Abstract
RsRGA4 (for Ralstonia solanacearum-responsive gene A4) encodes a polypeptide similar to S-locus glycoprotein (SGP) from Brassica rapa and SGP-like proteins from Ipomoea trifida and Medicago truncatula. Therefore, we designated RsRGA4 as NtSGLP (for Nicotiana tabacum SGP-like protein) and NbSGLP (its Nicotiana benthamiana ortholog). NbSGLP is expressed in root, leaf, petal, gynoecium, and stamen. Expression of NbSGLP was strongly induced by inoculation with an avirulent strain of R. solanacearum (Rs8107) and slightly enhanced by inoculation with virulent R. solanacearum (RsOE1-1). Expression of NbSGLP was induced by inoculation with an hrpY-deficient mutant of RsOE1-1 and Rs8107. Expression was also induced by aminocyclopropane carboxylic acid and salicylic acid. Virus-induced gene silencing of NbSGLP enhanced the growth of Rs8107. Growth of RsOE1-1 and appearance of wilt symptoms were also accelerated in silenced plants. Expression of PR-1a and EREBP was reduced, and markers for basal defense, such as callose deposition and reduced vascular flow, were compromised in NbSGLP-silenced plants. Moreover, growth of Pseudomonas cichorii, Pseudomonas syringae pv tabaci, and P. syringae pv mellea was also enhanced in the silenced plants. On the other hand, silencing of NbSGLP did not interfere with the appearance of the hypersensitive response. NbSGLP was secreted in a signal peptide-dependent manner. Agrobacterium tumefaciens-mediated expression of NbSGLP induced PR-1a and EREBP expression, callose deposition, and reduced vascular flow. NbSGLP-induced callose deposition and reduced vascular flow were not observed in salicylic acid-deficient N. benthamiana NahG plants. Taken together, SGLP might have a role in the induction of basal defense in Nicotiana plants.
Plants have a variety of active defense mechanisms to protect themselves from microbial pathogen infection. These responses include the hypersensitive response (HR; Levine et al., 1994), production of phytoalexins (Kuc, 1972), activation of pathogenesis-related proteins (Mauch and Staehelin, 1989), induction of an oxidative burst (Baker and Orlandi, 1995), cross-linking of cell wall glycoproteins (Bradly et al., 1992; Brisson et al., 1994), and lignification (Vance et al., 1980). A common feature of plant active defense responses is the transcriptional activation of a large number of genes upon pathogen infection or treatment with pathogen elicitors (Rushton and Somssich, 1998). Some of the pathogen-induced genes encode proteins with direct antimicrobial activities or enzymes involved in biosynthesis of antimicrobial compounds. Other pathogen-induced genes encode proteins with regulatory functions in signal transduction pathways of plant defense responses (Yang et al., 1997).
Ralstonia solanacearum is a devastating, soil-borne pathogen with a global distribution and a wide host range (Hayward, 1991). It causes bacterial wilt on several economically important solanaceous crops. R. solanacearum generally invades through wounded roots or natural openings, from which secondary roots subsequently emerge, and then proliferates in the intercellular spaces of the inner cortex and vascular parenchyma, before invasion into xylem vessels (Hayward, 1991; Vasse et al., 1995; Seile et al., 1997). R. solanacearum has evolved a highly specialized protein secretion system, known as the type III secretion system, whose main function is to deliver proteins from the bacterial cytoplasm into the host cell cytosol. The hrp genes encode the type III secretion system proteins and are required for the induction of the HR, a rapid localized programmed death of plant cells at the infection site, in resistant cultivars and in nonhost plants, and for pathogenicity to host plants (Boucher et al., 1992; Van Gijsegem et al., 1995; Kanda et al., 2003a).
In the tomato (Solanum lycopersicum), resistance to R. solanacearum is controlled by several loci (Thoquet et al., 1996a, 1996b). In Arabidopsis (Arabidopsis thaliana), resistance is monogenic and is conferred by the RRS1-R gene that encodes a novel R protein. This resistance is dependent upon salicylic acid (SA) and the NDR1 signaling pathway (Deslandes et al., 2002). Recently, PopP2, the cognate Avr protein for RRS1-R, was identified and shown to interact with the R protein (Deslandes et al., 2003). Although genetic identification of R genes has been extensively employed to analyze R. solanacearum-plant interactions, little is known regarding the molecular events in plants during the establishment of resistance or susceptibility to R. solanacearum. Previously, we isolated more than 50 gene fragments that were regulated in tobacco (Nicotiana tabacum) plants by inoculation with R. solanacearum (R. solanacearum-responsive genes [RsRGs]; Kiba et al., 2007). Many RsRGs showed no similarity with any other known genes and thus might represent novel genes related to plant defense responses. To identify RsRGs that are essential for defense responses, potato virus X (PVX) vector-mediated virus-induced gene silencing was performed in Nicotiana benthamiana. We previously reported that RsRGA6, encoding a small heat shock protein, was found to be essential for basic defense against R. solanacearum (Maimbo et al., 2007). In this study, we focused on clone RsRGA4, which encoded a protein with similarity to the S-locus glycoprotein (SGP). SGPs have been reported to be involved in the determination of self-incompatibility of Brassica plants (Takayama and Isogai, 2003). However, the role of proteins with similarity to SGP (SGP-like protein [SGLP]) in plant species other than Brassica is unknown. Moreover, little is known about the role of SGLP in plant defenses. Therefore, we carried out expression profiling and functional analysis of SGLP using N. benthamiana. We also discuss a possible mechanism by which this SGLP affects disease resistance.
RESULTS
Full-Length Sequence of RsRGA4
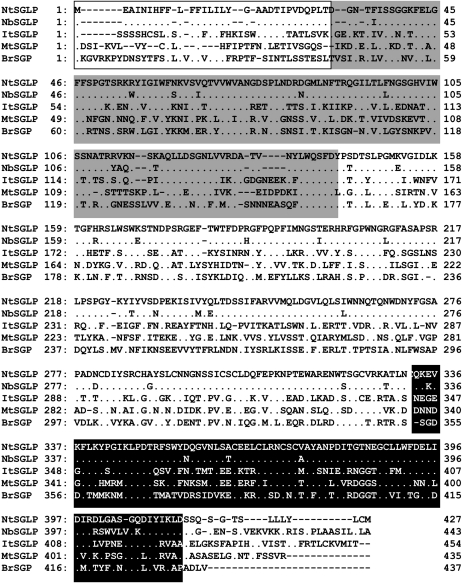
As shown in Figure 1, the deduced amino acid sequence of full-length RsRGA4 contains a secretion signal peptide, a bulb lectin domain, and a PAN apple motif. A protein database search showed 99.4%, 30.4%, 25.5%, and 18.1% amino acid identity with its orthologs from N. benthamiana, SGLP from Ipomoea trifida (AAA97903), Medicago truncatula (ABE82226), and SGP from Brassica rapa (BAB21000), respectively. Therefore, we designated RsRGA4 as NtSGLP (for N. tabacum SGLP), and its ortholog from N. benthamiana was designated as NbSGLP.
Figure 1.
Deduced amino acid sequences of NtSGLP and NbSGLP. Alignment of the deduced amino acid sequences of S-locus glycoprotein-like proteins from N. tabacum (NtSGLP), N. benthamiana (NbSGLP), Ipomoea trifida (ItSGLP), and Medicago truncatula (MtSGLP) and S-locus glycoprotein from Brassica rapa (BrSGP). Dots indicate identical amino acids. Dashes show amino acids that are not present in the sequences. The white box shows the signal peptide for extracellular secretion. The gray box indicates the bulb lectin domain. The black box indicates the PAN apple domain.
