Abstract
Pathogen-associated molecular patterns (PAMPs) trigger plant defenses when perceived by surface-localized immune receptors. PAMP-triggered immunity (PTI) plays a vital role in the resistance of plants to numerous potential pathogens. MicroRNA (miRNA) biogenesis is known to be important for PTI, but miRNA species involved in this process have not been fully explored. Here we show that the Arabidopsis (Arabidopsis thaliana) miRNA effector protein, Argonaute1 (AGO1), is required for a number of PTI responses including PAMP-induced callose deposition, gene expression, and seedling growth inhibition. Deep sequencing of AGO1-bound small RNAs led to the identification of a number of miRNAs that are up- or down-regulated by flg22, a well-studied PAMP. Overexpression of selected miRNAs in stable transgenic plants demonstrated that miR160a positively regulate PAMP-induced callose deposition, whereas miR398b and miR773 negatively regulate PAMP-induced callose deposition and disease resistance to bacteria, suggesting a complexity of the miRNA regulation in plant innate immunity.
Plants are equipped to detect conserved molecular features of microbes, termed pathogen-associated molecular patterns (PAMPs), and trigger defenses (Zipfel and Felix, 2005). PAMP-triggered immunity (PTI) allows plants to fend off a large number of potential pathogens (Li et al., 2005). For example, flg22, a conserved peptide derived from Pseudomonas syringae flagellin (Felix et al., 1999), is perceived by the receptor FLS2 at the plasma membrane (Gómez-Gómez and Boller, 2000; Chinchilla et al., 2007; Heese et al., 2007) and subsequently activates mitogen-activated protein kinases (MPKs), a transient oxidative burst (reactive oxygen species; Felix et al., 1999), callose (β-1,3-glucan) deposition at the cell wall (Brown et al., 1998; Gómez-Gómez et al., 1999), and the expression of defense-related genes (Zipfel et al., 2004; Zhang et al., 2007). Many plant pathogens can deliver a variety of effector proteins into the host cell to inhibit PTI signaling (Göhre and Robatzek, 2008; Zhou and Chai, 2008). To counteract, plants have evolved resistance proteins to sense the activity of some of these effectors to activate a second layer of inducible defenses called effector-triggered immunity (ETI; Chisholm et al., 2006; Jones and Dangl, 2006).
In plants, small RNAs including microRNAs (miRNAs) and small interfering RNAs (siRNAs) regulate diverse processes including development (Jones-Rhoades et al., 2006; Mallory and Vaucheret, 2006), abiotic stress tolerance (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Fujii et al., 2005), and antiviral defenses (Mourrain et al., 2000; Dalmay et al., 2001; Morel et al., 2002). Several recent studies indicate that small RNAs also participate in plant disease resistance to bacterial pathogens. For example, flg22 induces the accumulation of miR393, which contributes to plant resistance against bacteria by negatively regulating the mRNA level of F-box auxin receptors TIR1, AFB2, and AFB3 (Navarro et al., 2006). Induced accumulation of a natural antisense transcript-associated siRNA, nat-siRNAATGB2 (Katiyar-Agarwal et al., 2006), and a long siRNA, AtlsiRNA-1 (Katiyar-Agarwal et al., 2007), is required specifically for RPS2-mediated ETI, but not basal resistance to compatible P. syringae bacteria. Consistent with a role of these small RNAs in plant immunity, proteins required for small RNA biogenesis and function have been shown to be required for disease resistance to bacterial pathogens. For example, Dicer-Like1 (DCL1) and Hua Enhancer1, which are required for the biogenesis of both miRNAs and long siRNAs, are required for PTI resistance (Navarro et al., 2008). Likewise, AGO7 is required for the accumulation of AtlsiRNA-1 and RPS2 resistance (Katiyar-Agarwal et al., 2007). In addition, AGO4, which is required for RNA-directed DNA methylation, contributes to resistance nonspecifically to both adapted and nonadapted P. syringae through an unknown mechanism (Agorio and Vera, 2007).
A key component in the miRNA pathway is Argonaute1 (AGO1), which predominately binds mature miRNAs to form a RNA-induced silencing complex in cytoplasm and cleaves the target mRNA through miRNA-mRNA base pairing (Okamura et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005) or represses translation through an association with polysomes (Lanet et al., 2009). AGO1 contains three characteristic domains: PAZ, MID, and PIWI (Song and Joshua-Tor, 2006). PIWI domain adopts the structure of RNase H that contains the catalytic site formed by three residues (Asp, Asp, and His), and provides a slicer activity that executes the miRNA-guided cleavage of target RNA (Liu et al., 2004; Song et al., 2004; Rivas et al., 2005). Several studies have shown the involvement of AGO1 in plant antiviral defense (Morel et al., 2002; Qu et al., 2008). However, the role for AGO1 in plant defenses to bacterial infection has not been fully explored. More importantly, miRNA species regulating plant disease resistance remain largely unknown.
