoeur et al., 1995; Tardieu, 2006), mineral content (Snir and Neumann, 1997; Yu and Rengel, 1999), and pH of the substrate (Wilkinson, 1999). Any deviation in one of these factors from one laboratory to another may have affected the leaf growth phenotype at any of the levels analyzed in our experiment. Several of these factors were identical for all laboratories because of common materials (pots, substrate). Strict guidelines for micrometeorological conditions were provided, and in general, the prescribed temperature, incident light, daylength, and nutrient solution pH were reached. Although some deviations were recorded because of the constraints specific to each laboratory (Table I), they did not always result in detectable differences. For example, L2 reported a different watering regime (subirrigation to capacity instead of daily top pipetting to fixed weight), but this factor alone did not have an apparent effect on the leaf growth phenotype, since L2 belongs to the core group.
A primary source of variability may have been the evaporative demand in the growth chamber (Ben-Haj-Salah and Tardieu, 1996). Indeed, distance between lamps and rosettes varied greatly, from 200 cm down to 29 cm (L5), the smallest distance being set to reach the required incident light intensity at the plant level. Such a range may have resulted in different leaf surface temperatures and thereby varying evaporative demand. A second source of variability may have been light quality, which could differ significantly between growth chambers equipped with different lamps (Table I) and between lamps of varying age (Ohashi-Kaneko et al., 2006; Poudel et al., 2008). Furthermore, the fact that the sensitivity of leaf growth to such environmental factors is genotype dependent (Ober and Luterbacher, 2002; Reymond et al., 2004; Aguirrezabal et al., 2006; Khan et al., 2007; Laperche et al., 2007) may explain why the ranking of genotypes was not constant across all laboratories. The different slope of correlations between rosette fresh weight and RA observed in some laboratories (L4 and L7) could be linked to differences in light quality (Supplemental Fig. S1), considering the different lamps used by each laboratory in its growth chamber (Table I). Contrastingly, results reported by L1 with higher RA for a given fresh weight could result from either shading (although the light intensities measured by this laboratory are not the lowest values of all laboratories) or more probably different light quality.
The minutia of material sampling protocols can also influence results at the molecular level. Because of the rapidity of enzymatic responses, this is particularly true for metabolite profiling, for which small timing differences in the handling of leaves analyzed separately (i.e. between harvesting and freezing) can have a major impact. The difficulty of distinguishing genotypes on the basis of metabolite profiles when analyzing all samples collectively, therefore, might be explained by procedures used in each laboratory to dissect plants and freeze fifth leaves. Additionally, large differences in the harvested leaf size may have resulted in a deviation from standard protocols when extracting metabolites from given sample amounts, possibly contributing to technical variation. Nevertheless, in most cases, metabolite profiles did cluster according to genotypes when only considering samples grown in the same environmental conditions and harvested by the same team.
The Molecular Level Revealed a Stronger Sensitivity to Genotype × Environment Interaction (Including Sampling) Than Higher Organizational Levels
The comparison of genotype clustering based on separate laboratory data revealed that phenotypic traits (leaf growth) better discriminated genotypes than metabolic traits. For example, RA distinguished Col-4 from Ler and Ws and epidermal CD Ler from Col-4 and Ws. However, metabolite profile comparison showed a stronger genotype × environment interaction. This is consistent with a model in which a biological system responds to the environment first by modulating its metabolic activity, while phenotypic changes at the higher organizational level may eventually appear in the longer term. This strong genotype × environment interaction for metabolic traits is supported by the observation that the genotypic effect becomes more prominent when metabolite profiles are compared within one laboratory, in which noncontrollable differences of environmental conditions between laboratories were minimized. The results also suggest that laboratory-specific harvesting procedures may have altered the metabolic profiles despite observance of a standardized operating protocol. Together, our results confirm that metabolite profiling is a very sensitive technique to discriminate plants according to environment, genotype, or developmental stage under defined conditions (Fernie et al., 2004). Metabolites could be assigned to those determined by the genotype and those determined by the environment. Metabolites common to both of these categories included many from the central metabolism, including tricarboxylic acid cycle intermediates and the majority of the amino acids that significantly adjust to new environmental conditions such as changing rates of photosynthesis driven by different light intensities. In contrast, some sugars and the majority of lipids show no significant changes, suggesting a stronger buffering (in the case of sugars) or a stronger structural function (in the case of lipids). There were only a few genotype-specific metabolites, including three unknowns, an organic acid, and Suc, while there were 21 environment-specific significant differences. These included known stress metabolites such as raffinose, Pro, and 4-aminobutyric acid as well as the starch breakdown product maltose, which are also broadly consistent with the observed transcriptional differences.