Organ-Specific Expression of NbSGLP
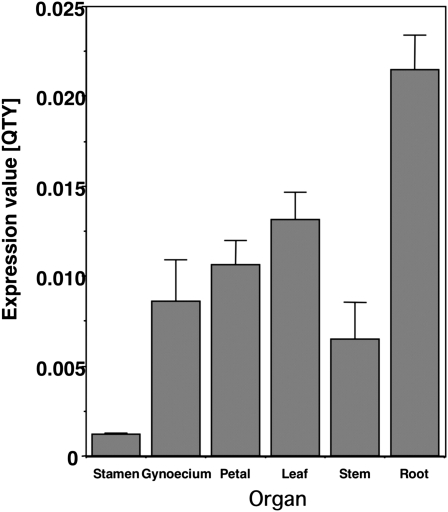
SGPs are components of the self-incompatibility mechanism and are expressed in the gynoecium (Takayama and Isogai, 2003). To determine the organ-specific expression of NbSGLP, total RNAs were isolated from the stamen, gynoecium, petal, leaf, stem, and root of N. benthamiana. Expression of NbSGLP was observed in all plant organs tested. The relative strength of expression in the tissues was as follows: root >> leaf > petal > gynoecium > stamen (Fig. 2). Therefore, expression of NbSGLP was not restricted to the gynoecium, indicating that NbSGLP plays a role in N. benthamiana other than in pollen-gynoecium interaction.
Figure 2.
Organ-specific expression of NbSGLP. Total RNA was isolated from root, leaf, stem, petal, gynoecium, and stamen of N. benthamiana. Quantitative real-time PCR was carried out as described in “Materials and Methods.” Expression values for NbSGLP are expressed as Qty values after normalization with the actin value. Values represent means and sd from triplicate experiments.
Expression of NbSGLP Is Induced by Inoculation with R. solanacearum
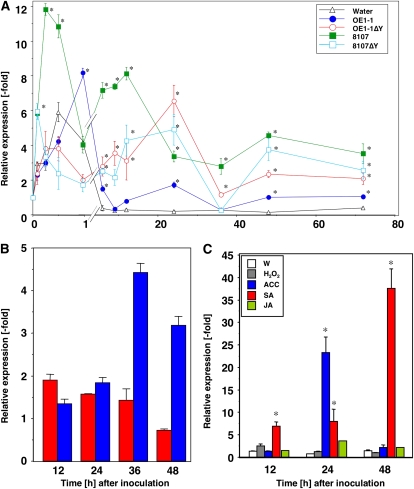
RsRGA4 (NtSGLP) was isolated as an R. solanacearum-responsive gene, which was induced in N. tabacum plants inoculated with a virulent strain of R. solanacearum (RsOE1-1) and with an avirulent strain of the bacterium (Rs8107), which induces HR on Nicotiana plants (Supplemental Fig. S1). To determine the expression profile of NbSGLP in response to R. solanacearum, RNA samples were isolated from leaves taken from N. benthamiana plants that had been inoculated with virulent strain RsOE1-1 of R. solanacearum and with an avirulent strain of the bacterium (Rs8107). The total RNAs were isolated from N. benthamiana leaves at 5, 15, and 30 min and at 1, 6, 9, 12, 24, 36, 48, and 72 h after inoculation (HAI) with RsOE1-1 and Rs8107. Expression analysis of NbSGLP by quantitative reverse transcription (qRT)-PCR showed that strong induction of NbSGLP was observed in Rs8107-inoculated N. benthamiana, and the strong expression was observed at 15 to 30 min after inoculation and 12 and 48 HAI. Expression of NbSGLP was slightly induced by inoculation with RsOE1-1 at 1 and 24 HAI (Fig. 3A).
Figure 3.
Induction pattern of NbSGLP expression. Expression values of NbSGLP are relative to the absolute nontreated control level and are normalized against actin values. Values represent means and sd from triplicate experiments. A, Total RNA was isolated from N. benthamiana leaves infiltrated with water, RsOE1-1 (OE1-1), hrpY mutant of RsOE1-1 (OE1-1ΔY), Rs8107 (8107), or hrpY mutant of Rs8107 (8107ΔY) after incubation at 25°C for the indicated times. B, Total RNA was isolated from N. benthamiana leaves infiltrated with INF1-expressing (red bars) or GUS-expressing (blue bars; control) A. tumefaciens at the indicated time points. C, Total RNA was isolated from N. benthamiana leaves at 12, 24, and 48 h after infiltration with water (W), SA, JA, H2O2, or ACC. Quantitative real-time PCR was carried out as described in “Materials and Methods.” Asterisks denote values significantly different from nontreated control leaves (P < 0.05).
Relationship between Expression of NbSGLP and Induction of Cell Death
Expression of NbSGLP was significantly induced in tobacco leaves inoculated with the HR-causing bacterial strain (Rs8107). To examine the relationship between cell death and induction of NbSGLP expression, the effect of a cell death-triggering agent, INF1-expressing Agrobacterium tumefaciens (Katou et al., 2003), was determined. We also examined the expression pattern of NbSGLP induced by inoculation with an hrpY (encoding the Hrp pilus) mutant of Rs8107 (Rs8107ΔY) that is not able to induce an HR and an hrpY mutant of RsOE1-1 (RsOE1-1ΔY). Expression of NbSGLP was induced in N. benthamiana plants inoculated with GUS-expressing control A. tumefaciens. INF1-expressing A. tumefaciens induced NbSGLP expression similarly to the GUS-expressing control at 12 and 24 HAI (Fig. 3B). NbSGLP was also up-regulated in N. benthamiana leaves inoculated with Rs8107ΔY, with kinetics similar to that in N. benthamiana leaves inoculated with the wild-type Rs8107. In addition, RsOE1-1ΔY also induced NbSGLP expression in N. benthamiana leaves (Fig. 3C). Therefore, expression of NbSGLP was induced in N. benthamiana by A. tumefaciens and by an hrp mutant of R. solanacearum. These results suggested that expression of the NbSGLP gene could be induced without cell death.
Effect of Intracellular Signaling Molecules on NbSGLP Expression
To elucidate the signaling pathways related to NbSGLP expression, well-known intracellular signaling molecules were infiltrated into N. benthamiana leaves. The signaling molecules used in this study were SA, methyl jasmonate (JA), the ethylene (ET) precursor aminocyclopropane carboxylic acid (ACC), and hydrogen peroxide (H2O2). Total RNA was isolated at 12, 24, and 48 h after treatment. Expression of NbSGLP was barely affected by the treatment with JA and H2O2. NbSGLP was induced at 24 h after treatment with ACC. Among these chemicals, SA was the most effective at inducing the expression of NbSGLP. Expression of NbSGLP increased at 12 to 48 h after SA treatment, and the expression level increased by a factor of 40 in comparison with nontreated tobacco plants at 48 h after treatment (Fig. 3C). These results suggested that induction of NbSGLP was mediated by both the ET and SA pathways.
Virus-Induced Gene Silencing of NbSGLP in N. benthamiana
Expression of NbSGLP was observed in N. benthamiana plants inoculated with R. solanacearum and in those treated with well-known signaling molecules, such as ACC and SA (Fig. 4). This information prompted us to test the function of NbSGLP in plant defense. We carried out a virus-induced gene silencing approach in N. benthamiana using the PVX vector. Three weeks after inoculation, there were no phenotypic differences between plants infected with A. tumefaciens carrying an empty pPVX201 vector and those with bacteria carrying pPVX-SGLP (data not shown). Analysis by qRT-PCR confirmed that the NbSGLP gene was silenced (Fig. 4).
