Abstract
Plant metabolism is highly coordinated with development. However, an understanding of the whole picture of metabolism and its interactions with plant development is scarce. In this work, we show that the deficiency in the plastidial glycolytic glyceraldehyde-3-phosphate dehydrogenase (GAPCp) leads to male sterility in Arabidopsis (Arabidopsis thaliana). Pollen from homozygous gapcp double mutant plants (gapcp1gapcp2) displayed shrunken and collapsed forms and were unable to germinate when cultured in vitro. The pollen alterations observed in gapcp1gapcp2 were attributed to a disorganized tapetum layer. Accordingly, the expression of several of the genes involved in tapetum development was down-regulated in gapcp1gapcp2. The fertility of gapcp1gapcp2 was rescued by transforming this mutant with a construct carrying the GAPCp1 cDNA under the control of its native promoter (pGAPCp1::GAPCp1c). However, the GAPCp1 or GAPCp2 cDNA under the control of the 35S promoter (p35S::GAPCp), which is poorly expressed in the tapetum, did not complement the mutant fertility. Mutant GAPCp isoforms deficient in the catalytic activity of the enzyme were unable to complement the sterile phenotype of gapcp1gapcp2, thus confirming that both the expression and catalytic activity of GAPCp in anthers are necessary for mature pollen development. A metabolomic study in flower buds indicated that the most important difference between the sterile (gapcp1gapcp2, gapcp1gapcp2-p35S::GAPCp) and the fertile (wild-type plants, gapcp1gapcp2-pGAPCp1::GAPCp1c) lines was the increase in the signaling molecule trehalose. This work corroborates the importance of plastidial glycolysis in plant metabolism and provides evidence for the crucial role of GAPCps in pollen development. It additionally brings new insights into the complex interactions between metabolism and development.
Glycolysis is a primary metabolic pathway whose main function is to oxidize hexoses to generate ATP, reducing power and pyruvate, and to produce precursors for anabolism (Plaxton, 1996). The major substrates fuelling glycolysis in plants are Suc and starch (Plaxton, 1996). Both metabolites are subjected to large diurnal changes in their concentrations that need to be coordinated with the rate of glycolysis to provide metabolic flexibility that can respond to differential demands of plant development and environmental stress acclimation. In plants, glycolysis occurs in both the cytosol and plastids that complicates our global understanding of this pathway and its interactions with development.
The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reversibly converts the glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate by coupling with the reduction of NAD+ to NADH. In mammals, glycolytic GAPDH has been extensively characterized where, besides its pivotal role in energy production, it has been implicated in transcriptional regulation, DNA repair, signal transduction cascades, and apoptosis (Hara et al., 2005; Kim and Dang, 2005; Hara and Snyder, 2006; Colell et al., 2007; Min et al., 2007; Lee et al., 2009). These additional functions of glycolytic enzymes suggest that links between metabolic sensors and development could be established directly through enzymes that participate in metabolism (Kim and Dang, 2005). Nonglycolytic functions of plant GAPDHs have also been suggested either because of the nuclear localization of some isoforms or by the suppression of H2O2-mediated cell death in protoplasts (Baek et al., 2008; Holtgrefe et al., 2008). However, whether these postulated signaling functions of plant GAPDHs rely on their glycolytic activity remains unproven.
A family of four genes encoding putative phosphorylating glycolytic GAPDHs (GAPC1, GAPC2, GAPCp1, and GAPCp2) is present in Arabidopsis (Arabidopsis thaliana; http://www.arabidopsis.org). GAPC1 and GAPC2 are cytosolic isoforms, while GAPCp1 and GAPCp2 are located in the plastids (Muñoz-Bertomeu et al., 2009). In spite of their low gene expression level compared to cytosolic GAPDHs, GAPCp down-regulation leads to drastic changes in the sugar and amino acid balance of the plant and causes arrested root development and sterility (Muñoz-Bertomeu et al., 2009). The arrested root development of gapcp mutants was attributed to a Ser deficiency, concluding that the major function of GAPCps in roots is to provide precursors for Ser biosynthesis (Muñoz-Bertomeu et al., 2009).
In this study, we investigated the role of GAPCps in pollen development. In flowering plants, pollen formation depends on differentiation and interaction of two cell types in the anther: the reproductive microsporocyte cells and the somatic cells that form the tapetum (Yang and Sundaresan, 2000). Here, we show that homozygous gapcp double mutants (gapcp1gapcp2) are male sterile. Pollen obtained from gapcp1gapcp2 has collapsed shapes and is unviable. This is attributed to alterations in the tapetum layer as a result of the lack of GAPCp catalytic activity in anther tissues. The mechanism responsible for male sterility is studied, especially its relationships with sugar signaling. Our work corroborates the importance of plastidial glycolysis in metabolism and provides further evidence of the clear links between metabolism and development.
RESULTS
GAPCp Expression Is Increased in Reproductive Organs
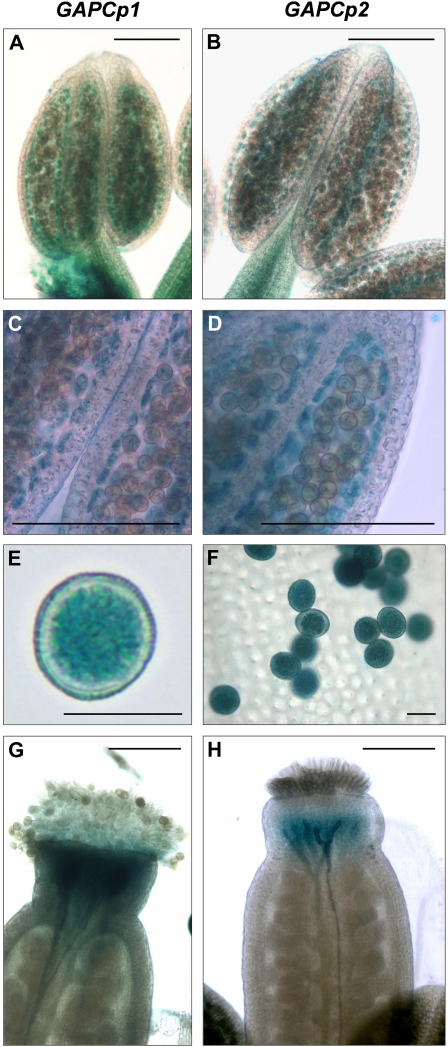
The expression of the four Arabidopsis phosphorylating glycolytic GAPDH genes in roots, leaves, shoots, and flowers was investigated by quantitative reverse transcription-PCR (Q-RT-PCR; Table I). The expression of cytosolic GAPC1 (At3g04120) and GAPC2 (At1g13440) isoforms was lower in flowers than the calculated mean expression value of all organs studied. However, the expression of the plastidial GAPCp1 (At1g79530) and GAPCp2 (At1g16300) was higher in roots and flowers compared to the other organs. Consequently, the relative abundance of GAPCp1 and GAPCp2 in flowers was increased between 2 and 6 times compared to both GAPC1 and GAPC2. Our data are in agreement with those found in the microarray databases where the relative expression of both GAPCp1 and GAPCp2 is increased by >4-fold in flowers compared to other glycolytic GAPDHs (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Specifically, GAPCp1 expression is higher in early stages of flower development (stage 9) than in older stages (stage 15), especially in nongreen organs, such as petals, stamens, and carpels (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Besides, GAPCp1 is more highly expressed in the earlier stages of pollen development (uninucleate microspores and bicellular pollen) compared to mature pollen (Honys and Twell, 2004). Histochemical GUS analyses clearly corroborated that both GAPCp1 and GAPCp2 were expressed in stamens and carpels (Muñoz-Bertomeu et al., 2009; Fig. 1). In stamens, the GUS expression was specifically observed in the tapetum and at an early stage of pollen development (Fig. 1, A–F). An in silico analysis of the GAPCp1 and GAPCp2 promoters using the promomer tool (Winter et al., 2007) revealed that promoter regions of both genes were significantly enriched in consensus binding sequences present in important genes controlling anther development, such as the floral homeotic gene AGAMOUS (Supplemental Table S1). These data indicate that GAPCp1 and GAPCp2 promoters have relevant information required for anther gene expression.
