Abstract
Heterologous regulatory elements and flanking sequences have been used in chloroplast transformation of several crop species, but their roles and mechanisms have not yet been investigated. Nucleotide sequence identity in the photosystem II protein D1 (psbA) upstream region is 59% across all taxa; similar variation was consistent across all genes and taxa examined. Secondary structure and predicted Gibbs free energy values of the psbA 5′ untranslated region (UTR) among different families reflected this variation. Therefore, chloroplast transformation vectors were made for tobacco (Nicotiana tabacum) and lettuce (Lactuca sativa), with endogenous (Nt-Nt, Ls-Ls) or heterologous (Nt-Ls, Ls-Nt) psbA promoter, 5′ UTR and 3′ UTR, regulating expression of the anthrax protective antigen (PA) or human proinsulin (Pins) fused with the cholera toxin B-subunit (CTB). Unique lettuce flanking sequences were completely eliminated during homologous recombination in the transplastomic tobacco genomes but not unique tobacco sequences. Nt-Ls or Ls-Nt transplastomic lines showed reduction of 80% PA and 97% CTB-Pins expression when compared with endogenous psbA regulatory elements, which accumulated up to 29.6% total soluble protein PA and 72.0% total leaf protein CTB-Pins, 2-fold higher than Rubisco. Transgene transcripts were reduced by 84% in Ls-Nt-CTB-Pins and by 72% in Nt-Ls-PA lines. Transcripts containing endogenous 5′ UTR were stabilized in nonpolysomal fractions. Stromal RNA-binding proteins were preferentially associated with endogenous psbA 5′ UTR. A rapid and reproducible regeneration system was developed for lettuce commercial cultivars by optimizing plant growth regulators. These findings underscore the need for sequencing complete crop chloroplast genomes, utilization of endogenous regulatory elements and flanking sequences, as well as optimization of plant growth regulators for efficient chloroplast transformation.
Over the expanse of time since the endosymbiotic events that led to the establishment of plant organelles, plant cells have evolved elaborate mechanisms to coordinate the expression of plastid genes with the changing developmental and functional requirements of the cell. In addition to the nucleus-encoded, plastid-localized RNA polymerase, the nucleus controls the expression of a suite of σ-factors required for the active transcription of photosynthetic genes by the plastid-encoded RNA polymerase (PEP), with PEP itself being transcribed by nucleus-encoded, plastid-localized RNA polymerase (Allison et al., 1996; Hess and Borner, 1999). Nuclear control over translation of plastid mRNA is exerted through the activities of numerous plastid-localized RNA-binding proteins (RBPs). RBPs appear to have a tight affinity for their cognate sequences in plastid mRNAs, and studies have demonstrated that their interactions are specific for particular genes (Nakamura et al., 1999, 2001; Shen et al., 2001; Meierhoff et al., 2003; Schmitz-Linneweber et al., 2005, 2006).
Plastid mRNAs, expressed as monocistrons or polycistrons, contain 5′ and 3′ untranslated regions (UTRs). Detailed analyses have demonstrated that within these UTRs lie cis-elements, often forming secondary structures, which facilitate the interaction with nucleus-encoded RBPs (Yang et al., 1995; Hirose and Sugiura, 1996; Klaff et al., 1997; Alexander et al., 1998; Zou et al., 2003; Merhige et al., 2005). RBPs display an array of functions, including processing of polycistronic transcription units, RNA maturation and editing, transcript stability and turnover, and the recruitment of additional protein factors involved in the initiation of translation in response to the requirements of the cell (Nickelsen, 2003; Schmitz-Linneweber and Barkan, 2007). In contrast to the high level of conservation found within protein-coding regions and ribosomal RNAs, intergenic and UTRs are highly variable in chloroplast genomes (Daniell et al., 2006; Saski et al., 2007; Timme et al., 2007). For example, comparison of nine Poaceae chloroplast genomes did not identify a single intergenic region that had 100% sequence identity (Saski et al., 2007).
Chloroplast transformation strategies have utilized both endogenous and heterologous regulatory elements to facilitate high levels of foreign gene expression. Hybrid systems comprising a modified tobacco (Nicotiana tabacum) ribosomal operon promoter (Prrn) in conjunction with a translational control region derived from the tobacco plastid-encoded rbcL gene or from bacteriophage T7 gene 10 (g10) to express foreign genes have been utilized in numerous species (Guda et al., 2000; Staub et al., 2000; Kuroda and Maliga, 2001; Ruhlman et al., 2007). Another approach incorporates the endogenous PSII protein D1 (psbA) 5′ and 3′ UTRs into transformation constructs (Verma et al., 2008). The potential of psbA 5′ UTR stems from its important role in plastids (Staub and Maliga, 1994). The PSII core protein D1 is a polytopic thylakoid membrane constituent with five membrane-spanning helices encoded by the plastid psbA gene (Marder et al., 1987). Expression of D1 is predominantly regulated at the level of translation and requires the participation of RBPs imported into plastids posttranslationally from the cytoplasm. PSII is highly susceptible to excessive light, and the primary target of the damage is D1. If the core protein is not efficiently removed and replaced, the result is impairment of electron transport, known as photoinhibition (Yamamoto, 2001). It is this cycle of turnover that makes the psbA 5′ UTR an attractive tool to enhance the level of foreign gene expression in transplastomic lines. The use of endogenous psbA regulatory elements has facilitated the generation of transplastomic tobacco lines with enhanced expression of a large number of soluble (Verma et al., 2008) and membrane (Singh et al., 2008) proteins, generating transplastomic lines conferring desired agronomic traits (Daniell et al., 2005; Bock, 2007; Verma and Daniell, 2007) or expressing biopharmaceutical proteins and vaccine antigens (Daniell et al., 2009b; Davoodi-Semiromi et al., 2009).
However, the majority of foreign proteins have been expressed in tobacco chloroplasts. Although heterologous sequences, commonly the tobacco psbA 5′ UTR, have been used to generate transplastomic plants from diverse genera, most cases thus far have been limited to first reports expressing only selectable marker genes, without useful agronomic traits or satisfactory levels of foreign protein accumulation (Verma and Daniell, 2007). In order to advance this field, efficient transformation of chloroplast genomes of several crop species should be accomplished. In addition, rapid and reproducible oral delivery systems expressing vaccine antigens, autoantigens, or biopharmaceuticals should be developed. There are two major limitations in accomplishing these goals. Development of a direct organogenesis system is key for the establishment of efficient and reproducible plastid transformation systems in crops regenerated via organogenesis. In addition, improvement in our understanding of the role of endogenous or heterologous regulatory sequences in transgene expression in plastids is essential. In this paper, we investigate both of these aspects. By optimizing different plant growth regulators (PGRs), we have developed a rapid and reproducible regeneration system for lettuce (Lactuca sativa). We used a bioinformatic approach to evaluate nucleotide variability in the regions upstream of chloroplast genes that comprise promoters and UTRs. We examined coding and noncoding sequences across 20 crop species representing most major clades of angiosperms, including four grasses and three legumes. We have used lettuce and tobacco transplastomic lines regulating transgenes with endogenous or heterologous regulatory elements to investigate RNA-protein interaction, foreign transcript accumulation and polyribosome association (polysome assay), and foreign protein accumulation. Our findings show that species-specific optimization of plastid transformation vectors and optimization of the growth hormone requirement will have a significant impact on foreign gene expression and transformation efficiency. These findings also underscore the need for sequencing complete crop chloroplast genomes.
RESULTS
Cultivars of Lettuce Respond Differently to PGRs and Regeneration Conditions
Five different lettuce commercial cultivars, Evola (LE), Great Lake (LG), Lentissima (LL), Romaine (LR), and Simpson Elite (LS), were evaluated for their organogenesis potential using the PGRs naphthaleneacetic acid (NAA) and 6-benzylaminopurine (BAP) either alone or in different combinations. Six leaf explants of 1 cm2 were placed on each petri plate, and five such plates were evaluated for each treatment. Direct shoots emerged from the leaf explants 21 d after culture in optimized regeneration medium. The shoot regeneration response was variable with different combinations of auxin (NAA) and cytokinin (BAP; Table I). We observed that auxin or cytokinin alone did not initiate shoot regeneration. A combination of auxin (NAA) and cytokinin (BAP) was necessary to induce direct shoot regeneration in different cultivars. Maximal direct shoot initiation was observed from the leaf explants cultured in medium supplemented with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP. Other combinations of PGRs induced callus formation and decreased shoot regeneration in all cultivars. Regardless of the cultivar studied, the medium supplemented with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP induced the most number of shoots. Therefore, we conducted three different sets of experiments in all the cultivars with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP to confirm the optimal levels of PGRs for direct shoot regeneration. LS showed the greatest regeneration potential (average 4.3 shoots per explant), whereas LL had the least response (0.43 shoots per explants). Regardless of the cultivar studied, the medium supplemented with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP induced the greatest number of shoots (Table I). The regeneration efficiency of other cultivars was LR (2.5), LG (1.5), and LE (1.2). Therefore, LS cultured on regeneration medium with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP was chosen for all of our transformation experiments.