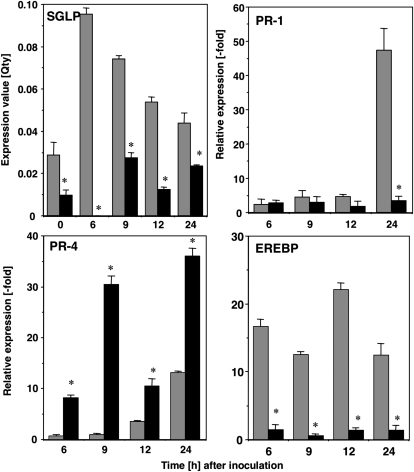
Figure 4.
Virus-induced gene silencing of NbSGLP and effect of silencing of NbSGLP on the expression of defense-related genes. N. benthamiana plants were infected with A. tumefaciens carrying either PVX (gray bars) or PVX:SGLP (black bars). Three weeks later, the fourth leaves above the primary A. tumefaciens-infected leaves were infiltrated with Rs8107, and total RNA was isolated at the indicated time points. The relative abundances of NbSGLP, PR-1a, PR-4, and EREBP transcripts were analyzed using quantitative real-time PCR with primer combinations described in Supplemental Table S1. Quantitative real-time PCR was carried out as described in “Materials and Methods.” Expression values of NbSGLP are expressed as Qty values after normalization with the actin value. Expression values of PR-1a, PR-4, and EREBP are relative to the absolute nontreated control level and are normalized against the actin values. Values represent means and sd of results from triplicate experiments. Asterisks denote values significantly different from empty PVX controls (P < 0.05).
Silencing of NbSGLP Compromised the Expression of Defense-Related Genes
To test the influence of the silencing of NbSGLP on expression of defense-related genes, total RNA was extracted from NbSGLP-silenced and control leaves at 6, 9, 12, and 24 HAI with Rs8107. As shown in Figure 4, expression of PR-1a (a marker gene for the SA signal pathway) was induced in control plants at 24 HAI with Rs8107, whereas PR-1a expression was compromised in NbSGLP-silenced plants. Expression of EREBP (a marker gene for the ET signal pathway) was observed in control plants at 6 to 24 HAI with Rs8107. However, the expression of EREBP was greatly reduced in NbSGLP-silenced leaves throughout the experiment. In contrast, the expression level of PR-4 (a marker gene for the JA signal pathway) was enhanced in NbSGLP-silenced leaves. The reductions in PR-1a and EREBP expression levels were consistent with the NbSGLP responses to SA and ACC (Fig. 3C). These results suggested that NbSGLP might have an important role(s) in the expression of defense-related genes through the SA and ET pathways.
Silencing of NbSGLP Accelerates Growth of R. solanacearum and Disease Development of Bacterial Wilt
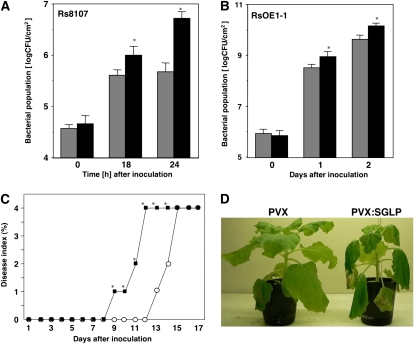
A reduction of defense-related gene expression was observed in NbSGLP-silenced plants, which raised the possibility that disease resistance to nonpathogenic bacteria is compromised and disease susceptibility to pathogenic bacteria might increase in the silenced plants. To address whether silencing of NbSGLP affected the growth of Rs8107, the bacterial suspension was inoculated into NbSGLP-silenced leaves and control leaves. The bacterial population was determined at 18 and 24 HAI. As shown in Figure 5A, growth of Rs8107 was significantly enhanced in NbSGLP-silenced plants at 24 HAI, showing an approximately 10-fold increase in comparison with control plants. We then confirmed the effect of NbSGLP silencing on the growth of RsOE1-1. Enhancement of the growth of RsOE1-1 was also observed in NbSGLP-silenced plants at 1 and 2 d after inoculation (Fig. 5B). However, acceleration of the RsOE1-1 growth (approximately 5-fold) was not remarkable in comparison with the growth of Rs8107 (approximately 10-fold). These results might reflect the expression level of NbSGLP in response to the two bacterial strains. We also observed the phenotypes of NbSGLP-silenced and control plants challenged with RsOE1-1. In control plants, bacterial wilt was first observed at 13 d, and the plants were completely wilted at 15 d after inoculation with RsOE1-1. When challenged with RsOE1-1, NbSGLP-silenced plants started to wilt at 9 d and were completely wilted at 12 d (Fig. 5, C and D).
Figure 5.
Effects of NbSGLP silencing on growth of R. solanacearum and development of bacterial wilt. N. benthamiana leaves were preinoculated with A. tumefaciens either carrying empty PVX (gray bars) or PVX:SGLP (black bars). A, Control and NbSGLP-silenced leaves were infiltrated with a bacterial suspension of Rs8107. B, Three weeks later, the fourth leaves above the primary A. tumefaciens-infected leaves were infiltrated with a bacterial suspension of RsOE1-1. The bacterial population was determined by plating at specified time points. Values are means of four replicate experiments with sd. Asterisks denote values significantly different from empty PVX controls (P < 0.05). C, Disease development of bacterial wilt was rated daily on a 0 to 4 disease index in empty PVX (white circles) or PVX:SGLP (black boxes). Each point represents the mean disease index of 10 plants combined from three separate experiments. Asterisks denote values significantly different from empty PVX controls (P < 0.05). D, Characteristic symptoms of bacterial wilt in control (PVX) and NbSGLP-silenced (PVX:SGLP) N. benthamiana. The photograph was taken 12 d after inoculation with RsOE1-1. [See online article for color version of this figure.]
Silencing of NbSGLP Accelerates Growth of Phytopathogenic Bacteria
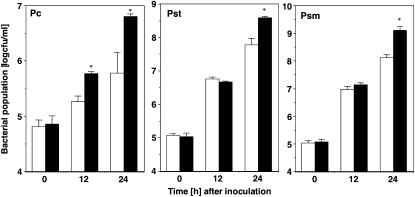
The growth of R. solanacearum and appearance of bacterial wilt were accelerated in NbSGLP-silenced plants. We then tested the role of NbSGLP in defense responses of N. benthamiana against bacterial pathogens other than R. solanacearum. As shown in Figure 6, growth of Pseudomonas cichorii, which is nonpathogenic and induces HR to N. benthamiana, was significantly enhanced in NbSGLP-silenced plants at 12 and 24 HAI, and an approximately 10-fold increase was observed in comparison with control plants at 24 HAI. Intriguingly, enhancement of the growth of Pseudomonas syringae pv tabaci and P. syringae pv mellea, both of which are pathogenic to N. benthamiana, was also observed in the silenced plants. However, acceleration of the growth of P. syringae pv tabaci and P. syringae pv mellea was only observed in later stages of infection (24 HAI) and was not remarkable (about 5-fold increase) in comparison with P. cichorii (10-fold increase).
Figure 6.
Effects of NbSGLP silencing on growth of phytopathogenic bacteria. N. benthamiana leaves were preinoculated with A. tumefaciens either carrying empty PVX (white bars) or PVX:SGLP (black bars). Control and NbSGLP-silenced leaves were infiltrated with a bacterial suspension of P. cichorii (Pc), P. syringae pv tabaci (Pst), or P. syringae pv mellea (Psm). The bacterial population was determined by plating at specified time points. Values are means of four replicate experiments with sd. Asterisks denote values significantly different from empty PVX controls (P < 0.05).
