Abstract
Carotenoid pigments in fruits are indicative of the ripening process and potential nutritional value. Papaya (Carica papaya) fruit flesh color is caused by the accumulation of lycopene or β-carotenoids in chromoplasts. It is a distinct feature affecting nutritional composition, fruit quality, shelf life, and consumer preference. To uncover the molecular basis of papaya flesh color, we took map-based cloning and candidate gene approaches using integrated genetic and physical maps. A DNA marker tightly linked to flesh color colocalized on a contig of the physical map with a cDNA probe of the tomato (Solanum lycopersicum) chromoplast-specific lycopene β-cyclase, CYC-b. Candidate gene sequences were obtained from amplified fragments and verified by sequencing two bacterial artificial chromosomes containing the two alleles. Sequence comparison revealed a 2-bp insertion in the coding region of the recessive red flesh allele resulting in a frame-shift mutation and a premature stop codon. A color complementation test in bacteria confirmed that the papaya CpCYC-b is the gene controlling fruit flesh color. Sequence analysis of wild and cultivated papaya accessions showed the presence of this frame-shift mutation in all red flesh accessions examined. Evaluation of DNA markers near CpCYC-b revealed a recombination hot spot, showing that CpCYC-b is located in a gene-rich region with a recombination rate at 3.7 kb per centimorgan, more than 100-fold higher than the genome average at 400 kb per centimorgan. Conserved microsynteny of the CpCYC-b region is indicated by colinearity of two to four genes between papaya, Arabidopsis (Arabidopsis thaliana), grape (Vitis vinifera), and tomato. Our results enhanced our understanding of papaya flesh color inheritance and generated new tools for papaya improvement.
Papaya (Carica papaya) is an important fruit crop grown throughout the tropics and subtropics for local market and temperate climate export markets. Consumer preferences for fruit traits such as size, shape, and flesh color vary widely by region and population demographics. In recent years, papaya fruit flesh color has received increased attention from breeders and consumers as an indicator of antioxidant activity and vitamin A nutrition. Papaya is one of the crops targeted for nutrient enrichment to be used in sustainable grass-roots programs to combat vitamin A deficiency in developing nations (Rodriguez-Amaya, 2003; World Health Organization, 2007). Papaya flesh color is the result of the accumulation of carotenoids in fruit cell chromoplasts, primarily lycopene in red flesh and β-carotenoids in yellow flesh, which provide antioxidant activity and vitamin A nutrition, respectively. Red flesh papaya softens faster and has a shorter shelf life, but some consumers prefer red flesh papaya, often called “strawberry papaya” in the market.
Papaya breeders recognized that fruit flesh color segregated in a simple Mendelian pattern, indicating a single major gene controlling this trait, with yellow flesh color dominant over red. While the basic pathway for carotenoid biosynthesis in plants was established in the 1960s, the genes encoding most of the enzymes in the pathway were not identified and characterized until the 1990s (Cunningham and Gantt, 1998; Hirschberg, 2001; DellaPenna and Pogson, 2006). The carotenoid composition profiles of red- and yellow-fleshed Hawaiian Solo papayas (Yamamoto, 1964) showed a strong accumulation of lycopene (approximately 63% of the total carotenoid content) in red-fleshed fruit, while none was detected in yellow-fleshed fruit. The profile of yellow-fleshed fruit showed mostly β-cryptoxanthin and β-carotene derivatives, up to 75% of the total carotenoid content, while red-fleshed fruit contained about half that amount. No α-carotenoids (e.g. lutein) were detected in either red- or yellow-fleshed fruit (Yamamoto, 1964).
Our group constructed a high-density genetic map of papaya using amplified fragment length polymorphism (AFLP) markers, and the flesh color locus was mapped near the end of linkage group 7 (LG7) and the two flanking markers were 3.4 and 3.7 centimorgan (cM), respectively (Ma et al., 2004). These closely linked AFLP markers were converted to sequence-characterized amplified region (SCAR) markers, but they were not polymorphic in our mapping populations (Z. Liu, P. Moore, Q. Yu, and R. Ming, unpublished data). We then cloned a papaya lycopene β-cyclase, CpLCY-b, the enzyme product of which mediates the conversion of lycopene (red) to β-carotene (yellow; Skelton et al., 2006). However, CpLCY-b was not differentially expressed between red- and yellow-fleshed papaya fruit and it showed 7-fold higher expression in leaves than in fruit, indicating its role as a chloroplast-specific lycopene β-cyclase. More recently, we constructed a high-density genetic map of papaya using simple sequence repeat markers, and the flesh color locus was mapped to the end of LG5 with the closest marker 13 cM away (Chen et al., 2007). In both papaya genetic maps, linkage group designations were assigned by genetic map length except for LG1, which contains the male-specific region of the Y chromosome.