Table I. Regeneration efficiency in lettuce cultivars.
Mean number of regenerated shoots from lettuce leaf explants cultured in media containing different combinations of the PGRs NAA and BAP. All concentrations of plant growth regulators are in μg mL−1. EFF, Number of regenerated shoots per explant.
| PGR | Evola (LE) |
Great Lake (LG) |
Lentissima (LL) |
Romaine (LR) |
Simpson Elite (LS) |
|||||
| Mean ± se | EFF | Mean ± se | EFF | Mean ± se | EFF | Mean ± se | EFF | Mean ± se | EFF | |
| NAA, 0.1 | 0 | 0 | 0 | 0 | 0 | |||||
| NAA, 0.05; BAP, 0.1 | 3.8 ± 0.9 | 0.63 | 3.8 ± 0.6 | 0.63 | 0.4 ± 0.12 | 0.06 | 7.2 ± 0.1 | 1.20 | 9.6 ± 0.1 | 1.60 |
| NAA, 0.05; BAP, 0.2 | 4.4 ± 0.0 | 0.73 | 4.2 ± 0.2 | 0.70 | 1.0 ± 0.21 | 0.16 | 8.4 ± 0.2 | 1.40 | 11.2 ± 0.4 | 1.86 |
| NAA, 0.1; BAP, 0.1 | 2.0 ± 0.1 | 0.33 | 5.0 ± 0.0 | 0.83 | 1.2 ± 0.2 | 0.20 | 6.0 ± 0.5 | 1.00 | 6.6 ± 0.1 | 1.10 |
| NAA, 0.1; BAP, 0.2 | 7.2 ± 0.4 | 1.20 | 8.8 ± 0.3 | 1.46 | 2.6 ± 0.1 | 0.43 | 15.2 ± 0.0 | 2.53 | 25.8 ± 0.9 | 4.30 |
| NAA, 0.2; BAP, 0.1 | 1.6 ± 0.1 | 0.26 | 1.4 ± 0.1 | 0.23 | 0 | 3.8 ± 0.0 | 0.63 | 3.6 ± 0.0 | 0.60 | |
| NAA, 0.2; BAP, 0.2 | 1.2 ± 0.9 | 0.20 | 1.8 ± 0.2 | 0.30 | 0 | 4.6 ± 0.4 | 0.76 | 4.8 ± 0.9 | 0.80 | |
| BAP, 0.1 | 0 | 0 | 0 | 0 | 0 | |||||
Upstream Sequence Variation Predominates in Plastid Genes Across Taxa
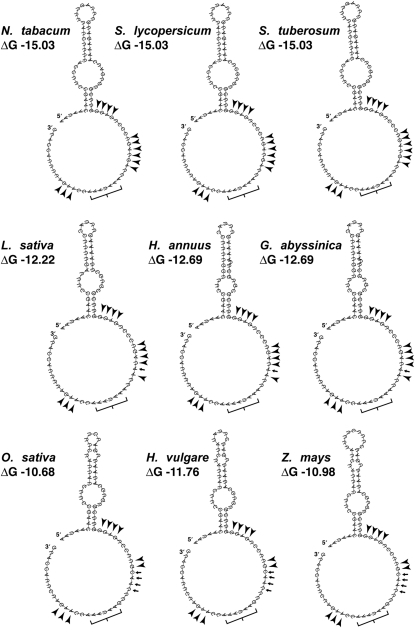
Sequences of intergenic spacer regions upstream from genes representing different functional groups were extracted from 20 complete plastid genomes representing most major clades of angiosperms, including four grasses and three legumes. Alignments were anchored by the inclusion of 100 bases from the coding regions of adjacent genes. Sequence identity was calculated for the region encompassed by 200 bases upstream of the translation start codon (i.e. promoters and UTRs). We observed that coding regions across all genera and genes display sequence identity of 80% to 97%, whereas the noncoding regions are 45% to 79%. In keeping with our findings throughout this study employing the psbA 5′ UTR as an experimental model system, we found that despite 95.0% identity in the coding region, identity in the psbA upstream region is 59% across all taxa (Fig. 1). The stem loop feature located at positions −49 to −71 in relation to the tobacco start codon has been shown experimentally to be an important determinant of translation efficiency (Eibl et al., 1999; Zou et al., 2003). Across all taxa, sequence identity for this element is 61%; tobacco and lettuce have 54.2% identity for this region.
Figure 1.
Nucleotide alignment of the psbA 5′ UTR. Intergenic spacers were extracted from complete plastid genomes of 20 species of angiosperm. Colored bases indicate agreements. Colored annotations below the tobacco sequence indicate regulatory elements (from 5′ to 3′): promoter and transcription start, purple; 5′ UTR terminal stem loop region, white; RBS, yellow (RBS3, R…; RBS2, full label; RBS1, R); AU box, green; translational start, blue (D). Identity across taxa is indicated by the histogram shown under the consensus sequence. Complete data from genomic analyses are available at http://chloroplast.cbio.psu.edu.
To determine how the nucleotide sequence variability in this region affects the secondary structure of the UTR, we used RNAfold (Gruber et al., 2008) to generate two-dimensional structure predictions based on minimum Gibbs free energy (ΔG; Fig. 2). Within families (Asteraceae, Solanaceae, Poaceae), stem and loop structures were highly similar, and predicted ΔG values showed little or no variation. However, among these three families, differences in nucleotide sequences result in structural and corresponding ΔG value variations. Relative to tobacco, the lettuce terminal stem and loop are reduced by one pair and three bases, respectively. Also, the base composition and conformation of the pairs within the stem are not conserved. Likewise, the stem bulge is reduced by two bases on each side, and the composition in this region is highly dissimilar. All alignments have been deposited in the Chloroplast Genome Database (http://chloroplast.cbio.psu.edu/) for public access.
Figure 2.
Theoretical predictions of secondary structure within the psbA 5′ UTR. The Vienna RNA Websuite tool, RNAfold, was used to produce two-dimensional structures for the psbA 5′ UTR for three representatives each from Solanaceae (top row), Asteraceae (middle row), and Poaceae (bottom row), based on minimum free energy. Unpaired bases upstream and downstream of the predicted stem and loop region are shown as circular, with 5′ and 3′ ends labeled accordingly; this representation is arbitrary and software dependent and is not intended as an indication of structure. ΔG values are given below the species names for each structure. Arrowheads indicate potential ribosome-binding sites (RBS). Small arrows indicate base changes in RBS relative to tobacco; these sites are identical within the families shown. Brackets indicate the AU box.
Plastid Transformation Vectors
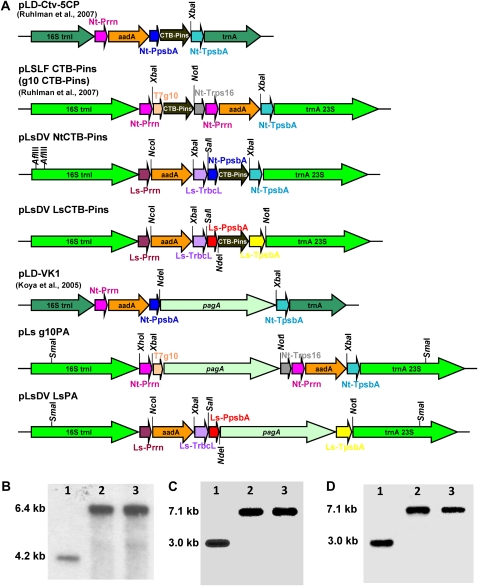
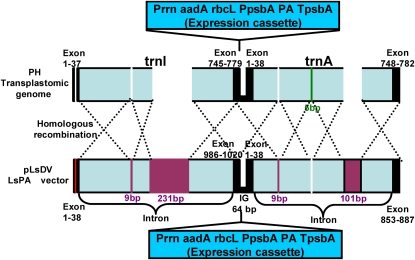
The pUC-based lettuce long flanking sequence vector was used to integrate foreign genes into the intergenic spacer region between the trnI and trnA genes as described previously (Ruhlman et al., 2007). The lettuce endogenous 16S Prrn and 3′ rbcL were amplified from the lettuce plastid genome and used to regulate the expression of the aadA gene from the GGAG ribosome-binding site for spectinomycin resistance. The aadA expression cassette was inserted into the spacer region between trnI and trnA and resulted in pLsDV vector. The pLsDV NtCTB-Pins (Fig. 3A) and pLsDV LsCTB-Pins plasmids were constructed by transferring the cholera toxin B-subunit-human proinsulin (CTB-Pins) expression cassette with tobacco or lettuce psbA regulatory sequences to pLsDV. The pLs g10PA (Fig. 3A) was constructed by replacing CTB-Pins with anthrax-protective antigen gene (pagA) in pLSLF CTB-Pins vector. The pLsDV LsPA (Fig. 3A) construct was made by cloning the pagA expression cassette with lettuce psbA regulatory sequences into the pLsDV vector. Abbreviations and regulatory elements for plants generated from different plasmids are given in Table II. The plasmids pLD VK1, pLD-Ctv-5CP, and pLSLF CTB-Pins used to generate Nt-Nt-PA, Nt-Nt-CP, and Ls-g10-CP plants have been reported previously (Koya et al., 2005; Ruhlman et al., 2007). In pLD-VK1 (Koya et al., 2005) and pLD-Ctv-5CP (Ruhlman et al., 2007), pagA and CTB-Pins, respectively were regulated by the tobacco psbA promoter and 5′ and 3′ UTRs. In pLSLF CTB-Pins (Ruhlman et al., 2007), CTB-Pins was driven by tobacco plastid Prrn from the 5′ translation control element of bacteriophage T7 g10.
