Abstract
Galactoglycerolipids are major constituents of photosynthetic membranes in chloroplasts. At least three parallel sets of enzymes are involved in their biosynthesis that must be coordinated in response to changing growth conditions. A potential candidate for a protein affecting the activity of different galactoglycerolipid pathways is the recently described digalactosyldiacylglycerol1 (dgd1) SUPPRESSOR1 (DGS1) protein of Arabidopsis (Arabidopsis thaliana) localized in the mitochondrial outer membrane. It was discovered based on a specific gain-of-function point mutation allele, dgs1-1, that causes a partial restoration of chloroplast galactoglycerolipid deficiency in the dgd1 mutant, which is defective in the lipid galactosyltransferase, DGD1. The dgs1-1 allele causes the accumulation of hydrogen peroxide that leads to an activation of an alternative, DGD1-independent galactoglycerolipid biosynthesis pathway in chloroplasts. Analysis presented here shows that the DGS1 protein is a component of a large protein complex, which explains the previously observed dominant negative phenotype following the expression of the dgs1-1 allele. The dgs1-1 allele causes the loss of mitochondrial alternative oxidase (AOX) protein that might be related to the accumulation of hydrogen peroxide in the dgs1-1 mutant background. This effect was posttranscriptional because mRNA levels for the major form of AOX were not affected in dgs1-1 mutant seedlings. Unlike dgs1-1, a loss-of-function allele, dgs1-2, had no effect on plant growth, AOX, and lipid composition to the extent tested, leaving the quest for a possible molecular function of DGS1 open. Apparently, the DGS1 wild-type protein does not directly affect lipid metabolism in mitochondria or chloroplasts.
In plant cells, the lipid composition of membranes in different organelles is quite distinct (Jouhet et al., 2007) and can even change in response to environmental cues such as phosphate limitation (Essigmann et al., 1998; Härtel et al., 2000). Biogenic membranes are defined as those capable of de novo lipid biosynthesis and are restricted to a few membranes such as the chloroplast envelopes or the endoplasmic reticulum (Benning, 2009). This raises the question for mechanisms of lipid trafficking between organelles and the control of membrane lipid diversity and lipid homeostasis of organelles. Following the discovery of an alternative galactoglycerolipid biosynthetic pathway induced by phosphate limitation in the digalactosyldiacylglycerol (DGDG)-deficient digalactosyldiacylglycerol1 (dgd1) mutant of Arabidopsis (Arabidopsis thaliana; Härtel et al., 2000) we set out to find factors controlling this alternative pathway. A suppressor mutant screen in the dgd1 mutant background was conducted under the assumption that repressors of the alternative galactoglycerolipid biosynthetic pathway could be identified (Xu et al., 2008). Consequently a dgd1 suppressor1 (dgs1) mutant was isolated that was represented by a single-point mutant allele (dgs1-1). The affected protein, DGS1, had two predicted membrane-spanning domains and was localized to the outer mitochondrial membrane. Orthologs of this protein were absent from mammals but present in plant and fungal mitochondria. However, mechanistic data on these orthologs were insufficient to provide clues toward the molecular or biochemical function of the Arabidopsis DGS1 protein (Xu et al., 2008).
The dgs1-1 point mutant allele caused a dominant negative growth phenotype following overexpression in wild-type plants and the mode of inheritance of the dgs1-1 allele was semidominant. Reduced growth following the overexpression of the dgs1-1 mutant allele was correlated with the accumulation of hydrogen peroxide (reactive oxygen species [ROS]). In turn, exposure of dgd1 plants to hydrogen peroxide led to the activation of the alternative DGD1-independent galactoglycerolipid biosynthetic pathway (Xu et al., 2008), explaining the suppression of the dgd1 DGDG deficiency in the presence of the dgs1-1 suppressor allele. However, the function of the DGS1 wild-type protein could not be explained by this observation. Moreover, the mechanism by which the dgs1-1 point mutant allele causes elevated levels of hydrogen peroxide remained unclear. Here we continue to probe the mechanism of action of the dgs1-1 mutant allele. Furthermore, we critically test our hypothesis that the DGS1 wild-type protein is directly involved in the regulation of the alternative galactoglycerolipid biosynthetic pathway and that hydrogen peroxide formation is a component in the signal transduction pathway connecting phosphate deprivation and the expression of the DGD1-independent galactoglycerolipid biosynthetic pathway. If this hypothesis were correct, a DGS1 loss-of-function mutant should be impaired in the activation of the alternative galactoglycerolipid biosynthetic pathway following phosphate deprivation. Furthermore, the effect of the apparent dgs1-1 gain-of-function allele should be epistatic to the effect of phosphate deprivation, but not additive, which would indicate a parallel mechanism. Here we provide a comparative analysis of a loss-of-function dgs1-2 T-DNA insertion allele and the apparent dgs1-1 gain-of-function point mutant allele that led to the originally described dgd1 suppressor phenotype.