Table I. Expression analysis of glycolytic GAPDHs genes of Arabidopsis by Q-RT-PCR.
Values represent the relative expression (± sd) of each GAPDH in different organs compared to the mean expression value of all organs assayed. The ratio represents the relative abundance of plastidial GAPDHs in different organs compared to the cytosolic isoforms.
| Relative Expression |
Ratio |
|||||||
| GAPC1 | GAPC2 | GAPCp1 | GAPCp2 | GAPCp1/GAPC1 | GAPCp1/GAPC2 | GAPCp2/GAPC1 | GAPCp2/GAPC2 | |
| Root | 1.34 ± 0.28 | 1.37 ± 0.20 | 6.90 ± 0.50 | 4.38 ± 1.03 | 5.14 | 5.02 | 2.55 | 2.49 |
| Leaf | 0.93 ± 0.13 | 1.01 ± 0.00 | 0.12 ± 0.01 | 0.16 ± 0.04 | 0.13 | 0.12 | 0.27 | 0.25 |
| Shoot | 1.55 ± 0.10 | 0.71 ± 0.00 | 0.41 ± 0.05 | 0.95 ± 0.05 | 0.27 | 0.58 | 0.50 | 1.08 |
| Flower | 0.37 ± 0.00 | 0.74 ± 0.07 | 2.38 ± 0.06 | 1.67 ± 0.01 | 6.37 | 3.22 | 4.18 | 2.11 |
Figure 1.
Expression analysis of GAPCp1 and GAPCp2 in flowers. Expression of GUS under the control of GAPCp1 and GAPCp2 promoters in stamens (A–D), pollen (E and F), and carpels (G and H). Bars = 0.15 mm (A–D), 20 μm (E and F), and 0.25 mm (G and H).
T-DNA Insertions in GAPCp1 and GAPCp2 Trigger Male Sterility
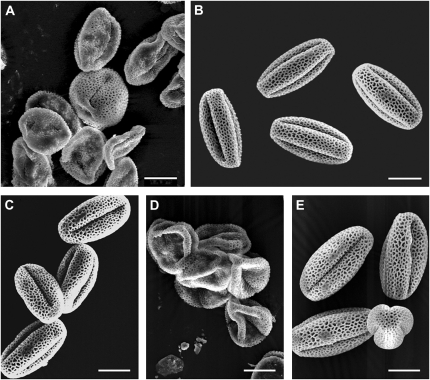
Multiple independent T-DNA insertion mutant lines affecting GAPCp1 and GAPCp2 were obtained in a previous study (Muñoz-Bertomeu et al., 2009). Although none of the single mutants of either GAPCp1 or GAPCp2 presented a clear visual phenotype, different homozygous gapcp double mutants (gapcp1gapcp2) showed growth defects resulting in dwarf phenotypes and sterility (Muñoz-Bertomeu et al., 2009). The causes of this gapcp1gapcp2 sterility were investigated. The stamens and pistils of gapcp1gapcp2 look macroscopically normal when compared to wild-type plants (Fig. 2A). However, gapcp1gapcp2 produced small siliques with no seeds (Fig. 2B). Crosses of wild-type pollen with gapcp1gapcp2 ovules rescued the fertile phenotype, producing normal seeds and siliques (Fig. 2B). Yet, crosses of gapcp1gapcp2 pollen with wild-type ovules produced small siliques with no seeds like those observed in the self-crossed gapcp1gapcp2 (Fig. 2B). These findings indicate that gapcp1gapcp2 mutations trigger male sterility. Scanning electron microscopy revealed that most of wild-type pollen had a uniform shape, but pollen from gapcp1gapcp2 had shrunken and collapsed shapes (Fig. 3, A and B). In vitro germination assays demonstrated that pollen from three different homozygous gapcp1gapcp2 alleles (gapcp1.1/gapcp1.1 gapcp2.1/gapcp2.1, gapcp1.1/gapcp1.1 gapcp2.2/gapcp2.2, and gapcp1.1/gapcp1.1 gapcp2.3/gapcp2.3) were unable to germinate (Table II), thus confirming that male sterility is genetically linked to both the GAPCp1 and GAPCp2 loci.
Figure 2.
The gapcp1gapcp2 double mutation causes male sterility. A, Side and top views of wild-type (left) and gapcp1gapcp2 (right) flowers. Bars = 1.5 mm. B, Siliques obtained from (1) gapcp1gapcp2 plants fertilized with wild-type pollen, (2) wild-type plants fertilized with gapcp1gapcp2 pollen, (3) gapcp1gapcp2 plants, (4) heterozygous gapcp1GAPCp2 plants, and (5) wild-type plants. Bar = 1.5 cm.
Figure 3.
gapcp1gapcp2 pollen shows aberrant phenotypes. Scanning electron micrographs of pollen isolated from gapcp1gapcp2 (A), the wild type (B), gapcp1gapcp2 transformed with a genomic GAPCp1 construct (C; pGAPCp1::GAPCp1geno), gapcp1gapcp2 transformed with GAPCp1 cDNA under the control of the 35S promoter (D; p35S::GAPCp1c), and gapcp1gapcp2 transformed with GAPCp1 cDNA under the control of its native promoter (E; pGAPCp1::GAPCp1c). Bars = 10 μm.
Table II. Germination rate of pollen from the wild type, heterozygous (g1.1/g1.1 G2/g2.1, G1/g1.1 g2.1/g2.1), double mutants (g1.1/g1.1 g2.1/g2.1, g1.1/g1.1 g2.2/g2.2, g1.1/g1.1 g2.3/g2.3), and transformed gapcp1 gapcp2 double mutant lines.