The results from different laboratories were considerably more consistent for transcript profiles than for metabolite profiles and developmental variables. Transcript profiles separated well between genotypes. This apparent robustness of transcript profiles is consistent with genotype and environment affecting just a subset of the total gene component. Although genotype × environment interactions are based on signaling cascades and regulatory networks and such regulation often affects transcript abundance, it is also relayed by processes such as translation rate, protein stability, and enzyme activity, thereby explaining why effects on transcript levels were less pronounced than effects on metabolite and cell or organ phenotypes. This may reflect also that processes operate at different temporal scales: metabolism is regulated over milliseconds to minutes; transcription is regulated over minutes to hours; while cell division takes place over several hours to tens of hours. Nevertheless, differences in microenvironment conditions did affect the transcript profiles, as shown by the GO analysis of the most differentially expressed gene classes, and strict control of growth conditions is required for successful standardization of transcript profiling experiments.
Interestingly, despite the clear control of growth conditions and harvesting time via the standard protocol, the most significantly overrepresented GO categories with respect to interlaboratory variation were related to rhythmic processes. Consistent with this, several of the metabolites that showed high environmental dependency have also been reported to be diurnally regulated (Gibon et al., 2006). As we have no record for deviations in the harvest time between laboratories, this leads to the interesting possibility that the temporal profiles of metabolites and of transcripts varied between environments. Although this has not been explicitly studied, there is clear evidence indicating that light quality can affect rhythmic processes, for example the nucleocytoplasmic partitioning of photoreceptors (Kircher et al., 2002), which in turn provide input signals for the circadian clock. This notion is supported by the finding that the GO categories “red or far-red light signaling pathway” and “response to red light” also were enriched among the genes with high interlaboratory variance (Table II). Together, these data indicate that the light quality dependency of molecular rhythms warrants further study and highlight the challenges of monitoring molecular changes in the presence of even slightly divergent environmental conditions.
What Is the Leaf Phenotype of a Genotype?
This study stresses that there is no simple answer to this question. First, by definition, the phenotype expresses a particular genotype × environment interaction. Our results revealed substantial phenotypic plasticity in three Arabidopsis genotypes, suggesting that stabilizing phenotypic values is difficult across different laboratories. Even small variations in experimental conditions can strongly affect the leaf growth phenotype and metabolite profiles. Therefore, it is crucial that phenotypic values are completed with a precise and comprehensive description of the genotype studied and its specific environment.
Second, the definition of a stable phenotypic value depends on the level of observation (as shown by the differences between molecular, cellular, and even whole plant phenotype clustering) and the time of sampling (as potentially shown by differential expression of circadian genes). Strikingly, the Arabidopsis genotypes differed at the whole rosette and cellular levels, but individual leaf area at the same nodal position was similar for all three. The absence of phenotypic differences at one level may be the result of compensatory mechanisms at others. For example, the smaller epidermal CA in Ler leaves is compensated by a higher number of epidermal cells (Tisné et al., 2008), as is also the case in so-called compensation mutants (Ferjani et al., 2007). Similarly, the absence of changes in final leaf area can be explained by compensation between the rate and duration of leaf expansion (Aguirrezabal et al., 2006). Our multilevel phenotype analysis provides new evidence that differences at one level do no necessarily reflect differences at other levels. It confirms that a phenotype observed at a specific organizational level cannot be easily inferred from another organizational level (Granier and Tardieu, 2009). Our experiment supports the view that the leaf system has a complex status as defined by Wu and Marceau (2002) for ecological systems (i.e. a system is complex when it is not completely reducible to its components).
CONCLUSION
Our findings underscore that the challenge of describing, monitoring, and precisely controlling environmental plant growth conditions is too often underestimated. Growth phenotypes and molecular profiles should be interpreted with caution when comparing independent experiments, even when obtained from plants of the same genotype. However, we also demonstrate that dedicated efforts can result in coherent and reproducible data from different laboratories when produced with particular attention to standard environmental conditions and following a detailed protocol. Our results also show that the conformity between experimental platforms should be rigorously tested prior to engaging in large-scale multisite projects.