HR Cell Death Caused by R. solanacearum, P. cichorii, and INF1 Are Not Affected by Silencing of NbSGLP
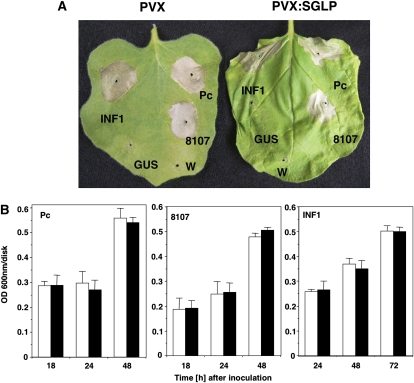
The HR is one of the best-characterized plant defenses against pathogens. To examine the response of NbSGLP-silenced plants to HR triggering, nonpathogenic bacteria Rs8107 and P. cichorii and HR-like cell death-inducing INF1-expressing A. tumefaciens were inoculated into NbSGLP-silenced plants and control plants. An HR lesion developed in both the control and NbSGLP-silenced plants at 48 HAI with Rs8107, P. cichorii, and INF1-expressing A. tumefaciens (Fig. 7A). Cell death timing was similar in control and NbSGLP-silencing plants by all HR-triggering infiltrations, as detected by Evans blue staining (Fig. 7B). These results suggested that NbSGLP might not act upstream of cell death induction.
Figure 7.
Effects of NbSGLP silencing on induction of HR. N. benthamiana plants were infected with A. tumefaciens carrying either PVX or PVX:SGLP. Control and silenced leaves were infiltrated with Rs8107 (8107), P. cichorii (Pc), or A. tumefaciens harboring GUS (control; GUS) or INF1 (INF1). A, Photographs were taken 4 d after infiltration of each bacterium. W, Water. B, Cell death was estimated in control (white bars) and NbSGLP-silenced (black bars) plants inoculated with P. cichorii, Rs8107, or A. tumefaciens harboring INF1 by Evans blue staining, as described in “Materials and Methods.” [See online article for color version of this figure.]
Silencing of NbSGLP-Compromised Basal Defenses
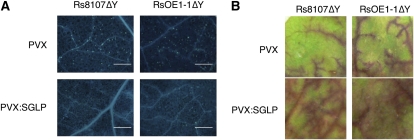
Expression of NbSGLP was induced by the inoculation of an hrp mutant of R. solanacearum, and growth of several phytopathogenic bacteria was also accelerated in NbSGLP-silenced plants. These results suggested a role for NbSGLP in basal defenses. To check whether silencing of NbSGLP affected the induction of basal defenses, hrp mutants of R. solanacearum (Rs8107ΔY and RsOE1-1ΔY) were inoculated into NbSGLP-silenced leaves and control leaves. Callose deposition, a well-known marker of basal defense, was induced in control plants inoculated with both Rs8107ΔY and RsOE1-1ΔY. In contrast, callose deposition was reduced in NbSGLP-silenced plants (Fig. 8A). As another marker of basal defense, we also analyzed vascular flow in NbSGLP-silenced leaves and control leaves. Reduction of vascular flow was observed in control plants inoculated with both Rs8107ΔY and RsOE1-1ΔY. On the other hand, the reduction of vascular flow was compromised in NbSGLP-silenced plants (Fig. 8B). These results suggested that NbSGLP might have a role in the induction of basal defenses.
Figure 8.
Effects of NbSGLP silencing on induction of basal defenses. N. benthamiana plants were infected with A. tumefaciens carrying either PVX or PVX:SGLP. Control and NbSGLP-silenced leaves were infiltrated with the hrp mutant of Rs8107 (Rs8107ΔY) and RsOE1-1 (RsOE1-1ΔY) and incubated for 24 h. A, Callose deposition was detected by aniline blue staining. Bars = 100 μm. B, Vascular flow was visualized by staining with Safranin O. [See online article for color version of this figure.]
NbSGLP Is an Extracellular Secreted Protein
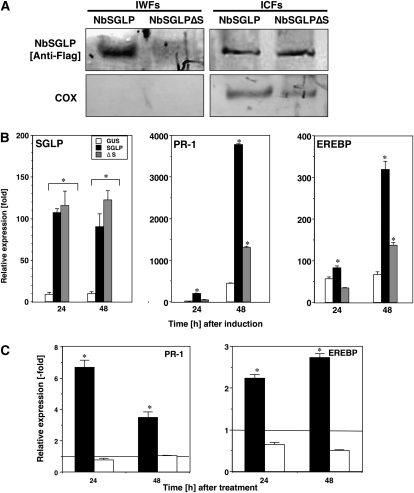
The deduced amino acid sequence of NbSGLP contained a signal peptide for extracellular secretion. To determine whether NbSGLP protein is secreted outside the cell or not, the full-length open reading frame (ORF) of NbSGLP and an ORF encoding a signal peptide-deleted NbSGLP (NbSGLPΔS) were transiently expressed in N. benthamiana leaves under the control of the 35S promoter. Immunoblot analysis was carried out with intercellular washing fluids (IWFs) and intracellular fractions (ICFs) from NbSGLP- or NbSGLPΔS-expressing N. benthamiana. As shown in Figure 9A, immunoreactive protein was detected in ICFs from both NbSGLP- and NbSGLPΔS-expressing N. benthamiana by immunoblot analysis with anti-Flag antibody, confirming the synthesis of NbSGLP and NbSGLPΔS proteins in N. benthamiana leaves. Immunoblot analysis of IWF from NbSGLP-expressing leaves showed the presence of an immunoreactive protein band. On the other hand, there was no immunoreactive protein detected in the IWF from NbSGLPΔS-expressing N. benthamiana leaves. An antibody against cytochrome oxidase (an intracellular marker protein) recognized a protein in the ICFs but not in the IWFs. These results suggested that NbSGLP protein was secreted outside plant cells mediated by the function of the N-terminal signal peptide.
Figure 9.
Transient expression of NbSGLP and induction of defense-related genes in N. benthamiana. A, The IWFs and ICFs were prepared from N. benthamiana leaves inoculated with full-length NbSGLP- and signal peptide-deleted NbSGLPΔS- expressing A. tumefaciens. Both IWFs and ICFs were separated by 15% SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes. The blots were subjected to western-blot analyses with monoclonal antibodies raised against the Flag tag sequence (NbSGLP [Anti-Flag]) and cytochrome oxidase (COX). B, Total RNA was isolated from N. benthamiana leaves inoculated with GUS, full-length NbSGLP-, and signal peptide-deleted NbSGLPΔS-expressing A. tumefaciens at the indicated time points. C, Total RNA was isolated from N. benthamiana leaves treated with the IWF fraction from NbSGLP-expressing (black bars) or NbSGLPΔS-expressing (white bars) leaves. The relative abundances of PR-1a and EREBP transcripts were analyzed using quantitative real-time PCR with primer combinations described in Supplemental Table S1. Expression values of NbSGLP, PR-1a, and EREBP are relative to the absolute nontreated control level and are normalized against the actin values. Lines showed expression levels of respective genes in nontreated control plants. Values represent means and sd from triplicate experiments. Asterisks denote values significantly different from nontreated controls (P < 0.05).
