Abstract
Flavonoids synthesized by the phenylpropanoid pathway participate in myriad physiological and biochemical processes in plants. Due to the diversity of secondary transformations and the complexity of the regulation of branched pathways, single gene strategies have not been very successful in enhancing the accumulation of targeted molecules. We have expressed an Arabidopsis (Arabidopsis thaliana) transcription factor, AtMYB12, in tobacco (Nicotiana tabacum), which resulted in enhanced expression of genes involved in the phenylpropanoid pathway, leading to severalfold higher accumulation of flavonols. Global gene expression and limited metabolite profiling of leaves in the transgenic lines of tobacco revealed that AtMYB12 regulated a number of pathways, leading to flux availability for the phenylpropanoid pathway in general and flavonol biosynthesis in particular. The tobacco transgenic lines developed resistance against the insect pests Spodoptera litura and Helicoverpa armigera due to enhanced accumulation of rutin. Suppression of flavonol biosynthesis by artificial microRNA reversed insect resistance of the AtMYB12-expressing tobacco plants. Our study suggests that AtMYB12 can be strategically used for developing safer insect pest-resistant transgenic plants.
More than 7,000 diverse flavonoids have been identified in different plant species. These are synthesized by the phenylpropanoid pathway (Ververidis et al., 2007) and impart pigmentation to flowers (Tanaka et al., 2008), provide protection against damage by UV radiation (Izaguirre et al., 2007) and insects (Thoison et al., 2004; Diaz Napal et al., 2009), as well as act as signaling molecules (Kobayashi et al., 2004). About 20% of the carbon fixed in photosynthesis is directed into this pathway, which produces the majority of phenolic compounds found in nature. The biosynthesis of naringenin, the central molecule of the pathway, involves the enzymes Phe ammonia lyase (PAL), cinnamate 4-hydroxylase, 4-coumarate:CoA ligase, chalcone synthase, and chalcone isomerase. The intermediates or naringenin subsequently lead to the biosynthesis of lignins, flavones, flavonols, and anthocyanins. Structurally diverse compounds are formed due to the action of enzymes that bring about regiospecific condensation, cyclization, aromatization, hydroxylation, glycosylation, acylation, prenylation, sulfation, and methylation reactions.
Flavonols, among flavonoids, are also considered as health-protective components in functional foods and provide protection against cardiovascular diseases, certain types of cancers and degenerative disorders (Nijveldt et al., 2001; Korkina, 2007), platelet aggregation (Graf et al., 2005), and osteoporosis (Trivedi et al., 2009). Flavonols, especially rutin, have been shown to have insecticidal properties (Simmonds, 2003). It has also been reported that soybean (Glycine max) genotype PI 227687, which accumulates rutin, has been used widely in breeding programs as a source of insect resistance (Hoffmann-Campo et al., 2006). Biosynthesis and accumulation of these flavonoids are species specific and show tight spatial and temporal regulation. There has been growing interest to engineer plants for higher flavonol content by manipulating the expression of genes of the biosynthetic pathway. Manipulation of the expression of individual genes has provided limited success for the enhancement in flavonol biosynthesis and accumulation, due to the complex regulation of various branches of the phenylpropanoid pathway and poor substrate availability (Muir et al., 2001). The increase of flux into one branch of the pathway will normally decrease the flux into other branches, unless total flux into the pathway is enhanced.
Transcription factors that regulate the expression of genes involved in metabolic pathways have been discussed as promising tools for engineering the levels of metabolites (Broun, 2004). The transcription factors are often present as gene families and regulate target genes in tissue- and species-related patterns (Bovy et al., 2002). Specificity of the regulatory genes has not been studied in detail in most cases. Analysis of changes in transcriptomes and metabolomes should provide clues related to regulation by transcription factors in heterologous systems. AtMYB12 has been identified as a flavonol-specific transcription factor in Arabidopsis (Arabidopsis thaliana; Mehrtens et al., 2005; Stracke et al., 2007). It has been heterologously expressed in plants, like tobacco and tomato (Solanum lycopersicum), leading to high-level accumulation of polyphenolic compounds, especially flavonols (Luo et al., 2008). AtMYB12 expression and enhanced accumulation of flavonols in transgenic lines have not been studied for global changes in transcriptomes or metabolomes leading to increased flux availability to the flavonol-specific pathway. In this study, we report changes in genome-wide expression and the metabolite profile in AtMYB12-expressing transgenic tobacco lines and show that apart from regulating the phenylpropanoid pathway, this transcription factor modulates other metabolic pathways as well, which lead to increased flux availability to this pathway. Most importantly, the high-level accumulation of the flavanol rutin in AtMYB12-expressing tobacco lines provided resistance against Spodoptera litura and Helicoverpa armigera insects. This resistance was observed through an insect bioassay performed using plant extract or purified rutin from transgenic lines as well as in planta. When AtMYB12-expressing lines were suppressed for the expression of the tobacco flavonol synthase (NtFLS) gene using artificial microRNA (miRNA), the decreased accumulation of rutin reversed the insect resistance in transgenic lines.
RESULTS
Enhanced Flavonol Content in AtMYB12-Expressing Plants
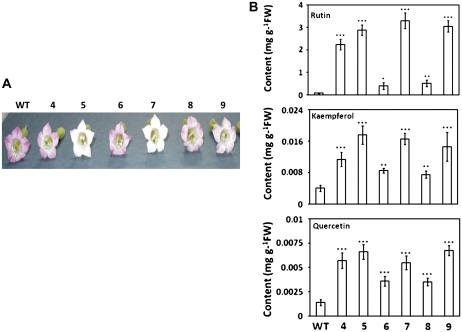
AtMYB12-expressing T0 tobacco transgenic lines displayed reduced floral pigmentation (Fig. 1A) due to reduction in anthocyanin content when compared with wild-type flowers (Supplemental Fig. S1A). The total flavonoid content of these lines was highly elevated in terms of quercetin equivalents (Supplemental Fig. S1B). The flower color phenotype segregated in the T1 generation. Based on kanamycin resistance, we selected homozygous transgenic lines for further analysis. In homozygous transgenic lines, rutin (up to 3.2 mg g−1 fresh weight) was the most dominant flavonol in nonhydrolyzed methanolic extract of transgenic lines (Fig. 1). This increase was more than 40-fold in comparison with the wild-type plants. Kaempferol and quercetin increased up to 3.2- and 5-fold, respectively (Fig. 2). In hydrolyzed methanolic extract of leaf samples, while rutin was undetectable, quercetin (up to 3.3 mg g−1 fresh weight) and kaempferol (up to 0.82 mg g−1 fresh weight) were major flavonols, and their levels were much higher than those of their nonhydrolyzed counterparts (Supplemental Fig. S2). This may result from the acid hydrolysis, resulting in the release of sugar moieties in quercetin and kaempferol backbones, leading to the conversion of rutin and kaempferol glycosides into quercetin and kaempferol, respectively.