Figure 3.
Schematic representation of expression cassettes and confirmation of homoplasmy by Southern hybridization. A, Schematic representation of the chloroplast transformation vectors. Ls, L. sativa; Nt, N. tabacum; Prrn, rRNA operon promoter; aadA, aminoglycoside 3′-adenylytransferase gene; TrbcL, 3′ UTR of rbcL; Trps16, 3′ UTR of rps16; g10, 5′ translation control element of bacteriophage T7 gene10; CTB-Pins, coding sequence of CTB subunit fused to human proinsulin; pagA, coding sequence of Bacillus anthracis protective antigen gene; PpsbA, promoter and 5′ UTR of psbA gene; TpsbA, 3′ UTR of psbA gene. B to D, Homoplasmic transformants generated for this study: Ls-Nt-CP (B), Ls-Ls-PA (C), Ls-g10-PA (D). Lane 1, The wild type (B, 4.2 kb; C and D, 3.0 kb); lanes 2 and 3, independent transplastomic lines (B, 6.4 kb; C and D, 7.1 kb). Five micrograms of total DNA was digested completely with AflIII for the Ls-Nt-CP blot and with SmaI for the Ls-Ls-PA and Ls-g10-PA blots.
Table II. Transplastomic lines utilized in various analyses.
Citations for lines that have been published previously are given in the right column. T7, Bacteriophage T7; T7 g10, T7 gene 10 translational control element.
| Line Name/Gene of Interest | Plasmid Name | Plant Background and Regulatory Elements for Gene of Interest | Regulatory Elements for Selectable Marker aadA | First Reported |
| Nt-Nt-CP/CTB-Pins | pLD-Ctv-5CP | Tobacco background with endogenous psbA 5′ and 3′ UTRs | Endogenous ribosomal operon promoter and GGAG ribosome-binding site | Ruhlman et al. (2007) |
| Ls-Ls-CP/CTB-Pins | pLsDV LsCTB-Pins | Lettuce background with endogenous psbA 5′ and 3′ UTRs | Endogenous ribosomal operon promoter, GGAG ribosome-binding site, and endogenous 3′ rbcL (TrbcL) | This work |
| Ls-Nt-CP/CTB-Pins | pLsDV NtCTB-Pins | Lettuce background with tobacco psbA 5′ and 3′ UTRs | Endogenous ribosomal operon promoter, GGAG ribosome-binding site, and endogenous 3′ rbcL (TrbcL) | This work |
| Ls-g10-CP/CTB-Pins | pLSLF CTB-Pins | Lettuce background with tobacco ribosomal operon promoter, T7 g10, and tobacco 3′ rps16 (Trps16) | Tobacco ribosomal operon promoter, GGAG ribosome-binding site, and tobacco psbA 3′ UTR | Ruhlman et al. (2007) |
| Nt-Nt-PA/PA | pLD-VK1 | Tobacco background with endogenous psbA 5′ and 3′ UTRs | Endogenous ribosomal operon promoter and GGAG ribosome-binding site | Koya et al. (2005) |
| Nt-Ls-PA/PA | pLsDV LsPA | Tobacco background with lettuce psbA 5′ and 3′ UTRs | Lettuce ribosomal operon promoter, GGAG ribosome-binding site, and lettuce 3′ rbcL (TrbcL) | This work |
| Ls-Ls-PA/PA | pLsDV LsPA | Lettuce background with endogenous psbA 5′ and 3′ UTRs | Endogenous ribosomal operon promoter, GGAG ribosome-binding site, and endogenous 3′ rbcL (TrbcL) | This work |
| Ls-g10-PA/PA | pLs g10PA | Lettuce background with tobacco ribosomal operon promoter, T7 g10, and tobacco 3′ rps16 (Trps16) | Tobacco ribosomal operon promoter, GGAG ribosome-binding site, and tobacco psbA 3′ UTR | This work |
Generation of Transplastomic Lines and Confirmation of Homoplasmy
Plastid transformation of lettuce and tobacco was carried out as described (Daniell et al., 2005; Ruhlman et al., 2007), with modifications of regeneration media as described above. In lettuce, six to seven 21-d-old, fully expanded leaves (approximately 4 cm2) were placed per plate for bombardment. On average, five to six primary regenerants were obtained per 10 bombardments. Most importantly, shoots were formed from the leaf explants without the formation of callus. Primary regenerants identified by PCR were subjected to an additional round of regeneration followed by rooting in selective media. Site-specific integration and homoplasmy of the transplastome were confirmed by Southern hybridization with flanking sequence probe specific for lettuce or tobacco. Generation and confirmation of homoplasmy for CTB-Pins lines Nt-Nt-CP and Ls-g10-CP and PA line Nt-Nt-PA have been reported previously (Koya et al., 2005; Ruhlman et al., 2007). All newly generated transplastomic lines were found to be homoplasmic, containing no detectable wild-type copies of the plastome (Fig. 3, B–D).
Homologous Recombination between Lettuce and Tobacco DNA Sequences
We analyzed the nucleotide sequence in the regions flanking the transgene cassette integration site from both of the Nt-Ls-PA transplastomic lines obtained. The tobacco transformants were generated using the pLsDV LsPA construct containing the lettuce flanking sequence. Comparison of tobacco wild-type plastid DNA sequence in the trnI and trnA exon regions corresponding to vector pLsDV LsPA uncovered a single base-pair deletion. Absent in the transplastomic lines were DNA stretches of 101, 231, and two of 9 bp that were present in the vector DNA. Alignment of this region to the lettuce chloroplast genome identified the unique sequences in the trnI and trnA introns (Fig. 4, purple bars) as those that were eliminated in tobacco transplastomic lines. Even though the chloroplast vector used for transformation contained these additional sequences, we did not detect any of the sequences unique to the lettuce trnI exon (19th base), trnI intron (9 and 231 bp), or trnA intron (9 and 101 bp) in the Nt-Ls-PA transplastomic lines. All unique sequences were deleted by looping out during recombination. Looping out of unique lettuce sequences in transplastomic lines was supported by observed identical DNA sequences in both lines. The nonhomologous species-specific nucleotide sequence deletions in transplastomic lines were flanked on either side by regions sharing complete homology. These differences were generated by up to eight recombination events (Fig. 4). Even though all sequences unique to the lettuce chloroplast genome were eliminated, a 5-bp sequence unique to the tobacco chloroplast genome was not looped out during the recombination process. Nucleotide sequence analysis did not show any illegitimate recombination in the regions analyzed. Use of lettuce flanking sequence for transformation of the tobacco chloroplast genome significantly reduced the transformation efficiency, probably due to complex homologous recombination events. While the use of pLD vectors with tobacco flanking sequence under optimal conditions generated up to 15 independent events in 10 bombardments, only two independent transplastomic lines were obtained with 30 bombardments when the pLsDV LsPA vector was used for bombardment. However, the nucleotide sequences of the expression cassette in the bombarded chloroplast expression vector and the Nt-Ls-PA transplastomic chloroplast genome were identical, confirming the absence of any recombination or modification of the psbA regulatory elements (data not shown).
Figure 4.
Schematic representation of homologous recombination between the tobacco transplastomic genome and the lettuce transformation vector. Total DNA was isolated from the tobacco Nt-Ls-PA transplastomic lines, sequenced using appropriate primers, and aligned with tobacco and lettuce sequences from the National Center for Biotechnology Information database. A unique lettuce intron sequence of the transformation vector is indicated by purple bars. A unique tobacco intron sequence of the plastome is indicated by the green bar. Blank indicates looped out sequence(s). Black bars indicate the exon region.