Here we show that AGO1 positively regulates PAMP-induced callose deposition, defense gene expression, and seedling growth inhibition, and contributes to PAMP-induced disease resistance to P. syringae. Deep sequencing of AGO1-bound small RNAs identified a small number of miRNA species whose accumulation was up- or down-regulated by flg22. Overexpression of selected miRNAs in stable transgenic plants indicated that miR160a positively regulates PAMP-induced callose deposition, whereas miR398b and miR773 negatively regulate PAMP-induced callose deposition. Furthermore, miR398b and miR773 overexpression plants showed enhanced susceptibility to both virulent and nonpathogenic strains of P. syringae, indicating an important role of these miRNAs in disease resistance.
RESULTS
AGO1 Contributes to flg22-Induced Disease Resistance
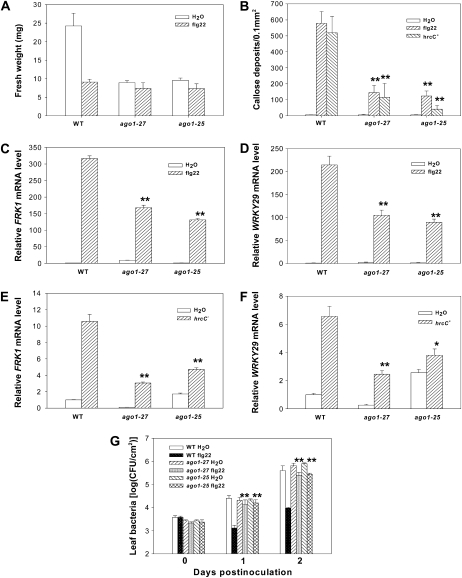
Flg22 treatment causes a strong reduction in Arabidopsis (Arabidopsis thaliana) seedling growth (Gómez-Gómez et al., 1999). The ago1-25 and ago1-27 mutants carrying a point mutation in PIWI domain are impaired in posttranscriptional gene silencing and viral resistance (Morel et al., 2002), but do not affect miRNA accumulation (Vaucheret et al., 2004). We tested the effect of ago1-25 and ago1-27 mutations on flg22-mediated growth inhibition. After growing in the one-half Murashige and Skoog liquid medium containing 10 μm flg22 for 5 d, the wild-type seedlings displayed a significant reduction (60%) in fresh weight compared to control seedlings grown without flg22. The growth of ago1-25 and ago1-27 mutants was reduced only slightly (15% to approximately 20%) by flg22 treatment (Fig. 1A), indicating that AGO1 is required for flg22-mediated seedling growth inhibition. An examination of flg22-induced callose deposition showed that the two mutants had significantly reduced callose deposition compared to wild type (Fig. 1B). Similarly, callose deposition induced by a nonpathogenic P. syringae mutant hrcC−, which lacks a functional type III secretion apparatus but contains a collection of PAMPs (Yuan and He, 1996), was also compromised in the ago1 mutants. We examined the expression of FRK1 and WRKY29, two PAMP-response genes (Asai et al., 2002), in ago1-25 and ago1-27 plants treated with flg22 and hrcC− mutant bacteria. The two ago1 mutants accumulated 50% to 70% less transcripts compared to that in wild type (Fig. 1, C–F), indicating that AGO1 is partially required for flg22-induced gene expression. We also tested if MAP kinase activation and transient oxidative burst, two early events in PTI signaling, were affected in ago1 mutants. Supplemental Figure S1 shows that the ago1-25 mutant had normal MAP kinase activation and oxidative burst in response to flg22 treatment, indicating that early and late PTI signaling events were differentially impacted by the ago1 mutations. To determine whether AGO1 plays a role in plant resistance to bacteria, we conducted flg22-mediated protection assay on ago1-27 and ago1-25 (Zipfel et al., 2004). While pretreatment of wild-type plants with flg22 inhibited the growth of virulent DC3000 bacteria by approximately 100-fold 2 d after inoculation, it only slightly inhibited bacterial growth in the two ago1 mutants (Fig. 1G), indicating that AGO1 plays an important role in flg22-induced resistance to bacteria.
Figure 1.
AGO1 contributes to PTI. A, ago1 mutants are partially insensitive to flg22-induced seedling growth inhibition. Wild-type (WT) and ago1 mutant seedlings were treated with or without 10 μm flg22 for 5 d, and fresh weight was measured. B, ago1 mutants are compromised in flg22- and hrcC−-induced callose deposition. Wild-type and ago1 plants were infiltrated with water, 1 μm flg22, or 2 × 107 cfu/mL hrcC− bacteria for 12 h before stained for callose. C to F, ago1 mutants are compromised in PAMP-induced gene expression. Wild-type and ago1 mutants were syringe infiltrated with 2 μm flg22, 2 × 107 cfu/mL hrcC− bacteria, or water for 4 h, and RNA was extracted for quantitative RT-PCR analysis. mRNA level was normalized to that in water-treated wild-type plants. G, AGO1 is required for flg22-induced resistance to P. syringae DC3000. Wild-type and ago1 mutants were infiltrated with 1 μm flg22 or water for 24 h before infiltrated with 5 × 105 cfu/mL DC3000 bacteria. Leaf bacterial population was determined at the indicated times. Error bars indicate sd. Student's t test was carried out to determine the significance of difference between wild-type and mutant plants following flg22 or hrcC− treatment. Asterisks (* and **) indicate significant difference at a P value <0.05 and <0.01, respectively. The data shown are representative of three independent experiments.