Induction Pattern of Defense-Related Genes by Transient Expression of NbSGLP
To analyze the effect of transient expression of NbSGLP on defense-related gene expression, full-length NbSGLP and signal peptide-deleted NbSGLPΔS were transiently expressed in N. benthamiana leaves under the control of the 35S promoter (Fig. 9B). qRT-PCR confirmed high-level expression of NbSGLP and NbSGLPΔS in N. benthamiana leaves in comparison with GUS-expressing control leaves, and similar levels of expression were achieved for both constructs. Expression of PR-1a and EREBP were significantly up-regulated in N. benthamiana leaves that transiently expressed the full-length NbSGLP. These results suggested that NbSGLP has a role in the regulation of SA- and ET-dependent signaling pathways. The expression level of both PR genes was drastically reduced in NbSGLPΔS-expressing leaves in comparison with the full-length NbSGLP-expressed leaves. In addition to these experiments, we analyzed the induction of these defense-related genes by treatment with the IWF fraction from the full-length NbSGLP- and NbSGLPΔS-expressing leaves. As shown Figure 9C, both PR-1a and EREBP expression was induced by the treatment with IWF fraction from the full-length NbSGLP-expressed leaves. In contrast, no significant induction was observed in N. benthamiana leaves treated with IWF fraction from the NbSGLPΔS-expressed leaves. These results suggested that induction of these defense-related genes requires an extracellularly secreted NbSGLP.
Induction of Basal Defenses by Transient Expression of NbSGLP and Its Dependency on SA
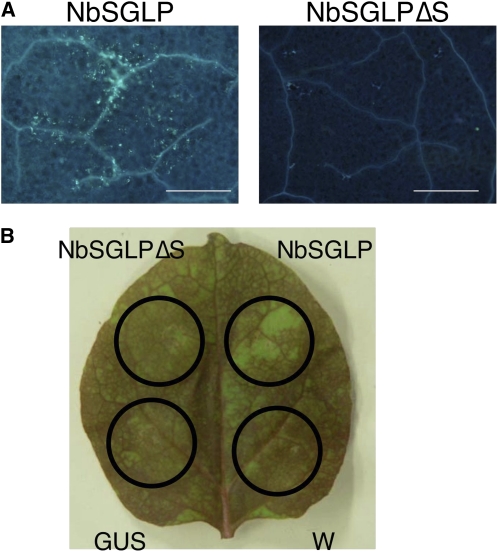
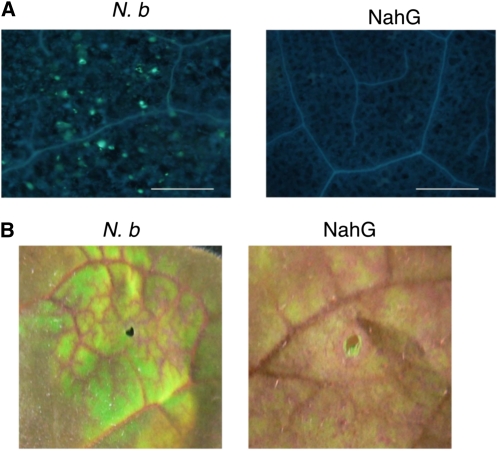
The callose deposition and reduction of vascular flow were compromised in NbSGLP-silenced plants (Fig. 8), indicating the role of NbSGLP in the induction of basal defenses. Transient expression of NbSGLP induced callose deposition and reduction of vascular flow, whereas these basal defenses were not induced in N. benthamiana by transient expression of NbSGLPΔS (Fig. 10). Intriguingly, NbSGLP-induced callose deposition and reduction of vascular flow were not observed in SA-deficient N. benthamiana NahG plants (Fig. 11). These results suggested that NbSGLP has an ability to induce basal defenses, and SA is required for induction of the basal defenses.
Figure 10.
Transient expression of NbSGLP and induction of basal defenses in N. benthamiana. A, N. benthamiana plants were inoculated with full-length NbSGLP- and signal peptide-deleted NbSGLPΔS-expressing A. tumefaciens. Callose deposition was detected by aniline blue staining. Bar = 100 μm. B, N. benthamiana plants were infiltrated with water (W), GUS, and full-length NbSGLP- and signal peptide-deleted NbSGLPΔS-expressing A. tumefaciens. Vascular flow was visualized by staining with Safranin O. [See online article for color version of this figure.]
Figure 11.
Role of SA in NbSGLP-induced basal defenses in N. benthamiana. The full-length NbSGLP-expressing A. tumefaciens was inoculated into N. benthamiana (N. b) and N. benthamiana NahG. A, Callose deposition was detected by aniline blue staining. Bars = 100 μm. B, Vascular flow was visualized by staining with Safranin O. [See online article for color version of this figure.]
DISCUSSION
In this study, we isolated genes from N. tabacum and N. benthamiana that were similar to SGP from Brassica rapa (NtSGLP and NbSGLP). SGPs were originally reported to be encoded by the S-locus, which is well known to be involved in the determination of self-incompatibility of Brassica plants (Takayama and Isogai, 2003). Database searching showed that SGLPs exist not only in Nicotiana plants but also in Medicago and Ipomoea plants. Therefore, SGLPs are evolutionally conserved in plants. However, little is known about the role of SGLP in plant species other than Brassica. The organ-specific expression patterns of NbSGLP indicated a different role of SGLP than in pollen-stigma interaction (Fig. 2). Expression analysis of NtSGLP and NbSGLP showed greatly increased expression in Nicotiana plants in response to an avirulent strain of R. solanacearum (Rs8107) in comparison with the virulent strain of R. solanacearum (RsOE1-1; Fig. 3A; Supplemental Fig. S1). These results suggested that SGLP might have a role in plant defense responses in Nicotiana plants against R. solanacearum.
Plant defense responses are initiated by the recognition of pathogen-derived molecules (Nurnberger and Brunner, 2002; Schulze-Lefert, 2004) and are divided into two categories: effectors-triggered immunity (ETI) and pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI; Jones and Dangl, 2006). In the case of pathogenic bacteria, the type III secretion apparatus encoded by the hrp genes enables effector proteins to be injected into plant cells. Effector proteins are recognized by plant cells, after which ETI develops. hrp-deficient bacterial pathogens lose the ability to induce ETI but still induce PTI. PTI is triggered by the recognition of generally conserved elicitors (PAMPs). PTI effectively restricts the growth of a majority of potential pathogens (Ma and Guttman, 2008). Rapid induction of NbSGLP expression was induced in N. benthamiana leaves within 5 min after inoculation of Rs8107. The expression of NbSGLP was also induced in N. benthamiana leaves inoculated with hrp mutants of R. solanacearum (RsOE1-1ΔY and Rs8107ΔY). Moreover, GUS-expressing A. tumefaciens also enhanced NbSGLP expression (Fig. 3). Therefore, NbSGLP might be induced by PAMPs recognition and be related to induction of PTI.
The growth of Rs8107 was accelerated in N. benthamiana plants in which NbSGLP was silenced (Fig. 5A), whereas Rs8107 did not systemically spread, was restricted to the infiltrated area, and did not cause bacterial wilt in the silenced plants (data not shown). Reduction in expression of defense-related genes, including PR-1a and EREBP, was observed in NbSGLP-silenced plants challenged with Rs8107 (Fig. 4). Generally, plant resistance responses to avirulent bacterial pathogens are extremely complex and are likely to involve myriad cellular processes, including expression of PR proteins (Maleck et al., 2000; Mysore et al., 2002; Tao et al., 2003). Therefore, reduction of disease resistance seems to be caused by a reduction in the expression of defense-related genes. In contrast, induction of HR by Rs8107 and INF1 was scarcely affected by NbSGLP silencing (Fig. 7). Therefore, disease resistance is at least partially mediated by NbSGLP, but HR-mediated defense seems not to be related to NbSGLP.