Decrease of Foreign Protein Accumulation with Heterologous Regulatory Elements
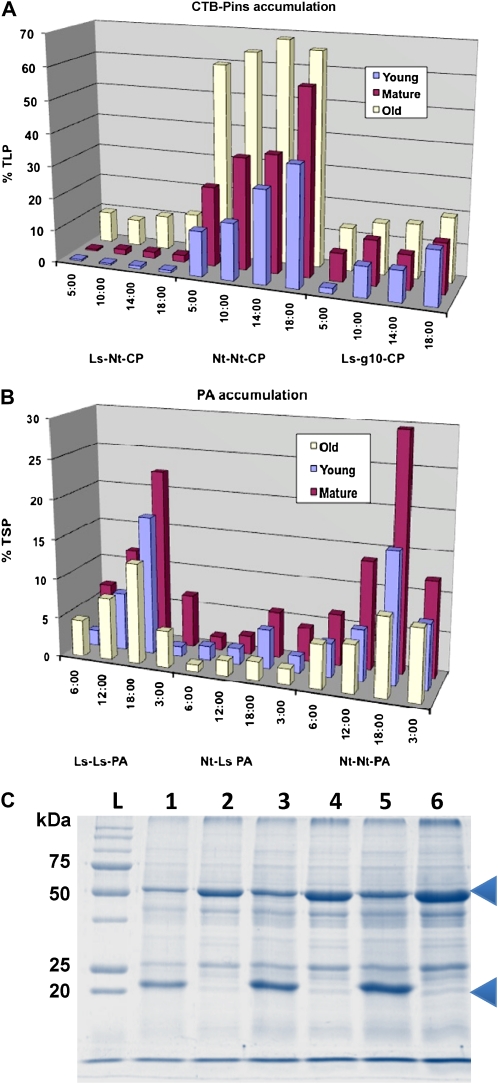
Second generation (T1) transplastomic homoplasmic plants of lettuce and tobacco expressing CTB-Pins or PA were grown in the greenhouse for 8 to 10 weeks. Leaves of different developmental stages were harvested at four time points during the light cycle to evaluate foreign protein accumulation. Expression of CTB-Pins was quantified by densitometric analysis of crude homogenates against known quantities of CTB standard (Fig. 5A). For these quantitative studies, homogenates have been used, as we found that up to 90% of CTB-Pins protein is retained in the pelleted fraction following centrifugation. We modified our protein extraction and SDS sample loading buffer to include 100 mm dithiothreitol and increased the buffer-to-tissue ratio to enhance the solubility of CTB-Pins. In all three lines analyzed, the older leaves showed the highest levels of CTB-Pins accumulation, up to 72% of total leaf protein (TLP) in the tobacco line Nt-Nt-CP, 12.3% of TLP in Ls-Nt-CP, and approximately 25% of TLP in Ls-g10-CP. To confirm this estimate for tobacco, we quantified samples by densitometric analysis of Coomassie Brilliant Blue-stained gels. We estimated that in Nt-Nt-CP lines, CTB-Pins accumulation exceeds that of Rubisco by a factor of up to 2 (Fig. 5C), yet no deleterious phenotype was observed, likely as this level of accumulation was attained over time (Bally et al., 2009; Oey et al., 2009). It appeared that the observed reduction in Rubisco large subunit was concomitant with increased levels of CTB-Pins accumulation, as wild-type tissue sampled at the same light and developmental stages showed no such reduction in Rubisco. In mature leaves harvested at 6 pm, the lettuce line accumulated CTB-Pins to 2.2% ± 0.8% of TLP, whereas the tobacco line reached 57% ± 2.1%, a 96% reduction in foreign protein, despite the fact that both of these lines express CTB-Pins from the tobacco psbA 5′ UTR. CTB-Pins accumulation was determined for a mature T0 lettuce line (Ls-Ls-CP) at the same point in the light cycle. This line, which has the endogenous psbA regulatory elements, expressed CTB-Pins to 24.0% ± 3.6% of TLP, 11-fold more than the Ls-Nt-CP line. The expression pattern observed in Nt-Nt-CP young and mature leaves was consistent with developmental and light regulation, with increasing levels of foreign protein as maturing leaves synthesize and accumulate proteins throughout the light cycle. Minimal differences observed in the Ls-Nt-CP and Ls-g10-CP lettuce lines were not consistent with developmental stage or harvest time and therefore were attributed to the overall physiological condition of the plants and experimental variation.
Figure 5.
Accumulation of foreign protein in transplastomic lettuce and tobacco. A and B, CTB-Pins accumulation estimated by densitometry (A) and PA accumulation estimated by ELISA (B) presented as a function of light and developmental stage. Highest accumulation is shown in the back rows. The order of young, mature, and old is different in A and B because of the accumulation of higher CTB-Pins in older leaves and PA in mature leaves. The bars of the histograms represent means of at least three independent determinations. C, SDS-PAGE stained with Coomassie Brilliant Blue. Lanes 1, 3, and 5, 10, 20, and 30 μg of TLP from Nt-Nt-CP older leaf at 6:00 pm; lanes 2, 4, and 6, corresponding amounts of wild-type protein extract; lane L, molecular mass standards. Arrowheads indicate positions of CTB-Pins (22 kD) and Rubisco (53 kD).
Expression of PA in transplastomic tobacco and lettuce was determined by ELISA of young, mature, and older leaves. PA was mostly observed in the soluble fraction; therefore, quantitation by ELISA was accurate. The maximum PA expression was observed in mature leaves compared with young or older leaves in all the lines examined. In mature leaves harvested at 6 pm from lettuce and tobacco lines, PA accumulation reached 22.4% ± 1.0% and 29.6% ± 0.9% of the total soluble protein (TSP), respectively, when PA expression was regulated by endogenous regulatory elements. In Nt-Ls-PA, the foreign protein represented 5.8% ± 0.1% of TSP in mature leaves at 6 pm, an 80% reduction of expression in tobacco (Fig. 5B). In order to investigate whether this reduction in expression level was the result of any deletion or modification of the regulatory elements or coding sequence of the PA expression cassette, DNA from both of the independent Nt-Ls-PA lines was isolated and the nucleotide sequence of the expression cassette was determined. As mentioned above, the nucleotide sequence of the PA expression cassette in the bombarded chloroplast expression vector and the Nt-Ls-PA transplastomic chloroplast genome was identical (data not shown), confirming the absence of any recombination or modification of the psbA regulatory elements or the PA coding sequence. Like the CTB-Pins lines, PA accumulation followed predictable developmental and light-regulated patterns when endogenous psbA regulated transgene expression. CTB-Pins and PA accumulated foreign gene products differently in older leaves. While CTB-Pins was present at the highest levels, PA expression was the lowest in older leaves. This difference was probably due to protection from proteolytic degradation conferred by the aggregation of CTB-Pins.
Decrease of Foreign Gene Transcripts with Heterologous Regulatory Elements
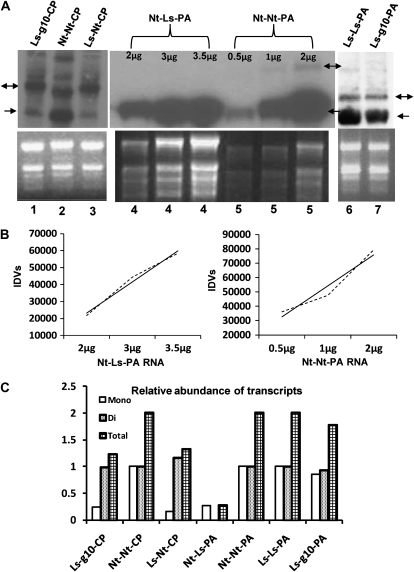
Total RNA was isolated from different transplastomic lines, and northern blots were prepared to examine the transcript populations generated from various regulatory elements. Northern blots were probed with radiolabeled coding regions for CTB-Pins (Fig. 6A, lanes 1–3, for Ls-g10-CP, Nt-Nt-CP, and Ls-Nt-CP) and PA (Fig. 6A, lanes 4–7, for Nt-Ls-PA, Nt-Nt-PA, Ls-Ls-PA, and Ls-g10-PA). The relative abundances of CTB-Pins and PA transcript species were assessed by densitometry (Fig. 6C). In order to ensure the validity of quantitation using densitometry, different concentrations of RNA were loaded and linearity was observed between loaded RNA concentration and observed transcript signals. Plots of integrated density values with increasing concentrations of RNA are shown in Figure 6B to show the linearity and validity of the densitometric quantitation. Ethidium bromide staining of RNA gels is also shown in the lower frame of Figure 6A as an indicator of equal loading. All of the plants under analysis carry two engineered promoters, one upstream of aadA and one upstream of the gene of interest, allowing for transcription of monocistrons and also dicistrons arising from RNA polymerase read-through of the upstream 3′ UTR element where present. While the accumulation of dicistronic mRNA for CTB-Pins varied minimally (Fig. 6A, lanes 1–3), tobacco plants with endogenous psbA UTR (Fig. 6A, lane 2) expressed 84% more monocistronic mRNA and 66% total mRNA for CTB-Pins than lettuce plants with the heterologous tobacco psbA UTR (Fig. 6A, lane 3). As seen in transplastomic lines, where genes are integrated within the ribosomal operon, a number of larger species were detectable using the CTB-Pins probe but were not quantified for this analysis. In PA lines where endogenous 5′ psbA was used, monocistronic transcript (Fig. 6A, lane 5 with 2 μg of RNA) was elevated by 72% over those where heterologous psbA UTR was used as the regulatory element (Fig. 6A, lane 4 with 2 μg of RNA). In lettuce PA lines, accumulation of dicistronic mRNA varied minimally, whereas lettuce plants with endogenous psbA UTR expressed 14% more monocistronic mRNA than lettuce Ls-g10-PA plants (Fig. 6A, lanes 6 and 7).