AGO7 was reported to be required for RPS2-specified ETI resistance (Katiyar-Agarwal et al., 2007), but a potential role in PTI defenses has not been investigated. We therefore tested if the ago7 mutant also impact PTI. The ago7 mutant (zip-1; Hunter et al., 2003), a likely null allele, was completely normal in PTI responses when flg22-induced callose deposition, FRK1 expression, seedling growth inhibition, and oxidative burst were measured (Supplemental Fig. S2, A–D). Consistent with the normal PTI responses, hrcC− mutant bacteria multiplied normally in ago7 (Supplemental Fig. S2E), and flg22 pretreatment provided similar protection against the virulent DC3000 bacteria in ago7 and wild-type plants (Supplemental Fig. S2F). In contrast, ago7 plants showed reduced resistance to DC3000 (avrRpt2), confirming previous report (Supplemental Fig. S2G). Together these results are consistent with a specific role of AGO7 in RPS2 resistance to bacteria.
DCL1 is required for miRNA biogenesis. It has been shown that dcl1-9 mutant is compromised in resistance to hrcC− mutant bacteria (Navarro et al., 2008). An examination of PTI defenses showed that, like ago1 mutants, callose deposition was reduced by approximately 50% to 70% in dcl1-9 compared to wild-type control (Supplemental Fig. S3A), and flg22-induced resistance to DC3000 bacteria was profoundly diminished in dcl1-9 plants (Supplemental Fig. S3F). Unlike ago1 mutants, FRK1 and WRKY29 expression was not significantly altered in dcl1 (Supplemental Fig. S3, B–E). These results confirm an important role of DCL1 in PTI resistance. Like ago1 mutants, dcl1-9 was not affected in flg22-induced MAP kinase activation and transient oxidative burst (Supplemental Fig. S4). Taken together, these data indicate that both AGO1 and DCL1 are required for flg22-induced plant resistance to P. syringae bacteria, although their roles in specific PTI responses differ.
Characterization of AGO1-Bound Small RNAs during PTI Defenses
As an effector protein, AGO1 must act through its bound small RNAs in PTI responses. We therefore examined AGO1-associated small RNA species in flg22- or water-treated plants by Illumina deep sequencing. Consistent with previous reports (Mi et al., 2008), AGO1-bound small RNAs displayed a strong bias for sequences beginning with a 5′-terminal uridine and a length of 21 nt (Supplemental Fig. S5, A and B), and this was not altered by flg22 treatment. Small RNAs mapped to the Arabidopsis genome were categorized based on their genomic locations and functions (Supplemental Fig. S5C; Supplemental Table S1). A total of 1,477,337 and 1,385,186 genome-matched small RNA reads were obtained from water-treated and flg22-treated AGO1 complexes, respectively. These represent 46,555 and 62,387 unique small RNA sequences for water and flg22 treatment, respectively, suggesting that flg22 treatment induces the biogenesis of significant number of unique AGO1-bound small RNAs. Most of the flg22-induced small RNA reads belong to non-miRNAs.
The majority of AGO1-bound small RNA reads are miRNAs in both treatments. Among them, 89 and 91 known miRNA sequences were identified in water and flg22 treatments, respectively. Together these constitute most of the known miRNA species, indicating that our sequencing had a robust coverage of miRNAs (Table I; Supplemental Table S1). Most of the 67 miRNAs reported by a previous report (Mi et al., 2008) were identified in this study, except for a few low-copy miRNA species.
Table I. miRNAs up- or down-regulated by flg22.
miRNAs with greater than 100 reads and showing >30% increase or decrease in flg22 treatment are selected.
| ID | Normalization Reads |
Fold-Change |
|
| Water | Flg22 | Flg22/Water | |
| Up-regulated | |||
| miR158a | 13,243 | 31,311 | 2.36 |
| miR158b | 55 | 160 | 2.88 |
| miR160a | 311 | 413 | 1.33 |
| miR160b | 287 | 375 | 1.31 |
| miR160c | 311 | 413 | 1.33 |
| miR161.2 | 591 | 1,153 | 1.95 |
| miR167d | 9,975 | 20,168 | 2.02 |
| miR169a | 947 | 1,774 | 1.87 |
| miR391 | 772 | 1,453 | 1.88 |
| miR393a | 617 | 1,307 | 2.12 |
| miR393b | 617 | 1,307 | 2.12 |
| miR396a | 37,169 | 52,352 | 1.41 |
| miR399f | 457 | 1,378 | 3.01 |
| miR822 | 2,999 | 4,530 | 1.51 |
| miR824 | 1,703 | 3,102 | 1.82 |
| miR1888 | 131 | 204 | 1.55 |
| Down-regulated | |||
| miR156a | 2,901 | 1,906 | 0.66 |
| miR156b | 2,928 | 1,926 | 0.66 |
| miR156c | 2,901 | 1,906 | 0.66 |
| miR156d | 3,856 | 2,567 | 0.67 |
| miR156e | 2,889 | 1,899 | 0.66 |
| miR156f | 2,889 | 1,899 | 0.66 |
| miR168a | 15,772 | 8,553 | 0.54 |
| miR168b | 15,766 | 8,541 | 0.54 |
| miR398b | 1,036 | 733 | 0.71 |
| miR398c | 1,036 | 733 | 0.71 |
| miR773 | 225 | 114 | 0.51 |
In an effort to identify small RNA species important to PTI responses, the reads encompassing the defined miRNA sequence ±2 nts on each side were calculated. We focused on flg22-regulated miRNAs in this study. miRNAs with at least 100 reads and >30% increase or decrease in flg22 treatment were selected. In total, 16 up-regulated and 11 down-regulated miRNAs were identified (Table I; Supplemental Table S2). RNA-blot analyses were carried out for nine selected miRNAs (Fig. 2A). Flg22 treatment induced miR393 accumulation to approximately 1.6-fold compared to the water control in an RNA-blot analysis, which is consistent with our sequencing data (approximately 2-fold) and a previous report (Navarro et al., 2006). Likewise, miR158a, miR160a, miR167, miR169, miR391, and miR396 were induced by flg22 to 1.6-, 2-, 2-, 1.6-, 1.5-, and 1.4-fold, respectively, compared to the water control. These miRNAs accumulated similarly in ago1-25 and wild-type plants, and this is in agreement with previous report that mutation in PIWI domain does not affect miRNA binding with AGO1 protein (Vaucheret et al., 2004). Contrary to above miRNAs, miR398b abundance was slightly reduced upon flg22 treatment. miR773 expressed at a level below the detection limit of RNA-blot analysis.