Mutant lines were transformed with the following constructs: GAPCp1 cDNA fused to the 35S promoter (p35S::G1c), GAPCp1 cDNA fused to the GAPCp1 native promoter (pG1::G1c), GAPCp1 genomic fragment fused to the GAPCp1 native promoter (pG1::G1geno), and mutated GAPCp1 cDNAs on the indicated amino acids (C236G, H263A, R318E, and K311A) fused to the GAPCp1 native promoter. Values are the percentage of germination (± sd) compared to wild-type plants. The germination rate of wild-type plants is shown in parentheses. For simplicity, g stands for gapcp and G stands for GAPCp.
| Genotype | Germination (%) |
| Wild type | 100 (75%) |
| g1.1/g1.1 G2/g2.1 | 101 ± 12 |
| G1/g1.1 g2.1/g2.1 | 95 ± 7 |
| g1.1/g1.1 g2.1/g2.1 | 0 |
| g1.1/g1.1 g2.2/g2.2 | 0 |
| g1.1/g1.1 g2.3/g2.3 | 0 |
| g1.1/g1.1 g2.1/g2.1 p35S::G1c | 0 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1c | 58 ± 8 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1geno | 93 ± 36 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1c (C236G) | 0 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1c (H263A) | 0 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1c (R318E) | 0 |
| g1.1/g1.1 g2.1/g2.1 pG1::G1c (K311A) | 97 ± 7 |
The transmission of the T-DNA insertion in self-fertilized GAPCp1/gapcp1 gapcp2/gapcp2 or gapcp1/gapcp1 GAPCp2/gapcp2 heterozygous plants was normal (1:2:1 wild type:heterozygous:double mutant Mendelian ratio; Muñoz-Bertomeu et al., 2009), indicating that gapcp1gapcp2 pollen obtained from heterozygous plants germinate normally. In addition, the germination percentage of pollen from heterozygous plants (GAPCp1/gapcp1.1 gapcp2.1/gapcp2.1 or gapcp1.1/gapcp1.1 GAPCp2/gapcp2.1) did not differ from that of wild-type plants (Table II), confirming that male sterility in gapcp1gapcp2 is caused by sporophytic effects. Thus, a defect in the anther development of the gapcp1gapcp2 is probably the responsible for the pollen sterility observed in these plants.
Tapetum Development Is Impaired in gapcp1gapcp2
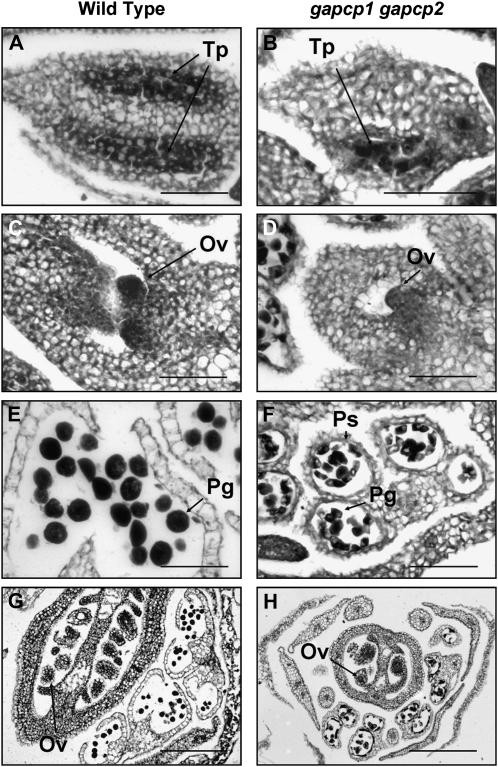
In order to explore the causes of pollen dysfunction in gapcp1gapcp2, we next investigated anther development. Sections of anthers and carpels of the wild type and gapcp1gapcp2 were obtained from flowers at the same developmental stage (Fig. 4). At an early developmental stage (stages 5–6; Smyth et al., 1990), the tapetum layer in gapcp1gapcp2 was completely disorganized (Fig. 4B). We also observed a delay in the ovule development of gapcp1gapcp2 (Fig. 4D). At a later stage of flower development (stages 8–10), pollen grains were clearly observed in both the wild type and gapcp1gapcp2, although the malformations noted in the mutant pollen could already be distinguished (Fig. 4F). At this developmental stage, the ovules of gapcp1gapcp2 developed normally, corroborating that this mutant has fertile ovules (Fig. 4H).
Figure 4.
The tapetal cell layer is disorganized in gapcp1gapcp2. The micrographs show transverse sections of anthers and carpels at early (top) and late (bottom) developmental stages (stages 5 and 6 and stages 8–10). A and E, Wild-type anthers; B and F, gapcp1gapcp2 anthers; C and G, wild-type carpels; D and H, gapcp1gapcp2 carpels. Ov, Ovule; Pg, pollen grain; Ps, pollen sac; Tp, tapetum. Bars = 50 μm for A to D and F and 250 μm for G and H.
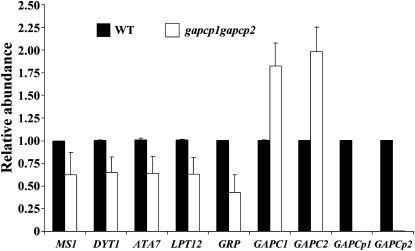
To further confirm that tapetum development was impaired in gapcp1gapcp2, the expression of a set of genes reported to be important in the tapetum and/or pollen wall development was studied. The following mRNA levels of both the wild type and gapcp1gapcp2 were analyzed in closed buds (stages 5–10) by Q-RT-PCR: MALE STERILITY1 (MS1; Wilson et al., 2001; Ito and Shinozaki, 2002), DYSFUNCTIONAL TAPETUM1 (DYT1; Zhang et al., 2006), ARABIDOPSIS THALIANA ANTHER7 (ATA7; Rubinelli et al., 1998), LIPID TRANSFER PROTEIN12 (LPT12; Yang et al., 2007), and GLYCINE RICH PROTEIN At4g37900 (Yang et al., 2007). In agreement with the microscopy analyses, the expression of these genes was down-regulated in the gapcp1gapcp2 inflorescences (Fig. 5). The expression of the glycolytic GAPDHs GAPCp1, GAPCp2, GAPC1, and GAPC2 was also analyzed (Fig. 5). As expected, the expression of GAPCp1 and GAPCp2 was nearly undetectable in gapcp1gapcp2. Interestingly, the expression of the cytosolic GAPC1 and GAPC2 was up-regulated in the mutant, probably reflecting a compensatory effect for the lack of plastidial GAPDH activities.
Figure 5.
Expression of important genes in tapetum development is impaired in gapcp1gapcp2. Q-RT-PCR analysis of MS1, DYT1, ATA7, LPT12, GLYCINE-RICH PROTEIN (GRP), GAPC1, GAPC2, GAPCp1, and GAPCp2 in wild-type (WT) and gapcp1gapcp2 flower buds (stages 5–10).
Expression and Catalytic Activity of GAPCp in Anthers Is Necessary for Mature Pollen Development
Since GAPCp mutations have pleiotropic effects on the plant, it might be argued that GAPCp deficiency could have an indirect effect on gametophyte development as a consequence of a general alteration of metabolism at the whole-plant level in gapcp1gapcp2. To answer this question, gapcp1gapcp2 were transformed with GAPCp under the control of different promoters. The 35S promoter has a very low or no expression in the tapetum (Skirycz et al., 2007; Grienenberger et al., 2009). For this purpose, a GAPCp1 or GAPCp2 cDNA construct under the control of this 35S promoter (p35S::GAPCp1c or p35S::GAPCp2c) complemented all phenotypes except the male sterility in gapcp1gapcp2 (Fig. 6, A and B). However, all the phenotypes of gapcp1gapcp2, including male sterility, could be complemented with a genomic construct (pGAPCp1::GAPCp1geno) of the GAPCp1 locus (Fig. 6). When the GAPCp1 cDNA was expressed under the control of its native promoter (pGAPCp1::GAPCp1c), full complementation of gapcp1gapcp2 was once again achieved (Fig. 6).