MATERIALS AND METHODS
Plant Material
Seeds of the Arabidopsis (Arabidopsis thaliana) accessions Col-4 (N933), Ler (NW20), and Ws (N2360) were distributed to all laboratories, named L1 to L10 hereafter. All seeds originated from the same batch provided by the Nottingham Arabidopsis Stock Centre.
Plant Growth Conditions
The reference experiment was performed by L3 with the PHENOPSIS automated platform (Granier et al., 2006) that also served to define the common experimental protocol for all laboratories. Pots, soil substrate, and nutrient solution were provided by L3 to all other sites, together with a detailed protocol (for complete documentation, see Supplemental Protocol S1). Seeds were stored at 4°C and imbibed in water 24 h before sowing. Each genotype was grown in 30 independent cylindrical pots for the reference experiment (L3) and in 10 independent pots for the comparative experiment (all 10 laboratories, including L3). Pots were filled with a mixture (1:1, v/v) of a loamy soil and organic compost at a humidity of 0.30 g water g−1 dry soil. Seeds and water were aspirated with a pipette, and three seeds were sown at the center of each pot. Ten milliliters of a modified one-tenth-strength Hoagland solution (Hoagland and Arnon, 1950) was added to the pot surface just before sowing. The pots were transferred to a growth chamber and covered with aluminum foil during 48 h. Daylength in the growth chamber was fixed at 16 h using a mix of cool-white fluorescent tubes, sodium, and hydrargyrum quartz iodide (HQI) lamps. The other growth conditions were as follows: air temperature at 20°C during the day and 21.5°C during the night; air humidity between 70% and 75%; and incident light measured at the plant level approximately 150 μmol m−2 s−1. During the germination phase, water was sprayed at regular intervals on the pots to maintain sufficient humidity at the soil surface. Beginning at plant germination, each pot was processed daily to calculate the soil water content by weighing and to adjust it to 0.40 g water g−1 dry soil by addition of an appropriate volume of nutrient solution. The adjustment of soil water content was done automatically with the PHENOPSIS automaton in the L3 growth chamber during the reference and the comparative experiments, while it was done manually in the other laboratories during the comparative experiment.
Measurement of Leaf Growth Variables in the Reference Experiment
Rosette LN
The LN was recorded for 10 plants per genotype every 2 to 3 d from stage 1.0 (two cotyledons visible) until stage 6.0 (first flower open), when plants were harvested. Developmental stages were defined according to Boyes et al. (2001).
RA
The PHENOPSIS automaton imaged the plants daily from stages 1.0 to 6.0 using a Sony SSC-DC393P camera. The total projected leaf area (RA; mm2) was determined for 10 rosettes per genotype every 2 to 3 d using image-analysis software (Bioscan-Optimas version 4.10).
Individual Leaf Area
From stages 1.0 to 6.0, five rosettes per genotype were dissected every 2 to 3 d. The AL6 (mm2) was measured after imaging with a binocular magnifying (×160) glass for leaves smaller than 2 mm2 or with a flatbed scanner for larger ones.
Epidermal CD, CN, and CA
A negative film of the adaxial epidermis of the same sixth leaf was obtained after evaporation of a varnish spread on its surface. These epidermal imprints were analyzed using a microscope (Leitz DM RB; Leica) supported by the image-analysis software Optimas. Mean epidermal CD (cells mm−2) was estimated by counting the number of epidermal cells in three zones (at the tip, middle, and base) of each leaf. Total epidermal CN was estimated from epidermal CD and leaf area. Mean epidermal CA (μm2) was measured from 25 epidermal cells in a central zone of each leaf.
Estimation of Dynamic Variables
For each genotype, RA, LN, AL6, CN, and CA were plotted as a function of time (days after stage 1.0). Sigmoid curves (Eq. 1) were fitted to the data to estimate the duration and rate of processes.
| (1) |
Durations (d) of the phases of leaf production, rosette leaf expansion, sixth leaf expansion, cell production, and cell expansion were calculated from the respective sigmoid curves as:
| (2) |
Maximal rates (Rmax) of leaf production, rosette leaf expansion, sixth leaf expansion, cell production, and cell expansion were calculated from the respective sigmoid curves as:
| (3) |
The mean values of d and Rmax and associated sd values were estimated by bootstrapping the nonlinear model for 1,000 replicates using the “boot” function in R 2.9.0 (R Development Core Team, 2008; http://www.r-project.org/). Mean values were compared between genotypes with a t test at 5%.