Figure 1.
Change in flower color and comparative analysis of flavonols in transgenic tobacco lines expressing AtMYB12 under the control of the cauliflower mosaic virus 35S promoter. A, Flower-specific phenotypes of the wild type (WT) and six independent (T4–T9) transgenic lines. B, Contents of rutin, kaempferol, and quercetin in leaves of the wild type and different transgenic lines. Compounds were quantified by separating methanolic extracts from leaves of the wild type and transgenic lines using HPLC. The graph shows values ± sd of four leaves from each independent transgenic line. Asterisks indicate values that differ significantly from the wild type according to Student's paired t test: * P < 0.05, ** P < 0.01, *** P < 0.001. FW, Fresh weight. [See online article for color version of this figure.]
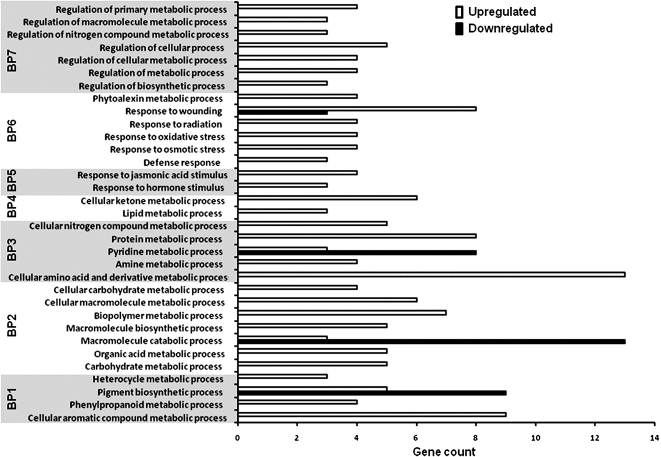
Figure 2.
Biological processes related to differentially regulated genes in AtMYB12 in transgenic tobacco lines. Arabidopsis homologs of the differentially expressed genes in AtMYB12-expressing tobacco lines were identified and grouped with respect to their predicted roles in different biological processes using WEGO (level 4). Different biological processes were further grouped under secondary plant product (BP1), carbohydrate metabolism (BP2), amino acid metabolism (BP3), lipid metabolism (BP4), hormone action (BP5), stress and defense responses (BP6), and regulation (BP7).
Transcriptome Analysis in AtMYB12-Expressing Transgenic Lines
AtMYB12 has been shown to be a flavonol-specific transcription factor targeting genes related to the phenylpropanoid pathway in Arabidopsis (Mehrtens et al., 2005) and heterologous system (Luo et al. 2008). Microarray analysis on RNA extracted from the leaves of wild-type and AtMYB12-expressing tobacco plants (line 7) identified a number of differentially expressed genes. A combined criterion of 2-fold or greater change and P < 0.05 in the t tests showed 205 and 257 probe sets as up- and down-regulated, respectively. These probes sets represented 57 and 92 up- and down-regulated unigenes, respectively (Supplemental Tables S1 and S2). Arabidopsis homologs of all the differentially expressed genes in AtMYB12-expressing tobacco lines were identified and grouped with respect to their predicted roles in different biological processes using WEGO (Fig. 2) as well as with respect to the interactive network leading to the modulation of different processes (Supplemental Fig. S4). Our analysis suggests that different biological processes related to secondary plant products, carbohydrate metabolism, amino acid metabolism, lipid metabolism, hormone action, stress and defense responses, and regulation of metabolic processes are differentially expressed in AtMYB12-expressing transgenic tobacco lines.
Expression of Genes Related to Phenylpropanid Pathway-Related Genes
Most of the structural genes of the phenylpropanoid pathway present on the arrays were up-regulated in the transgenic lines. The up-regulation of these genes as well as others involved in different pathways was further confirmed by reverse transcription (RT)-PCR (Supplemental Fig. S3). Although the expression of PAL, chalcone synthase, chalcone isomerase, flavonone-3-hydroxylase, and FLS increased in the leaves and flowers, expression of two dihydroflavonol 4-reductase (DFR) genes, DFR1 and DFR2, is flower specific and not regulated by AtMYB12 in transgenic lines. The DFR and PAL expression has been shown not to be regulated by AtMYB12 in Arabidopsis (Stracke et al., 2007). However, in AtMYB12-expressing tobacco, the genes originally identified as targets of AtMYB12 (Mehrtens et al., 2005; Stracke et al., 2007) in Arabidopsis as well as PAL were up-regulated. While the expression of putative ferulate-5-hydroxylase and cinnamoyl-CoA reductase involved in lignin biosynthesis was up-regulated, expression of two methyltransferases, caffeoyl-CoA 3-O-methyltransferase and catechol O-methyltransferase, was down-regulated. An EST encoding for a putative Rha biosynthetic gene (Arabidopsis Rha synthase [AtRHM1], a homolog), responsible for the formation of UDP-Rha residues from Glc, was up-regulated in the transgenic lines. A similar RHM gene coexpresses with flavonol biosynthetic genes in Arabidopsis, suggesting its involvement in rhamnosylation of flavonols (Yonekura-Sakakibara et al., 2008). ESTs showing homology with Petunia hybrida anthocyanin 3-O-glucoside:rhamnosyltransferase and cinnamate-4-hydroxylase were also up-regulated in AtMYB12-expressing tobacco lines.
Expression of Genes Related to Amino Acid Metabolism
Apart from the phenylpropanoid pathway, expression of genes involved in other pathways was also significantly modulated in the transgenic lines. Two genes, encoding 3-dehydroquinate dehydratase/shikimate dehydrogenase and 5-enolpyruvylshikimate-3-phosphate synthase, were up-regulated in AtMYB12 transgenic lines. These two enzymes are involved in the shikimic acid pathway responsible for the production of aromatic amino acids, the carbon flux for the phenylpropanoid pathway. Asn biosynthesis appears to be highly influenced by AtMYB12, as Asn synthase showed up-regulation while putative asparaginase, involved in the catabolism of Asn, was down-regulated in the transgenic lines. Expression of Gln synthetase, Asp aminotransferase, Glu synthase, and Arg decarboxylase, having roles in amino acid biosynthesis and related biosynthetic processes, were also up-regulated in the transgenic lines.
Expression of Genes Related to Carbohydrate and Lipid Metabolism
Enhanced expression of a putative transaldolase and enoyl-CoA hydratase/isomerase in tobacco transgenic lines indicated that AtMYB12 expression affects the pentose phosphate pathway and β-oxidation of fatty acids also. In addition, genes like glyceraldehyde-3-phosphate dehydrogenase, pyruvate decarboxylase, and Glc-6-P/phosphate transporter were differentially regulated.