Figure 6.
Phenotype of gapcp1gapcp2 and transformed lines. A, Phenotype of wild-type plants, gapcp1gapcp2 transformed with a genomic GAPCp1 construct (pGAPCp1::GAPCp1geno), gapcp1gapcp2 transformed with GAPCp1 cDNA under the control of its native promoter (pGAPCp1::GAPCp1c), gapcp1gapcp2 transformed with GAPCp1 cDNA under the control of the 35S promoter (p35::GAPCp1c), gapcp1gapcp2 transformed with GAPCp2 cDNA under the control of the 35S promoter (p35S::GAPCp2c), and gapcp1gapcp2. Bar = 10 cm. B, Close-ups to show differences in the siliques obtained from the different lines. Bar = 5 cm.
Pollen morphology of gapcp1gapcp2 transformed either with pGAPCp1::GAPCp1geno or pGAPCp1::GAPCp1c was similar to the wild type (Fig. 3, C and E). However, pollen from gapcp1gapcp2-p35S::GAPCp1c line presented the same morphological alterations as gapcp1gapcp2 (Fig. 3D). The same was true for pollen viability. Only pollen from gapcp1gapcp2 transformed either with pGAPCp1::GAPCp1geno or pGAPCp1::GAPCp1c was viable in an in vitro germination assay (Table II).
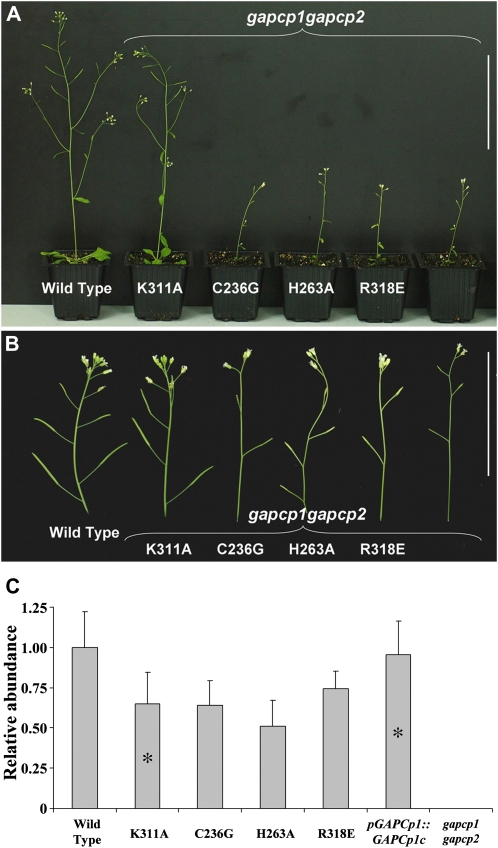
Several nonglycolytic functions for GAPDHs have been demonstrated in mammals and yeast (Hara and Snyder, 2006). To separate glycolytic functions from other additional putative functions of the GAPCps in anther development, amino acid substitutions were made to GAPCp1. GAPDHs are one of the most conserved proteins in living organisms. Sequence alignment of GAPCps with other GAPDHs from Arabidopsis identified several conserved amino acids already described in mammals as being essential for catalytic activity (Cys-236 and His-263), binding to the substrate glyceraldehyde-3-P (Arg-318) or putatively interacting with other regulatory proteins (Lys-311; Supplemental Fig. S1). Mutated versions of GAPCp1 were used to transform gapcp1gapcp2. Fifteen (C236G), 10 (H263A), and 32 (R318E) independent T2 transgenic lines expressing the mutated isoforms that affect the glycolytic activity of GAPCp1 were studied. None of them was able to complement any of the phenotypes observed in the mutant (Fig. 7, A and B). Only the construct that carried a mutation that enables the catalytic function of GAPCp1 (K311A) was able to complement the male sterility phenotype and all the other phenotypes present in gapcp1gapcp2 (Fig. 7, A and B). The in vitro germination assays confirmed that the K311A transformed lines were the only ones with viable pollen (Table II). Seedlings from a pool of three single insertion T3 homozygous lines were used to verify the expression of all the mutant constructs in Arabidopsis (Fig. 7C). This expression was compared to that of GAPCp1 in the wild type and in gapcp1gapcp2 complemented with pGAPCp1::GAPCp1c. The expression of all the mutated GAPCp1 versions was similar but slightly lower than that found in the lines transformed with the wild-type pGAPCp1::GAPCp1c. No correlation between expression level of the mutated GAPCp1 versions and complementation of the sterile phenotype was found.
Figure 7.
Mutated versions of GAPCp1 do not complement gapcp1gapcp2 phenotypes. A, From left to right: the wild type, gapcp1gapcp2 transformed with mutated versions of GAPCp1 (K311A, C236G, H263A, and R318E), and gapcp1gapcp2. Bar = 10 cm. B, Close-ups to show the differences in the siliques obtained from the different lines. Bar = 5 cm. C, Q-RT-PCR analysis of GAPCp1 expression level in 3-week-old seedlings from the wild type, gapcp1gapcp2 transformed with mutated versions or wild-type versions of GAPCp1 (K311A, C236G, H263A, R318E, and pGAPCp1::GAPCp1c), and gapcp1gapcp2. Asterisk indicates line with fertile phenotype.
Ser Supplementation Rescues All the Phenotypes Studied in gapcp1gapcp2 But Not Male Sterility
gapcp1gapcp2 roots display a deficiency in Ser that causes root developmental arrest (Muñoz-Bertomeu et al., 2009). This was attributed to a limitation of 3-phosphoglycerate, the substrate for the phosphorylated pathway of Ser biosynthesis, in the mutant plastids. However, Ser levels in flower buds of gapcp1gapcp2 were not reduced compared to control plants (Table III). There was even a clear tendency of an increase in the Ser content of the mutants. In accordance with these results, Ser supplementation rescued general growth but did not rescue the male sterility phenotype of gapcp1gapcp2 (Fig. 8).
Table III. Most important metabolite changes in flower buds of gapcp1gapcp2, gapcp1gapcp2 transformed with GAPCp1, or GAPCp2 cDNA under the control of the 35S promoter (p35S::GAPCp1c; p35S::GAPCp2c) and gapcp1gapcp2 transformed with a genomic GAPCp1 construct (pGAPCp1::GAPCp1geno) compared to the wild type.