Measurement of Leaf Growth Variables in the Multilaboratory Comparative Experiment
In all laboratories, 10 plants per genotype were harvested at the middle of the light period (i.e. after 8 h of light) when plants had reached stage 6.0 (i.e. the end of the growth period). Rosettes were dissected, individual leaves were detached, and their lamina were separated from the petiole. The fifth leaf lamina was immediately frozen in liquid nitrogen for metabolome and transcriptome analyses. All other laminas were fixed onto a sheet of paper with double-sided adhesive tape in order of leaf emergence on each individual rosette. The montage was scanned for measurements of final individual leaf area. The area of the fifth leaf was estimated as the mean area of the fourth and sixth leaves. For practical reasons, the molecular profiles were extracted from the fifth leaf but cellular variables were measured on the sixth leaf of the same plant. The imprint of the adaxial epidermis was obtained for all sixth leaves. The following leaf growth variables were measured by L3 from scans and imprints as described above: LN (leaves), area of individual leaves (ALi; mm2), RA (mm2), mean epidermal CD in the sixth leaf (CD; cells mm−2), total epidermal CN in the sixth leaf (CN; cells), and mean epidermal CA in the sixth leaf (CA; μm2).
All leaf growth variables measured in the reference and comparative experiments are summarized in Supplemental Table S1.
Metabolite Profiling
Five biological replicates (i.e. five samples of the fifth leaf, each derived from an independent plant; see above) were analyzed for each genotype and laboratory by L4. Samples that were not delivered frozen to L4 (from L7, L8, and L10) were discarded.
GC-MS Analysis
Leaf samples were homogenized and extracted as described by Lisec et al. (2006). Derivatized samples were run in an Agilent 7683 series autosampler (Agilent Technologies) coupled to an Agilent 6890 gas chromatograph coupled to a Leco Pegasus 2 time-of-flight mass spectrometer (LECO). Chromatogram acquisition parameters were those described previously (Weckwerth et al., 2004). Chromatograms were exported from Leco ChromaTOF software (version 3.25) to R software. Peak detection, retention time alignment, and library matching were performed with an in-house R script. Completely randomized GC-MS samples were measured on 5 consecutive days and normalized for the measurement-day effect by dividing each metabolite value by the median of all values for this metabolite measured in the same batch followed by a log2 transformation to center the values around zero (Lisec et al., 2006). The median from the five biological replicates per genotype and laboratory was determined for each of the 89 metabolites. An ANOVA was conducted using the statistical framework R. The model contained two factors, genotype and laboratory. P values were corrected for multiple testing applying the method of Benjamini and Hochberg (1995).
Transcript Profiling
Two or three biological replicates (i.e. samples of the fifth leaf, each derived from an independent plant; see above) were measured per genotype and laboratory by L6. RNA extraction was done on a Qiagen QiaCube robot with the Qiagen RNA plant extraction kit. After discarding samples that did not arrive in a frozen state or yielded RNA of insufficient quality, 41 samples from L1 to L6 were selected for analysis. RNA was amplified and labeled with the GeneChip Expression 3′ Amplification One-Cycle Target Labeling kit (Affymetrix). Labeled RNA was hybridized to AGRONOMICS1 microarrays. The AGRONOMICS1 array is a custom-made Arabidopsis Col-0 tiling array that contains the complete paths of both genome strands with on average one 25mer probe per 35-bp genome sequence window. The microarray enables reliable expression profiling of more than 31,000 Arabidopsis genes and gives very similar results to the widely used ATH1 GeneChip for the set of common probes (Rehrauer et al., 2010). The arrays were scanned using an Affymetrix 3000 7G confocal scanner. All data processing was performed using R (version 2.8.1). Signal values were derived from Affymetrix *.CEL files using Robust Multichip Average. This analysis yielded expression summaries for 31,455 Arabidopsis genes. When hybridizing Ws or Ler samples to arrays based on the Col sequence, sequence polymorphisms could decrease hybridization efficiency and thus cause reduced signals. To avoid such artifacts, 547 genes (i.e. 1.7%) were excluded that always gave lower apparent expression signals in Ler or Ws than in Col-4. Identification of enriched functional categories was done using the ATCOECIS tool (http://bioinformatics.psb.ugent.be/ATCOECIS; Vandepoele et al., 2009). The enrichment analysis focused on genes with intermediate to high expression levels by excluding any signals that were smaller than the median of all signals. Divisive hierarchical clustering was performed using the “diana” algorithm implemented in the “cluster” R package. Microarray raw and processed data will be available via ArrayExpress and the AGRON-OMICS data repositories.