Expression of Genes Related to the Auxin Response
Expression of certain ESTs homologous to auxin-repressed genes, such as Drm3-like protein and putative auxin-repressed protein, was up-regulated in the transgenic lines, while that of the auxin-induced genes, such as parA (Takahashi and Nagata, 1992; Takahashi et al., 1995), was down-regulated. In addition, an auxin efflux carrier was down-regulated. Taken together, the microarray data suggest that an auxin-depleted state may prevail in tobacco leaves expressing AtMYB12 transcription factor.
Expression of Genes Related to Stress and Defense Responses
Interestingly, a set of ESTs up-regulated in AtMYB12-expressing tobacco lines are related to genes involved in biotic and abiotic stresses. These stress-responsive genes include β-1,3-glucanases (Calo et al., 2006), osmotin (Castillo Ruiz et al., 2005), harpin-induced1 (Takahashi et al., 2004), snakin (Berrocal-Lobo et al., 2002), and thionins (Pelegrini and Franco, 2005).
Modulation of Metabolite Profile by AtMYB12
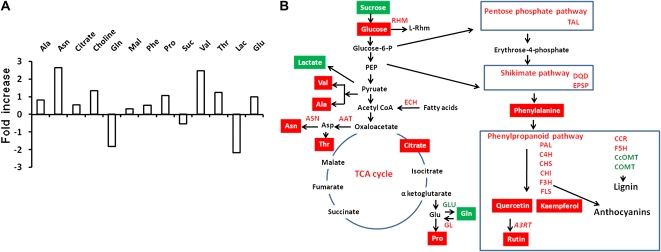
The microarray analyses of transgenic lines suggested that AtMYB12 expression in tobacco affects genes belonging to diverse aspects of plant metabolism. Limited metabolite profiles of leaves of transgenic plants also showed a marked shift (Fig. 3A; Table I). One-dimensional 1H NMR spectra of aqueous methanolic extracts of wild-type and AtMYB12-expressing transgenic plants were obtained using one-pulse sequence with suppression of water resonance by presaturation following a standard protocol (Sobolev et al., 2005). Our analyses showed changes in a number of metabolites in transgenic plants in comparison with the wild type (Fig. 3A). Enhanced accumulation of most of the amino acids, like Val, Ala, Phe, Thr, Pro, and Asn, as well as citrate, choline, and Glc was observed in AtMYB12-expressing transgenic tobacco lines. Out of these, Asn and Val accumulated more than 2.5-fold. Gln, Suc, and lactate accumulation in these lines decreased in comparison with the wild type. Increased accumulation of Phe suggests that AtMYB12 expression not only up-regulates the phenylpropanoid pathway but also leads to enhanced biosynthesis of aromatic amino acids to meet the increasing demand of flux in the pathway.
Figure 3.
Effects of AtMYB12 expression on different metabolites and metabolic sectors of tobacco leaves. A, Modulation of different metabolites as identified using a 400-MHz 1H NMR spectrum of aqueous methanolic extracts of leaves of wild-type and transgenic tobacco plants in D2O buffer. Data were recorded for at least three independent leaves (Table I) and plotted as fold change in each transgenic line with respect to the wild type. B, Schematic representation based on limited metabolite profiling and transcriptome analysis of the metabolic sectors altered in tobacco plants expressing AtMYB12. Red and green boxes show metabolites with increased and decreased contents, respectively, in AtMYB12-expressing lines in comparison with the wild type; enzymes encoded by different genes are shown in italics either in red or green representing up- or down-regulation in AtMYB12-expressing lines. Different metabolites are as follows: Lac, lactose; Mal, malate; PEP, phosphoenolpyruvate; Rhm, UDP-l-Rha. Enzymes encoded by differentially regulated genes are as follows: A3RT, anthocyanin 3-O-glucoside rhamnosyltransferase; AAT, Asp aminotransferase; ASN, Asn synthase; C4H, cinnamate 4-hydroxylase; CcOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CHI, chalcone isomerase; CHS, chalcone synthase; COMT, catechol O-methyltransferase; DQD, 3-dehydroquinate dehydratase/shikimate dehydrogenase; ECS, enoyl-CoA hydratase/isomerase; EPSP, 5-enoylpyruvylshikimate-3-phosphate synthase; F3H, flavonone-3-hydroxylase; F5H, ferulate-5-hydroxylase; FLS, flavonol synthase; GL, Glu synthase; GLU, Gln synthetase; PAL, Phe ammonia lyase; RHM, Rha synthase; TAL, transaldolase. [See online article for color version of this figure.]
Table I. Identified metabolites and their content as analyzed by 1H NMR spectrum.
1H chemical shift is reported with respect to TSP signal (δ = 0.00 ppm) in CD3OD and a binary mixture of CD3OD-D2O (8:2), respectively. Data were recorded for at least three independent leaves. sd values of metabolite contents for independent measurement are given in parentheses. ND, Not detected.
| Metabolites | 1H (ppm) and Multiplicity | Content |
||
| Control | Transgenic | P | ||
| mg g−1dry wt | ||||
| Ala | 1.48 (d) | 0.272 (0.002) | 0.479 (0.006) | 0.001 |
| Asn | 2.85 (dd), 2.93 (dd) | 0.199 (0.005) | 1.256 (0.006) | 0.001 |
| Citrate | 2.65 (dd), 2.80 (dd) | 6.715 (0.239) | 9.867 (0.719) | 0.002 |
| Choline | 3.19 (s) | 0.143 (0.003) | 0.368 (0.005) | 0.001 |
| Glc | 3.25 (dd), 4.64 (d), 5.22 (d) | 1.502 (0.160) | 2.971 (0.212) | 0.001 |
| Gln | 2.14 (m), 2.46 (m), 3.65 (t) | 1.328 (0.312) | 0.372 (0.003) | 0.006 |
| Lactate | 1.33 (d), 4.14 (q) | 0.196 (0.002) | 0.044 (0.003) | 0.001 |
| Malate | 2.39 (dd), 4.33 (dd) | 3.330 (0.269) | 4.023 (0.183) | 0.013 |
| Phe | 7.32 (m), 7.37 (m), 7.41 (m) | 0.731 (0.003) | 1.041 (0.051) | 0.001 |
| Pro | 2.07 (m), 2.35 (m), 4.14 (m) | 5.167 (0.283) | 10.874 (1.033) | 0.001 |
| Rutin | 3.80 (d), 6.20 (s), 6.87 (d) | ND | 2.832 | 0.001 |
| Suc | 5.42 (d) | 7.024 (0.164) | 4.897 (0.175) | 0.001 |
| Thr | 1.32 (d), 4.26 (m) | 0.689 (0.009) | 1.611 (0.085) | 0.001 |
| Val | 0.99 (d), 1.03 (d), 2.27 (m) | 0.162 (0.002) | 0.904 (0.013) | 0.001 |
On the basis of genome-wide expression analysis as well as metabolic profiling, metabolic sectors altered in tobacco plants expressing AtMYB12 have been generated (Fig. 3B). These pathways are interconnected to each other and help in increasing substrate flux for biosynthesis of flavonols.