RESULTS
Introduction of dgs1-2 Does Not Suppress the dgd1 Growth Phenotype
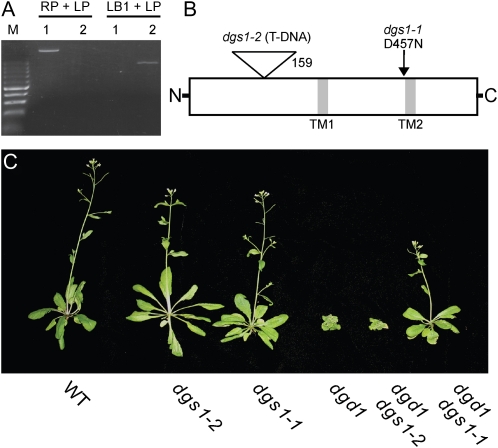
To further explore the function of DGS1 (At5g12290), we identified a potential loss-of-function allele caused by the insertion of a T-DNA into the N-terminal portion of At5g112290 (SAIL_391_F040; Sessions et al., 2002). A homozygous line was obtained following selfing as confirmed by PCR-based genotyping (Fig. 1A). The insertion was also confirmed by DNA sequencing and the insertion site corresponds to amino acid 159 in the peptide sequence (Fig. 1B). As will be shown below, this disruption leads to a loss of DGS1 protein in leaf mitochondria isolated from mutant plants. Therefore, this dgs1-2 allele described here is also referred to as a loss-of-function allele. This is in contrast to the previously described semidominant dgs1-1 allele, which here is referred to as a gain-of-function allele, because its overexpression leads to drastic phenotypes increasing in severity as more protein is present (Xu et al., 2008). The growth phenotype of the two dgs1 alleles in the wild-type and the dgd1 mutant background is shown in Figure 1C. There are no apparent growth abnormalities for either mutant allele in the otherwise wild-type background under standard growth conditions. Moreover, contrary to the dgs1-1 allele, the dgs1-2 allele does not affect growth in the dgd1 background. This result agrees with the notion that dgs1-1 is a gain-of-function allele, while dgs1-2 is a loss-of-function allele. The lack of an apparent growth phenotype of the dgs1-2 allele raises the stakes in determining the molecular or biochemical function of the wild-type DGS1 protein that led to its maintenance during evolution.
Figure 1.
Comparison of the dgs1-2 T-DNA null allele and the dgs1-1 point mutant gain-of-function allele in wild-type (WT) and dgd1 backgrounds. A, Genotyping of wild-type (1) and dgs1-2 homozygous (2) plants using the PCR primers RP, LP, and LB1 described under “Materials and Methods.” B, Diagram showing the relative positions of the dgs1-1 and dgs1-2 mutations in the DGS1 protein. The relative positions of two predicted transmembrane-spanning domains are also indicated (TM1 and TM2). C, Growth phenotypes of 30-d-old plants sown directly on soil. The respective genotypes are indicated along the bottom.
DGS1 Is Found in a Mitochondrial High Molecular Weight Complex
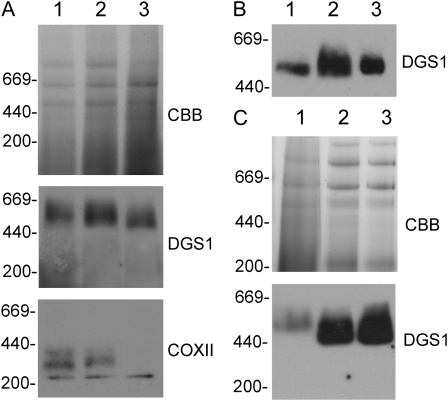
One of the simplest explanations for the mechanism of a dominant negative mutation is the poisoning of a protein complex by the mutated protein (Veitia, 2007). To query this mechanism for the possible action of the dgs1-1 allele, we explored whether the DGS1 protein is present in a higher Mr complex. To obtain sufficient material for further analysis, we switched to isolated mitochondria from cauliflower (Brassica oleracea) florets, which can be easily prepared in high quantity without contamination of chloroplast membranes. Following solubilization of mitochondrial proteins using increasing amounts of Triton X-100 and separation of complexes by blue-native PAGE, a high Mr complex migrating between the 440 and 669 kD markers (approximately 500 kD) containing a peptide reacting with anti-Arabidopsis DGS1 antibody serum is present (Fig. 2A). This complex is stable up to 1 mg Triton-X100 per mg protein, the highest concentration tested. For comparison, antiserum against subunit two of complex IV (COX II) of the cytochrome c oxidase detected a high Mr complex (approximately 300 kD) presumably representing complex VIa of the respiratory electron transport chain as described by Eubel et al. (2003). Contrary to the DGS1 immunogenic complex, this complex was not stable at 1 mg Triton-X100 per mg protein (Fig. 2A). The high Mr DGS1 immunogenic complex was also present following solubilization with two other detergents, dodecylmaltoside and Tween 20 (Fig. 2B). Isolated leaf mitochondria from Arabidopsis also showed the presence of a DGS1-immunogenic high molecular complex that was indistinguishable in wild-type and dgs1-1 mitochondria (Fig. 2C). While we have no evidence at this time to conclude whether DGS1 forms a complex with other proteins, our data consistently suggest that wild-type DGS1 and dgs1-1 mutant proteins are present in a higher Mr complex that could provide a mechanism for the function of the dominant negative dgs1-1 allele.