Data are normalized to the mean response calculated for the wild type. Values are represented as the mean ± se of seven to 10 independent determinations from three different transgenic lines. Values in parentheses are absolute values (μmol g fresh weight−1). Asterisk indicates that the absolute value was not quantified. Those values that are significantly different to the wild type are set in bold, P < 0.05. For simplicity, g stands for gapcp and G stands for GAPCp.
| Wild Type | g1g2 | g1g2 p35S::G1c | g1g2 p35S::G2c | g1g2 pG1::G1geno | |
| Amino acids | |||||
| Ile | 1.00 ± 0.12 (0.13) | 2.09 ± 0.35 | 1.64 ± 0.28 | 1.60 ± 0.23 | 1.47 ± 0.20 |
| Ser | 1.00 ± 0.15 (2.49) | 1.68 ± 0.30 | 1.66 ± 0.22 | 1.58 ± 0.16 | 1.29 ± 0.14 |
| Trp | 1.00 ± 0.15 (0.03) | 2.50 ± 0.41 | 2.03 ± 0.37 | 2.01 ± 0.11 | 1.95 ± 0.29 |
| Sugars and sugar alcohols | |||||
| Cellobiose | 1.00 ± 0.09 (*) | 2.53 ± 0.48 | 1.69 ± 0.14 | 1.79 ± 0.25 | 1.66 ± 0.16 |
| Myoinositol | 1.00 ± 0.04 (0.96) | 2.05 ± 0.25 | 1.48 ± 0.10 | 1.78 ± 0.11 | 1.21 ± 0.07 |
| Raffinose | 1.00 ± 0.00 (0.01) | 6.50 ± 0.85 | 2.03 ± 0.04 | 4.72 ± 0.09 | 1.76 ± 0.03 |
| Suc | 1.00 ± 0.04 (5.25) | 1.14 ± 0.05 | 1.14 ± 0.10 | 1.04 ± 0.09 | 0.96 ± 0.04 |
| Trehalose | 1.00 ± 0.09 (0.16) | 4.68 ± 0.45 | 2.64 ± 0.26 | 3.92 ± 0.55 | 1.37 ± 0.05 |
Figure 8.
Ser supplementation does not rescue the male sterility phenotype of gapcp1gapcp2. A, gapcp1gapcp2 seeds were germinated in plates supplemented with Ser, which were then transplanted to pots in a greenhouse. Plants were either irrigated or sprayed with 0.1 mm Ser. From left to right: the wild type and gapcp1gapcp2 supplemented with Ser and gapcp1gapcp2. Bar =10 cm. B, Close-ups of the plants to show the differences in the siliques obtained from the different lines. Bar = 5 cm.
Metabolite Analysis in Flowers of gapcp1gapcp2 and Transformed Lines
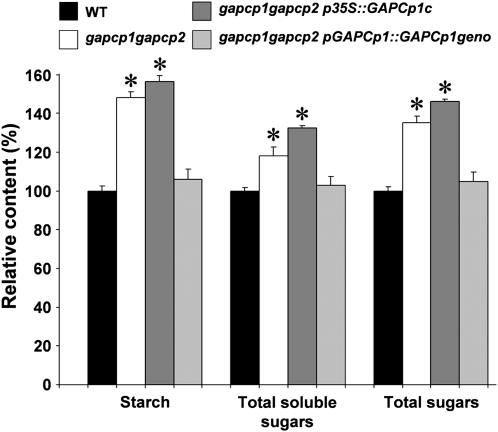
To investigate the effect of GAPCp deficiency on anther metabolism, flower buds of wild-type, gapcp1gapcp2, and gapcp1gapcp2 transformed lines were harvested and the carbohydrate content measured. The starch and total soluble sugar contents in gapcp1gapcp2 significantly increased by 48% and 18%, respectively, compared to the wild type (Fig. 9). Accordingly, the total sugars in gapcp1gapcp2 buds significantly increased by 35% compared to the wild type. The carbohydrate contents also increased in buds of gapcp1gapcp2 transformed with the p35S::GAPCp1c construct (sterile line), but they did not increase in those mutants transformed with the genomic construct (pGAPCp1::GAPCp1geno) of the GAPCp1 locus (fertile line).
Figure 9.
Starch and soluble sugar content in closed flower buds of gapcp1gapcp2 and transformed lines. Carbohydrate content was measured in the wild type (WT), gapcp1gapcp2, gapcp1gapcp2 transformed with GAPCp1 cDNA under the control of the 35S promoter (p35S::GAPCp1c), and gapcp1gapcp2 transformed with a genomic GAPCp1 construct (pGAPCp1::GAPCp1geno). Values (relative content ± sd) were normalized to the mean value of the wild type in mg g fresh weight−1 (starch: 6.54 ± 0.16, soluble sugars 5.1 ± 0.1, total sugars 11.55 ± 0.25). Asterisk indicates significance at a P value <0.05.
To further investigate the metabolic alterations in gapcp1gapcp2, a detailed analysis of the metabolites in flower buds was performed by a gas chromatography-mass spectrometry method (Table III; Supplemental Table S2). Although the content of Suc was slightly increased (14%) in gapcp1gapcp2, some other less abundant sugars, such as cellobiose, myoinositol, raffinose, and trehalose, were increased by >2-fold compared to the wild type (Table III). The most important changes noted in gapcp1gapcp2 were the increases in raffinose (6.5-fold increase) and in the signaling molecule trehalose (4.7-fold increase) compared to the wild type (Table III). The levels of the amino acids Ile and Trp also significantly increased by >2-fold in gapcp1gapcp2 compared to the wild type.
The metabolite profiles of the sterile gapcp1gapcp2-p35S::GAPCp1c and gapcp1gapcp2-p35S::GAPCp2c lines were similar but not identical (Table III; Supplemental Table S2). However, both lines displayed significantly higher levels of cellobiose, myoinositol, raffinose, trehalose, and Trp than the wild type, as found in gapcp1gapcp2 (Table III). In general, the metabolite changes measured in the gapcp1gapcp2-p35S::GAPCp1c line were weaker than those found in the gapcp1gapcp2-p35S::GAPCp2c line, especially in the raffinose and trehalose levels (2.0- versus 4.7-fold increase in raffinose and 2.6- versus 3.9-fold increase in trehalose in gapcp1gapcp2-p35S::GAPCp1c and gapcp1gapcp2-p35S::GAPCp2c, respectively, compared to the wild type). The fertile gapcp1gapcp2-pGAPCp1::GAPCp1geno line did not fully phenocopy the metabolite profile of the wild type, although a clear trend of a similar profile to the wild type was observed (Table III; Supplemental Table S2). None of the metabolites measured varied >2-fold in the gapcp1gapcp2-pGAPCp1::GAPCp1geno line compared to the wild type. By comparing this fertile complemented line with the sterile lines (gapcp1gapcp2, gapcp1gapcp2-p35S::GAPCp1c, and gapcp1gapcp2-p35S::GAPCp2c), the most important change in the measured metabolites was the reduction of the trehalose content in the fertile line to levels resembling those observed in the wild type (only a 1.37-fold increase compared to 4.7-, 2.6-, and 3.9-fold increase in the sterile lines, respectively).
DISCUSSION
GAPCp Disruption Affects Tapetum Organization and Impairs Pollen Development
Mutations in the GAPCp genes cause male sterility. However, gapcp1gapcp2 gametes obtained from heterozygous mothers are viable, indicating that the pollen sterility is not due to an alteration in the gamete itself, but to alterations in the cells participating in pollen development in gapcp1gapcp2. Several of the genes controlling tapetum development have been found to affect pollen viability and/or development (Colcombet et al., 2005; Mizuno et al., 2007; Yang et al., 2007; Zhu et al., 2008). The tapetum plays a major role in sporogenesis and is also critical in pollen wall and pollen coat formation (Paxson-Sowders et al., 2001; Dong et al., 2005; Yang et al., 2007). Pollen from gapcp1gapcp2 presented severe alterations of the pollen wall that may suggest that the tapetum is not functioning properly in these mutants. The disorganization of the tapetum layer, along with the down-regulation of the genes involved in tapetum development in gapcp1gapcp2, would support this idea.