It is presumed that red fruit flesh in papaya represents a disruption in the carotenoid biosynthesis pathway during fruit development and ripening; however, we do not know whether this disruption is due to a mutation or pretranscriptional or posttranscriptional regulatory mechanisms. Similar expression levels of the chloroplast-specific CpLCY-b in both red- and yellow-fleshed papaya fruit were detected, implying either posttranscriptional control or the presence of a second, chromoplast-specific lycopene β-cyclase (Skelton et al., 2006). Developmental regulation of carotenoid biosynthesis has been previously reported, but few of the precise regulatory factors have been fully described. The tomato (Solanum lycopersicum) chromoplast-specific lycopene β-cyclase, CYC-b, was identified by characterization of the Beta (B) mutation that accumulates high levels of β-carotene in its fruit and shows dramatically increased expression at the breaker stage that remained high during fruit ripening (Ronen et al., 2000; Bramley, 2002). The wild-type b allele did not show increased expression, and the authors suggest a pretranscriptional regulatory mechanism based on the sequence differences identified in the 2.5-kb region upstream of the coding region. In ripening pepper fruit, induction of gene expression for four carotenoid biosynthesis genes (phytoene synthase [Psy], phytoene desaturase [Pds], β-carotene hydroxylase [Chy-b], and capsanthin-capsorubin synthase [Ccs]) occurred at the onset of fruit ripening except in white pepper fruit, which lack accumulated carotenoids (Ha et al., 2007). Nonred pepper varieties were originally thought to lack Ccs, which mediates the cyclization of xanthophylls (yellow) to ketocarotenoids (red), as it was previously shown to be absent in two yellow pepper varieties (Bouvier et al., 1994; Hugueney et al., 1995). However, Ccs was identified in two different yellow pepper varieties, and expression of Ccs in these varieties is not induced during fruit ripening (Ha et al., 2007). A nonsense-mediated posttranscriptional gene silencing (i.e. nonsense-mediated mRNA decay) mechanism was suggested, since no cis-acting regulatory sequences or mutations were identified. The Orange (Or) mutation in cauliflower causes the accumulation of β-carotene in normally nonpigmented tissues by regulation of chromoplast formation, and up-regulation of the carotenoid biosynthesis genes is not required for carotenoid accumulation in Or mutants (Li et al., 2001; Lu et al., 2006). The functional role of the Or allele might be regulation of the cellular process that initiates proplastid differentiation into carotenoid sink structures (i.e. chromoplasts).
Papaya genomic resources have been developed in recent years, including the draft genome sequence and the integrated physical and genetic map (Ming et al., 2008; Yu et al., 2009). Papaya belongs to the order Brassicales and has no whole genome duplication (WGD) event since the paleohexaploidy event that is common among all Rosid lineages (Ming et al., 2008). Contrarily, the model plant species Arabidopsis (Arabidopsis thaliana) has experienced two WGD events since its divergence with papaya (Bowers et al., 2003). Although Arabidopsis is an excellent model species, papaya can be a useful outgroup for comparative genomics and a good model system for fruit trees, especially for agronomic traits such as fleshy fruit development and tree-like growth habit. The lack of recent WGD events makes papaya more amenable to studies of gene conservation and synteny (Freeling et al., 2008; Lyons et al., 2008). We report here the cloning and characterization of the papaya chromoplast-specific lycopene β-cyclase, CpCYCb, and genomic analysis of the surrounding region that includes a recombination hot spot in papaya.
RESULTS
Positional Cloning and Sequence Characterization of the Candidate Gene
After papaya fruit flesh color was mapped in our high-density AFLP map (Ma et al., 2004), we used map-based cloning and candidate gene approaches to clone the target gene. The SunUp hermaphrodite bacterial artificial chromosome (BAC) library was hybridized by a SCAR probe derived from an AFLP marker 3.4 cM away from papaya fruit flesh color and a tomato CYC-b cDNA probe. Five BACs were identified by the SCAR marker and two by the tomato CYC-b cDNA probe. These seven BACs were mapped to a single fingerprint contig FPC-1648 of the papaya physical map (Supplemental Fig. S1; Yu et al., 2009). Colocalization of the flesh color-linked SCAR marker and the CYC-b cDNA probe on a single contig indicated that the papaya CYC-b orthologous gene was a strong candidate for controlling fruit flesh color. The papaya draft genome sequence of SunUp female integrated with FPC-1648 was subjected to Genescan, which identified a gene with homology to citrus Ccs and tomato CYC-b. Lycopene (red) was the major carotenoid identified in red-fleshed papaya, but no ketocarotenoids (red), which are converted from violaxanthins (yellow) by the CCS enzyme, were identified in the red-fleshed papaya carotenoid profile. For that reason, we named our orthologous gene CpCYC-b based on its function in papaya fruit. Allelic sequences of the 1,485-bp coding region of CpCYCb were sequenced from genomic DNA of the dominant yellow-fleshed Kapoho and recessive red-fleshed SunUp cultivars by primer walking on amplified DNA fragments from both directions. Results were confirmed by sequencing of two BACs, SH18O09 and DM105M02, containing the two alleles (for details, see below). Three synonymous substitutions and one 2-bp insertion were identified within the SunUp CpCYC-b predicted coding region. The 5′ region of Kapoho and SunUp CpCYC-b showed 100% amino acid sequence identity up to the 2-bp insertion, which produces a frame-shift mutation resulting in a premature stop codon and a truncated coding region in SunUp CpCYC-b allele (Fig. 1).
Figure 1.
Sequence comparison of CpCYC-b in Kapoho and SunUp. A, DNA sequence comparison of CpCYC-b in Kapoho (yellow fleshed) and SunUp (red fleshed) revealed a 2-bp insertion in SunUp causing a frame-shift mutation. B, Amino acid sequence of the 2-bp indel region showing a truncated protein-coding region following the 2-bp insertion in SunUp.
A collection of 15 wild and cultivated papaya accessions was used to test for the candidate CpCYC-b gene. A pair of primers was designed to amplify and sequence a fragment containing the 2-bp insertion in the red flesh allele. Sequence analysis showed that the 2-bp insertion is present in all red-fleshed genotypes and absent from all yellow-fleshed genotypes tested (Supplemental Table S1).
Phylogenetic analysis of the CpCYC-b amino acid sequence with known and putative lycopene cyclase sequences obtained from GenBank grouped Kapoho CpCYC-b with other known chromoplast-specific lycopene cyclases and away from the chloroplast-specific lycopene β-cyclases (Supplemental Table S2; Supplemental Fig. S2). Amino acid sequences of CpCYC-b and CpLCY-b shared 60% identity over a 369-amino acid region, whereas CpCYC-b shared the highest homology (77% over 436 amino acids) with citrus Ccs. The initial Genescan of the CpCYC-b region identified by map-based cloning indicated a citrus Ccs gene homolog. Cluster analysis clearly separated CpCYC-b from CpLCY-b.