Figure 2.
Blue-native PAGE analysis of the DGS1 complex. A, Cauliflower mitochondrial proteins solubilized with varying amounts of Triton X-100 (0.2, 0.33, and 1 mg Triton X-100 per mg mitochondrial protein in lanes 1, 2, and 3, respectively) and separated by blue-native PAGE. The Coomassie Brilliant Blue-stained proteins (CBB) and corresponding immunoblots with the DGS1 antibody (DGS1) or the COX II antibody (COXII) are shown. The relative positions of protein standards are indicated (thyroglobulin at 669 kD, ferritin at 440 kD, and catalase at 200 kD). B, Blue-native PAGE of cauliflower mitochondrial proteins solublized with different detergents Triton X-100 (lane 1), dodecylmaltopyranoside (lane 2), or Tween 20 (lane 3), followed by immunodetection of DGS1. C, Blue-native PAGE separation of Triton X-100 solubilized crude cauliflower mitochondria (lane 1), and from isolated Arabidopsis mitochondria from wild-type (lane 2) and homozygous dgs1-1 plants (lane 3).
Phosphate Deprivation and dgs1-1 Mutation Have Additive Effects on Lipid Metabolism
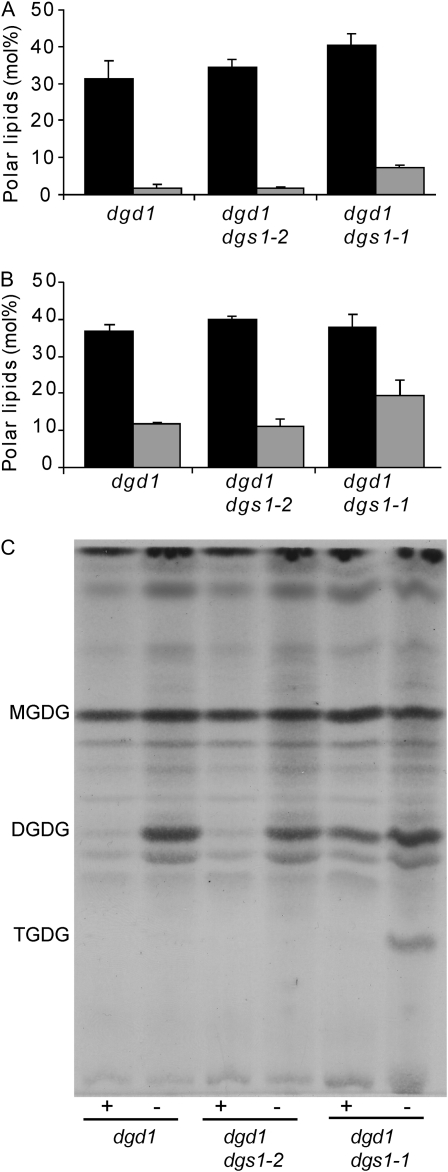
Phosphate deprivation and introduction of the dgs1-1 allele into the dgd1 mutant background lead to the accumulation of DGDG (Fig. 3), respectively. To test whether induction of DGDG biosynthesis following phosphate deprivation requires DGS1, we compared galactoglycerolipid contents in the dgd1 single mutant and the dgd1 dgs1-1 and dgd1 dgs1-2 homozygous double mutants under normal growth conditions (Fig. 3A) and under conditions of phosphate deprivation (Fig. 3B). Introducing the dgs1-2 loss-of-function allele had no effect on the accumulation of DGDG compared to the dgd1 homozygous single mutant following phosphate deprivation, suggesting that DGS1 is not required for phosphate-dependent activation of the DGD1-independent pathway. Phosphate deprivation enhanced the accumulation of DGDG in the dgd1 dgs1-1 homozygous double mutant in an additive manner. Based on this result one must conclude that phosphate deprivation and introduction of the dgs1-1 mutation act independently and represent different mechanisms of activating the DGD1-independent pathway of galactoglycerolipid biosynthesis.
Figure 3.
Galactoglycerolipid content in the dgd1 single homozygous mutant and the dgd1 dgs1-1 and dgd1 dgs1-2 double homozygous mutants under normal and phosphate-deprived growth conditions. MGDG (black bars) and DGDG (gray bars) relative amounts present in leaves of 10-d-old plants grown on MS agar plates (A), and 10-d-old plants transferred to MS agar plates lacking phosphate (B) for an additional 10 d. Relative amounts are expressed on a % mol basis of polar glycerolipid and the average and sd of three independent measurements is shown. C, A TLC plate stained with α-naphthol to reveal the separated galactoglycerolipids. Plants were grown in the presence (+) or absence of phosphate (−) as described for A and B.