Increased Rutin Accumulation Leads to Insect Resistance in Plants
Accumulation of certain secondary plant products (Kappers et al., 2005) including flavonoids (Thoison et al., 2004) has been reported to confer insect resistance in plants. Insect bioassay using S. litura on detached leaves of control and AtMYB12-expressing tobacco suggested that the transgenic lines were distinctly insect resistant. More than 70% mortality was noted. The surviving insects showed marked weight loss (Fig. 4A; Supplemental Table S3). Release of these insects on potted transgenic plants for 10 d showed 75% mortality with more than 90% weight loss of the surviving insects (Fig. 4B; Supplemental Table S4). To study whether enhanced content of the flavonols in the transgenic lines provided insect resistance, the ethanolic fraction of the control and transgenic lines was fed to the larvae of S. litura and H. armigera mixed with the synthetic diet. Fifty percent and 100% mortality of S. litura and H. armigera insects, respectively, was observed after feeding for 10 d. Significant weight loss (more than 95%) was observed for the surviving S. litura insects (Table II; Fig. 4C; Supplemental Tables S5–S8). Since the dominant flavonol in transgenic lines was rutin, we studied whether enhanced accumulation of rutin leads to insect mortality. Rutin was isolated from the transgenic lines and fed to the larvae of S. litura and H. armigera by mixing in synthetic diet (1 and 2 mg mL−1). At both concentrations, more than 65% mortality and severe weight loss of the surviving insects (Fig. 4, C and D; Supplemental Tables S5–S9) were noted, suggesting that high rutin in AtMYB12 transgenic lines leads to insect resistance.
Figure 4.
AtMYB12 expression leads to insect resistance in tobacco plants. A, Insect bioassay on detached leaves of wild-type (WT) and different AtMYB12-expressing transgenic tobacco plants. Two-day-old S. litura larvae were released on detached leaves of the wild type and selected transgenic lines (T4–T7), followed by measurements taken after 3 d. More than 70% mortality and severe weight loss of surviving insects were observed in different transgenic lines. B, Twenty-five neonate S. litura larvae were released on potted wild-type and AtMYB12-expressing transgenic plants (T5 and T7). Photographs of plants were taken 10 d after release of insects. C and D, Growth reduction of S. litura (C) and H. armigera (D) larvae 10 d after release on semisynthetic diet containing solvent (SL), extract from control plant (CE), extract from AtMYB12-expressing transgenic plant (TE), and rutin at 1 mg mL−1 (R1) and 2 mg mL−1 (R2) purified from the transgenic plants. Observations on mortality (percentage) and weight of surviving larvae were made on different days. In the case of H. armigera, the larvae were released individually to avoid cannibalism. For details, see Supplemental Tables S3 to S9. [See online article for color version of this figure.]
Table II. Percentage larval mortality and weight loss in control and AtMYB12-expressing tobacco lines.
NA, Not available.
| Extract/Compound | S. litura | H. armigera |
| % mortality (% weight loss) | ||
| Control extract (1 mg mL−1) | 0.0 (0.53) | 10.0 (62.39) |
| Transgenic extract (1 mg mL−1) | 50.00 (95.45) | 100 (NA) |
| Rutin (1 mg mL−1) | 46.66 (95.32) | 86.66 (98.08) |
| Rutin (2 mg mL−1) | 66.66 (97.35) | 100 (NA) |
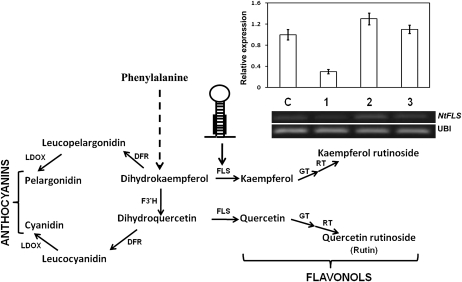
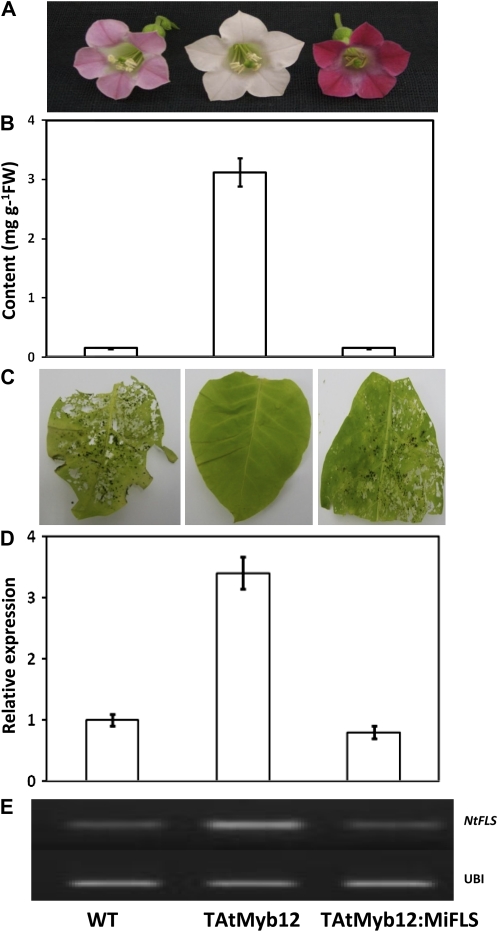
Our study suggests that AtMYB12 overexpression in tobacco modulates different metabolic sectors (Fig. 3B), leading to enhanced rutin content that gives insect resistance in tobacco. To further validate whether rutin was responsible in planta for insect resistance, its synthesis in a transgenic line (line 7) was blocked using artificial miRNA (Fig. 5). Artificial miRNA for tobacco NtFLS was prepared using the hairpin structure of Arabidopsis miRNA159a as a backbone (Niu et al. 2006). Three constructs, using different regions of NtFLS, were prepared and analyzed for efficacy using agroinfiltration in leaves of the transgenic lines (Fig. 5). The miRNA that showed maximum inhibition (amiFLS1) of NtFLS expression (as noted by RT-PCR) was utilized to raise stable transgenic lines in the AtMYB12 background. Transgenic plants harboring both AtMYB12 and amiFLS miRNA showed increased floral color in comparison with AtMYB12 transgenic lines as well as the wild type (Fig. 6A). Intense red floral color suggested diversion of the pathway to synthesize excess anthocyanins, following the inhibition of flavonols by the amiFLS miRNA. On the other hand, rutin content was drastically reduced in the double transgenic lines and was comparable to the wild type (Fig. 6B). Feeding by S. litura larvae on the double transgenic lines showed reversion of insect resistance (Fig. 6C), substantiating the involvement of flavonols, specifically rutin, in insect resistance.