Differential Expression of Carotenoid Biosynthesis Genes during Fruit Development and Ripening
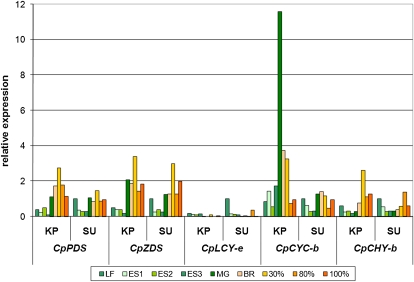
Five genes along the carotenoid biosynthesis pathway (Supplemental Fig. S3) were selected from papaya EST or genomic sequences to verify the role of CpCYC-b in the expression of fruit flesh color. These genes were examined for differential gene expression between yellow- and red-fleshed papaya during fruit development and ripening (Fig. 2; Supplemental Table S3). The CpPDS and CpZDS genes, whose enzyme products mediate the conversion of phytoene to lycopene, were similarly up-regulated over the expression levels in leaf tissue during ripening of both yellow- and red-fleshed papaya fruit. Expression of CpLCY-e was almost nondetectable in any stage of developing and ripening fruit. Expression of both CpCYC-b and CpCHY-b was up-regulated above baseline levels in leaf tissue in yellow-fleshed Kapoho papaya fruit, which showed 11.5-fold higher CpCYC-b expression in mature green fruit and 2.6-fold higher CpCHY-b expression in 30% ripe fruit (Fig. 3).
Figure 2.
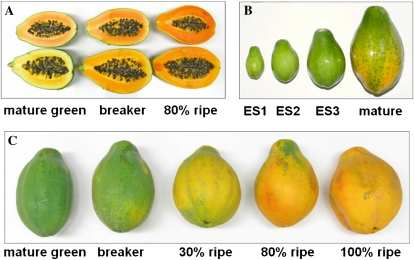
Papaya fruit expansion and ripening stages for candidate gene expression analysis. A, Color development in fruit flesh of SunUp (top row) and Kapoho (bottom row). B, Expansion size (ES) of developing papaya fruit compared with mature fruit. C, Fruit ripening stages based on change in fruit skin color.
Figure 3.
Differential expression of carotenoid biosynthesis genes in developing and ripening papaya fruit. Data are normalized to actin gene expression in SunUp leaf tissue. KP, Kapoho; SU, SunUp; LF, leaf; ES, expansion size; MG, mature green; BR, breaker. Fruit ripeness stages are listed as percentages (30%, 80%, and 100%).
Color Complementation
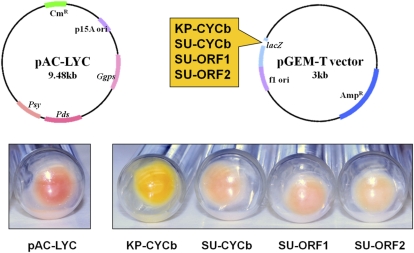
The full-length CpCYC-b coding regions from Kapoho and SunUp as well as two short open reading frames from SunUp were cloned for transformation into bacteria that lack lycopene β-cyclase activity (Supplemental Table S4). The gene product of the full-length CpCYC-b cloned from Kapoho was able to mediate the conversion of lycopene (red) to β-carotene (yellow). None of the SunUp CpCYC-b constructs exhibited lycopene β-cyclase activity, as indicated by the lack of color change (Fig. 4). Color complementation with the Kapoho CpCYC-b construct demonstrated functional activity of the gene product as a lycopene β-cyclase.
Figure 4.
Color complementation of a pAC-LYC bacterial line with four CpCYC-b constructs. The pAC-LYC plasmid (Cunningham and Gantt, 2007) enables lycopene production and accumulation in E. coli. pGEM-T expression vectors (Promega) carrying a CpCYC-b construct (Supplemental Table S3) from Kapoho (KP; yellow flesh) or SunUp (SU; red flesh) were transformed into pAC-LYC cells. Functional activity of the CpCYC-b construct is indicated by a color shift from red (lycopene) to yellow (β-carotene).
The carotenoids accumulated from each cell line were extracted with ethanol to confirm β-carotene production. The SunUp CpCYC-b absorption spectra corresponded to a lycopene standard, while the Kapoho CpCYC-b absorption spectra showed an admixture of lycopene and β-carotene peaks (Supplemental Fig. S4). Plasmid DNA from each transformed line was isolated to verify the presence of both plasmids by SacI digestion and PCR amplification of the CpCYC-b insert and Pds gene from the pAC-LYC plasmid (data not shown).
Sequence Analysis and Gene Content of Two BACs Containing the Two CpCYC-b Alleles
Two BAC clones, one from red flesh SunUp and the other from yellow flesh AU9, were completely sequenced using a shotgun method to reveal the genomic features of the CpCYC-b alleles and its neighboring regions. SH18O09 was identified from the SunUp hermaphrodite BAC library using a tomato CYC-b probe before the CpCYC-b had been cloned. DM105M02 was identified by Southern hybridization from the AU9 male BAC library using PCR-amplified fragments of CpCYC-b from Kapoho and SunUp genomic DNA as a probe. The shotgun sequencing reads were assembled into single contigs spanning 78,726 bp for DM105M02 and 65,684 bp for SH18O09 (GenBank accession nos. GQ478572 and GQ478573). The gapless comparison of these two BACs revealed 99.0% sequence identity over the 64,996-bp overlapping region, excluding two large insertions of 1,805 and 2,256 bp in nontransgenic AU9 or deletions in the transgenic SunUp (Supplemental Fig. S5). Analysis with the papaya repeat database revealed one Ty1/Copia-type retroelement and less than 2% low-complexity DNA (Nagarajan et al., 2008; Wang et al., 2008). The BAC sequences were searched against the papaya whole genome shotgun sequence and aligned to supercontig_195 of the C. papaya Core Annotation database. The draft genome covered 92.7% of the 65,684-bp SunUp BAC SH18O09.