Two different enzymes are known that could be responsible for DGD1-independent biosynthesis of DGDG: First, the paralog of DGD1, DGD2, is known to be induced following phosphate deprivation (Kelly and Dörmann, 2002). Second, a processive galactolipid:galactolipid galactosyltransferase discovered in preparations of plastids (van Besow and Wintermans, 1978) but not yet defined at the molecular level might be activated in dgs1-1 as well. Indeed, analysis of leaf lipid crude extracts by thin-layer chromatography (TLC) revealed the accumulation of what appeared to be—based on staining and migration—the oligogalactoglycerolipid trigalactosyldiacylglycerol in the homozygous dgd1 dgs1-1 double mutant following phosphate deprivation (Fig. 3C). This oligogalactoglycerolipid is a product of the above-mentioned processive galactolipid:galactolipid galactosyltransferase and is also present in endoplasmic reticulum-to-plastid lipid-trafficking mutants (Xu et al., 2003). It also accumulates in ozone-treated spinach (Spinacea oleracea) leaves (Sakaki et al., 1990). The latter may be relevant here, because dgs1-1 mutant plants accumulate ROS (Xu et al., 2008), which would also be present following ozone treatment.
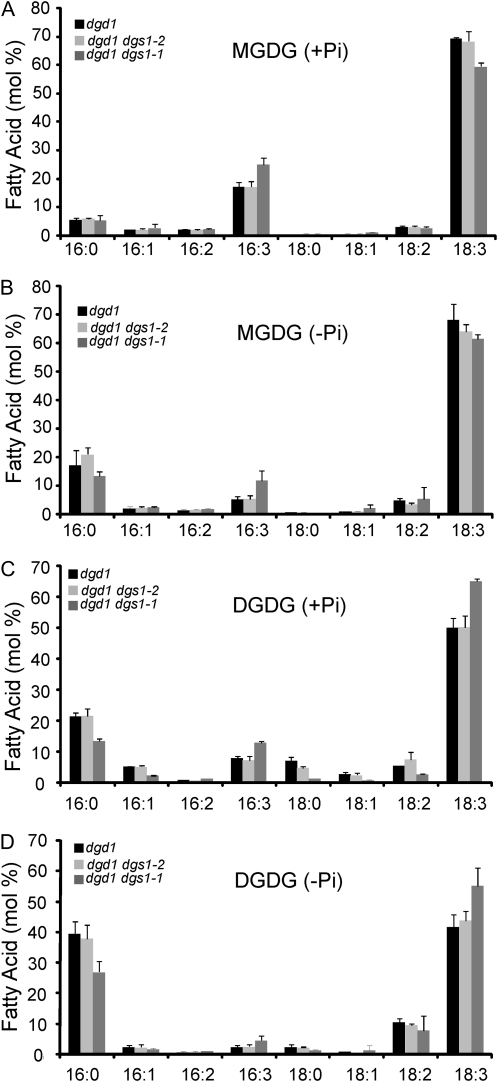
Close examination of the fatty acid profile of monogalactosyldiacylglycerol (MGDG) and DGDG in the three different mutant lines again showed no difference between the dgd1 homozygous mutant and the dgd1 dgs1-2 homozygous double mutant line under phosphate replete and depleted growth conditions (Fig. 4). The dgd1 dgs1-1 homozygous double mutant stood slightly out with a relative increase in 16:3 (carbons: position of double bonds) and decrease in 18:3 fatty acids in MGDG and lowered relative amounts of 16:0 and increased 18:3 fatty acids in DGDG independent of the growth conditions.
Figure 4.
Fatty acid profiles of MGDG (A and B) and DGDG (C and D) in the dgd1 single homozygous mutant and the dgd1 dgs1-1 and dgd1 dgs1-2 double homozygous mutants under normal (A and C) and phosphate-limited (B and D) growth conditions. The data are presented on a % mol basis and represent the average and sd from three independent experiments. Fatty acids are designated with carbon numbers: number of double bonds.
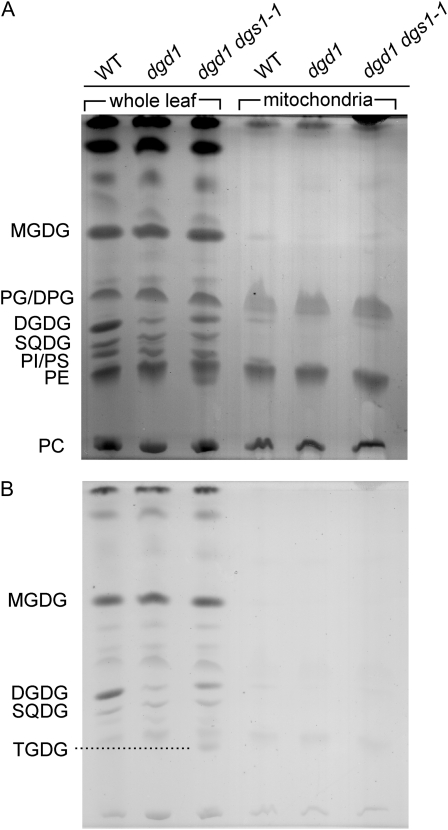
Because it was reported that phosphate deprivation leads to accumulation of DGDG in mitochondria of plant cells (Jouhet et al., 2004) and because DGS1 is a mitochondria-localized protein (Xu et al., 2008), we tested whether the presence of the dgs1-1 mutant allele could affect the lipid composition of mitochondria. As shown in Figure 5 no differences in lipid composition were observed between mitochondria isolated from wild-type, dgd1, or dgd1 dgs1-1 mutant leaves from plants grown under normal conditions. Thus, the DGDG accumulating in the dgs1-1 mutant is associated with nonmitochondrial membranes, again distinguishing the process of DGDG accumulation in the presence of the dgs1-1 allele from DGDG accumulation under phosphate-limiting growth conditions.