Figure 5.
Inhibition of rutin biosynthesis in an AtMYB12-expressing transgenic line by amiFLS. Artificial miRNA was designed and cloned in plant expression vector under the control of the cauliflower mosaic virus 35S promoter. Part of the phenylpropanoid pathway synthesizing flavonols and anthocyanin as well as associated enzymes is represented. The hairpin structure represents amiFLS to inhibit NtFLS expression in AtMYB12-expressing tobacco lines. The constructs expressing three different amiFLS proteins under the control of the cauliflower mosaic virus 35S promoter were agroinfiltrated in tobacco leaves. Relative expression of NtFLS was studied through semiquantitative RT-PCR analysis to select the most potent construct inhibiting NtFLS expression. Expression of ubiquitin was used for normalization. Bars C, 1, 2, and 3 represent the control and three different amiFLS-expressing constructs. Construct 1 reduced expression of NtFLS approximately 3-fold and was further used for stable transformation. F3′H, Flavonoid 3′-hydroxylase; GT, glucosyltransferase; LDOX, leucoanthocyanidin dioxygenase; RT, rhamnosyltransferase.
Figure 6.
Effect of amiFLS in AtMYB12-expressing transgenic lines. A and B, Flower color phenotype (A) and decrease in rutin content by amiFLS (B) in AtMYB12-expressing transgenic lines. FW, Fresh weight. C, Insect bioassay in the wild type (WT), AtMYB12-expressing (TAtMyb12), and AtMYB12-expressing but NtFLS-suppressed (TAtMyb12:MiFLS) transgenic line 7. Two-day-old S. litura larvae were released on detached leaves of the plants followed by observations after 3 d. Decreased rutin content in AtMYB12-expressing transgenic lines through amiFLS shows reversion of insect resistance, suggesting the involvement of rutin in insect resistance. D and E, RT-PCR analysis and relative expression of NtFLS in the wild type, AtMYB12-expressing, and amiFLS-suppressed transgenic line 7. Expression of NtFLS decreased due to the expression of amiFLS in the AtMYB12-expressing transgenic lines. Expression of ubiquitin was used for normalization. [See online article for color version of this figure.]
DISCUSSION
Transcription factors of the MYB family regulate multiple steps in metabolic pathways and can have dramatic effects on the accumulation of flavonoids (Tohge et al., 2005; Butelli et al., 2008). Our study demonstrates that one of the flavonol-specific transcription factors, AtMYB12 (Stracke et al., 2007; Luo et al., 2008), not only up-regulates the expression of genes involved in the phenylpropanoid pathway but also leads to several other changes in the transcriptome, leading to additional carbon flux for enhanced accumulation of flavonols and several other metabolites (Fig. 3B). Interestingly, enhancement in the content of flavonols in tobacco is much higher in comparison with that reported in Arabidopsis overexpressing AtMYB12 (Mehrtens et al., 2005). The higher accumulation in tobacco may be due to additional targets of AtMYB12. For example, enhanced expression of PAL in tobacco can contribute to increased flux into the phenylpropanoid pathway. Enhanced expression of ESTs encoding RHM1 and rhamnosyltransferase in microarray analysis suggests their possible roles in Rha residue formation and rhamnosylation of flavonol aglycones. Our HPLC data also suggest that most flavonol accumulation in transgenic lines is in the rhamnosylated form, posing additional demand for UDP-Rha residues.
Limited metabolite profiling by NMR revealed reduced accumulation of Suc and enhanced content of amino acids. Similar correlation between Suc and amino acid content has been observed in other systems, suggesting that de novo biosynthesis of amino acids is regulated by Suc level (Roessner-Tunali et al., 2003; Broeckling et al., 2005; Sánchez-Sampedro et al., 2007). Higher accumulation of flavonols by AtMYB12 in our study appears to utilize additional carbon flux and energy, leading to a decrease in Suc content. Enhanced accumulation of citrate and malate could be due to increased respiration to meet extra demand for energy as well as carbon flux for increased de novo synthesis of the amino acids by enzymes involved in amino acid biosynthesis (Roessner-Tunali et al., 2003). Decreased Gln content in AtMYB12 transgenic lines may be due to down-regulation of the gene encoding a Gln synthetase. The decrease in Gln content may lead to the increased accumulation of Asn and Pro observed in our study.
The microarray and NMR analyses suggest that the shikimate pathway, responsible for the biosynthesis of aromatic amino acids, is up-regulated in the AtMYB12-expressing transgenic tobacco lines. Two genes of this pathway, 3-dehydroquinate dehydratase/shikimate dehydrogenase and 3-phosphoshikimate 1-carboxyvinyltransferase, were up-regulated (Fig. 3B). Up-regulation of a gene encoding a putative transaldolase, an enzyme involved in the pentose phosphate pathway, may also be involved in increasing flux to the shikimate pathway. Modulated expression of only a few enzymes of these pathways could be concluded due to limited transcriptome data available on the tobacco genome. The possibility of up-regulation of other genes involved in the shikimate pathway and/or Phe biosynthesis cannot be ruled out.
Up-regulation of putative enoyl-CoA hydratase/isomerase by AtMYB12 is indicative of the impact of the β-oxidation pathway, which might lead to increases in carbon flux in the form of acetyl-CoA. This can be correlated with the depletion of lactate due to reduced anaerobic utilization of pyruvate over efficient aerobic conversion into acetyl-CoA to enter into tricarboxylic acid cycle. In addition, down-regulation of a gene encoding a putative pyruvate decarboxylase also indicates efficient preferential aerobic breakdown of pyruvate. Altered expression of certain auxin-responsive genes in transgenic plants suggests lower auxin levels in the leaves, which might be due to the inhibition of auxin transport by flavonoids, particularly aglycone flavonols like quercetin and kaempferol (Peer and Murphy, 2007).
One of the significant observations of our transcriptome analysis is up-regulation of genes involved in biotic and abiotic stress response in plants. Genes up-regulated in transgenic lines may either be a direct target of AtMYB12 or regulated by the accumulation of flavonols, leading to a stress-related signaling cascade. Flavonoids have been known to act as signaling molecules in stresses and root nodule formation (Kobayashi et al., 2004). Components of the signal transduction machinery through flavonoids leading to resistance against stresses within plants are not yet known. Modulated expression of putative MYB and WRKY transcription factors as well as a calmodulin-like protein in AtMYB12-expressing transgenic tobacco suggests a possible involvement of such a signaling machinery. In addition, up-regulation of β-1,3-glucanase may be due to the auxin-depleted state of leaves, as this is known to be negatively regulated by auxin in tobacco (Mohnen et al., 1985). Another stress-responsive gene, Snakin2, an antimicrobial peptide, is up-regulated in AtMYB12-expressing lines. This gene is known to be positively regulated by pathogen attack, wounding, and abscisic acid (Berrocal-Lobo et al., 2002). Enhanced choline levels in transgenic plants are also suggestive of a stress response, as its increased accumulation has been observed after elicitor treatments (Sánchez-Sampedro et al., 2007). These observations suggest that AtMYB12 expression and subsequent accumulation of flavonols may lead to metabolic and transcriptional alterations that may mimic the stress response. This stress response may be generated by hormonal cross talk or metabolic alterations, resulting in activation of defense-related proteins/compounds.