Figure 2.
Expression of selected miRNAs and their target genes in response to flg22 treatment. A, RNA-blot analysis of miRNAs. Wild-type (WT) and ago1-25 leaves were infiltrated with flg22 (F) or water (H) for 1 h before RNA extraction. Fifteen micrograms small RNA was loaded. RNA blots were hybridized with DNA oligonucleotide probes complementary to the indicated miRNAs. U6 was used as loading control. Values below each section represent relative abundance of miRNA normalized to U6 control. B to E, Quantitative RT-PCR analyses of mRNA levels for genes targeted by miR167 (ARF6 and ARF8), miR160 (ARF10, ARF16, and ARF17), miR773 (MET2), and miR398 (COX5b.1, CSD1, and CSD2). Error bars indicate sd. Student's t test was used to determine the significance of difference between water and flg22 treatments. Asterisks (* and **) indicate significant difference at a P value <0.05 and <0.01, respectively. The experiments were repeated two times with similar results.
Real-time reverse transcription (RT)-PCR was used to determine if the flg22-regulated expression of miRNAs correlated with the expression of their putative target genes (protein coding). miR167 targets auxin response factors ARF6 and ARF8 (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004), which is supported by the down-regulation of ARF6 and ARF8 mRNA in the 35S:miR167 transgenic plants (Wu et al., 2006), whereas miR160a targets ARF10, ARF16, and ARF17 (Mallory et al., 2005). The ARF genes encode auxin response factors (Mallory et al., 2005). Flg22 treatment repressed the expression of ARF10, ARF16, and ARF17, but did not significantly alter ARF6 and ARF8 (Fig. 2, B and C). miR398 targets COX5b.1, CSD1, and CSD2 (Jones-Rhoades and Bartel, 2004), which, respectively, encode a cytochrome c oxidase and two copper superoxide dismutase. Flg22 treatment enhanced the accumulation of COX5b.1, CSD1, and CSD2 transcripts (Fig. 2D). Although miR773 RNA level was not detectable by RNA-blot analysis, flg22 treatment resulted in greater expression of its target gene MET2 (Fig. 2E; Fahlgren et al., 2007), which encodes a DNA methyltransferase. The expression of the genes targeted by six other flg22-induced miRNAs was not significantly altered by flg22 treatment, with the exception of At3g03580, which was reduced by flg22 treatment (Supplemental Fig. S6). Target genes for miR156, which was identified as down-regulated by flg22, showed reduced expression in response to flg22 for reasons unknown (Supplemental Fig. S6).
miR160a, miR398b, and miR773 Regulate PTI Defenses
To further study the function of miR160a, miR398b, miR773, and miR158a, we generated stable transgenic plants overexpressing the four miRNAs. Three independent T2 transgenic lines overexpressing these miRNAs were identified by RNA-blot analyses (Figs. 3A–5A; Supplemental Fig. S7A). We next determined if the overexpression of the miRNAs reduced their target transcripts. The expression of ARF16 and ARF17 was reduced to 10% to 20% in the three miR160a overexpression plants compared to the wild-type control, whereas ARF10 was not significantly altered (Fig. 3B). Consistent with previous reports that transgenic plants expressing miR160-resistant forms of ARF10 and ARF17 (arf10 and arf17) display serrated leaves (Mallory et al., 2005; Liu et al., 2007), our 35S:miR160a plants exhibited a loss of leaf serration. We examined callose deposition induced by flg22 and the hrcC− mutant bacteria. Figure 3C shows that miR160a overexpression led to greater callose deposition in both treatments, indicating that miR160a positively regulates PAMP-induced callose deposition. However, miR160a overexpression plants were not significantly altered in basal resistance to DC3000 bacteria (Fig. 3D).
Figure 3.