Figure 5.
Mitochondria of dgd1 dgs1-1 homozygous mutant plants do not accumulate galactoglycerolipids. Lipid extracts were prepared from leaves (on an equal fresh weight basis) and isolated mitochondria (on an equal mg protein basis) separated by TLC and stained with iodine (A), and after destaining, with α-naphthol (B). WT, Wild type; lipids: DPG, diphosphatidylglycerol (cardiolipin); PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SQDG, sulfoquinovosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
Alternative Oxidase Protein Levels Are Specifically Reduced in the dgs1-1 Mutant Allele
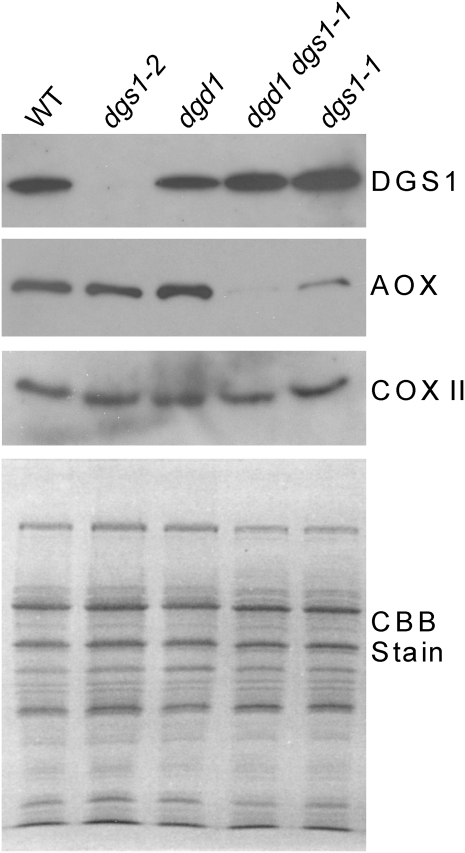
In search for molecular defects in mitochondria with altered DGS1 activity or levels, we compared the protein levels of the DGS1, alternative oxidase (AOX), and COX II of the respiratory electron transport chain in isolated mitochondria. Mitochondria were prepared from leaves of the wild-type, the dgs1-2, dgd1, dgs1-1, and the dgd1 dgs1-1 homozygous single and double mutants, respectively, and protein levels were compared using immunoblotting. As shown in Figure 6, the DGS1 protein is completely absent from the dgs1-2 mutant, confirming that this is a null or loss-of-function allele as already indicated above. Moreover, the presence of the dgs1-1 allele does not lead to a decrease in DGS1-immunogenic protein; if anything its protein level might actually be increased.
Figure 6.
AOX is reduced in dgs1-1 mutant mitochondria. Immunoblots of proteins from isolated mitochondria prepared from leaves of wild-type (WT), dgs1-1, dgs1-2, and dgd1 single homozygous mutants, and the dgd1 dgs1-1 double homozygous mutant are shown. The blots were probed with affinity-purified DGS1 antibody (DGS1), an AOX antibody (AOX), or a cytochrome c oxidase subunit II antibody (COX II). Equal amounts of protein were loaded as shown in the Coomassie Brilliant Blue-stained gel (CBB) at the bottom.
Using an AOX monoclonal antibody that detects all major forms of this protein (Finnegan et al., 1999), it was apparent that AOX levels are strongly reduced in the dgs1-1 gain-of-function mutant but unaffected in the loss-of-function mutant, dgs1-2 (Fig. 6). The observed loss of the AOX protein was not due to a general deterioration of mitochondrial complexes because COX II used here as a control was unaffected in any of the mutant backgrounds (Fig. 6). Moreover, the Coomassie Brilliant Blue-stained gel (Fig. 6, bottom section) did not show any differences in the mutant lines, suggesting no coarse aberration in mitochondrial protein patterns.
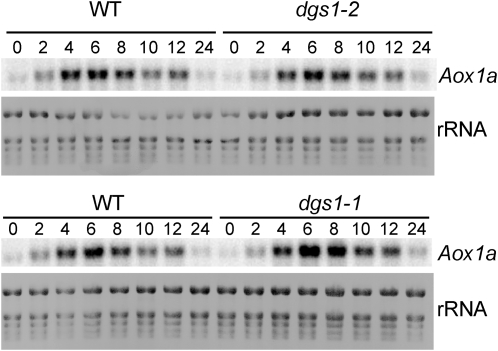
To begin to understand how the presence of the dgs1-1 allele could affect AOX protein levels, we examined the expression of the major AOX1a gene of Arabidopsis. To be able to readily detect AOX1a transcript levels in seedling extracts, we treated the seedlings with Antimycin A, which blocks respiratory electron transport at complex III and leads to a transient induction of AOX1a expression (Zarkovic et al., 2005). As shown in Figure 7, the presence of neither of the two dgs1 mutant alleles affected the expression of the AOX1a-encoding gene at the level of mRNA. Therefore, the observed reduction of AOX protein levels in the dgs1-1 mutant allele must be due to a posttranscriptional mechanism. Moreover, the wild-type DGS1 protein is not involved in regulating AOX protein or transcript levels, at least under the conditions tested, because the dgs1-2 loss-of-function allele had no effect.