A set of genes up-regulated in AtMYB12-overexpressing lines were related to plant stress and defense response. We analyzed transgenic lines for insect resistance against S. litura and H. armigera. The insect feeding experiment on detached leaves (Fig. 4A) as well as on plants (Fig. 4B) established that AtMYB12-overexpressing lines provided resistance against these insects. The larvae feeding studies on the chromatographic fraction of the leaf extract suggested that AtMYB12-expressing tobacco lines conferred insect resistance through increased accumulation of flavonols, especially rutin (Fig. 4, C and D). Inhibition of flavonol biosynthesis using artificial miRNA targeting NtFLS expression caused reversion of insect tolerance in AtMYB12-expressing tobacco to insect sensitivity and a concomitant decrease in rutin content. Rutin is an environmentally friendly insecticide. It interacts as a noncompetitive inhibitor with Arg kinase, a key enzyme in the cellular energy metabolism of insects (Wu et al., 2009), which may lead to the insect resistance in AtMYB12-expressing transgenic tobacco lines. Engineering crop plants to develop transgenic seeds with endogenous resistance to insect pests has been one of the major commercial successes of plant biotechnology. Genetically modified maize (Zea mays), potato (Solanum tuberosum), and cotton (Gossypium hirsutum) plants expressing genes encoding entomocidal δ-endotoxins from Bacillus thuringiensis have been cultivated globally. In spite of field success, uses related to the safety in deployment of bacterial genes in plants have been restricting the release of genetically modified crops in several countries, especially in the European Union. There is a need to develop new strategies for engineering insect resistance, preferably using genes of plant origin. Also, the pyramiding of independently acting genes can be a good guard against the development of resistance in insect pests. A few plant-based genes (Ferry et al., 2004) have been identified in recent years. We propose that AtMYB12 is a good candidate gene for use in developing insect-resistant crop plants.
MATERIALS AND METHODS
Plasmid Construction and Plant Transformation
The full-length open reading frame of the AtMYB12 cDNA from Arabidopsis (Arabidopsis thaliana) was isolated using oligonucleotide primers (AtMYB12F, 5′-ATAACCGCTCTAGAAAATGGGAAGAG-3′, and AtMYB12R, 5′-CGGATCAGAGCTCAATATCATCATGAC-3′) having recognition sites for XbaI and SacI restriction endonucleases. The PCR product was cloned in the binary vector pBI121 (Clontech) under the control of the cauliflower mosaic virus 35S promoter, replacing the GUS gene. Binary vectors harboring the desired construct were transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation. For tobacco (Nicotiana tabacum ‘Petit Havana’) transformation, a leaf disc cocultivation method (Horsch et al., 1985) was used for generating transgenic plants, which were selected on medium containing 100 mg L−1 kanamycin. Several transgenic tobacco lines constitutively expressing AtMYB12 were selected based on floral color phenotype and RT-PCR and grown in a glasshouse. Seeds were harvested, sterilized, and plated on solid half-strength MS medium (Murashige and Skoog, 1962) supplemented with kanamycin. Antibiotic-resistant plants were transferred in the glasshouse and grown until maturity. Wild-type and control plants were grown in pots at 26°C on a 16-h-day/8-h-night cycle.
Gene Expression Analysis
Total RNA was extracted from young leaves and flower petals of flowering tobacco plants using the Spectrum Plant Total RNA kit (Sigma-Aldrich), which was subsequently treated with RNase-free DNase (Fermentas Life Sciences). RNA was subjected to RT to generate first-strand cDNA using oligo(dT) primers (MBI Fermentas). RT-PCR analysis of a set of selected genes was carried out using 2× PCR Master mix (Fermentas Life Sciences). The lists of selected genes and oligonucleotide primers used in the study are provided as Supplemental Table S9. The primers for the tobacco ubiquitin gene were used as a loading control to ensure that equal amounts of cDNA were used in all the reactions. PCR was carried out using the following cycle conditions: an initial denaturation at 94°C for 2 min, 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final 5-min extension at 72°C.
Analysis of Anthocyanin and Flavonoid Content
For estimation of total anthocyanin, fresh tissue was ground into fine powder in liquid N2 and extracted with acidic (0.1% HCl, v/v) methanol for 18 h at room temperature. After extraction, samples were centrifuged for 20 min at 5,000g. Anthocyanins in the supernatants were determined spectrophotometrically through recording A530 and A657. Quantification of anthocyanin was carried out as follows: anthocyanin = (A530 − 0.25 × A657) M−1, where M is the weight of the plant material taken for the analysis. Total flavanoid content as quercetin equivalent was determined as described previously (Aslan et al., 2007). Total flavanoid content was expressed in milligrams of quercetin equivalents per gram of extract.
Individual flavanols in the plants were determined either as aglycones or as flavanol glycosides by preparing acid-hydrolyzed or nonhydrolyzed extracts, respectively. For preparation of acid-hydrolyzed extract, plant material was extracted with 80% ethanol overnight at room temperature with brief agitation. The filtrates were evaporated to 1.0 mL, and 3 volumes of HCl (1 m) was added followed by incubation at 94°C for 2 h to hydrolyze any conjugate forms of flavanoids. After hydrolyzation, samples were extracted with ethyl acetate, evaporated to dryness, and resuspended in 80% methanol. For nonhydrolyzed extracts, plant material was extracted in methanol:water (70:30) overnight at room temperature with brief agitation. Extracts were filtered through a 0.2-μm filter (Millipore) before separation and quantitation of flavonols using a liquid chromatograph (Waters) and a Merck Purospher star (250 × 4.6 mm, 5-μm pore size) C18 column with guard column of the same chemistry. Elution of flavonols was carried out at a flow rate of 0.8 mL min−1 with 0.5% phosphoric acid as solvent A and methanol as solvent B, using a gradient elution with 75% to 70% A (0–5 min), 70% to 50% A (5–10 min), 50% to 20% A (10–15 min), and 20% to 80% A (15–25 min). Flavonols were quantified by calculating the area of an individual peak and comparing this with a standard obtained from Sigma-Aldrich.