In addition to HR-mediated defense, plants show a symptomatic defense. The defense is defined as basal defense and is reportedly induced by hrp-deficient bacterial pathogens (Klement et al., 1999; Szatmari et al., 2005). Basal defense is also activated by virulent pathogens on susceptible hosts (Jones and Dangl, 2006). Our study showed that expression of NbSGLP was induced in an hrp gene-independent manner (Fig. 3B). Expression of NbSGLP was induced not only by avirulent Rs8107 but also by virulent RsOE1-1 (Fig. 3A). Growth of RsOE1-1, and development of bacterial wilt, were accelerated in NbSGLP-silenced plants (Fig. 5, C and D). In addition to changing the expression pattern of NbSGLP, hrp mutants of R. solanacearum, such as RsOE1-1ΔY and Rs8107ΔY, induced callose deposition and reduced vascular flow, both of which are markers for basal defense in N. benthamiana, whereas these basal defenses were reduced in NbSGLP-silenced plants (Fig. 8). The same phenomenon was observed by transient expression of NbSGLP (Fig. 10). Taken together, these results suggest that NbSGLP (and NtSGLP) might have a role in the regulation of basal defense responses.
During the initiation of defense responses, signal transduction cascades are triggered and endogenous signaling compounds are produced. In tobacco, SA is a critical signaling molecule leading to the expression of acidic types of PR genes (Ohashi and Ohshima, 1992; Pontier et al., 1994; Guo et al., 2000). JA also mediates the expression of basic types of PR genes. It has also been shown that SA and JA antagonistically inhibit SA and JA production and SA- and JA-dependent PR gene expression, respectively (Niki et al., 1998). In addition to SA and JA, ET also has a crucial role in plant defense (Block et al., 2005). An ET-responsive factor, TSRF1, is reportedly up-regulated by ET, SA, or R. solanacearum infection. Overexpression of TSRF1 in tobacco and tomato constitutively activates the expression of PR genes and subsequently enhances transgenic plant resistance to the bacterial wilt caused by R. solanacearum (Zhang et al., 2004). Our previous study demonstrated that expression of PR-1a was induced by the infiltration of Rs8107, and Rs8107-induced acquired resistance was mediated by the SA-dependent signaling pathway (Kiba et al., 2003). Therefore, SA- and ET-dependent defense responses might have crucial roles in plant defense against R. solanacearum. In this study, NbSGLP was induced by treatment with SA and ACC (Fig. 3). Transient expression of NbSGLP showed significant induction of PR-1a and EREBP but not PR-4 (Fig. 9B; Supplemental Fig. S2). In contrast, a reduction in expression of PR-1a and EREBP was observed in NbSGLP-silenced plants, whereas expression of PR-4 was enhanced in the silenced plants (Fig. 4). Therefore, NbSGLP might have a role in the regulation of SA- and ET-dependent defense and might antagonistically suppress the JA-dependent signaling pathway. Intriguingly, NbSGLP-induced basal defenses were compromised in SA-deficient NahG plants. Recent reports indicate that SA has a critical role in the induction of basal defense in potato against Phytophthora infestans (Halim et al., 2007). SA-dependent basal immunity was also reported to be required for plant defense against bacterial pathogens, such as Erwinia amylovora, P. syringae, and Pantoea stewartii in Arabidopsis plants (DebRoy et al., 2004). Taken together, these results suggest that NbSGLP (and NtSGLP) might have a role in the regulation of basal defense responses through the SA-dependent pathway.
Structural analysis of the deduced amino acid sequence of NbSGLP showed two conserved motifs: a bulb lectin domain (B-type lectin) and a PAN apple domain. The most characterized proteins containing B lectin and the PAN domain are S receptor kinases and the SGP encoded by the S-locus of Brassica plants, which is well known to be involved in the determination of self-incompatibility of Brassica plants (Takayama and Isogai, 2003). Recently, the dominant resistance gene, Pi-d2, which confers resistance to rice (Oryza sativa) blast, has been shown to encode a receptor-like kinase protein with a predicted extracellular domain comprising B-lectin and PAN domains (Chen et al., 2006). Taken together, there are at least two hypothetical models for the function of SGLP in Nicotiana plants. A possible hypothetical model is that SGLP might act as a component of a receptor complex and recognize pathogen-derived molecules (PAMPs/elicitors). Another possible model is that SGLP might be involved in intercellular (infected cell to peripheral cells) signal transduction. Further analysis will be required to determine the cognate receptor or elicitor molecule(s) to clarify the SGLP-mediated signaling cascade leading to plant defense.
MATERIALS AND METHODS
Plant and Bacterial Material
Nicotiana tabacum ‘Samsun NN’, Nicotiana benthamiana, and N. benthamiana (NahG) were grown in a growth room with a 16-h/8-h photoperiod at a light intensity of 10,000 lux at 25°C (Kiba et al., 2003).
Bacterial strains used in this study are listed in Supplemental Table S2. Ralstonia solanacearum strains OE1-1 (RsOE1-1) and 8107 (Rs8107) were grown for 16 h at 30°C in peptone-yeast extract (PY) medium. Pseudomonas cichorii SPC9018, Pseudomonas syringae pv tabaci 6605, and P. syringae pv mellea MAFF302303 were cultured in PY medium containing 20 μg mL−1 rifampicin (Marutani et al., 2005; Kiba et al., 2006b). hrpY mutants of R. solanacearum 8107 (Rs8107ΔY) and OE1-1 (RsOE1-1ΔY) were cultured in PY medium containing 50 μg mL−1 spectinomycin or 50 μg mL−1 kanamycin, respectively (Kanda et al., 2003a; Maimbo et al., 2007). The bacterial population was measured spectrophotometrically at an optical density at 600 nm, and the suspension was adjusted to 108 colony-forming units (cfu) mL−1 for inoculation. Inoculation of bacteria was carried out by leaf infiltration with the bacterial suspension using a syringe. The leaf-infiltration method produces the same phenotype in tobacco plants against R. solanacearum strains when compared with the root-inoculation method (Kanda et al., 2003a, 2003b; Shinohara et al., 2005). Reproducible expression of defense-related genes was also observed in tobacco leaves inoculated with R. solanacearum isolates RsOE1-1, Rs8107, and a mutant strain of the bacteria (Kanda et al., 2003a, 2003b; Kiba et al., 2003; Maimbo et al., 2007).
Treatment with Intracellular Signaling Molecules
N. benthamiana plant leaves were treated by leaf infiltration using a syringe by the method described previously (Maimbo et al., 2007). Concentrations of chemicals used in the experiment were as follows: 0.3 mm H2O2 (Nacalai Tesque), 50 μm SA (sodium salicylate; Sigma), 50 μm JA (Nacalai Tesque), and 100 μm ACC (Sigma).
Isolation of RNA and cDNA Synthesis
Total RNA was isolated from N. tabacum ‘Samsun NN’ and N. benthamiana leaves by a previously described method (Kiba et al., 2003). RNA samples were treated with DNase I (RNase free; Takara) to degrade contaminating genomic DNA, according to the manufacturer's instructions. RT was carried out with 1 μg of total RNA and the oligo(dT) primer (Supplemental Table S1) using Moloney murine leukemia virus reverse transcriptase (Takara) according to the manufacturer's instructions.
Isolation of Full-Length cDNA
For isolation of the complete cDNA of RsRGA4, a modified RACE method was performed (Frohman et al., 1988). PCR amplification was performed with a primer combination of A4-S and oligo(dT)-AD, as listed in Supplemental Table S1. cDNAs from N. tabacum and N. benthamiana were used as templates. Cycling parameters were as follows: 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Amplified cDNA fragments were cloned into the vector pGEM T-Easy (Promega) creating pGEMNtA4 (full-length RsRGA4 from N. tabacum) and pGEMNbA4 (its ortholog from N. benthamiana; Supplemental Table S3).
Sequencing
Sequence analysis was performed using M4 and RV primers (Supplemental Table S1) with the reagents for the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and an Applied Biosystems 3100 Avant Automated Sequencer according to the manufacturer's instructions. The sequence analysis was carried out using DNASIS (version 3.6; Hitachi) and the BLAST network service from the National Center for Biotechnology Information (Altschul et al., 1990).