Figure 6.
Northern blotting of total RNA. To examine foreign transcript abundance in transplastomic lines, 2 μg of total RNA (except for Nt-Ls-PA and Nt-Nt-PA) was separated by electrophoresis, blotted to nylon membranes, and probed with radiolabeled CTB-Pins or PA fragment. A, Top panel, autoradiographs; bottom panel; ethidium bromide-stained rRNA. Monocistrons are indicated by one-headed arrows, and dicistrons are indicated by two-headed arrows. B, Plots of integrated density values (IDVs). Broken lines show data points, and solid lines show trend lines. C, Densitometric estimation of signal intensity was used to calculate relative abundance of foreign transcripts in lines with heterologous regulatory elements when compared with those with endogenous elements. Lines with endogenous elements were assigned a value of 1. Mono, CTB-Pins or PA monocistron; Di, aadA and CTB-Pins or PA dicistron; Total, Mono and Di combined values.
Foreign Gene Transcripts with Endogenous 5′ UTR Were Stabilized in Nonpolysomal Fractions
Total RNA was prepared from fractions separated through Suc gradients to evaluate the polysome association of foreign gene transcripts in lettuce transformants that express CTB-Pins from the endogenous psbA UTR (Ls-Ls-CP) versus the tobacco psbA 5′ UTR (Ls-Nt-CP). Fractions were collected as 12 aliquots of 500 μL each from the bottom of the gradient and numbered 1 through 12 accordingly. Polysome-associated transcripts are expected to migrate with higher molecular mass translation complexes into the lower fractions, while transcripts not associated with polysomes should remain in the upper fractions. The number of ribosomes loaded per mRNA at a given time is variable, such that polysome association may be ascribed to several fractions collected from the lower portion of the gradient. For our analysis, we have designated fractions 2 through 7 as polysome-associated and fractions 8 through 12 as nonpolysomal. Ethidium bromide-stained gels showing ribosomal RNA profiles indicated that at the time of sedimentation, polyribosome integrity was intact. It was seen that fractions from the lower portion of the gradients are well distributed with rRNA and that all fractions showed proportions of rRNA that would be expected in the various fractions of the density gradient (Fig. 7). The rRNAs may migrate this depth in the gradient (15%–55% Suc) only when complexed as polyribosomes.
Figure 7.
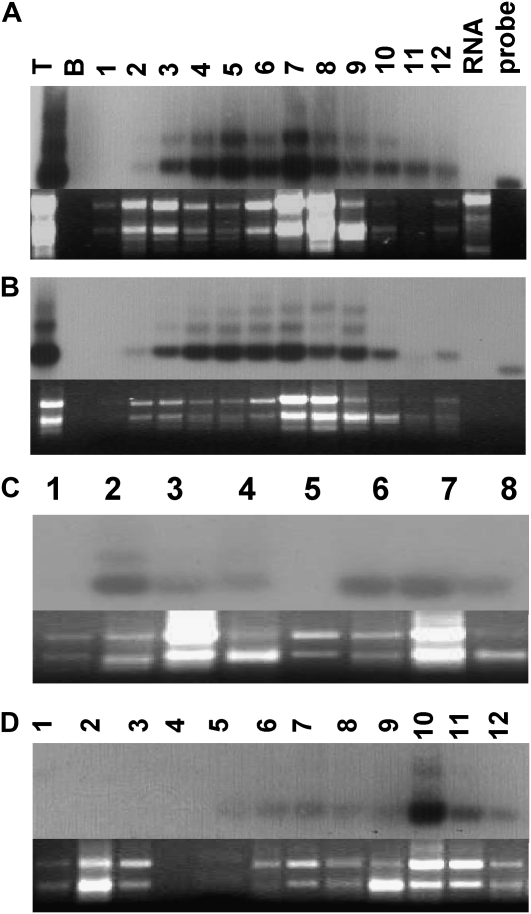
Polysome assay. Suc gradient fractions were separated through 1.2% agarose and transferred for northern blotting with the CTB-Pins probe. Lanes are numbered above A for both A (Ls-Ls-CP) and B (Ls-Nt-CP). Lane T, Total RNA; lane B, blank; lanes 1 to 12, fractions 1 to 12 collected from the bottom of the gradient; lane RNA, RNA standards (in A; bands are 0.5, 1, 2, and 3 kb); lane probe, CTB-Pins probe. C, Lanes 1 to 4 and 5 to 8, fractions 1, 4, 8, and 11 from Ls-Nt-CP and Ls-Ls-CP, respectively. D, Controls. Lanes 1 to 3, Pooled fractions from the wild-type sample; lane 4, blank; lanes 5 to 7, each lane contains two pooled factions from 2 to 7 of puromycin-treated sample (Ls-Ls-CP); lanes 8 to 12, fraction corresponds to lane number. Transcripts identified in fractions 2 to 7 are considered to be polysome associated; fractions 8 to 12 are considered nonpolysomal. Ethidium bromide-stained agarose gels are shown below each blot.
The distribution of ribosomal RNAs in gradient fractions was similar between these two lines. Northern blots of 12 fractions collected from the bottom of the gradient were probed with the radiolabeled, full-length coding sequence for CTB-Pins to localize foreign transcripts. Blots prepared from all fractions of the gradient for Ls-Ls-CP or Ls-Nt-CP lines show that the CTB-Pins transcript present in all fractions was predominantly monocistronic, although the dicistron was readily detectable and in some fractions abundant (Fig. 7, A and B). One larger CTB-Pins mRNA species was observed only in Ls-Nt-CP lines; this likely corresponds to a processed intermediate originating from the endogenous Prrn upstream of the insertion site. Foreign mRNA is more abundant in Ls-Ls-CP lines in the upper fractions, suggesting that the transcript pool not associated with polysomes is stabilized in these lines (Fig. 7A). To account for the variation in transfer and hybridization efficiency that can confound blot-to-blot comparisons, selected fractions were prepared from an independent isolation of these two lines and were blotted together to confirm this observation (Fig. 7C). Also, the ethidium bromide-stained gel image showed that the rRNAs were equally abundant in the fractions of the Ls-Ls-CP and Ls-Nt-CP lines, facilitating a direct comparison of the samples and supporting the results observed in Figure 7, A and B. Samples from untransformed plants and puromycin-treated controls are shown in Figure 7D. Puromycin is known to cause disruption of polyribosomes by premature chain termination, leading to a shift of transcripts out of the polyribosome complex. This antibiotic is commonly used as a control to demonstrate that the signal detected in lower (polysome-associated) fractions is accurate; the addition of puromycin should cause the same sample to fractionate all transcripts, including the gene of interest, into the upper (polysome-free) region of the gradient after high-speed centrifugation. Autoradiographs were used for densitometry. In the Ls-Nt-CP line, 22% to 37% of total signal was associated with the two nonpolysomal fractions, whereas in the Ls-Ls-CP line, 40% to 65% of the total signal was found in these fractions.