Transcriptome Analysis
The quality of total RNA isolated from leaves of wild-type and AtMYB12-expressing tobacco (line 7) was validated using a nanodrop spectrophotometer as well as a Bioanalyzer (Agilent 2100). Microarray-based gene expression profiling was performed using Agilent Platform with the Agilent tobacco whole genome microarray with 44,000 probe sets as per standard protocol. Normalization and statistical analysis were carried out using Agilent's GeneSpring GX version 10.0 software. To identify statistically significantly differentially expressed genes, a combined criterion of 2-fold or greater change and P < 0.05 in the t tests was adopted. To obtain annotations for the differentially regulated probe sets, target sequences from the sequence information file of the tobacco genome array were searched using BLASTx against the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Arabidopsis (The Arabidopsis Information Resource; www.arabidopsis.org) databases. Arabidopsis homologs of all the differentially expressed genes in AtMYB12-expressing tobacco lines were identified and grouped with respect to their predicted roles in cellular components, molecular functions, and biological processes using the Web Gene Ontology Annotation Plotting tool (WEGO; http://wego.genomics.org.cn) as well as with respect to an interactive network leading to modulation of different processes using Tools for Mining Pathway Collections and Ontologies, Pathway Studio (Ariadne Genomics).
Limited Metabolite Profiling by NMR
One-dimensional 1H NMR spectra of aqueous methanolic extracts of leaves of wild and transgenic tobacco plants were obtained using one-pulse sequence with suppression of water resonance by presaturation. In each experiment, the sample was dissolved in 500 μL of deuterated binary mixture of MeOD-D2O (8:2) and taken in a 5-mm NMR tube for analysis. A coaxial tube containing 30 μL of 0.375% trisilylpropionic acid (TSP) in deuterium oxide was inserted into the NMR tube before recording the NMR spectra. TSP served as a chemical shift reference as well as an internal standard for quantitative estimation. Typical parameters for both the extractions were as follows: spectral width, 6,000 Hz; time domain data points, 32,000; flip angle, 45°; relaxation delay, 5 s; spectrum size, 32,000 points; line broadening for exponential window function, 0.3 Hz. To confirm the assignments, two-dimensional correlation spectroscopy (COSY) and total COSY (TOCSY) were carried out using the Bruker's standard pulse program library. The spectral width of COSY was 6,000 Hz in both dimensions, with 512 t1 increments for each t1, and 16 transients using 2.5-s relaxation delay were added with 2,048 complex data points. The phase-sensitive data were obtained by the time-proportional phase incrementation method. The resulting data were zero filled up to 1,024 in the t1 dimension and were weighted with 90° squared sine window functions in both dimensions prior to double Fourier transformation. In the case of TOCSY, the spin-locking time was set to 60 ms, which included duration of 2.5 ms for the trim pulses. The assignments of metabolites of wild and transgenic tobacco leaves was carried out by the use of one-dimensional and two-dimensional COSY spectra. The assignments were further reinstated based on the existing literature values obtained in lettuce (Lactuca sativa) leaves by Sobolev et al. (2005). Differential accumulation of metabolites was analyzed and plotted as log of fold change.
Insect Toxicity Bioassay
The genetically homozygous larvae of Spodoptera litura and Helicoverpa armigera were maintained in the laboratory on semisynthetic diet at 26°C ± 2°C and 70% relative humidity on a 14-h-day/10-h-night cycle. In the case of detached leaf insect bioassays, leaves from the fourth to sixth nodes from the base of 2-month-old wild-type and transgenic tobacco were washed thoroughly in distilled water, blotted dry and placed in a plastic beaker, and fed to first instar larvae of S. litura for 3 d. For on-plant bioassay, 25 neonate larvae were released on 2-month-old plants and observation was made after 10 d. For experiments with leaf extracts and purified rutin, stock solutions of different extracts or rutin were mixed separately with semisynthetic diet before solidifying. Control treatment consisted of ethanol mixed with diet. Medium was poured into the petri dishes (60 mm), which were kept open overnight to evaporate the solvent. One-day-old, 30 larvae of S. litura or H. armigera were fed to the preparation for 10 d. In the case of H. armigera, the larvae were released individually to avoid cannibalism. Observations for mortality (percentage) and surviving larvae weights were made on 3, 5, 7, and 10 d after start of the experiment. Each treatment was replicated three times.
Artificial Pre-amiRNA Construct and Analysis of Transgenic Lines
Tobacco FLS nucleotide sequence (gi:164454784) was downloaded from the National Center for Biotechnology Information for selection of putative artificial miRNA. Putative artificial miRNA for NtFLS was identified using online software (http://wmd.weigelworld.org/cgi-bin/mirnatools.pl). A total of 59 putative artificial miRNA sequences for NtFLS were obtained and analyzed for their specificity using http://bioinfo3.noble.org/psRNATarget and BLASTn. Three putative sequences were selected for consideration for amiRNA preparation based on their specificity to the target. A 545-bp fragment containing the entire sequence of the Arabidopsis miR159a was cloned by PCR amplification using primers miR159-F1 (5′-ATATCTCCTTCATAGCTCTAATG-3′) and miR159R1 (5′-AAATAACACGCTAAACATTGCTTCG-3′) and utilized for mutagenesis to yield amiRNA for NtFLS using oligonucleotide primers provided in Supplemental Table S9. Mutagenesis of pre-miR159a was performed by PCR. Each primary amiRNA construct, having 235 nucleotides consisting miRNA159a fold-back structure, was cloned at XbaI and SacI sites in pIG121 binary vector having hygromycin selection. The pre-amiFLS constructs were tested for their effectiveness through agroinfiltration in tobacco leaves (Voinnet et al., 2003). amiFLS1 inhibited expression of NtFLS and was used for transformation in AtMYB12-expressing tobacco transgenic plants. Transgenic plants were selected on the basis of resistance to hygromycin. Expression of NtFLS, rutin content, as well as insect toxicity in double transgenic plants expressing AtMYB12 and amiFLS1 were studied as per the procedures given above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_130314.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of AtMYB12 expression on flower anthocyanin and leaf flavonoid contents.
Supplemental Figure S2. Effects of AtMYB12 expression on leaf quercetin and kaempferol contents.
Supplemental Figure S3.Semiquantitative RT-PCR analysis of genes up-regulated in AtMYB12-expressing tobacco transgenic lines.
Supplemental Figure S4. Functional network predicted from the genes differentially regulated by AtMYB12 in transgenic tobacco lines.
Supplemental Table S1. List of up-regulated genes in AtMYB12-expressing tobacco lines.
Supplemental Table S2. List of down-regulated genes in AtMYB12-expressing tobacco lines.
Supplemental Table S3. Insect bioassay on detached leaves of wild-type and AtMYB12-expressing transgenic tobacco plants.
Supplemental Table S4. Insect bioassay on wild-type and AtMYB12-expressing transgenic tobacco lines.