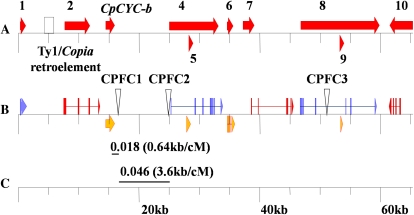
The aligned BAC sequence covered a 65.3-kb region and contained 10 predicted genes. Seven predicted genes are supported by papaya EST data, with a minimum alignment of 167 amino acids and an e-value less than 1e-50 (Fig. 5). Reverse transcription (RT)-PCR results confirmed the expression of the remaining three predicted genes 5, 6, and 9 (Supplemental Table S5; Supplemental Fig. S6). RT-PCR products were sequenced to determine intron positions of the predicted gene sequences. Papaya EST or cDNA sequences were searched against the Arabidopsis nonredundant protein database to identify orthologous genes and to examine gene order (Supplemental Table S6). Homologues to these 10 papaya genes were spread across all five Arabidopsis chromosomes and included one collinear block corresponding to a 12.7-kb region of Arabidopsis chromosome 5 containing AT5G22350 to AT5G22380. The grape (Vitis vinifera) and tomato genome databases were searched for orthologous genes (Supplemental Table S7), and colinear blocks were identified on chromosomes 8 and 11, respectively (Fig. 6). Gene order is conserved in all four genomes for genes 1 and 4, although the orientation of gene 1 is reversed in tomato. Papaya shows conserved gene order with tomato for genes 7 and 10, with grape for genes 6 and 8, and with Arabidopsis for genes 1, 2, and 4.
Figure 5.
Consensus map of DM105M02 and SH18O09 BAC sequences. A, Positions and orientations of 10 predicted papaya genes. B, Alignment of EST (top) or gene expression data (bottom) for nine predicted papaya genes and CpCYC-b. Inverted triangles indicate the positions of CPFC1, CPFC2, and CPFC3 indel polymorphisms. C, Recombination frequency detected in the KD × 2H94 F2 population between CpCYC-b, CPFC1, and CPFC2. The local recombination rate is indicated in parentheses by the physical-to-genetic distance ratio.
Figure 6.
Colinear genomic regions in papaya, tomato, Arabidopsis, and grape. Gray boxes indicate orthologous gene positions and orientations, and shaded areas connect conserved genes. Gene numbers are assigned based on start position on papaya BAC clone SH18O09; gene 3 is CpCYC-b. Orthologous gene information is described in Supplemental Table S5.
Recombination Frequency in the CpCYC-b Region
Multiple attempts to utilize the 2-bp insertion/deletion polymorphism (indel) for differentiating between the yellow- and red-fleshed papaya fruit using agarose gels or by designed primers at the insertion site were unsuccessful. A codominant marker, CPFC1 (for C. papaya flesh color 1), was developed based on a 36-bp indel detected between Kapoho and SunUp approximately 580 bp downstream of the CpCYC-b coding region (Supplemental Table S8). CPFC1 genotypes of 15 different papaya accessions were determined using standard PCR and gel electrophoresis equipment, and three recombinants for the CPFC1 and flesh color loci were identified (Supplemental Fig. S7). Subsequent screening of the 219 hermaphrodite F2 progeny of the Khaek Dum (KD) × 2H94 mapping population identified four recombinants, indicating a genetic distance of 0.9 cM between CPFC1 and CpCYC-b.
In addition to the CPFC1 marker, two SCAR markers, CPFC2 and CPFC3, based on indels between DM105M02 and SH18O09 were screened in two F2 mapping populations segregating for papaya fruit flesh color. CPFC2 is a dominant PCR marker based on a 1,805-bp indel located 9.0 kb downstream of the CpCYC-b coding region. CPFC3 is a codominant PCR marker based on a 54-bp indel located 36.9 kb downstream of CpCYC-b. Ten recombinants between CPFC1 and CPFC2 were identified in the KD × 2H94 mapping population, giving an estimated genetic distance of 2.3 cM between the two markers. The KD and 2H94 parents are not polymorphic for the CPFC3 marker, and the AU9 and SunUp parents are not polymorphic for the CPFC1 marker. Two recombinants between CpCYC-b and CPFC2 were identified in the AU9 × SunUp mapping population of 54 F2 individuals, giving an estimated genetic distance of 1.9 cM. No F2 individuals carrying the AU9 allele for CPFC3 were detected in either dominant or codominant fashion in the AU9 × SunUp mapping population. The average recombination rate in this region was 3.7 kb cM−1, more than 100-fold higher than the genome average of 400 kb cM−1.
DISCUSSION
Papaya fruit flesh color is indicative of the nutritional benefit provided by the carotenoids responsible for flesh color and is a phenotypic trait important to consumer preference. Cloning the gene controlling fruit flesh color has been a long pursuit by our research group. Previous attempts via map-based cloning were hindered by the lack of polymorphic markers in this region, and a candidate gene approach did not yield positive clones from our papaya cDNA libraries. It turned out that our EST collection of 16,362 unigenes contains no CpCYC-b or CpCHY-b2 (Ming et al., 2008). Our failure to capture these two genes is likely because we constructed our papaya cDNA libraries of developing fruit from the red fruit flesh variety SunUp (Paull et al., 2008; Q. Yu, P.H. Moore, and R. Ming, unpublished data). Colocalization on the physical map of the flesh color-linked SCAR marker and the tomato CYC-b cDNA probe is the result of a 100-fold increase of recombination rate in the CpCYC-b region. Although the genetic distances are inflated in our AFLP genetic map due to the use of dominant markers, we did not expect the SCAR marker, which is located 3.4 cM away from the flesh color locus, to hybridize to the neighboring region of the target gene (Ma et al., 2004; Chen et al., 2007). Three of the five BACs hybridized by the SCAR marker overlapped with BACs hybridized by the tomato CYC-b probe. Tomato CYC-b and SunUp CpCYC-b share just 75% sequence identity over a 682-bp genomic sequence length; thus, differences in hybridization specificity or autoradiography signal strength for each probe may account for why the same BACs were not identified by both probes and why the other two SCAR marker BACs do not overlap with the tomato CYC-b BACs.