Quantitative Real-Time PCR
Quantitative real-time PCR was carried out by the method of Maimbo et al. (2007). RT was carried out with 1 μg of total RNA and the oligo(dT) primer (Supplemental Table S1) using Moloney murine leukemia virus reverse transcriptase (Takara) according to the manufacturer's instructions. Real-time PCR was carried out with a 20-μL reaction mixture containing 1 μL of the cDNA stock and 10 pm of the respective primers (Supplemental Table S1) using the SYBR premix ExTaq (Takara) with an Applied Biosystems 7300 real-time PCR system. Cycling parameters were the same for all primers: an initial 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 1 min. Melting curve runs were also performed at the end of each PCR to verify the specificity of the primers by the presence of a single product. Relative quantification of gene expression was carried out according to the instructions for the Applied Biosystems 7300 real-time PCR system, using the comparative cycle threshold method for the calculation of Qty value. All values were normalized to the expression values of the actin gene as an internal standard in each cDNA stock. Expression analyses were carried out with at least two biological replications to ensure that expression patterns were reproducible. We have shown characteristic data in the figures. sd values and differences between expression ratios of controls and other samples were tested for statistical significance using the t test.
Plasmid Construction for Agrobacterium tumefaciens-Mediated Transient Expression
For detection of NbSGLP protein by immunoblot analysis, a Flag tag sequence was added to the C-terminal region of both constructs and expressed under the control of the cauliflower mosaic virus 35S promoter. A full-length ORF of NbSGLP was amplified with primers A4ORF-S and A4Flag-A (Supplemental Table S1) using pGEMNbA4 as a template. A cDNA fragment containing a signal peptide-deleted ORF of NbSGLP was amplified with primers A4dS-ATG and A4Flag-A (Supplemental Table S1) using pGEMNbA4 as a template. These cDNA fragments were subcloned into the TA cloning site of pGEM T-Easy, creating pGEMSGLP and pGEMSGLPdelS. The pGEMSGLP and pGEMSGLPdelS plasmids were digested with BamHI and SacI (Takara) and ligated into the pBI121 vector (Clontech) digested with the same enzymes. The constructs containing these inserts were designated pBI-SGLP and pBI-SGPLdelS, respectively. We also used the binary vector p35S-INF1 containing a fusion between the signal peptide of N. tabacum PR-1a and the Phytophthora inf1 gene driven by the 35S promoter of Cauliflower mosaic virus (Huitema et al., 2005). The binary vector p35S-GUS containing the GUS gene driven by the 35S promoter of the Cauliflower mosaic virus (Katou et al., 2003) was used as a control. These binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101, which harbors the transformation helper plasmid pSoup (Hellens et al., 2000), and inoculated into N. benthamiana leaves as described previously (Katou et al., 2003; Supplemental Table S3).
Preparation of IWFs and ICFs, and Immunoblot Analysis
The IWFs from N. benthamiana leaves from full-length NbSGLP- and signal peptide-deleted NbSGLPΔS-expressing A. tumefaciens were extruded by the method of De Wit and Spikeman (1982). After isolation of IWFs, leaf samples were homogenized with 10 mm Tris-HCl (pH 7.6) containing a protease inhibitor cocktail (Nacalai Tesque). The homogenates were centrifuged at 800g for 10 min. The supernatants were used as the ICFs. The IWF (10 μg) and ICF (20 μg) were separated by 10% SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes (Bio-Rad). The blots were subjected to western-blot analyses with a monoclonal antibody raised against the Flag-tag sequence (Sigma) or with a cytochrome oxidase monoclonal antibody (Abnova). Cross-reacting proteins were visualized with an alkaline phosphatase-conjugated secondary antibody raised in goat against mouse or rabbit IgG (Bio-Rad) with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (Nacalai Tesque).
DNA Constructs and Seedling Infection for Virus-Induced Gene Silencing
A 189-bp cDNA fragment representing the 3′ end sequence of NbSGLP was amplified with primers PVX:A4U and PVX:A4L (Supplemental Table S1) using pGEMNbA4 as a template. This cDNA fragment was subcloned into the TA cloning site of pGEM T-Easy, and pGEMA4PVX was created. The pGEMA4PVX plasmid was digested with PstI and SalI (Takara) and ligated into pPVX201 (Maimbo et al. 2007) digested with Sse8387I and SalI (Takara). The construct containing this insert in the antisense orientation was designated pPVX-SGLP. Plasmid pPVX201 that did not contain any insert was used as a control. These binary plasmids were transformed into A. tumefaciens strain GV3101 and inoculated into N. benthamiana leaves as described previously (Katou et al., 2003). Three weeks after the initial A. tumefaciens inoculation, R. solanacearum, P. cichoriim, P. syringae pv mellea, P. syringae pv tabaci, and A. tumefaciens were inoculated into a N. benthamiana leaf three to four leaves above the A. tumefaciens-inoculated leaf as a challenge inoculation.
Evaluation of Cell Viability
For evaluation of viability, leaf discs were stained with Evans blue (Wako) by the method of Kiba et al. (2006a). Bacterial suspensions were adjusted to give an initial optical density at 600 nm of 0.1 (1.0 × 108 cfu mL−1). Fifty microliters of water or bacterial suspension was infiltrated into a N. benthamiana leaf. Leaf discs (1 cm2) were punched out and submerged in 1 mL of 0.25% Evans blue and incubated for 20 min. They were then washed several times with water to remove excess and unbound dye. The discs were homogenized, and bound dye was extracted with 1% SDS. The extracted dye was measured spectrophotometrically at 600 nm.
Inoculation of Bacteria and Disease Index
Inoculation of bacteria was carried out by leaf infiltration with the bacterial suspension at 108 cfu mL−1 using a syringe. The plants were coded and inspected daily for wilting symptoms for 17 d. Each assay was repeated in at least six successive trials, and the disease index was recorded as described previously (Shinohara et al., 2005).
Vascular Dye Accumulation Assay and Detection of Callose
R. solanacearum or A. tumefaciens was infiltrated on N. benthamiana leaves. Each leaf was detached with a razor at the base of the petiole, and the petiole was directly placed in a 0.5% (w/v) Safranin O solution for 12 h (Oh and Collmer, 2005). Detection of callose deposition was determined by the method of Adam and Somerville (1996). Leaves were cleared in alcoholic lactophenol. Cleared leaves were rinsed with 50% ethanol, rinsed with water, and then stained for 30 min in 150 mm K2HPO4 (pH 9.5) containing 0.01% aniline blue. Samples were mounted in 50% glycerol, and callose deposition was detected under epifluorescent illumination (Olympus BX-51).
Sequence data from this article have been submitted to GenBank, EMBL, and the DNA Data Bank of Japan under the accession numbers AB479030 and AB479031 (NtSGLP and NbSGLP, respectively).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression pattern of NtSGLP in response to inoculation with R. solanacearum.
Supplemental Figure S2. Transient expression of NbSGLP and induction of defense-related genes in N. benthamiana.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. Bacterial strains used in this study.
Supplemental Table S3. List of plasmids used in this study.
Supplementary Material
Acknowledgments
We thank Dr. David C. Baulcombe of the Sainsbury Laboratory, John Innes Centre, for providing the PVX vector. We also acknowledge Dr. Jonathan D.G. Jones of the Sainsbury Laboratory, John Innes Centre, for providing construct for NahG transgenic N. benthamiana. Additionally, we thank Dr. Yuki Ichinose of Okayama University and Kasumi Takeuchi of the National Institute of Agrobiological Science for providing P. syringae pv tabaci and P. syringae pv mellea, respectively.