GAPCp Activity Is Essential for Viable Pollen Development
GAPCp1 or GAPCp2 cDNA under the control of the 35S promoter could complement all the gapcp1gapcp2 phenotypes, except the male sterile phenotype. It is known that the 35S promoter has poor expression in the tapetum layer of Arabidopsis (Skirycz et al., 2007; Grienenberger et al., 2009). As in our case, the Arabidopsis mutant in the SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) and SERK2 genes, which are essential for tapetum development, could not be complemented when the SERK open reading frame was expressed under the control of the 35S promoter, but could be complemented with a genomic clone (Colcombet et al., 2005). These authors suggested that the noncoding sequences of the SERK locus might contribute to SERK function. By using the GAPCp1 native promoter fused to the GAPCp1 cDNA, we concluded that the GAPCp1 cDNA expression is sufficient to complement the gapcp1gapcp2 male sterility phenotype. Therefore, these results indicate that GAPCp expression in those cells participating in pollen development is necessary to achieve fertile pollen. They also suggest that the male sterility phenotype is not the result of a general alteration of the plant metabolism in gapcp1gapcp2, but the consequence of the lack of expression of GAPCp in anther specific tissues.
Recent works using mutant lines in other plastidic glycolytic enzymes downstream of GAPCp, such as pyruvate kinase (Andre and Benning, 2007; Baud et al., 2007) or enolase (Prabhakar et al., 2009), have not reported the drastic phenotype observed with gapcp1gapcp2, especially the male sterility phenotype. We wondered if GAPCps could have additional nonglycolytic functions like those described for hexokinase (Rolland et al., 2006). Because of this, we transformed gapcp1gapcp2 with GAPCps inactivated in their catalytic domain. Cys-236 has been described as being essential for catalytic activity (Didierjean et al., 2003). This Cys residue has also been reported to be important in signaling mechanisms such as S-nitrosylation-dependent apoptotic cell death cascades (Hara et al., 2005). To ensure that only the glycolytic functions of GAPCp were altered, other important amino acids for GAPCp1 glycolytic activity were also substituted (H263A and R318E; Didierjean et al., 2003). None of these mutant constructs was able to complement the male sterile phenotype of gapcp1gapcp2, thus confirming that the glycolytic activity of GAPCp is essential for correct pollen development. Therefore, GAPCps do not seem to have other functions other than catalytic activity in plants, or at least they are not involved in the phenotypes studied in gapcp1gapcp2.
Role of GAPCp in Flower Metabolism and Pollen Development
The metabolite profile of gapcp1gapcp2 flower buds is clearly different from that of the wild type. These metabolite changes seem to be specific of GAPCp activity in flower organs since the gapcp1gapcp2-p35S::GAPCp lines, with a wild-type phenotype except for pollen viability, showed a similar metabolite profiling to gapcp1gapcp2, while that of the gapcp1gapcp2-pGAPCp1::GAPCp1geno line, with a fertile phenotype, was more similar to the wild type.
GAPCp disruption leads to alterations of pollen development that could be associated with changes in the metabolite profile of gapcp1gapcp2 anthers. Since metabolite analysis was performed in flower buds, it could be argued that the changes observed do not represent those that may be occurring in the anther. If the only difference between the sterile gapcp1gapcp2-p35S::GAPCp and the fertile gapcp1gapcp2-pGAPCp1::GAPCp1geno lines is the differential expression of GAPCp in anthers, it could then be assumed that the metabolite differences between these two lines are a good indication of the changes occurring in this organ.
The main function of GAPCps in roots is to provide precursors for Ser biosynthesis (Muñoz-Bertomeu et al., 2009). Although Ser deficiency in some of the cellular types of anthers cannot be ruled out, the high levels of Ser in flowers of gapcp1gapcp2, as well as the noncomplementation of the male sterility by Ser supplementation, make this possibility unlikely. Thus, GAPCp main function in anthers seems to be different from that found in roots.
Carbohydrates not only play a critical role in anther and pollen development as an energy source, but are also necessary for pollen wall biosynthesis (Clement and Audran, 1995; Goetz et al., 2001; Woo et al., 2008). Starch and Suc levels increased in gapcp1gapcp2 compared to controls. The impaired carbohydrate homeostasis in gapcp1gapcp2 could affect pollen development. In this way, accumulation of starch in the pollen grain may well lead to reduced male fertility in tomato (Solanum lycopersicum; Nashilevitz et al., 2009). It has also been shown that the timing of callose wall formation and degradation is crucial for the normal pollen development (Izhar and Frankel, 1971; Warmke and Overman, 1972; Toller et al., 2008). The metabolism of callose could be affected in the gapcp1gapcp2 tapetum as a consequence of the general alteration of the carbohydrate metabolism. In this case, the effect of GAPCp would be indirect.
The trehalose content in gapcp1gapcp2 flowers increased by almost 5-fold compared to the control. The levels of this metabolite remained high in the sterile lines (gapcp1gapcp2, gapcp1gapcp2-p35S::GAPCp1c, and gapcp1gapcp2-p35S::GAPCp2c), but drastically dropped to approach wild-type levels in the fertile complemented line (gapcp1gapcp2-pGAPCp1::GAPCp1geno) where the starch and Suc levels were also similar to the wild type. The trehalose pathway is a central metabolic regulator of Suc and starch metabolism in plants (Wingler et al., 2000; Schluepmann et al., 2003; Paul, 2008; Paul et al., 2008; Schluepmann and Paul, 2009). Wingler et al. (2000) showed a strong activation of ADP-Glc pyrophosphorylase and accumulation of starch in response to trehalose feeding. Accordingly, the activities of enzymes involved in the synthesis of Suc and starch, such as ADP-Glc pyrophosphorylase, increased in gapcp1gapcp2 (Muñoz-Bertomeu et al., 2009). Thus, the high levels of trehalose in gapcp1gapcp2 could be the direct consequence of GAPCp inactivity and might well lead to the observed changes in the main pools of carbohydrates. On the other hand, consensus is emerging in that trehalose also plays a central role in coordinating metabolism with development. Numerous effects on plant development have been observed as result of alterations of the trehalose pathway, which range from embryo and leaf development to inflorescence architecture (Eastmond et al., 2002; Pellny et al., 2004; Satoh-Nagasawa et al., 2006; Chary et al., 2008). Although trehalose has never been directly related to gametophyte development, overexpression of trehalose synthesis genes in maize (Zea mays) produced plants with sterility and seed formation-related problems (Almeida et al., 2007). In spite of the low expression of 35S promoter in anther tissues of Arabidopsis, expression of the trehalose-6-P synthase with the cauliflower mosaic virus 35S promoter also produced mature plants with poor seed set (Schluepmann et al., 2003). Also, Arabidopsis plants treated with trehalose and Validamycin A, a strong inhibitor of the enzyme trehalase, accumulated high levels of trehalose and showed alterations in fruiting having much less siliques and no seeds (Muller et al., 2001). All these fertility problems could be attributed to male sterility, although in no instance was this possibility investigated by the authors.