Figure 7.
Analysis of AOX1a transcript levels in seedlings treated with Antimycin A. Two week-old seedlings of the wild-type (WT) and the dgs1-1 and dgs1-2 homozygous mutants were sprayed with 25 μm Antimycin A and samples of aerial tissue were taken at the indicated times (0–24 h). Total RNA (2.5 μg) from these samples were isolated, processed for RNA blotting, and specifically probed with for AOX1a mRNA. Methylene blue-stained total RNA (rRNA) is shown as a loading control.
The dgs1-1 Mutant Shows Phenotypes of an AOX1a-Deficient Mutant
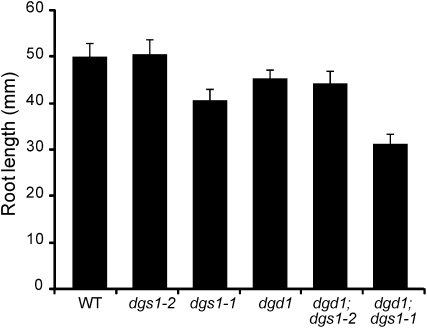
AOX-deficient mutants of Arabidopsis in general have subtle phenotypes unless exposed to multiple environmental stresses (Umbach et al., 2005; Giraud et al., 2008; Strodtkotter et al., 2009). One robust and easily observable phenotype is reduced root growth of AOX1a-deficient lines (Giraud et al., 2008). Comparing root length of seedlings between the different Arabidopsis lines used in this study, it is apparent that only for those lines carrying the dgs1-1 allele root length is decreased (Fig. 8). Thus, the dgs1-1 root phenotype repeats the root phenotype of the AOX1a-deficient lines consistent with the reduction of AOX protein in the dgs1-1 line only. However, a specific difference between the described AOX1a-deficient lines and the dgs1-1 mutant allele is the previously observed accumulation of hydrogen peroxide (Xu et al., 2008), which was reported not to be the case for the AOX1a-deficient lines (Giraud et al., 2008).
Figure 8.
The dgs1-1 but not the dgs1-2 mutant displays a root growth phenotype. Seedlings were grown for 10 d on vertically placed MS agar plates supplemented with 1% Suc, and root length was measured. The data represent the average and sd from five individual plants. WT, Wild type.
DISCUSSION
The primary goal of this study was to determine whether the DGS1 protein of Arabidopsis plays a direct role in the regulation of the DGD1-independent pathway of DGDG biosynthesis. Until now, only a gain-of-function mutant allele, dgs1-1, was available that causes a severe growth phenotype when overexpressed (Xu et al., 2008). The original connection between DGS1 and lipid metabolism derived from the fact that the dgs1-1 allele partially suppresses the lipid phenotype of the DGDG-deficient dgd1 mutant by activating a DGD1-independent pathway similar to the effect of phosphate deprivation of the dgd1 mutant or wild type. Given that ROS levels are increased in dgs1-1, AOX protein levels are reduced, and the DGD1-indepdendent pathway is induced by ROS, one could conceive a simple hypothesis for the possible function of DGS1 in phosphate-mediated regulation of galactoglycerolipid biosynthesis. Phosphate deficiency would lead to a slowing of ATP synthesis in mitochondria, causing a backup of the electron transport chain as the two processes are tightly coupled. The resulting overreduction of the respiratory electron transport chain leads to ROS formation that can be compensated for by AOX activity. If DGS1 would affect AOX activity or abundance in response to phosphate deprivation, a mechanism by which ROS could serve as a signal for the activation of the DGD1-independent pathway can be envisioned. Unfortunately, two key observations disagree with this hypothesis: (1) Plants carrying the dgs1-2 allele completely lack DGS1 but are normal in their response to phosphate deprivation with regard to galactoglycerolipid metabolism (Fig. 3, A and B). (2) Induction of DGDG biosynthesis due to the presence of the dgs1-1 allele and phosphate deprivation are additive and therefore involve two independent mechanisms (Fig. 3, A and B). Based on these observations we conclude that DGS1 is not directly involved in the low phosphate induction of DGDG biosynthesis by the DGD1-indepdendent pathway.