Quantitative RT-PCR analysis and subsequent functional analysis in bacteria confirmed the role of CpCYC-b in controlling fruit flesh color in papaya. The elevated expression of CpCYC-b and CpCHY-b in yellow-fleshed Kapoho versus red-fleshed SunUp validated that there is activation of the carotenoid biosynthesis pathway in yellow fruit papaya. The disruption of this pathway in red flesh varieties is caused by a frame-shift mutation induced by a 2-bp insertion. This insertion of two Ts after five Ts could be the result of replication slippage during meiosis (Fig. 1; Karthikeyan et al., 1999; Ball et al., 2005; Kuo et al., 2006). The mRNA transcripts for CpCYC-b and CpCHY-b are detectable in low levels from developing red-fleshed papaya (Fig. 4). This may be a result of nonsense-mediated mRNA decay, which detects premature stop codons (or premature termination codons) to target the transcripts for destruction (Byers, 2002; Ha et al., 2007). Between the SunUp and AU9 BAC sequences, we detected 100% sequence identity in the 3,659 bp upstream of the CpCYC-b coding region to the predicted TATA box. We would predict similar gene transcription levels based on sequence identity in the promoter region, and extreme differences in the detection level of mRNA transcripts by quantitative RT-PCR would indicate a posttranscriptional regulatory mechanism.
Confirmation of this frame-shift mutation in 15 diverse accessions from both wild and cultivated germplasm indicates the prevalence of this mutation, if not the only mutation, in red-fleshed papaya varieties. The center of origin for papaya is southern Mexico, Belize, and Costa Rica. The earliest recorded transport of papaya was in 1525, when seeds were taken to Panama and Dominican Republic (Morton, 1987), and the single major introduction made to Hawaii was in 1910 from Barbados (Storey, 1969). The recessive CpCYC-b allele of Sunset, the progenitor of SunUp, is from Hawaiian Solo var Line 9 (Hamilton et al., 1993). The Taiwan cv Wang Peng is developed from Sunrise, a sister line of Sunset, and the red flesh allele is also from Line 9. Two Thai cultivars, KD and Khag Nam, have dramatically different fruit morphology with elongated large fruit up to 10 pounds compared with the Hawaiian Solo varieties that bear 1-pound fruit (hence the name Solo, meaning that one fruit can be consumed by one person). The red flesh allele of the two Thai varieties is likely from a different source. The Indian cv Coimbatore has large, oval-shaped fruit, and its red flesh allele might also be from another source. The wild germplasm UH918 collected from Costa Rica has not been used in any breeding program (Kim et al., 2002), and its red flesh allele is not ancestral to the commercial cultivars. It is possible that the frame-shift mutation reported here occurred multiple times in the wild germplasm and in commercial cultivars during the domestication and breeding processes.
Differential expression analysis of five genes along the carotenoid biosynthesis pathway between yellow- and red-fleshed papaya fruit showed evidence of developmental regulation of carotenoid biosynthesis in fruit tissues. Kapoho and SunUp both showed a slight up-regulation of CpPDS and CpZDS, which convert phytoene (colorless) to lycopene (red), during fruit development. Kapoho also showed a dramatic increase in expression of CpCYC-b during fruit ripening, facilitating the production of β-carotene (yellow) and its derivatives. The gene product encoded by the SunUp CpCYC-b allele is nonfunctional, as shown by color complementation. The presence of β-carotenoids in ripe red-fleshed papaya varieties is likely due to the activity of the chloroplast-specific CpLCY-b, whose expression is detectable at low levels during flower and fruit development (Skelton et al., 2006).
With the exception of CpLCY-e, the other carotenoid biosynthesis genes examined in papaya, including the chloroplast-specific CpLCY-b, are expressed at detectable levels throughout fruit development. In this study, CpPDS, CpZDS, and CpCYC-b each showed an increase in expression beginning around the mature green stage of fruit development. Unlike tomato, in which developing fruit are green so that accumulated carotenoids can be masked by chlorophyll, developing papaya fruit have white fruit flesh, indicating a void of both chlorophyll and carotenoids. It is likely that activity of the carotenoid biosynthesis gene products does not begin until the later stages of fruit development. Direct examination of the accumulated carotenoids in developing papaya fruit tissues would reveal when the carotenoid biosynthesis enzymes are active and clarify whether the developmental regulation of carotenoid biosynthesis is based on gene expression or the availability of the precursor substrate.
In support of marker-assisted selection in papaya breeding programs, a simple PCR-based screening test using agarose gels has been developed based on the CPFC1 marker to identify individuals in a segregating population with the desired flesh color. For any breeding population where the parental genotypes show polymorphism for this marker, this simple PCR test will be able to identify fruit flesh color genes with approximately 98% certainty based on the recombination frequency observed in the KD × 2H94 mapping population. It should be noted that this tightly linked marker, only 580 bp away from the target gene, is still not 100% accurate due to the extremely high recombination rate. A more precise test could be done at a higher cost and lower throughput using acrylamide sequencing gels by targeting the two-nucleotide insertion of the red flesh CpCYC-b.
Recombination is responsible for the creation of novel allele combinations and also serves a role in chromosome segregation and in the repair of damaged DNA. Many studies have shown that recombination does not occur randomly in a genome, and indeed, genomic regions with extremely high or low rates of recombination (hot spots and cold spots, respectively) have been identified in yeast, mammal, and plant models (Hey, 2004; Mézard, 2006; Li et al., 2007). In plants, recombination hot spots have been defined as DNA regions of a few kilobases in length that exhibit a higher rate of recombination than the surrounding DNA or more specifically as regions where recombination junctions are more likely to form (Weil, 2002; Hey, 2004; Mézard, 2006). Our data here, based on the KD × 2H94 F2 mapping population (n = 219), indicate that the local recombination rate in the CpCYC-b region (3.7 kb cM−1) is more than 100-fold higher than the genome average (400 kb cM−1). The smaller AU9 × SunUp F2 mapping population (n = 54) indicates a recombination rate between CpCYC-b and CPFC2 (4.8 kb cM−1) that is 82-fold higher than the genome average. While these data give only an estimation of local recombination rates, the “true” population recombination rate must also account for the molecular recombination process and the effective population size (De Iorio et al. 2005), they clearly indicate a recombination hot spot in the nongenic 9.0-kb region located between CpCYC-b and CPFC2.