The blockage in the plastidial glycolytic pathway in gapcp1gapcp2 could be short-circuited through the cytosolic pathway, where the highly active cytosolic GAPDHs could metabolize the triose-phosphates. The increased expression of the cytosolic GAPDH isoforms in gapcp1gapcp2 would support this idea. Then metabolites, such as pyruvate, 3-phosphoglycerate, or phosphoenolpyruvate, could reenter the plastidial pathway through highly selective transporters present in the inner plastid membrane (Weber et al., 2005). The question that arises in the light of our data is why a deficiency in a minor GAPDH catalyzing a reversible reaction in plastids is able to provoke such drastic developmental alterations. Our results demonstrate a link between carbohydrate metabolism and pollen development. They also emphasize the importance of the GAPCps as regulators of the plant primary metabolism and as key enzymes in the connections between metabolism and development. Further studies will be needed to elucidate the role of specific metabolites, such as trehalose, in the network connecting GAPCp with pollen development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds (ecotype Columbia-0) were supplied by the European Arabidopsis Stock Centre (Scholl et al., 2000) and by the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/index.html). gapcp1 and gapcp2 single and double T-DNA mutant isolation and characterization were described by Muñoz-Bertomeu et al. (2009). Unless stated otherwise, seeds were grown under greenhouse conditions in pots filled with a (1:1, v/v) mixture of vermiculite and fertilized peat (KEKILA 50/50; Kekkilä Iberia) and were irrigated with demineralized water as needed. After a 4-d treatment at 4°C, seeds were placed in the greenhouse at 24°C for a 16-h day and at 17°C for an 8-h night photoperiod and were supplemented with artificial light as needed. Because gapcp1gapcp2 are sterile, lines were maintained in heterozygosis (gapcp1/GAPCp1 gapcp2/gapcp2 or gapcp1/gapcp1 GAPCp2/gapcp2). Homozygous gapcp1gapcp2 plants were identified in segregating populations by the sterile phenotype. For Ser supplementation experiments, gapcp1gapcp2 was grown in plates for 20 to 22 d and then transplanted to pots. Seeds were sterilized and sown on 0.8% agar plates containing one-fifth-strength Murashige and Skoog with Gamborg vitamins buffered with 0.9 g L−1 MES (adjusted to pH 5.7 with Tris). After a 4-d treatment at 4°C, seeds were placed in a growth chamber (SANYO; MLR-351H) at 22°C for a 16/8-h day/night photoperiod (100 μmol m−2 s−1). Homozygous gapcp1gapcp2 was identified in segregating populations by the short root phenotype after 8 to 10 d and was then subcultured on media containing 0.1 mm Ser for an additional 8 to 12 d. At day 20 to 22, plantlets were transplanted to pots as described above and were either irrigated or sprayed with 0.1 mm Ser.
Primers
All primers used in this work are listed in Supplemental Table S3.
Plasmid Constructs, Site-Directed Mutagenesis, and Plant Transformation
Standard methods were used to make the gene constructs (Sambrook and Russell, 2001). The cDNAs corresponding to genes GAPCp1 (At1g79530) and GAPCp2 (At1g16300) were placed under the control of the 35S promoter (p35S::GAPCp1c and p35S::GAPCp2c) by cloning them into a modified pGreen II plant transformation vector (Hellens et al., 1993) as described (Muñoz-Bertomeu et al., 2009). The construct p35S::GAPCp1c was used to exchange the 35S promoter by a 1.5-kb fragment corresponding to the native GAPCp1 promoter (−1,521 to +18 relative to the GAPCp1 translation start). The GAPCp1 promoter was PCR amplified from BACs T8K14 (supplied by Arabidopsis Biological Resource Center) using PromAt1g79530FHindIII and PromAt1g79530RXhoI primers, introducing HindIII and XhoI sites and cloned in p35S::GAPCp1c, giving pGAPCp1::GAPCp1c. For the genomic complementation, a 5.5-kb fragment including 1,520 nucleotides upstream of the ATG of GAPCp1 was obtained and cloned into a plant transformation plasmid derived from pFP101 named pGAPCp1::GAPCp1geno as described (Muñoz-Bertomeu et al., 2009). For gene promoter-reporter fusions, the promoter regions of gene GAPCp1 and GAPCp2 were fused to the GUS gene in pCAMBIA1303 as described (Muñoz-Bertomeu et al., 2009). Site-directed mutagenesis was performed with the Quikchange II XL site-directed mutagenesis kit (Stratagene) using the plasmid pGAPCp1::GAPCp1c as a template. The mutagenic primers used were the following: GAPCp1.C236G.F, GAPCp1.C236G.R, GAPCp1.H263A.F, GAPCp1.H263A.R, GAPCp1.K311A.F, GAPCp1.K311A.R, GAPCp1.R318E.F, and GAPCp1.R318E.R. All PCR-derived constructs were verified by DNA sequencing.
gapcp1gapcp2 was transformed with the different constructs using the floral dipping method (Clough and Bent, 1998) with Agrobacterium tumefaciens carrying pSOUP. Because gapcp1gapcp2 was unable to produce seeds, we transformed the progeny of heterozygous plants (gapcp1.1/gapcp1.1 GAPCp2/gapcp2.1 or GAPCp1/gapcp1.1 gapcp2.1/gapcp2.1) with the different constructs. Transformants were selected by antibiotic selection and gapcp1gapcp2 verified by PCR genotyping using gene-specific primers (LP1 and RP1 for GAPCp1 and LP3 and RP3 for GAPCp2) and left border primers of the T-DNA insertions (LB1_SAIL for gapcp1.1 and LBa1 for gapcp2.1). Additional primers At1g79530FBProm and At1g79530R3Ex were used to differentiate genomic transgene insertion from the endogenous At1g79530. Single insertion homozygous T3 lines were selected for characterization.
Pollen Germination and GUS Activity Assays
In vitro pollen germination assays were done using the optimized solid medium described by Boavida and McCormick (2007). Experiments were performed with pollen from nine freshly opened flowers from three independent plants. In vitro pollen germination was examined under an Olympus BH-2 microscope. Images were collected from at least two different microscopy fields (about 200 pollen grains were scored) for each replicate. A pollen grain was classified as germinated if the pollen tube length was longer than the pollen grain diameter.
For GUS activity assays, plant organs were incubated in GUS buffer (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 2 mm 5-bromo-4-chloro-3-indonyl-β-d-GlcUA [X-GlcA]; Duchefa) overnight at 37°C. The plant material was cleared either in 70% ethanol or Hoyer's solution before microscopy observation. At least six independent transgenic lines showed identical GUS-staining patterns and only differed in the expression level of GUS. Images were acquired with a Leica DM1000 microscope and a Leica DC350 digital camera.
Microscopy
Floral buds of different developmental stages were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in phosphate buffer, pH 7, for 4 h at 4°C. After several washes with water, ethanol dehydration series were performed followed by washes in xylene. Afterward, samples were embedded in plasticized paraffin for 2 h at 60°C and subsequently the formed blocks were poured. Sections (8 μm maximum) were cut using a microtone (Microm), deparaffinized in xylene, and rehydrated and stained with Weigert's hematoxiline. A Leica DMLS microscope with a Leica DC300 digital camera was used for cytological observations.