Statistical Analysis of Leaf Growth Variables
Statistical analyses were done using R. Mean values and sds were calculated for each growth variable, after verifying that variance was homogenous and residuals distributed normally (data points falling at more than 2 sd from the mean were considered as outliers, which represented 2% of the data homogenously distributed among genotypes, laboratories, and variables). Results were compared between laboratories by one-way ANOVA, followed by Tukey's posthoc tests to identify the homogenous groups of laboratories at the 1% level. The mean values were also compared with two-way ANOVA, using laboratories and genotypes as factors, either between the different laboratories or only between the laboratories identified as having a similar phenotype.
Five leaf growth variables (LN, RA, AL6, CD, and CA), 89 metabolites, and 500 genes were included in PCA. Individual variables were centered and scaled to unit variance before extracting the principal components, using the “pca” package in R.
Data Sharing
Data sets and sample information were obtained from data analysts and individual laboratories in spreadsheet and file-archive formats. These data were parsed to normalize identifiers and to verbosely specify metadata terms and data sets organized by sample identifiers. These data were uploaded into the openBIS software (http://www.cisd.ethz.ch/software/openBIS) and validated for completeness to ensure long-term public accessibility of the data.
The microarray raw data can be found under the accession number E-TABM-917.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlation of rosette fresh weight and rosette leaf area.
Supplemental Figure S2. Core laboratory phenotypes per genotype.
Supplemental Figure S3. CV for gene expression values.
Supplemental Figure S4. Divisive hierarchical clustering of gene expression.
Supplemental Table S1. Measured leaf growth variables.
Supplemental Table S2. ANOVA of metabolite profiles.
Supplemental Protocol S1. Detailed protocol distributed to all laboratories.
Supplementary Material
Acknowledgments
We thank J.J. Thioux for the preparation of pots, substrate, and nutrients for the comparative experiment.
References
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C. (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Atkin OK, Loveys BR, Atkinson LJ, Pons TL. (2006) Phenotypic plasticity and growth temperature: understanding interspecific variability. J Exp Bot 57: 267–281 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haj-Salah H, Tardieu F. (1996) Quantitative analysis of the combined effects of temperature, evaporative demand and light on leaf elongation rate in well-watered field and laboratory-grown maize plants. J Exp Bot 47: 1689–1698 [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 57: 289–300 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Clerget B, Dingkuhn M, Goze E, Rattunde HFW, Ney B. (2008) Variability of phyllochron, plastochron and rate of increase in height in photoperiod-sensitive Sorghum varieties. Ann Bot (Lond) 101: 579–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G, Rouphael Y, Cardarelli M, Temperini O, Rea E. (2007) Optimization of substrate composition for organic lettuce transplant production. Adv Hortic Sci 21: 106–110 [Google Scholar]
- Cookson SJ, Chenu K, Granier C. (2007) Day length affects the dynamics of leaf expansion and cellular development in Arabidopsis thaliana partially through floral transition timing. Ann Bot (Lond) 99: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Granier C. (2006) A dynamic analysis of the shade-induced plasticity in Arabidopsis thaliana rosette leaf development reveals new components of the shade-adaptive response. Ann Bot (Lond) 97: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R. (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot (Lond) 82: 17–26 [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Granier C, Massonnet C, Turc O, Muller B, Chenu K, Tardieu F. (2002) Individual leaf development in Arabidopsis thaliana: a stable thermal-time-based program. Ann Bot (Lond) 89: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Tardieu F. (2009) Multi-scale phenotyping of leaf expansion in response to environmental changes: the whole is more than the sum of parts. Plant Cell Environ 32: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler Jd A, Bergounioux C, Montagu MV, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. (1950) The water culture method for growing plants without soil. Calif Agric Exp Stn Circ 347: 1–32 [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, et al. (2005) Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp Funct Genomics 6: 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan HUR, Link W, Hocking TJ, Stoddard FL. (2007) Evaluation of physiological traits for improving drought tolerance in faba bean (Vicia faba L.). Plant Soil 292: 205–217 [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Ádám E, Schäfer E, Nagy F. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A, Brancourt-Hulmel M, Heumez E, Gardet O, Hanocq E, Devienne-Barret F, Le Gouis J. (2007) Using genotype × nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor Appl Genet 115: 399–415 [DOI] [PubMed] [Google Scholar]
- Lecoeur J, Wery J, Turc O, Tardieu F. (1995) Expansion of pea leaves subjected to short water-deficit: cell number and cell size are sensitive to stress at different periods of leaf development. J Exp Bot 46: 1093–1101 [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Ober ES, Luterbacher MC. (2002) Genotypic variation for drought tolerance in Beta vulgaris. Ann Bot (Lond) 89: 917–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Kaneko K, Matsuda R, Goto E, Fujiwara K, Kurata K. (2006) Growth of rice plants under red light with or without supplemental blue light. Soil Sci Plant Nutr 52: 444–452 [DOI] [PubMed] [Google Scholar]
- Poudel PR, Kataoka I, Mochioka R. (2008) Effect of red- and blue-light-emitting diodes on growth and morphogenesis of grapes. Plant Cell Tissue Organ Cult 92: 147–153 [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
- Ray JD, Sinclair TR. (1998) The effect of pot size on growth and transpiration of maize and soybean during water deficit stress. J Exp Bot 49: 1381–1386 [Google Scholar]
- Rehrauer H, Aquino C, Gruissem W, Henz SR, Hilson P, Laubinger S, Naouar N, Patrignani A, Rombauts S, Shu H, et al. (2010) AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol 152: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Muller B, Tardieu F. (2004) Dealing with the genotype × environment interaction via a modelling approach: a comparison of QTLs of maize leaf length or width with QTLs of model parameters. J Exp Bot 55: 2461–2472 [DOI] [PubMed] [Google Scholar]
- Schilling M, Pfeifer AC, Bohl S, Klingmuller U. (2008) Standardizing experimental protocols. Curr Opin Biotechnol 19: 354–359 [DOI] [PubMed] [Google Scholar]
- Shipley B. (2000) Plasticity in relative growth rate and its components following a change in irradiance. Plant Cell Environ 23: 1207–1216 [Google Scholar]
- Snir N, Neumann PM. (1997) Mineral nutrient supply, cell wall adjustment and the control of leaf growth. Plant Cell Environ 20: 239–246 [Google Scholar]
- Tardieu F. (2006) Leaf growth under water-limited conditions. Ribaut JM, , Drought Adaptation in Cereals. Haworth Press, London, pp 145–169 [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jones AR, Zhu WM, Apweiler R, Aebersold R, Deutsch EW, et al. (2007) The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol 25: 887–893 [DOI] [PubMed] [Google Scholar]
- Tisné S, Reymond M, Vile D, Fabre J, Dauzat M, Koornneef M, Granier C. (2008) Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis. Plant Physiol 148: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME. (2003) Remodelling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O. (2004) Process for the integrated extraction identification, and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83 [DOI] [PubMed] [Google Scholar]
- Wilkinson S. (1999) pH as a stress signal. Plant Growth Regul 29: 87–99 [Google Scholar]
- Wu JG, Marceau D. (2002) Modeling complex ecological systems: an introduction. Ecol Modell 153: 1–6 [Google Scholar]
- Yu Q, Rengel Z. (1999) Micronutrient deficiency influences plant growth and activities of superoxide dismutases in narrow-leafed lupins. Ann Bot (Lond) 83: 175–182 [Google Scholar]
- Zimmermann P, Schildknecht B, Craigon D, Garcia-Hernandez M, Gruissem W, May S, Mukherjee G, Parkinson H, Rhee S, Wagner U, et al. (2006) MIAME/Plant: adding value to plant microarray experiments. Plant Methods 2: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.