Supplemental Table S5. Larval growth reduction caused by feeding S. litura on diet supplemented with AtMYB12-expressing tobacco leaf extract.
Supplemental Table S6. Larval mortality caused by feeding S. litura on diet supplemented with AtMYB12-expressing tobacco leaf extract.
Supplemental Table S7. Larval growth reduction caused by feeding H. armigera on diet supplemented with AtMYB12-expressing tobacco leaf extract.
Supplemental Table S8. Larval mortality caused by feeding H. armigera on diet supplemented with AtMYB12-expressing tobacco leaf extract.
Supplemental Table S9. The primers used for semiquantitative analysis of selected genes by RT-PCR.
Supplemental Materials and Methods S1. Oligonucleotide primers used for mutagenizing the scaffold of miRNA159a for designing artificial miRNAs for NtFLS.
Supplementary Material
Acknowledgments
We thank Profs. Raja Ray and C.L. Khetrapal (Centre of Biomedical Magnetic Resonance, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, India) for providing the NMR facility to carry out our experiments.
References
- Aslan M, Deliorman Orhan D, Orhan N, Sezik E, Yesilada E. (2007) In vivo antidiabetic and antioxidant potential of Helichrysum plicatum ssp. plicatum capitulums in streptozotocin-induced-diabetic rats. J Ethnopharmacol 109: 54–59 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al. (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW. (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Broun P. (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 2: 202–209 [DOI] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Calo L, García I, Gotor C, Romero LC. (2006) Leaf hairs influence phytopathogenic fungus infection and confer an increased resistance when expressing a Trichoderma α-1,3-glucanase. J Exp Bot 57: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Castillo Ruiz RA, Herrera C, Ghislain M, Gebhardt C. (2005) Organization of phenylalanine ammonia lyase (PAL), acidic PR-5 and osmotin-like (OSM) defence-response gene families in the potato genome. Mol Genet Genomics 274: 168–179 [DOI] [PubMed] [Google Scholar]
- Diaz Napal GN, Carpinella MC, Palacios SM. (2009) Antifeedant activity of ethanolic extract from Flourensia oolepis and isolation of pinocembrin as its active principle compound. Bioresour Technol 100: 3669–3673 [DOI] [PubMed] [Google Scholar]
- Ferry N, Edwards MG, Gatehouse JA, Gatehouse AM. (2004) Plant-insect interactions: molecular approaches to insect resistance. Curr Opin Biotechnol 15: 155–161 [DOI] [PubMed] [Google Scholar]
- Graf BA, Milbury PE, Blumberg JB. (2005) Flavonols, flavones, flavanones, and human health: epidemiological evidence. J Med Food 8: 281–290 [DOI] [PubMed] [Google Scholar]
- Hoffmann-Campo CB, Neto JAR, de Oliveira MCN, Oliveira LJ. (2006) Detrimental effect of rutin on Anticarsia gemmatalis. Pesquisa Agropecu Bras 41: 1453–1459 [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eicholtz D, Rogers SG, Fraley RT. (1985) A simple method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Izaguirre MM, Mazza CA, Svatos A, Baldwin IT, Ballaré CL. (2007) Solar ultraviolet-B radiation and insect herbivory trigger partially overlapping phenolic responses in Nicotiana attenuata and Nicotiana longiflora. Ann Bot (Lond) 99: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappers IF, Aharoni A, van Herpen Teun WJM, Luckerhoff Ludo LP, Dicke M, Bouwmeester HJ. (2005) Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 309: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Naciri-Graven Y, Broughton WJ, Perret X. (2004) Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol Microbiol 51: 335–347 [DOI] [PubMed] [Google Scholar]
- Korkina LG. (2007) Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell Mol Biol 53: 15–25 [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C. (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenols. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D, Shinshi H, Felix G, Meins F. (1985) Hormonal regulation of beta1,3-glucanase messenger RNA levels in cultured tobacco tissues. EMBO J 4: 1631–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric De Vos CH, van Tunen AJ, Verhoeyen ME. (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19: 470–474 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog FA. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nijveldt RJ, van Nood E, van Hoorn DE, Boelens PG, van Norren K, van Leeuwen PA. (2001) Flavonoids: a review of probable mechanisms of action and potential applications. Am J Clin Nutr 74: 418–425 [DOI] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Pelegrini PB, Franco OL. (2005) Plant gamma-thionins: novel insights on the mechanism of action of a multi-functional class of defense proteins. Int J Biochem Cell Biol 37: 2239–2253 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Urbanczyk-Wochniak E, Czechowski T, Kolbe A, Willmitzer L, Fernie AR. (2003) De novo amino acid biosynthesis in potato tubers is regulated by sucrose levels. Plant Physiol 133: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Sampedro A, Kim HK, Choi YH, Verpoorte R, Corchete P. (2007) Metabolomic alterations in elicitor treated Silybum marianum suspension cultures monitored by nuclear magnetic resonance spectroscopy. J Biotechnol 130: 133–142 [DOI] [PubMed] [Google Scholar]
- Simmonds MS. (2003) Flavonoid-insect interactions: recent advances in our knowledge. Phytochemistry 64: 21–30 [DOI] [PubMed] [Google Scholar]
- Sobolev P, Brosio E, Gianferri R, Segre AL. (2005) Metabolic profile of lettuce leaves by high-field NMR spectra. Magn Reson Chem 43: 625–638 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Berberich T, Yamashita K, Uehara Y, Miyazaki A, Kusano T. (2004) Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Mol Biol 54: 613–622 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Hasezawa S, Kusaba M, Nagata T. (1995) Expression of the auxin-regulated parA gene in transgenic tobacco and nuclear localization of its gene products. Planta 196: 111–117 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Nagata T. (1992) parB: an auxin-regulated gene encoding glutathione S-transferase. Proc Natl Acad Sci USA 89: 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. (2008) A biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Thoison O, Sévenet T, Niemeyer HM, Russell GB. (2004) Insect antifeedant compounds from Nothofagus dombeyi and N. pumilio. Phytochemistry 65: 2173–2176 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Trivedi R, Kumar A, Gupta V, Kumar S, Nagar GK, Romero JR, Dwivedi AK, Chattopadhyay N. (2009) Effects of Egb 761 on bone mineral density, bone microstructure, and osteoblast function: possible roles of quercetin and kaempferol. Mol Cell Endocrinol 302: 86–91 [DOI] [PubMed] [Google Scholar]
- Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N. (2007) Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I. Chemical diversity, impacts on plant biology and human health. Biotechnol J 2: 1214–1234 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wu XQ, Zhu WJ, Lü JR, Xia Y, Yang JM, Zou F, Wang XY. (2009) The effect of rutin on arginine kinase: inhibition kinetics and thermodynamics merging with docking simulation. Int J Biol Macromol 44: 149–155 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.