Overexpression of miR160a enhances PAMP-induced callose deposition. A, Accumulation of miR160a in 35S:miR160a plants. Twenty micrograms small RNA was loaded for RNA-blot analysis. B, Overexpression of miR160a down-regulates ARF16 and ARF17 mRNA in transgenic plants. RNA was extracted from T2 generation of 35S:miR160a plants for quantitative real-time RT-PCR analyses. Error bars indicate sd. C, Flg22- and hrcC−-induced callose deposition in 35S:miR160a plants. D, 35S:miR160a plants were not affected in resistance to DC3000 bacteria. Wild-type (WT) and 35S:miR160a transgenic plants were infiltrated with 5 × 105 cfu/mL DC3000 bacteria, and leaf bacterial population was determined at the indicated times. Error bars indicate sd. The experiment was repeated three times with similar results. Student's t test was carried out to determine the significance of difference between 35S:miR160a and wild-type plants within each treatment. Asterisks (* and **) indicate significant difference at a P value of <0.05 and <0.01, respectively.
Figure 4.
miR398b negatively regulates PTI. A, RNA-blot analysis of miR398b in transgenic plants. B, Quantitative RT-PCR analyses of CSD1, CSD2, and COX5b.1 transcripts in 35S:miR398b transgenic plants. C, miR398b overexpression represses flg22- and hrcC−-induced callose deposition. D, miR398b overexpression enhances plant susceptibility to DC3000. Wild-type (WT) and 35S:miR398b transgenic plants were infiltrated with 5 × 105 cfu/mL DC3000 bacteria, and leaf bacterial population was determined at the indicated times. E, miR398b overexpression plants support growth of nonpathogenic DC3000 hrcC− mutant bacteria. Plants were spray inoculated with 5 × 108 cfu/mL DC3000 hrcC− mutant bacteria, and bacterial population in the leaf was determined at the indicated times. Error bars indicate sd. Student's t test was performed to determine the significance of difference between 35S:miR398b and wild-type plants within each treatment. Asterisks (* and **) indicate significant difference at a P value of <0.05 and <0.01, respectively. The experiments were repeated two (B, C, and E) and three (D) times with similar results.
Figure 5.
miR773 negatively regulates PTI. A, RNA-blot analysis of miR773 in transgenic plants. B, Quantitative RT-PCR analyses of MET2 transcripts in 35S:miR773 transgenic plants. C, miR773 overexpression represses flg22- and hrcC−-induced callose deposition. D, miR773 overexpression enhances plant susceptibility to DC3000. Wild-type (WT) and 35S:miR773 transgenic plants were infiltrated with 5 × 105 cfu/mL DC3000 bacteria. Leaf bacterial population was determined at the indicated times. E, miR773 overexpression plants support growth of nonpathogenic DC3000 hrcC− mutant bacteria. Plants were spray inoculated with 5 × 108 cfu/mL DC3000 hrcC− mutant bacteria, and bacterial population in the leaf was determined at the indicated times. Error bars indicate sd. Student's t test was done to determine the significance of difference between 35S:miR773 and wild-type plants within each treatment. Asterisks (* and **) indicate significant difference at a P value of <0.05 and <0.01, respectively. The experiments were repeated two times with similar results.
MiR398b transgenic plants displayed slightly yellowish leaves, but were otherwise normal in growth and development. COX5b.1 mRNA level in two miR398b transgenic lines (nos. 4 and 5) was reduced by 80% compared to wild-type control, whereas CSD2 transcript was completely abolished in these transgenic lines (Fig. 4B). Callose deposition induced by flg22 and hrcC− bacteria was decreased in the two transgenic lines (Fig. 4C). In agreement with reduced PTI defenses, 35S:miR398b transgenic plants were significantly more susceptible to DC3000 bacteria and supported 3- to approximately 5-fold more bacterial proliferation compared to wild type (Fig. 4D). In addition, the 35S:miR398b transgenic plants also supported DC3000 hrcC− bacteria by 7- to 10-fold (Fig. 4E), indicating that miR398b negatively regulates PAMP-triggered disease resistance.
MiR773 transgenic plants were morphologically indistinguishable from wild-type plants. The three 35S:miR773 transgenic lines examined all showed greatly reduced MET2 mRNA level (approximately 10%–20% of wild-type control; Fig. 5B). The transgenic plants displayed reduced callose deposition (Fig. 5C) and enhanced disease susceptibility to P. syringae DC3000 (Fig. 5D) and DC3000 hrcC− bacteria (Fig. 5E), indicating that, like miR398, miR773 also negatively regulates PTI resistance to P. syringae.
The two 35S:miR158a lines examined showed reduced expression of the target gene At3g03580 (Supplemental Fig. S7B). However, these plants were largely normal when PAMP-induced callose deposition was examined (Supplemental Fig. S7C). Furthermore, these plants supported normal growth to P. syringae DC3000 bacteria. These results did not support a role of miR158a in PTI resistance.
DISCUSSION
In this study, we systematically examined the role of AGO1, AGO7, and DCL1 in various PTI responses. ago1 and dcl1 mutants are compromised in PTI responses and flg22-induced disease resistance, indicating that overall AGO1 and DCl1 positively regulate PTI. In contrast, the ago7 mutant was completely normal in PTI resistance, suggesting a more specific role of AGO7 in RPS2 resistance. Thus AGO1 and AGO7 likely control distinct immune pathways in Arabidopsis.