References
- Adam L, Somerville SC. (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J 9: 341–356 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Baker C, Orlandi EW. (1995) Active oxygen in plant pathogenesis. Annu Rev Phytopathol 33: 299–321 [DOI] [PubMed] [Google Scholar]
- Block A, Schmelz E, O'Donnell P, Jones JB, Klee HJ. (2005) Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol 138: 1481–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher CA, Gough CL, Arlat M. (1992) Molecular genetics of pathogenicity determinants of Pseudomonas solanacearum with special emphasis on hrp genes. Annu Rev Phytopathol 30: 443–461 [Google Scholar]
- Bradly D, Kjellbom P, Lamb CJ. (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel rapid defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al. (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46: 794–804 [DOI] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99: 2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit PJG, Spikeman G. (1982) Evidence for the occurrence of race- and cultivar-specific elicitors of necrosis in intercellular fluids of compatible interaction of Cladosporium fulvum and tomato. Physiol Plant Pathol 21: 1–11 [Google Scholar]
- Frohman MA, Dush MK, Martin GR. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Salih G, Klessig DF. (2000) Activation of a diverse set of genes during the tobacco resistance response to TMV is independent of salicylic acid; induction of a subset is also ethylene independent. Plant J 21: 409–418 [DOI] [PubMed] [Google Scholar]
- Halim VA, Eschen-Lippold L, Altmann S, Birschwilks M, Scheel D, Rosahl S. (2007) Salicylic acid is important for basal defense of Solanum tuberosum against Phytophthora infestans. Mol Plant Microbe Interact 20: 1346–1352 [DOI] [PubMed] [Google Scholar]
- Hayward HC. (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29: 65–87 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards AE, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Huitema E, Vleeshouwers VGAA, Cakir C, Kamoun S, Govers F. (2005) Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol Plant Microbe Interact 18: 183–193 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kanda A, Ohnishi S, Tomiyama H, Hasegawa H, Yasukohchi M, Kiba A, Ohnishi K, Okuno T, Hikichi Y. (2003a) Type III secretion machinery-deficient mutants of Ralstonia solanacearum lose their ability to colonize resulting in loss of pathogenicity. J Gen Plant Pathol 69: 250–257 [Google Scholar]
- Kanda A, Yasukohchi M, Ohnishi K, Kiba A, Okuno T, Hikichi Y. (2003b) Ectopic expression of Ralstonia solanacearum effector protein PopA early in invasion results in loss of virulence. Mol Plant Microbe Interact 16: 447–455 [DOI] [PubMed] [Google Scholar]
- Katou S, Yamamoto A, Yoshioka H, Kawakita K, Doke N. (2003) Functional analysis of potato mitogen-activated protein kinase kinase, StMEK1. J Gen Plant Pathol 69: 161–168 [Google Scholar]
- Kiba A, Maimbo M, Kanda A, Tomiyama H, Ohnishi K, Hikichi Y. (2007) Isolation and expression analysis of candidate genes related to Ralstonia solanacearum-tobacco interaction. Plant Biotechnol 24: 409–416 [Google Scholar]
- Kiba A, Sangawa Y, Ohnishi K, Yao N, Park PY, Nakayashiki H, Tosa Y, Mayama S, Hikichi Y. (2006a) Induction of apoptotic cell death leads to the development of bacterial rot caused by Pseudomonas cichorii. Mol Plant Microbe Interact 19: 112–122 [DOI] [PubMed] [Google Scholar]
- Kiba A, Takata O, Ohnishi K, Hikichi Y. (2006b) Comparative analysis of induction pattern of programmed cell death and defense-related responses during hypersensitive cell death and development of bacterial necrotic leaf spots in eggplant. Planta 224: 981–994 [DOI] [PubMed] [Google Scholar]
- Kiba A, Tomiyama H, Takahashi H, Hamada H, Ohnishi K, Okuno T, Hikichi Y. (2003) Induction of resistance and expression of defense-related genes in tobacco leaves infiltrated with Ralstonia solanacearum. Plant Cell Physiol 44: 287–295 [DOI] [PubMed] [Google Scholar]
- Klement Z, Bozso Z, Ott PG, Kecskes ML, Rudolph K. (1999) Symptomless resistance response instead of the hypersensitive reaction in tobacco leaves after infiltration of heterologous pathovar of Pseudomonas syringae. J Phytopathol 147: 467–475 [Google Scholar]
- Kuc J. (1972) Phytoalexins. Annu Rev Phytopathol 10: 207–232 [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Ma W, Guttman DS. (2008) Evolution of prokaryotic and eukaryotic virulence effectors. Curr Opin Plant Biol 11: 412–419 [DOI] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. (2007) Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol 145: 1588–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 23–61 [DOI] [PubMed] [Google Scholar]
- Marutani M, Taguchi F, Shimizu R, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. (2005) Flagellin from Pseudomonas syringae pv. tabaci induced hrp-independent HR in tomato. J Gen Plant Pathol 71: 289–295 [Google Scholar]
- Mauch F, Staehelin A. (1989) Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3 glucanase in bean leaves. Plant Cell 1: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB. (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32: 299–315 [DOI] [PubMed] [Google Scholar]
- Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y. (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol 39: 500–507 [Google Scholar]
- Nurnberger T, Brunner F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5: 318–324 [DOI] [PubMed] [Google Scholar]
- Oh HS, Collmer A. (2005) Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J 44: 348–359 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Ohshima M. (1992) Stress-induced expression of gene for pathogenesis-related proteins in plants. Plant Cell Physiol 33: 819–826 [Google Scholar]
- Pontier D, Godiard L, Marco Y, Roby D. (1994) hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. (1998) Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol 1: 311–315 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P. (2004) Plant immunity: the origami of receptor activation. Curr Biol 14: R22–R24 [PubMed] [Google Scholar]
- Seile E, MacGarvey JA, Schell MA, Denny TP. (1997) Role of extra-cellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87: 1264–1271 [DOI] [PubMed] [Google Scholar]
- Shinohara R, Kanda A, Ohnishi K, Kiba A, Hikichi Y. (2005) The contribution of folate biosynthesis to Ralstonia solanacearum proliferation in the intercellular spaces. Appl Environ Microbiol 71: 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari A, Ott PG, Varga GJ, Besenyei E, Czelleng A, Klement Z, Bozso Z. (2005) Characterization of basal resistance (BR) by expression patterns of newly isolated representative genes in tobacco. Plant Cell Rep 25: 728–740 [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A. (2003) Molecular mechanism of self-recognition in Brassica self-incompatibility. J Exp Bot 54: 149–156 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interaction with bacterial pathogen Pseudomonas syringae. Plant Cell 11: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Olivier J, Sperisen C, Rogowsky P, Laterrot H, Grimsley N. (1996a) Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii 7996. Mol Plant Microbe Interact 9: 826–836 [Google Scholar]
- Thoquet P, Olivier J, Sperisen C, Rogowsky P, Prior P, Anaïs G, Mangin B, Bazin B, Nazer R, Grimsley N. (1996b) Polygenic resistance of tomato plants to bacterial wilt in the French West Indies. Mol Plant Microbe Interact 9: 837–842 [Google Scholar]
- Vance CP, Kink IK, Sherwood RT. (1980) Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18: 259–288 [Google Scholar]
- Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. (1995) The hrp gene cluster of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol 15: 1095–1114 [DOI] [PubMed] [Google Scholar]
- Vasse J, Frey P, Trigalet A. (1995) Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant Microbe Interact 8: 241–251 [Google Scholar]
- Yang Y, Shah J, Klessig DF. (1997) Signal perception and transduction in plant defense responses. Genes Dev 11: 1621–1639 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. (2004) Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol 55: 825–834 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.