However, we now have a better understanding of the mechanism by which the dgs1-1 allele causes the activation of the DGD1-indepdent pathway. As previously observed, this pathway is clearly induced by ROS produced in the mitochondria (Xu et al., 2008). Moreover, the point mutation in the DGS1 protein present in the dgs1-1 allele leads to an accumulation of ROS. What we discovered here is that the presence of the dgs1-1 allele also leads to a reduction in AOX protein in mitochondria. It would seem plausible that this reduction of AOX could be the cause of the observed accumulation of hydrogen peroxide in the dgs1-1 mutant as a role of AOX in the alleviation of mitochondrial ROS has been well documented (Maxwell et al., 1999; Fiorani et al., 2005; Umbach et al., 2005). This conclusion appears to be in conflict with a recent report that AOX1a-deficient mutants do not accumulate hydrogen peroxide (Giraud et al., 2008). However, a more recent report suggests that AOX1d is induced in the AOX1a mutant background and could partially compensate for the loss of the major AOX1a (Strodtkotter et al., 2009). Thus, while we have no solid proof at this time that the reduction of AOX protein observed in dgs1-1 could be the cause of hydrogen peroxide accumulation in this mutant line, it still seems to be a plausible scenario. This raises the question of how the dgs1-1 mutant protein affects AOX protein levels. Clearly, based on our AOX transcript analysis (Fig. 7) the effect must be posttranscriptional. Because DGS1 is localized in the outer membrane (Xu et al., 2008) while AOX is associated with the inner membrane, it is unlikely that the two proteins can directly interact. It is also not likely that the DGS1 wild-type protein is required for the processing or import of AOX protein into the mitochondria, because the presence of the dgs1-2 loss-of-function allele has no effect on AOX protein levels (Fig. 6). Moreover, orthologs of the DGS1 protein are present in organisms lacking AOX such as yeast (Saccharomyces cerevisiae; Xu et al., 2008). Thus, the effect of the dgs1-1 allele on AOX protein levels may be indirect and not directly related to the function of the DGS1 wild-type protein.
If the DGS1 wild-type protein is not directly involved in the induction of the alternative galactoglycerolipid biosynthetic pathway following phosphate deprivation and is not required to establish or maintain AOX protein levels, what is its molecular function? This question has become even more difficult to answer because we have not yet been able to discern any phenotype of the dgs1-2 loss-of-function allele from that of the wild type under all conditions tested. The biological function at the organism level of well-studied proteins such as AOX and many other Arabidopsis proteins is not clear as well, but it is increasingly apparent that many plant proteins are of conditional importance as they help sessile plants to cope with a changing environment. Clearly, we have not yet found the conditions under which the loss of the DGS1 protein becomes critical.
The DGS1 protein has come to our attention because a single-point mutation leads to suppression of a galactoglycerolipid biosynthesis defect in Arabidopsis and because this point mutation causes a dominant negative phenotype. This observation suggested that DGS1 is present in a protein complex because a defective protein can poison the complex's function as discussed for possible mechanisms of dominant negative mutations (Veitia, 2007). Indeed we have shown that DGS1 is present in a mitochondrial complex supporting this scenario. The phenotype of the dgs1-1 allele is clearly fascinating. However, a rigorous analysis of the dgs1-2 loss-of-function mutant submitted to a gauntlet of conditions, or the systematic biochemical analysis of the DGS1 protein complex will be required to provide more definitive clues toward the molecular function of DGS1 in the wild-type plant.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) plants (Columbia-2), and the dgs1-1 and dgd1 dgs1-1 mutants are as previously described (Xu et al., 2008). The dgs1-2 T-DNA insertion line (Sessions et al., 2002), SAIL_391_F040, was obtained from the Arabidopsis Biological Resource Center at Ohio State University. This line was selfed and tested for homozygosity (lack of segregation of the T-DNA marker). Homozygous dgd1 dgs1-2 double-mutant plants were obtained from the F2 seeds of a cross between dgd1 and dgs1-2 homozygous mutant plants. Genotyping for dgd1 homozygosity was performed by PCR with a derived cleaved amplified polymorphic sequence marker (Xu et al., 2003). Homozygosity for dgs1-2 was confirmed by the absence of a PCR amplicon when using the forward and reverse primers 5′-GAGGTGAGTGAACTAGAACATTCCATGAG-3′ and 5′-ACTCAACATAAAGCTAGAAAGCCCATTG-3′, respectively, which span the T-DNA insertion, and the presence of a PCR amplicon when the above reverse primer is used with a primer complementary to the left border of the T-DNA insertion (Alonso et al., 2003). In general, surface-sterilized seeds were grown on agar-solidified Murashige and Skoog (MS) medium or soil as previously described (Xu et al., 2003).
Isolation of Mitochondria and Immunoblotting
Mitochondria were isolated from leaf tissue of 3-week-old, soil-grown Arabidopsis plants and purified using Percoll gradients according to Keech et al. (1995), while crude cauliflower (Brassica oleracea) mitochondrial preparations were performed as previously described (Douce et al., 1972).