Stromal RBPs Preferentially Associate with Endogenous psbA 5′ UTR, Providing Transcript Stability
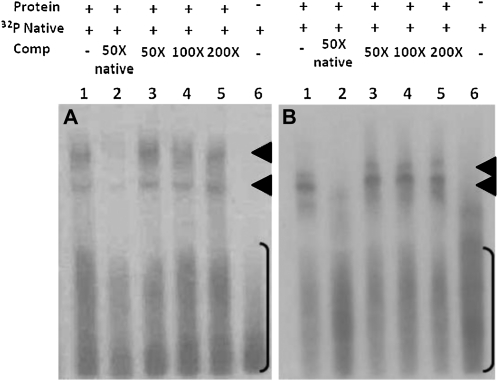
Extracts of plastid stromal proteins were prepared for lettuce and tobacco as described in “Materials and Methods.” Labeled, full-length psbA 5′ UTRs specific to each system were transcribed in vitro, as were the unlabeled competitors. Reactions were separated on native gels to evaluate the affinity of endogenous UTRs for stromal protein binding in the presence of up to 200-fold molar excess of competitor species. As shown in Figure 8, in the lettuce and tobacco systems, heterologous (unlabeled) psbA 5′ UTR was an ineffective competitor for the association with RBPs in the stromal extracts. Two bands were observed in positive control reactions (lane 1) that were absent in reactions that did not contain stromal proteins (lane 6). The complexes migrated to the apparent molecular mass of 150 and 100 kD in lettuce extracts and 100 and 80 kD in tobacco. The complexes formed between stromal proteins and labeled UTRs were disassociated by 50-fold molar excess of unlabeled endogenous UTR but not by 50-, 100-, or 200-fold excess of unlabeled heterologous psbA UTR. This result was consistent over three independent experiments in both backgrounds.
Figure 8.
RNA EMSA competition assay. Stromal proteins isolated from lettuce (A) or tobacco (B) were incubated with radiolabeled endogenous psbA 5′ UTR (lane 1). Competitions included 50× unlabeled endogenous psbA 5′ UTR (lane 2) and 50×, 100×, or 200× unlabeled heterologous psbA UTR (tobacco in A and lettuce in B; lanes 3–5, respectively). Lane 6 shows labeled probe. Brackets indicate free probe, and arrowheads indicate complexes associated with labeled RNA.
DISCUSSION
We report here a rapid and reproducible method of chloroplast transformation in lettuce. A plant system having direct shoot regeneration (without forming callus), a species-specific vector with endogenous regulatory elements, optimized DNA delivery, age of the explants, and culture conditions were crucial in the optimization of lettuce chloroplast transformation. We found that lettuce cv Simpson Elite had the highest direct shoot regeneration potential, with maximum number of shoots (4.3), among the five cultivars examined in this study. The ability to produce direct shoot regeneration from somatic cells varies among the different cultivars of lettuce (Xinrun and Conner, 1992). The induction of direct shoots was highly dependent on the auxin and cytokinin ratios. Auxin or cytokinin alone did not induce shoot formation in any of the analyzed cultivars of lettuce. Leaf explants incubated on medium supplemented with 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP induced the maximum number of shoots in all cultivars, confirming that the optimum level of auxin-cytokinin balance is essential to induce the maximum number of shoots in all the cultivars examined. Two types of organogenesis are known. In “direct organogenesis,” the explants undergo a minimum proliferation before forming an organ such as shoot or root, whereas in “indirect organogenesis,” explants undergo an extensive proliferation before it develops into shoot or root (Ovecka et al., 2000) and often requires redetermination of differentiated cells (dedifferentiation; George et al., 2008). The lettuce chloroplast transformation system reported here is established through direct organogenesis and therefore is more rapid than the indirect organogenesis established through callus reported previously (Lelivelt et al., 2005; Kanamoto et al., 2006). Regeneration and efficiency of this system are comparable to tobacco, five to six transplastomic shoots per 10 bombardments, the most efficient transplastomic system developed so far. Such a rapid and reproducible transformation system has already facilitated the expression of several vaccine antigens or autoantigens in lettuce chloroplasts (Davoodi-Semiromi et al., 2009).
A survey of the literature demonstrates that there is considerable interest in chloroplast transformation, both in the established system of tobacco and in crop species that have been more recalcitrant to transformation by this approach. Constructs designed and tested for tobacco transformation have been successfully used for potato (Solanum tuberosum) and tomato (Solanum lycospersicum) plastid transformation, but the plastid transformation frequency was considerably lower than in tobacco (Sidorov et al., 1999; Ruf et al., 2001). The lower transformation efficiency observed in potato could be the result of poor homologous recombination with the tobacco flanking sequences. A similar lower efficiency was observed when Petunia flanking sequences were utilized to transform the tobacco chloroplast genome (DeGray et al., 2001), confirming that lower homology reduces the transformation efficiency. In this study, use of lettuce flanking sequence resulted in much lower transformation efficiency of the tobacco chloroplast genome, probably due to the presence of stretches of heterologous sequences in the lettuce trnI/trnA region. None of the earlier studies on integration of foreign genes investigated the exchange of DNA sequences in transplastomic genomes using heterologous flanking sequences. In this study, we did not detect any of the sequences unique to the lettuce chloroplast genome in the trnI and trnA region of the transplastomic tobacco chloroplast genome, confirming their elimination during homologous recombination events. However, in another study, the Nicotiana plastid transformants obtained using Solanum plastid sequences showed the presence of mutations conferring spectinomycin and streptomycin resistance from the Solanum plastid genome, but the authors did not study foreign gene integration or determine nucleotide sequences of transplastomic genomes but used RFLP to study recombination (Kavanagh et al., 1999). In our investigations, all sequences unique to the lettuce chloroplast genome were eliminated, but a 5-bp sequence unique to the tobacco chloroplast genome was not looped out during the recombination process, suggesting that the recombination events are determined by the recipient chloroplast genome. The most important factor for homologous recombination is the extent and degree of sequence similarity between the recombining fragments and is mediated by recombination/repair enzymes. The RecA protein is mainly responsible for facilitating recombination, and its homolog has been identified in land plant plastids (Cerutti et al., 1992, 1993; Inouye et al., 2008). Studies demonstrate a log-linear relationship between recombination rate and the degree of sequence divergence, independent of the activity of the mismatch repair system (Shen and Huang, 1986; Majewski and Cohan, 1998). Chloroplast transformation studies in Chlamydomonas reinhardtii indicated that higher levels of homologous recombination require at least 150- to 200-bp sequence similarity (Newman et al., 1990, 1992; Suzuki et al., 1997). Furthermore, the ratio of heterologous and homologous regions between the recipient chloroplast genome and the donor insert influence the frequency of exchange events between chloroplast genomes (Newman et al., 1990). The stretches of heterologous DNA fragments in the recombining flanking sequences present a barrier to recombination and lead to lower chloroplast transformation efficiency.
More than 20 reports describe the generation of transplastomic crop plants through the use of species-specific flanking regions to facilitate homologous recombination between the shuttle vector and the plastid genome. Despite the recognized need for customized integration sequences, the expression cassettes used in these experiments more commonly contain heterologous regulatory elements. For example, rice (Oryza sativa) psbA 5′ and 3′ UTRs were used to express interferon-γ in transplastomic tobacco (Leelavathi and Reddy, 2003), resulting in expression levels similar to those reported for heterologous regulatory sequences in our study. Although a quantitative evaluation of Cry1Ab protein was not reported, only a single transplastomic event was achieved in soybean (Glycine max) plastid transformation using heterologous regulatory sequences (Dufourmantel et al., 2005). In both of these studies, the use of heterologous sequences was motivated by concern for potential recombination with endogenous regulatory elements; indeed, in these examples, there was no evidence of recombination between introduced sequences and the plastome. While these spurious rearrangements occasionally can occur and are interesting, they are not common enough to preclude the application of endogenous regulatory elements for the purpose of achieving the highest levels of transgene expression. In this study, we have utilized untransformed lettuce and tobacco plants along with a suite of transplastomic lines to investigate potential mechanisms that underlie the variation we have observed in foreign protein accumulation resulting from the use of endogenous or heterologous regulatory elements. Our findings suggest that these differences may be influenced at multiple levels, including transcription, mRNA processing, mRNA stability, and translation of foreign gene products.
We examined the relative abundance of foreign gene transcripts in total RNA extractions of transplastomic lines and found the greatest variation in the monocistron pools in the different CTB-Pins lines. Sequence alignments reveal that there is 100% identity between lettuce and tobacco in the region of the ribosomal operon P1 promoter required for full PEP activity (−64 to +17; Suzuki et al., 2003). This is supported by similar levels of dicistron accumulation in the lettuce and tobacco lines, where transcription of this mRNA species is driven by heterologous or endogenous Prrn. These data support the findings of Sriraman et al. (1998) that heterologous Prrn P1 can facilitate transcription in transplastomic plants. Although our observation was consistent for Ls-Ls-PA and Ls-g10-PA lines, in the Nt-Ls-PA line, which carries the lettuce Prrn, both the dicistron and monocistron were reduced. The core promoter elements shown to be sufficient for developmental regulation of psbA transcription are 90% identical between lettuce and tobacco (−42 to +9; Hayashi et al., 2003). Therefore, it seems unlikely that the differences we observed in the foreign transcript pool resulted directly from an inability to achieve PEP-mediated synthesis due to divergence in promoter sequence and structure in the heterologous system (i.e. Ls-Nt-CP).