For electron microscopy pollen was mounted on standard stubs and coated with gold-palladium prior to observation on a field emission microscope HITACHI S-4100.
Q-RT-PCR
Total RNA was extracted from roots, leaves, shoots, and closed buds using the RNeasy plant mini kit (Qiagen). RNA was treated with RNase free DNase (Promega). Then, 0.5 μg RNA was reverse transcribed using polyT primers and the first-strand cDNA synthesis kit for RT-PCR (Roche) according to the manufacturer's instructions. Real-time quantitative PCR reaction was performed using a 5700 sequence detector system (Applied Biosystems) with the Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. Each reaction was performed in triplicate with 1 μL of the first-strand cDNA in a total volume of 25 μL. The specificity of the PCR amplification was confirmed with a heat dissociation curve (from 60°C to 95°C). Efficiency of the PCR reaction was calculated and different internal standards were selected (Czechowski et al., 2005) depending of the efficiency of the primers. Relative mRNA abundance ± sd was calculated using the comparative Ct method according to Pfaffl (2001).
Metabolite Determination
Closed flower buds (stages 5–10) from plants grown in greenhouse conditions were used to determine metabolite content. Starch and total soluble sugars were determined with the ENZYTEC starch kit (ATOM). The levels of other metabolites were determined in derivatized methanol extracts by gas chromatography-mass spectrometry using the protocol defined by Lisec et al. (2006). The absolute concentrations of metabolites were determined by comparison with calibration standard curve response ratios of various concentrations of standard solutions as detailed by Roessner-Tunali et al. (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g79530 (GAPCp1), At1g16300 (GAPCp2), At3g04120 (GAPC1), and At1g13440 (GAPC2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment of GAPCps with other members of the GAPDH family in Arabidopsis.
Supplemental Table S1. Enriched consensus sequences in the promoter regions of GAPCp1 and GAPCp2.
Supplemental Table S2. Metabolite levels in flower buds of wild-type, mutant, and transformed lines.
Supplemental Table S3. Primers used in this work.
Supplementary Material
Acknowledgments
We thank the Servei Central de Suport a la Investigació Experimental at the Universitat de València for technical assistance. We also thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, Sabine Frietzs for helping in the design of pollen germination assays, and Prof. José Pertusa for valuable help with the microscopy.
References
- Almeida AM, Villalobos E, Araújo SS, Cardoso LA, Santos DM, Santos MA, Fevereiro PS, Torné JM. (2007) Electroporation of maize embryogenic calli with the trehalose-6-phosphate synthase gene from Arabidopsis thaliana. Acta Physiol Plant 29: 273–281 [DOI] [PubMed] [Google Scholar]
- Andre C, Benning C. (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol 145: 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Jin Y, Jeong JC, Lee HJ, Moon H, Lee J, Shin D, Kang CH, Kim DH, Nam J, et al. (2008) Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 69: 333–338 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C. (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S. (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Clement C, Audran JC. (1995) Anther wall layers control pollen sugar nutrition in Lilium. Protoplasma 187: 172–181 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, et al. (2007) GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129: 983–997 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. (2008) Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierjean C, Corbier C, Fatih M, Favier F, Boschi-Muller S, Branlant G, Aubry A. (2003) Crystal structure of two ternary complexes of phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus with NAD and D-glyceraldehyde 3-phosphate. J Biol Chem 278: 12968–12976 [DOI] [PubMed] [Google Scholar]
- Dong XY, Hong ZL, Sivaramakrishnan M, Mahfouz M, Verma DPS. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJH, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JDG, Smeekens SC, Graham IA. (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarc'h A, Kahmann U, Chriqui D, Roitsch T. (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA 98: 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674 [DOI] [PubMed] [Google Scholar]
- Hara MR, Snyder SH. (2006) Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol 26: 527–538 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Ellis TH, Lee D, Turner L. (1993) Repeated sequences as genetic markers in pooled tissue samples. Plant Mol Biol 22: 153–157 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R. (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Shinozaki K. (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol 43: 1285–1292 [DOI] [PubMed] [Google Scholar]
- Izhar S, Frankel R. (1971) Mechanism of male sterility in Petunia - relationship between ph, callase activity in anthers, and breakdown of microsporogenesis. Theor Appl Genet 41: 104–108 [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. (2005) Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30: 142–150 [DOI] [PubMed] [Google Scholar]
- Lee MN, Ha SH, Kim J, Koh A, Lee CS, Kim JH, Jeon H, Kim DH, Suh PG, Ryu SH. (2009) Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol 29: 3991–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Min J, Kim YK, Cipriani PG, Kang M, Khersonsky SM, Walsh DP, Lee JY, Niessen S, Yates JR, Gunsalus K, et al. (2007) Forward chemical genetic approach identifies new role for GAPDH in insulin signaling. Nat Chem Biol 3: 55–59 [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50: 751–766 [DOI] [PubMed] [Google Scholar]
- Muller J, Aeschbacher RA, Wingler A, Boller T, Wiemken A. (2001) Trehalose and trehalase in Arabidopsis. Plant Physiol 125: 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernandez E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Aharoni A, Kossmann J, Wolf S, Levy AA. (2009) The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato. Plant J 57: 1–13 [DOI] [PubMed] [Google Scholar]
- Paul MJ. (2008) Trehalose 6-phosphate: a signal of sucrose status. Biochem J 412: e1–e2 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang YH. (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441 [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Pellny TK, Ghannoum O, Conroy JP, Schluepmann H, Smeekens S, Andralojc J, Krause KP, Goddijn O, Paul MJ. (2004) Genetic modification of photosynthesis with E. coli genes for trehalose synthesis. Plant Biotechnol J 2: 71–82 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC. (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Prabhakar V, Lottgert T, Gigolashvili T, Bell K, Flugge UI, Hausler RE. (2009) Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett 583: 983–991 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rubinelli P, Hu Y, Ma H. (1998) Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol Biol 37: 607–619 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Paul MJ. (2009) Trehalose metabolites in Arabidopsis: elusive, active and central. Somerville CR, Meyerowitz EM, , The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0122, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-Roeber B. (2007) Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol 175: 425–438 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. (2008) Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J 54: 911–923 [DOI] [PubMed] [Google Scholar]
- Warmke HE, Overman MA. (1972) Cytoplasmic male sterility in sorghum. 1. Callose behavior in fertile and sterile anthers. J Hered 63: 103–108 [Google Scholar]
- Weber AP, Schwacke R, Flugge UI. (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56: 133–164 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wingler A, Fritzius T, Wiemken A, Boller T, Aeschbacher RA. (2000) Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol 124: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo MO, Ham TH, Ji HS, Choi MS, Jiang WZ, Chu SH, Piao RH, Chin JH, Kim JA, Park BS, et al. (2008) Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). Plant J 54: 190–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Sundaresan V. (2000) Genetics of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol 3: 53–57 [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun YJ, Timofejeva L, Chen CB, Grossniklaus U, Ma H. (2006) Regulation of Arabidopsis tapetum development and function by dysfunctional tapetum1 (dyt1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. (2008) Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.