For SDS-PAGE/immunoblot analyses, denatured crude leaf extracts were prepared as previously described (Martinez-Garcia et al., 1999), while protein fractions from purified organelles were denatured in a standard SDS-PAGE buffer (Laemmli, 1970). Protein concentration was quantified using the RCDC protein assay kit from Bio-Rad (Hercules) with bovine serum albumin as a standard. For blue-native PAGE, solubilized proteins from cauliflower or Arabidopsis mitochondria were prepared using Triton X-100, dodecylmaltopyranoside, or Tween 20 as described in Eubel et al. (2003) at 0.2, 0.33, and 1.0 mg detergent per mg protein (all at 25 mg protein mL−1). The supernatant fractions after a 16,000g centrifugation were removed and mixed with Coomassie Brilliant Blue R-250, and 50 μL from each sample was separated by blue-native PAGE (Schägger et al., 1994; Eubel et al., 2003). Immunoblotting was performed on polyvinylidene difluoride membranes using the CPS-1 chemiluminescent peroxidase substrate detection system from Sigma. DGS1-specific polyclonal antibodies were affinity purified as previously described (Xu et al., 2008). Monoclonal antibodies against a conserved epitope in plant AOXs (AOX mAb; Finnegan et al., 1999), and the cytochrome C oxidase subunit II (COX II mAb), were obtained from a Michigan State University-Department of Energy Plant Research Laboratory stock left by Lee McIntosh.
RNA Analysis
Total RNA from liquid N2-frozen leaf tissue of 2-week-old seedlings was isolated with the QIAGEN RNeasy plant mini-kit according to the manufacturer's instructions and separated by electrophoresis through a formaldehyde-denaturing agarose gel. RNAs were then transferred to a nylon filter that was processed for RNA blotting using ULTRAhyb hybridization buffer (Ambion). The probe used to specifically detect AOX1a mRNA was generated by PCR with the following primers: forward, 5′-TTTGTTCTTCGACGGTCCGTAC-3′; reverse, 5′-CGCGATTCCTTTATCTCCCTTG-3′, and labeled with 32P using the Megaprime DNA labeling system (GE Healthcare). Total RNAs were stained with methylene blue as previously described (Wilkinson et al., 1991).
Lipid Analysis
Lipids were extracted from leaves (Dörmann et al., 1995) on an equal fresh weight basis and separated by TLC on activated ammonium sulfate-impregnated silica gel TLC plates (Si250, Mallinckrodt Baker) with a solvent system of acetone/toluene/water (91:30:7.5, by volume). Gas chromatography-based quantification of fatty acid methyl esters was performed as previously described (Xu et al., 2005). MGDG and DGDG bands were scraped into individual vials, while the remaining polar lipids were collectively scraped into a single vial for fatty acid methyl ester derivatization (designated as the remainder of polar lipids). Lipids were extracted from mitochondria as described above, but loaded on TLC plates on an equal total mitochondrial protein basis. For visualization of total lipids separated by TLC, dried plates were placed in a chamber containing I2 vapors; for visualization of glycoglycerolipids, plates were stained with α-naphthol (Benning et al., 1995).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK118777 (DGS1 cDNA).
Acknowledgments
We would like to acknowledge the Arabidopsis Biological Resource Center at Ohio State University for providing seed stocks and Linda Danhof at the Michigan State University-Department of Energy Plant Research Laboratory, who provided us with monoclonal antibody sera for AOX and COX II left behind by the late Lee McIntosh.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Benning C, Huang ZH, Gage DA. (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys 317: 103–111 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Christensen EL, Bonner WD. (1972) Preparation of intact plant mitochondria. Biochim Biophys Acta 275: 148–160 [DOI] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jansch L, Braun HP. (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Wooding AR, Day DA. (1999) An alternative oxidase monoclonal antibody recognises a highly conserved sequence among alternative oxidase subunits. FEBS Lett 447: 21–24 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN. (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C. (2000) DGD1-independent biosynthesis of extraplastidic galactolipids following phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Marechal E, Baldan B, Bligny R, Joyard J, Block MA. (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Marechal E, Block MA. (2007) Glycerolipid transfer for the building of membranes in plant cells. Prog Lipid Res 46: 37–55 [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardstrom P. (1995) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Kelly AA, Dörmann P. (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T, Saito K, Kawaguchi A, Kondo N, Yamada M. (1990) Conversion of monogalactosyldiacylglycerols to triacylglycerols in ozone-fumigated spinach leaves. Plant Physiol 94: 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodtkotter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN. (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Besow A, Wintermans JFGM. (1978) Galactolipid formation in chloroplast envelopes. I. Evidence for two mechanisms in galactosylation. Biochim Biophys Acta 529: 44–53 [DOI] [PubMed] [Google Scholar]
- Veitia RA. (2007) Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19: 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, Doskow J, Lindsey S. (1991) RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res 19: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich J, Awai K, Benning C. (2005) Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Riekhof W, Froehlich JE, Benning C. (2003) A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J 22: 2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Moellering ER, Fan J, Benning C. (2008) Mutation of a mitochondrial outer membrane protein affects chloroplast lipid biosynthesis. Plant J 54: 163–175 [DOI] [PubMed] [Google Scholar]
- Zarkovic J, Anderson SL, Rhoads DM. (2005) A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol Biol 57: 871–888 [DOI] [PubMed] [Google Scholar]