The ago1 and dcl1 mutants investigated in this study showed defects in one or more of the late responses induced by PAMPs. However, these mutants displayed normal MAPK activation and transient oxidative burst, two events that occur less than 5 min after flg22 treatment. The data are consistent with the possibility that AGO1 and DCL1 act in later stages of PTI signaling. Because the ago1 mutants examined were partial loss-of-function alleles, we cannot rule out the possibility that the remaining AGO1 activity is sufficient to mediate MAPK activation and oxidative burst. It is also possible that PAMP-induced gene expression and callose deposition occur independent of MAPK activation and oxidative burst, as recently suggested by Lu et al. (2009) and Tsuda et al. (2009).
To date, only one miRNA (miR393) is known to be involved in the regulation of PTI defenses (Navarro et al., 2006). By using deep sequencing, we compared AGO1-bound small RNA in Arabidopsis plants after water and flg22 treatments. Our sequencing analyses led to the identification of 27 miRNAs that were either enriched or depleted in AGO1 upon flg22 treatment. Notably, miR160, miR167, miR393, miR396, and miR824 that were enriched in flg22-treated AGO1 had been shown to accumulate in plants treated with the DC3000 hrcC− bacteria (Fahlgren et al., 2007). RNA-blot analysis confirmed that the increased presence of at least some of theses miRNAs in flg22-treated AGO1 was likely caused by increased abundance of miRNAs. By constructing stable transgenic plants overexpressing miRNA genes, we further demonstrated that miR160, miR398, and miR773 play important role in regulating PTI defenses.
Flg22 is known to induce miR393 accumulation, which specifically targets TIR/AFB transcripts. The repression of TIR/AFB transcripts consequently down-regulates auxin signaling pathway and enhances plant resistance to DC3000 bacteria (Navarro et al., 2006). Our results showed that flg22 also induces miR160a accumulation and represses its target genes ARF16 and ARF17. ARF proteins bind auxin-responsive elements to activate or repress transcription of primary auxin-response genes (Hagen and Guilfoyle, 2002). Transgenic plants overexpressing miR160a exhibit enhanced callose deposition. Thus, multiple auxin pathway genes may be regulated by miRNAs during PTI defenses.
Intriguingly, some of the AGO1-bound miRNA apparently play a negative role in PTI resistance, although AGO1 overall positively regulates PTI resistance. Flg22 suppressed miR398b and miR773 accumulation. Consistent with this, flg22 treatment enhanced the expression of their target genes COX5b.1, CSD2, and MET1. miR398b and miR773 overexpression plants were compromised in PTI defenses exemplified by reduced callose deposition and supported greater DC3000 and DC3000 hrcC− proliferation, indicating that miR398b and miR773 negatively regulate plant disease resistance.
It was reported that inoculation of plants with incompatible strains DC3000 (avrRpm1) and DC3000 (avrRpt2) but not the compatible strain DC3000 represses miR398 levels (Jagadeeswaran et al., 2009). It is possible that miR398 is involved in both PTI and ETI defenses.
The findings that miR160a, miR398b, and miR773 regulate PTI raise interesting questions concerning the potential role of their target genes in PTI. CSD1 and CSD2 are copper- and zinc-containing superoxide dismutase enzymes that convert superoxide anion to hydrogen peroxide (Mori and Schroeder, 2004). It was shown previously that down-regulation of miR398 by oxidative stresses leads to the accumulation of CSD1 and CSD2 and elevated tolerance to a variety of stresses (Sunkar et al., 2006). MET2 is one of the seven known DNA methyltransferases in plants. A previous report showed that Arabidopsis DNA methyltransferases MET1 and MET2 are required for optimum root transformation by Agrobacterium (Crane and Gelvin, 2007). Future analyses of CSD1, CSD2, and MET2 functions may provide new insight into PTI regulation.
MATERIALS AND METHODS
Plants and Bacterial Strains
Arabidopsis (Arabidopsis thaliana) plants used in this study include the wild-type Columbia-0 (Col-0) and Landsberg erecta, and ago1-27, ago1-25, ago7, and dcl1-9 (Landsberg erecta background) mutants. Plants were grown in a growth room maintained at 23°C and 70% relative humidity with a 10/14 h day/night light. Bacteria used in this study include Pseudomonas strains Pseudomonas syringae pv tomato DC3000 and its nonpathogenic derivative hrcC−.
Construction of miRNA Overexpression Plants
To make the 35S:miRNA construct, miR160a genomic sequence containing 291 bp upstream and 133 bp downstream sequences, miR398b genomic sequence containing 163 bp upstream and 158 bp downstream sequences, and miR773 genomic sequence containing 76 bp upstream and 84 bp downstream sequence were PCR amplified from Col-0 genomic DNA. PCR products were cloned into 35S-pKANNIBAL vector between XhoI and KpnI. Constructs were transformed into Col-0 plants by Agrobacterium-mediated transformation. Transgenic plants were screened by spraying with 0.1% BASTA for two times.