Based on our sequence comparisons, the greatest variability upstream of the D1 coding region lies within the proposed stem loop structure of the 5′ UTR that has been shown to be involved in transcript stability and translation efficiency in land plants (Alexander et al., 1998; Eibl et al., 1999; Shen et al., 2001; Zou et al., 2003). This variation in the nucleotide sequence of the stem loop motif corresponds to variation in its structure and thermodynamic properties. Over our entire sample set, this region has a 61% sequence identity, and for lettuce and tobacco, the identity is 54.2%. Our results from polysome analyses indicated that foreign transcripts in lettuce lines that carried the endogenous 5′ UTR were more abundant in the upper fractions, giving some insight into the differences in foreign protein accumulation in the various lines used in this study.
It has been shown previously that the majority of psbA transcripts are not polysome associated in barley (Hordeum vulgare), spinach (Spinacia oleracea), and tobacco (Klein et al., 1988; Minami et al., 1988) and that this population is likely stabilized by RBPs in the stroma (Nakamura et al., 1999, 2001). A number of studies have investigated the cis-elements within the land plant psbA 5′ UTR to elucidate sequences that are required for the association of RBPs and the roles of these interactions in transcript stability, processing, and initiation of translation on plastid ribosomes. Using synthetic psbA 5′ UTRs with specific site mutations and internal deletions, it was determined that the stem loop region was a dispensable element, as its exclusion did not affect the translation of the fused lacZ reporter in 30S preparations from tobacco chloroplasts. The authors identified an AU-rich element (AU box) located between RBS1 and RBS2 that together were required and sufficient to initiate translation and proposed a role for the AU box as the primary target sequence for the binding of trans-acting factors (Hirose and Sugiura, 1996). Our sequence analysis, combined with the results from RNA electrophoretic mobility shift assay (EMSA), suggest that the AU box and adjacent RBS sites are not responsible for differences observed in our experimental system. Between lettuce and tobacco, there was sequence identity of 95% over this region (20 bp), the variation generated by a single base change in RBS2. We did find, however, that this region has diverged in a subset of species, including representatives from each of the clades in our analysis, which have variable TA (AU) insertions at the 3′ end of the AU box (Fig. 1).
Subsequent analyses have suggested the presence of an endonucleolytic cleavage site that is protected upon the binding of protein factors. This site is localized to the predicted stem loop region of the spinach psbA 5′ UTR (−49/−48 relative to the start of translation), and the binding interaction is sensitive to minute changes in secondary structure (Klaff et al., 1997; Alexander et al., 1998). Using both biochemical and theoretical approaches, Klaff and coworkers (1997) demonstrated that temperature-dependent structural transitions in the stem loop region either facilitated or ablated the binding of RBPs. The differences we have observed in this region between tobacco and lettuce result in a number of changes that could inhibit the interaction of RBPs, including base composition and size of the stem, bulge, and terminal loop. It is likely that these differences are iterated into changes in three-dimensional conformation as well.
Additional support for the role of the stem loop structure in the stabilization of transcripts that include the psbA 5′ UTR is provided from transplastomic studies. Tobacco plants were generated that express uidA (GUS) mRNA from chimeric genes where transcription was driven by Prrn with the full-length endogenous psbA 5′ UTR (85 bases) or mutation/deletion variants of the stem loop region. In all experimental constructs, mRNA abundance and corresponding translation products were reduced compared with the control, despite transcription from identical promoters (Zou et al., 2003). As in these examples, our data demonstrate that it is possible to accumulate foreign protein, albeit with reduced efficiency, with heterologous psbA UTR, due to the presence of ribosome-binding sites and the AU box downstream of the stem loop region.
RNA EMSA assays using wild-type stromal extracts demonstrated that the heterologous psbA 5′ UTR was not an effective competitor for binding factors that may be involved in transcript stability. We attribute this strongly preferential binding of the endogenous UTR to the stem loop structural element, as this is the primary region where significant sequence variation exists between lettuce and tobacco. Conceivably, in planta, the ability of foreign gene transcripts equipped with heterologous UTR elements to compete for stabilization factors would be hampered by the presence of abundant endogenous psbA transcripts, leading to rapid turnover of foreign RNA species in the plastid.
We have further evaluated the species-specific nature of protein factor binding to the psbA 5′ UTR as a mechanism influencing foreign protein accumulation in transplastomic lines. We found that exchange of the full-length UTR between lettuce and tobacco resulted in a reduction in CTB-Pins and PA expression of at least 97% and 80%, respectively. This effect was consistent in young and mature leaves sampled at four time points. CTB-Pins accumulated to much higher levels in older, but not senescent, leaves of Ls-Nt-CP plants, but this could not compensate for the differences in overall expression, as accumulation still showed a reduction of approximately 85% compared with tobacco plants with the endogenous UTR construct. PA lines did not accumulate foreign protein in older leaves to the same levels as CTB-Pins lines (Fig. 5). This is most likely due to differences in the solubility of the two proteins.
We estimate that CTB-Pins in fully expanded leaf tissue constituted 57% to 58% of the TLP when harvested near the end of the light period and reached as high as 72%. The use of endogenous psbA 5′ UTR has led to the accumulation of numerous gene products in transplastomic tobacco (Verma et al., 2008), including proteins that had been previously unattainable at satisfactory levels using this technology (Fernandez-San Millan et al., 2003; Dhingra et al., 2004; Singh et al., 2008). We have now generated transplastomic lettuce plants that accumulate abundant foreign protein. While the expression of the first therapeutic protein produced in lettuce was accomplished using the tobacco Prrn g10 system for the expression of the gene of interest and the selectable marker (Ruhlman et al., 2007), the implementation of lettuce-specific regulatory elements has contributed to the development of a highly reproducible transformation system that generates transplastomic lettuce plants expressing foreign proteins to high levels.
CONCLUSION
There are a great many factors to consider when designing transformation vectors for the generation of transplastomic lines, particularly when the target is a new species for which there is no standardized approach. The use of species-specific integration sequences (Verma and Daniell, 2007), codon optimization for plastid expression (Tregoning et al., 2003; Daniell et al., 2009a), and the inclusion of N-terminal stabilization sequences or fusion proteins have been essential to the development of transplastomic lines (Ye et al., 2001; Leelavathi and Reddy, 2003; Ruhlman et al., 2007). The emergent study of pentatricopeptide repeat proteins in addition to well-established research on RBPs have revealed that the interactions between protein factors and their cognate RNA sequences are highly specific. Our evaluation of UTR sequences from taxonomically diverse species for genes representing the various functional groups found in plastids combined with our experimental findings using the psbA 5′ UTR in particular argue for the use of species-specific regulatory elements for significant accumulation of foreign protein in transplastomic plants.
Similarly, the use of homologous flanking sequences significantly enhances transgene integration. However, comparison of intergenic spacer regions among four sequenced Solanaceae members revealed that only four of 150 spacer regions had 100% sequence identity (Daniell et al., 2006). Comparison of nine Poaceae chloroplast genomes revealed no intergenic spacer regions with 100% sequence identity (Saski et al., 2007). These studies point out the importance of using appropriate endogenous intergenic spacer regions for plastid transformation. While several hundred noncrop chloroplast genomes have been sequenced for phylogenetic studies or are in progress, no major effort has focused on sequencing crop chloroplast genomes for biotechnology applications or basic studies. This underscores the need for determining complete chloroplast genome sequences of crop species, because more than 45% of the chloroplast genome contains regulatory elements and intergenic spacer regions required for chloroplast transformation studies (Daniell et al., 2006; Saski et al., 2007). It is equally important to optimize regeneration and tissue culture conditions by examining growth hormone requirements to achieve reproducible and rapid transformation of crop species.
MATERIALS AND METHODS
Optimization of Direct Shoot Regeneration
Seeds of lettuce (Lactuca sativa) cultivars (New England Seed) were disinfected in a solution of 30% commercial bleach with 0.01% (v/v) Tween 20 for 5 min, rinsed five times in sterile water, and placed on half-strength Murashige and Skoog (MS; Murashige and Skoog, 1962) medium solidified with 0.6% (w/v) Phytablend (Caisson). After 21 d, fully expanded first leaves were dissected into 1 cm2 and placed adaxial side on the medium. The culture medium was composed of MS basal salts, 3% (w/v) Suc, and 0.6% (w/v) Phytablend. PGRs were added to the medium as shown in Table I, and pH was adjusted to 5.8 prior to autoclaving. Cultures were maintained at 26°C ± 2°C at 40 μE m−2 s−1 photon density, with 16 h of light and 8 h of dark.