Two chromosomal features commonly associated with plant recombination hot spots are regions of high gene density and telomeres (Mézard, 2006). The region encompassing CpCYC-b shows a gene density (6.5 kb gene−1) 2.3-fold higher than the genome average (15.0 kb gene−1), and high-density genetic mapping has placed the flesh color locus at or near the end of its linkage group, indicating its position near the telomere (Ma et al., 2004; Chen et al., 2007; Blas et al., 2009). Using the same AU9 × SunUp F2 mapping population analyzed here, Chen et al. (2007) mapped the flesh color locus to the end of LG5, with the nearest marker 13 cM away. Further enrichment of the AU9 × SunUp genetic map with AFLP markers (Blas et al., 2009) still placed the flesh color locus at the end of LG5, with the nearest marker 12 cM away, due to the 100-fold increase in recombination rate in this region.
Papaya is known to have limited genetic diversity among papaya germplasm based on DNA marker data (Kim et al., 2002), but it has not been quantified at the molecular level. Sequencing two homologous BACs from the uncultivated AU9 and commercial cv SunUp containing CpCYC-b alleles provided an opportunity to assess their DNA sequence variation. Genomic analysis revealed 99.0% gapless sequence identity, confirming the limited diversity due to the nature of self-pollination in hermaphrodite papaya and its coexistence and cross-breeding with dioecious varieties. Two large indels of 1,805 and 2,256 bp were found between these two BACs, and interestingly, these two large fragments are present in nontransgenic AU9 and absent in transgenic SunUp. The functional papaya ringspot virus coat protein gene was mapped to the same LG7 with the flesh color locus in the high-density AFLP map (Ma et al., 2004), and at least one of the three detected transgenic insertions in SunUp had shown rearrangements of DNA fragments caused by the particle bombardment (Suzuki et al., 2008). These two deletions in SunUp could have resulted from the double strand breakage-and-repair process that integrated the papaya ringspot virus coat protein gene.
MATERIALS AND METHODS
Plant Materials
Fruit and young leaf tissue of papaya (Carica papaya ‘SunUp’ and ‘Kapoho’) were collected from the Kunia Substation of the Hawaii Agriculture Research Center on Oahu. SunUp is a transgenic cultivar bearing red-fleshed fruit and containing the papaya ringspot virus coat protein gene. Kapoho is a nontransgenic cultivar with yellow-fleshed fruit. Fruits were collected at three expansion sizes and five ripening stages. Fruit flesh was cut away from the outer skin after removing seeds from the inner fruit cavity. Young leaves were collected before full leaf expansion; young seedlings were separated into root and shoot portions. All plant tissues for RNA isolation were flash frozen in liquid nitrogen immediately following collection.
Identification of the Candidate Gene Controlling Papaya Fruit Flesh Color
Papaya BAC libraries were screened with two probes, an AFLP-derived SCAR marker linked to papaya fruit flesh color and a tomato (Solanum lycopersicum) CYC-b probe provided by Dr. S. Tanksley (Cornell University). Positive BAC clones were localized on the papaya physical map, and the integrated draft genome sequence was screened with Genescan to identify candidate genes.
Phylogenetic Analysis
A multiple alignment of lycopene cyclase amino acid sequences obtained from GenBank was generated with ClustalW (http://asgpb.mhpcc.hawaii.edu/tools/) using a gap open penalty of 10 and gap extension penalty of 0.05. The alignments were analyzed with the Phylogeny Inference Package (PHYLIP) version 3.68 (Felsenstein, 2005). Distance measures of protein sequences were calculated using maximum likelihood estimates based on the Jones-Taylor-Thornton matrix model. An unrooted tree was generated using the neighbor-joining distance matrix method. Branch support was calculated by bootstrap resampling based on 1,000 replications.
Carotenoid Biosynthesis Gene Differential Expression
Five genes encoding enzymes of the carotenoid biosynthesis pathway were selected for characterization of their expression pattern during fruit development and ripening between yellow- and red-fleshed papayas: CpPDS, CpZDS, CpLCY-e, CpCYC-b, and CpCHY-b. Quantitative RT-PCR primers (Supplemental Table S2) were designed from conserved regions in the amino acid sequences of these enzymes obtained from GenBank. Briefly, amino acid sequences from several species were searched against the translated draft genome to identify papaya orthologs. A multiple alignment of the amino acid sequences was used to identify conserved regions that were targeted for primer design, and where possible, intron-spanning primer pairs were selected.
Color Complementation
The predicted open reading frames of the putative CpCYC-b were amplified from Kapoho and SunUp with primers incorporating the start and stop codons (Supplemental Table S3). Amplification reactions contained 1× Pfu PCR buffer, 0.16 mm deoxyribonucleotide triphosphates, 0.16 μm each primer, 1.25 units of PfuUltra DNA polymerase AD (Stratagene), and 12.5 ng of genomic DNA in a 50-μL total volume. PCR conditions were as follows: 95°C for 1 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s or 90 s; 72°C for 10 min; and hold at 4°C. Full-length CpCYC-b reactions received a 90-s extension time, while truncated CpCYC-b reactions received a 45-s extension time. For cloning purposes, 3′ A tails were added to the PCR products by incubation with 1 unit of GoTaq DNA polymerase (Promega) at 72°C for 10 min, then transferred to ice. A-tailed PCR products were ligated to pGEM-T vector (Promega) according to the manufacturer's recommendations. Five microliters of pGEM-T ligation mix was transformed into One-Shot TOP10 chemically competent Escherichia coli cells (Invitrogen) according to the manufacturer's protocol. Clones containing the CpCYC-b/pGEM-T constructs were selected on ampicillin selection medium. Plasmid DNA was isolated with the Zyppy Plasmid Miniprep kit (Zymo Research) according to the manufacturer's protocol. Orientation of the CpCYC-b inserts was confirmed by PCR amplification with SP6 and insert-specific primers.