Flg22-Protection Assay
Five- to 6-week-old plants were infiltrated with 1 μm flg22 or water 24 h before infiltrating 5 × 105 colony forming units (cfu)/mL DC3000 bacteria, and bacterial number was determined at indicated time points as described (Zipfel et al., 2004). Each data point consisted of at least four replicates.
Callose Staining
Five-week-old Arabidopsis leaves were infiltrated 1 μm flg22 for 12 h, and leaves were cleared, stained with 0.01% aniline blue for half an hour (Hauck et al., 2003). Callose deposition was captured with a fluorescence microscope and calculated by using the Image J software (Zhang et al., 2007). Each data consisted of at least six replicates.
Quantitative RT-PCR
RNA was extracted from leaves at indicated time points by Trizl reagent (Invitrogen) and reverse transcribed to obtain total cDNA using the SuperScript first-strand synthesis system (Invitrogen). SYBR Green Mix (TaKaRa) was used in real-time PCR to determine the abundance of mRNA. Gene expression level was normalized by using Actin 2 as a control. Primer used in real-time RT-PCR were: 5′-GGTGTCATGGTTGGTATGGGTC-3′ and 5′-CCTCTGTGAGTAGAACTGGGTGC-3′ for Actin2; 5′-TCTGAAGAATCAGCTCAAGGC-3′ and 5′-TGTTGGCTTCACATCTCTGTG-3′ for FRK1; 5′-AAGGATCTCCATACCCAAGG-3′ and 5′-ATCAGCGGATGGGATCATAGG-3′ for WRKY29; 5′-ATCCGCAGCTTATCTGTCAG-3′ and 5′-TATCATGCAGATCCCTCGCC-3′ for ARF6; 5′-ACGCGTATTTCAGTTGGGATG-3′ and 5′-ATGATGTGCCAGCATGCCATG-3′ for ARF8; 5′-AAAGGTTTAGTGGCTCGTGGG-3′ and 5′-TCCCGGTACACAACATGAGTC-3′ for ARF10; 5′-TGTCAAGGCATTAGATGCTCG-3′ and 5′-AACTCGCTTCACGTTTTGGAG-3′ for ARF16; 5′-TTATCAGGAGACCGGTCCATG-3′ and 5′-TATTGCCTGGCTCCCTGCATG-3′ for ARF17; 5′-TCCACATTTCAACCCCGATG-3′ and 5′-TTTCCAGTAGCCAGGCTGAG-3′ for CSD1; 5′-TCCTACAACTGTGAATGTTCG-3′ and 5′-AGGTCATCCTTAAGCTCGTG-3′ for CSD2; 5′-AGACCTGTCGGTTTCTATCTC-3′ and 5′-TTCGCTTGTCATAGTAGGAC-3′ for COX5b.1; 5′-ACCTGCCGGACGAAAATGTG-3′ and 5′-TCGTAGCTATCCGGAAACCC-3′ for MET2; 5′-ACCTGCCGGACGAAAATGTG-3′ and 5′-TGCGTCTCATGATCATGCTG-3′ for AGL16; 5′-TTTGTCCCTGAACCCACTTC-3′ and 5′-AAGAGAAGCTCTTGAGGAAC-3′ for GRF3; 5′-TTGTCGCAGAAGACTCGCTTG-3′ and 5′-AAACATTCATGTGTGGATGGG-3′ for SPL15; 5′-TTTGTCCCTGAACCCACTTC-3′ and 5′-AGAAACGTTGGTGAAGAC-3′ for SPL3; 5′-TCAAGGGGTGTCAATCGTGC-3′ and 5′-TCCTGTGGGGCATCTGATTG-3′ for AGO1; 5′-AGTGGAGATGATAACCCCTC-3′ and 5′-TTGCCTGTGGTAGGTAAGCC-3′ for AT1G17590; 5′-TGTCCCAAACCCATATCAGG-3′ and 5′-TTCTGTCCCGGATGACTTTC-3′ for AT5G12840; 5′-AAGCAACGGGTTCAGTGGAG-3′ and 5′-AAGCAGCCAAGGCTCTACAG-3′ for AT1G64100; 5′-TATAGCTGGATCGAAGTCGG-3′ and 5′-ACGCAATAGCAAGTCTCTCG-3′ for AT3G03580; 5′-TCAGTGCTTTGATCGATGTG-3′ and 5′-AATGTGTCACCATCCACTCC-3′ for AT1G06580; 5′-TAGAGATTGTGATAATGCCC-3′ and 5′-TAGGGCTCACTCCTTTAAGG-3′ for AT1G62910.
Growth Inhibition Assay
Arabidopsis seedlings were germinated for 8 d on one-half Murashige and Skoog agar plates and transferred to liquid one-half Murashige and Skoog medium containing 10 μm flg22 in a 24-well plate. Fresh weight was determined 5 d later.
Small RNA Gel-Blot Analysis
RNA-blot analysis for small RNAs from total extracts was performed as described (Qi et al., 2005; Mi et al., 2008). Leaves of 5-week-old plants were used for RNA extraction. miRNA probes were end labeled with γ-32P-ATP and T4 polynucleotide kinase.