The pAC-LYC plasmid that enables lycopene production and accumulation in E. coli (Cunningham and Gantt, 2007) was obtained from Dr. F.X. Cunningham, Jr., and Dr. E. Gantt (University of Maryland, College Park). The pAC-LYC plasmid contains bacterial genes encoding the geranylgeranyl pyrophosphatase, PSY, and PSD enzymes that, when expressed, sequentially modify the native isoprenoid substrates in E. coli to lycopene. pAC-LYC plasmid DNA was transformed into One-Shot TOP10 chemically competent cells according to the manufacturer's protocol and transformants selected with chloramphenicol. pAC-LYC cells were made competent according to Chung et al. (1989) and stored at −80°C. A 100-μL aliquot of pAC-LYC competent cells was transformed with 5 μL of Kapoho or SunUp CpCYC-b/pGEM-T construct. Transformants were grown in ampicillin + chloramphenicol selection medium. Plasmid DNA was isolated and amplified as above with insert-specific and PDS primers for pAC-LYC plasmid to confirm transformation.
Flesh Color Marker Evaluation
Primers targeting indel polymorphisms detected between Kapoho and SunUp or AU9 and SunUp in the region of CpCYCb were used to amplify genomic DNA. Fifteen papaya cultivars and breeding lines were screened with the CPFC markers. CPFC1, CPFC2, and CPFC3 flesh color markers were evaluated for segregation among a KD × 2H94 mapping population comprising 219 F2 hermaphrodite individuals or an AU9 × SunUp mapping population of 54 F2 individuals with known fruit flesh color phenotypes.
BAC Sequence Comparison and Gene Content
Two papaya BAC clones containing CpCYC-b were sequenced using a shotgun sequencing approach (GenBank accession nos. GQ478572 and GQ478573). BAC DNA was mechanically sheared using a Nebulizer (Invitrogen) to generate approximately 3-kb fragments. Sheared DNA was end repaired, size selected, purified, ligated into the pSMART cloning vector, and transformed into E. coli electrocompetent cells (Lucigen). BAC DNA inserts were cycle sequenced with ABI Big-Dye Terminator version 3.1 on a 3730XL DNA Analyzer (Applied Biosystems). Sequences were assembled with Vector NTI 10.3.1 ContigExpress (Invitrogen). Suspect and ambiguous bases were checked manually.
Predicted gene coding regions within the aligned BAC sequence were searched against the papaya EST database to confirm gene expression. Predicted genes without EST support were subjected to RT-PCR to confirm gene expression. Intron-spanning RT-PCR primers were designed from the papaya Core Annotation scaffold sequence (available at asgpb.mhpcc.hawaii.edu/papaya). RT-PCR conditions were as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s; and 72°C for 7 min. RT-PCR products were visualized by agarose gel electrophoresis and sequenced as above. Predicted genes were searched against the Arabidopsis (Arabidopsis thaliana) protein database to identify colinear blocks. The grape (Vitis vinifera) and tomato genome databases were searched for orthologous genes to examine microsynteny in the CpCYC-b region.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ478572 and GQ478573.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Position of BAC clones identified with papaya and tomato flesh color probes on FPC-1648 of the papaya physical map.
Supplemental Figure S2. Phylogenetic relationship among plant lycopene cyclases.
Supplemental Figure S3. Generalized model of the carotenoid biosynthesis pathway.
Supplemental Figure S4. Absorption spectra of extracted carotenoids from color complementation analysis.
Supplemental Figure S5. Sequence comparison of two papaya flesh color BACs.
Supplemental Figure S6. Gene expression of predicted papaya genes that lack EST support.
Supplemental Figure S7. Evaluation of the codominant CPFC1 marker in 15 papaya lines.
Supplemental Table S1. Sequence comparison of CpCYC-b alleles in 15 papaya lines.
Supplemental Table S2. GenBank accession numbers for amino acid sequences used in phylogenetic analysis of CpCYC-b.
Supplemental Table S3. Primer sequences for quantitative RT-PCR analysis.
Supplemental Table S4. Primer sequences for CpCYC-b cloning and color complementation testing.
Supplemental Table S5. Primer sequences for RT-PCR of predicted papaya genes without EST support.
Supplemental Table S6. Orthologous gene information for papaya, Arabidopsis, tomato, and grape genome comparisons.
Supplemental Table S7. Predicted function of 10 annotated genes on papaya flesh color BACs.
Supplemental Table S8. Primer sequences of SCAR markers linked to CpCYC-b.
Supplementary Material
Acknowledgments
We thank Andrea Gschwend and Anne-Florence Lava for technical assistance, Drs. Richard Manshardt and Maureen Fitch for providing papaya accessions D10 and Richter, Dr. Steve Tanksley for providing the tomato CYC-b probe used for BAC library screening, and Drs. Francis X. Cunningham, Jr., and Elizabeth Gantt for providing the pAC-LCY plasmid for color complementation tests.