Isolation of AGO1-Bound Small RNAs
Five- to 6-week-old Arabidopsis leaves were infiltrated with 2 μm flg22 or water and harvested 1 h later. One milliliter of Arabidopsis extract was incubated with Protein A-agarose for 60 min at 4°C. The precleared extracts were then incubated with 10 μL anti-AGO1 polyclonal antibodies at 4°C for 60 min, and 10 μL of Protein A agarose was added into the extracts and incubated for 2 h. Immunoprecipitates were washed three times (20 min each) in extraction buffer. AGO1-bound RNA was extracted from the immunoprecipitates with Trizol (Invitrogen). Small RNA library preparation, and sequencing of small RNAs were performed as described (Qi et al., 2005; Mi et al., 2008).
Bioinformatic Analysis of Small RNAs
The small RNA reads with length of 19 to 27 nt were mapped to the Arabidopsis nuclear, chloroplast, and mitochondrial genomes (http://www.arabidopsis.org/). The small RNAs with perfect genomic matches were used for further analysis. Annotation of small RNAs was performed using the following databases: TAIR7 annotations for coding sequences and noncoding RNAs (rRNAs, tRNAs, snoRNAs, snRNAs), and sequences from the intergenic regions (ftp://ftp.arabidopsis.org/Sequences/blast_datasets/TAIR7_blastsets/), Repbase (http://www.girinst.org) for transposons and repeats, ASRP for tasiRNA annotations (http://asrp.cgrb.oregonstate.edu/), and miRBase for miRNA annotations (http://microrna.sanger.ac.uk/sequences/). Annotations for the cis- or trans-natural antisense genes were extracted from published databases (Margulies et al., 2005; Wang et al., 2006). The abundance of small RNAs were calculated as reads per million.
Oxidative Burst
Leaves were sliced into 1 mm strips, and approximately 10 mm2 leaf strips were incubated in 200 μL water in a 96-well plate for 8 h prior to the addition of 1 μm flg22 in 200 μL buffer containing 20 mm luminol and 1 μg horseradish peroxidase (Sigma). Luminescence was determined with a Luminometer (Promega) for 30 to 40 min.
MAPK Activity Assay
Five-week-old plants were sprayed with 10 mm flg22 or water containing 0.02% Silwet L-77 for 10 min before protein extraction. Fifteen micrograms of total protein was electrophoresed on 10% SDS-PAGE gel, and the protein blot was reacted with anti-p-ERK antibody (cell signaling) to detect and determine phosphorylation state of MPK3, MPK4, and MPK6. A duplicate blot was reacted with anti-MPK6 antibodies (Sigma) to determine the amount of total MPK6.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GSE20448, GSM512702, and GSM512703.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ago1 mutants exhibit normal MAPK activation and transient oxidative burst.
Supplemental Figure S2. AGO7 differentially regulates PTI and RPS2 resistance.
Supplemental Figure S3. DCL1 contributed to PTI.
Supplemental Figure S4. dcl1-9 exhibits normal MAPK activation and transient oxidative burst.
Supplemental Figure S5. Flg22 treatment does not affect small RNA sorting to AGO1.
Supplemental Figure S6. Quantitative RT-PCR analysis of transcript levels of miRNA target genes in response to flg22.
Supplemental Figure S7. miR158 overexpression plants display normal PTI.
Supplemental Table S1. Category of AGO1-bound RNAs after flg22 treatment.
Supplemental Table S2. AGO1-bound miRNAs after flg22 treatment.
Supplementary Material
Acknowledgments
We thank Herve Vaucheret for providing ago1 mutants, Scott Poethig for ago7 mutant, and the Arabidopsis Biological Resource Center for dcl1-9 mutant.
References
- Agorio A, Vera P. (2007) ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I, Trethowan J, Kerry M, Mansfield J, Bolwell GP. (1998) Localization of components of the oxidative cross-linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non-pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J 15: 333–343 [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Crane YM, Gelvin SB. (2007) RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc Natl Acad Sci USA 104: 15156–15161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Juliana D, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez G, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS. (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–339 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin HL. (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Jr AV, Zhu JK, Staskawicz BJ, Jin HL. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103: 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crété P, Voinnet O, Robagliaa C. (2009) Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Lin HQ, Zhang WG, Zou Y, Zhang J, Tang XY, Zhou JM. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y. (2009) Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA 106: 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet (Suppl) 38: S31–S36 [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi SJ, Cai T, Hu YG, Chen YM, Hodges E, Ni FR, Wu L, Li S, Zhou HY, Long CZ, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Schroeder JI. (2004) Reactive oxygen species activation of plant Ca2+ channels: a signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol 135: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura A, Ishizuka A, Siomi H, Siomi MC. (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi YJ, Denli AM, Hannon GJ. (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Song JJ, Joshua-Tor L. (2006) Argonaute and RNA—getting into the groove. Curr Opin Struct Biol 16: 5–11 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chua NH, Wang XJ. (2006) Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol 7: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yuan J, He SY. (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol 178: 6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui HT, Chen LJ, Li HT, Zou Y, Long CZ, Lan LF, Chai JJ, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Chai JJ. (2008) Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 11: 179–185 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G. (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.