References
- Ball EV, Stenson PD, Abeysinghe SS, Krawczak M, Cooper DN, Chuzhanova NA. (2005) Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat 26: 205–213 [DOI] [PubMed] [Google Scholar]
- Blas AL, Yu Q, Chen C, Veatch O, Moore PH, Paull RE, Ming R. (2009) Enrichment of a papaya high-density genetic map with AFLP markers. Genome 52: 716–725 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Hugueney P, d'Harlingue A, Kuntz M, Camara B. (1994) Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J 6: 45–54 [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. (2003) Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Bramley PM. (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53: 2107–2113 [DOI] [PubMed] [Google Scholar]
- Byers PH. (2002) Killing the messenger: new insights into non-sense mediated mRNA decay. J Clin Invest 109: 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Yu Q, Hou S, Li Y, Eustice M, Skelton RL, Veatch O, Herdes RE, Diebold L, Saw J, et al. (2007) Construction of a sequence-tagged high-density genetic map of papaya for comparative structural and evolutionary genomics in Brassicales. Genetics 177: 2481–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CT, Niemela SL, Miller RH. (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA 86: 2172–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. (2007) A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth Res 92: 245–259 [DOI] [PubMed] [Google Scholar]
- De Iorio M, de Silva E, Stumpf MPH. (2005) Recombination hotspots as a point process. Philos Trans R Soc Lond B Biol Sci 360: 1597–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2005) PHYLIP (Phylogeny Inference Package) Version 3.6. http://evolution.genetics.washington.edu/phylip.html (March 4, 2010) [Google Scholar]
- Freeling M, Lyon E, Pedersen B, Alam M, Ming R, Lisch D. (2008) Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res 18: 1924–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SW, Kim JB, Park JS, Lee SW, Cho KJ. (2007) A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J Exp Bot 58: 3135–3144 [DOI] [PubMed] [Google Scholar]
- Hamilton RA, Ito PJ, Paull RE. (1993) ‘Sunset’ Solo Papaya. Hawaii Cooperative Extension Service, University of Hawaii, Honolulu [Google Scholar]
- Hey J. (2004) What's so hot about recombination hotspots? PLoS Biol 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Hugueney P, Badillo A, Chen HC, Klein A, Hirschberg J, Camara B, Kuntz M. (1995) Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J 8: 417–424 [DOI] [PubMed] [Google Scholar]
- Karthikeyan G, Chary KVR, Rao BJ. (1999) Fold-back structures at the distal end influence DNA slippage at the proximal end during mononucleotide repeat expansions. Nucleic Acids Res 27: 3851–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Moore PH, Zee F, Fitch MMM, Steiger DL, Manshardt RM, Paull RE, Drew RA, Sekioka T, Ming R. (2002) Genetic diversity of the Carica papaya as revealed by AFLP markers. Genome 45: 503–512 [DOI] [PubMed] [Google Scholar]
- Kuo HF, Olsen KM, Richards EJ. (2006) Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 173: 401–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hsia AP, Schnable PS. (2007) Recent advances in plant recombination. Curr Opin Plant Biol 10: 131–135 [DOI] [PubMed] [Google Scholar]
- Li L, Paolillo DJ, Parthasarathy MV, DiMuzio EM, Garvin DF. (2001) A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J 26: 59–67 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O'Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high-levels of B-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M. (2008) The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the Rosids. Trop Plant Biol 1: 181–190 [Google Scholar]
- Ma H, Moore PH, Liu Z, Kim MS, Yu Q, Fitch MMM, Sekioka T, Paterson AH, Ming R. (2004) High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166: 419–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mézard C. (2006) Meiotic recombination hotspots in plants. Biochem Soc Trans 34: 531–534 [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. (1987) Papaya. Morton J, , Fruits of Warm Climates. Julia F. Morton, Miami, pp 336–346 [Google Scholar]
- Nagarajan N, Navajas-Pérez R, Pop M, Alam M, Ming R, Paterson AH, Salzberg SL. (2008) Genome-wide analysis of repetitive elements in papaya. Trop Plant Biol 1: 191–201 [Google Scholar]
- Paull RE, Irikura B, Wu P, Turano H, Chen NJ, Blas A, Fellman JK, Gschwend AR, Wai CM, Yu Q, et al. (2008) Fruit development, ripening and quality related genes in the papaya genome. Trop Plant Biol 1: 246–277 [Google Scholar]
- Rodriguez-Amaya DB. (2003) Enhancing the Carotenoid Levels of Food through Agriculture and Food Technology. FoodAfrica. http://foodafrica.nri.org/nutrition/internetpapers/DeliaBRodriguez.pdf (March 4, 2010)
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. (2000) An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton RL, Yu Q, Srinivasan R, Manshardt R, Moore PH, Ming R. (2006) Tissue differential expression of lycopene β-cyclase gene in papaya. Cell Res 16: 731–739 [DOI] [PubMed] [Google Scholar]
- Storey WB. (1969) Papaya. Ferwerda FP, Wit F, , Outlines of Perennial Crop Breeding in the Tropics. H Veenman & Zonen, Wageningen, The Netherlands, pp 21–24 [Google Scholar]
- Suzuki JY, Tripathi S, Fermín GA, Jan FJ, Hou S, Saw JH, Ackerman CM, Yu Q, Schatz MC, Pitz KY, et al. (2008) Characterization of insertion sites in Rainbow papaya, the first commercialized transgenic fruit crop. Trop Plant Biol 1: 293–309 [Google Scholar]
- Wang J, Chen C, Na JK, Yu Q, Hou S, Paull RE, Moore PH, Alam M, Ming R. (2008) Genome-wide comparative analysis of microsatellites in papaya. Trop Plant Biol 1: 278–292 [Google Scholar]
- Weil CF. (2002) Finding the crosswalks on DNA. Proc Natl Acad Sci USA 99: 5763–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2007) Vitamin A Deficiency. World Health Organization. http://www.who.int/nutrition/topics/vad/en/ (March 4, 2010)
- Yamamoto HY. (1964) Comparison of the carotenoids in yellow- and red-fleshed Carica papaya. Nature 201: 1049–1050 [DOI] [PubMed] [Google Scholar]
- Yu Q, Tong E, Skelton RL, Bowers JE, Jones MR, Murray JE, Hou S, Guan P, Acob RA, Luo MC, et al. (2009) A physical map of the papaya genome with integrated genetic map and genome sequence. BMC Genomics 10: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.