Genome Analysis
Sequences of intergenic spacers upstream from genes representing different functional groups were extracted from complete plastid genomes on GenBank for 20 species representing most major clades of angiosperms (for taxa and genes, see Supplemental Tables S1 and S2). Alignments were anchored by the inclusion of 100 bases from the coding regions of adjacent genes. Sequences were aligned using MUSCLE (Edgar, 2004), followed by manual adjustment in Geneious (http://www.geneious.com/). Sequence identity was calculated for the region encompassed by 200 bases upstream of the translation start codon. For comparison, sequence identity was also determined for the aligned coding regions. Geneious was used to conduct motif searches for functional domains, promoters, and cis-translation elements and to calculate sequence identities for selected regions.
Vector Construction
The pUC-based lettuce long flanking plasmid (pLSLF) was used to integrate foreign genes into the intergenic spacer between tRNA-Ile and tRNA-Ala genes of the plastome inverted repeat region. Details of this vector, the g10 CTB-Pins expression cassette, and the pLD-CtV-5CP tobacco (Nicotiana tabacum) transformation vector were reported earlier (Ruhlman et al., 2007). The lettuce endogenous Prrn and 3′ rbcL for the expression of aadA from a GGAG RBS were amplified using total genomic lettuce DNA with sequence-specific primers and assembled in pBSSK+ vector. The aadA cassette was subcloned into pLSLF at the PvuII site, and the resulting vector is named as pLsDV. The lettuce endogenous psbA 5′ and 3′ UTRs were PCR amplified and cloned in pBSSK+ vector to make an intermediate vector with multiple cloning sites in between psbA 5′ and 3′ UTRs, resulting in pDVI-1. The CTB-Pins sequence and pagA sequence were cloned at NdeI and XbaI sites of pDVI-1 vector. The CTB-Pins and PA expression cassette with psbA 5′ and 3′ UTRs was released by digestion with SalI and NotI and ligated into the pLsDV vector. A transformation cassette for the generation of transplastomic lettuce plants that express CTB-Pins from tobacco psbA 5′ and 3′ UTRs was assembled by digestion of the pLD-CtV-5CP plasmid with SalI and XbaI to release the CTB-Pins coding region plus the tobacco psbA 5′ UTR. This fragment was ligated into a pUC intermediate plasmid upstream of the tobacco psbA 3′ UTR. The nucleotide sequence of the intermediate plasmid pUC-NtUTR-CTB-Pins was confirmed. The cassette was released by digestion with SalI and SnaBI and ligated into the pLsDV vector digested by SalI and EcoRV. The pagA coding sequence was released from pLD VK1 (Koya et al., 2005) plasmid by digesting with NdeI and NotI and cloned in pLSLF CTB-Pins vector. All cloning steps were carried out in Escherichia coli according to Sambrook and Russel1 (2001).
Bombardment and Selection of Transplastomic Lettuce
Seeds of lettuce cv Simpson Elite (New England Seed) were surface sterilized and germinated on MS medium solidified with 6 g L−1 Phytablend (Caisson). For each bombardment, six to seven young, fully expanded leaves of approximately 4 cm2 were placed adaxial side up on antibiotic-free modified lettuce regeneration medium (Kanamoto et al., 2006) containing 0.1 μg mL−1 NAA and 0.2 μg mL−1 BAP. Leaves were bombarded with 0.6-μm gold particles (Bio-Rad) coated with one of the plastid transformation vectors shown in Figure 3, and bombardments were carried out using the PDS-1000/He Biolistic device employing 900 p.s.i. rupture discs and a target distance of 6 cm as described by Kumar and Daniell (2004). Bombarded leaf samples were held in the dark at 25°C for 2 d, cut into 1-cm2 pieces, and then placed adaxial side down onto modified regeneration medium with 50 μg mL−1 spectinomycin dihydrochloride. Primary regenerants were screened by PCR for the transplastomic event, and positive shoots were subjected to an additional regeneration cycle on the same selective medium. Following the second cycle, regenerated shoots were rooted in half-strength MS medium containing 100 μg mL−1 spectinomycin. Plants were propagated by rooting of nodal sections in half-strength, hormone-free MS medium with spectinomycin. Rooted cuttings were hardened in Jiffy peat pots before transfer to the greenhouse for seed production. T1 seeds were harvested and plated on MS medium with 100 μg mL−1 spectinomycin along with wild-type lettuce to confirm maternal inheritance of plastid transgenes. Tobacco plants included in analysis are reported by Ruhlman et al. (2007) and Koya et al. (2005). Transplastomic lettuce plants expressing CTB-Pins from endogenous psbA 5′ and 3′ UTRs were contributed by D. Burberry. Transplastomic tobacco plants expressing PA from lettuce psbA 5′ and 3′ UTRs were generated by bombarding tobacco leaves with pLsDV LsPA lettuce transformation vector.
PCR Screening and Southern Blotting
Genomic DNA isolated from primary transformants was analyzed by PCR using primers 16SF (5′-CAGCAGCCGCGGTAATACAGAGGA-3′) and 3M (Singh et al., 2009). For Southern blotting, genomic DNA was isolated from young, in vitro-grown leaves ground in liquid N2 with chilled, sterile mortar and pestle, and extraction was carried out using a Qiagen DNeasy Plant Mini Kit (no. 69104). Five micrograms of total DNA was digested completely with AflIII for CTB-Pins Ls-Nt-CP line and SmaI for Ls-Ls-PA and Ls-g10-PA lines, and the resulting fragments were separated on 0.8% (w/v) Tris-acetate EDTA agarose gels and transferred to nylon membranes by capillary action. Plastid flanking sequence probe (1.3 kb) was amplified by PCR from lettuce genomic DNA. The PCR product was column purified, and labeled probe was generated by incubation with [α-32P]dCTP and Ready-To-Go DNA Labeling Beads (GE Healthcare). Blots were prehybridized for 1 h at 68°C in QuikHyb reagent (Stratagene). Blots were hybridized for 1 h at 68°C and washed twice at 37°C in 2× SSC (0.3 m sodium chloride and 30 mm sodium citrate, pH 7) and twice at 65°C in 0.1× SSC. Radiolabeled blots were exposed to film on intensifying screens at −80°C for 16 h.
Western-Blot and Densitometric Analyses
Second generation (T1) CTB-Pins transplastomic lettuce and tobacco were raised in the University of Central Florida greenhouse. Young, mature, and older fully expanded leaves from approximately 8-week-old plants were harvested in August at 5 am, 10 am, 2 pm, and 6 pm, ground in liquid N2, and stored at −80°C. Approximately 100 mg of leaf tissue was suspended in 5 volumes of protein extraction buffer (100 mm NaCl, 10 mm EDTA, 200 mm Tris-HCl, pH 8, 0.1% [v/v] Triton X-100, 100 mm dithiothreitol, 400 mm Suc, and 2 mm phenylmethylsulfonyl fluoride) and vortexed vigorously for 20 min at 4°C prior to determination of total protein using Bio-Rad Protein Assay Reagent. TLPs along with 100, 200, 400, and 600 ng of purified bacterial CTB (Sigma) were separated by SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting, according to Kumar and Daniell (2004). Immunoblotting with anti-CT primary antibody (1:3,500; Sigma) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:4,000; Southern Biotech) was employed for densitometric analysis. A SuperSignal West Pico HRP Substrate Kit (Pierce) was used for detection of chemiluminescence signal by exposure to film. Tissue collected at the latest developmental stage was prepared as above and subjected to SDS-PAGE along with CTB standards ranging from 0.5 to 3 μg. Gels were stained with Coomassie Brilliant Blue and used for densitometric quantitation.
Estimation of PA Protein Using ELISA
T1 PA transplastomic lettuce and tobacco were raised in the University of Central Florida greenhouse. Young, mature, and old fully expanded leaves from approximately 8-week-old plants were harvested at 6 am, 12 pm, 6 pm, and 3 am, ground in liquid N2, and stored at −80°C. ELISA of leaf extract supernatant was performed in duplicate on a 96-well EIA/RIA plate (Costar), along with purified PA as standard (courteously provided by Dr. Stephen H. Leppla, National Institutes of Health). The standard and samples were diluted in 15 mm Na2CO3, 35 mm NaHCO3, and 3 mm NaN3, pH 9.6. PA standard ranging from 5 to 400 ng mL−1 and sample dilutions of 1:10,000, 1:20,000, and 1:40,000 were loaded into the wells and incubated at 4°C overnight. The plate was blocked with 3% (w/v) fat-free milk in phosphate-buffered saline containing 0.05% (v/v) Tween 20 (PBST), incubated at 37°C for 1 h, and washed three times each with PBST and water. Primary polyclonal anti-PA antibody (1:3,000) in PBST was loaded into wells and incubated at 37°C for 1 h. The wells were then washed with PBST and water as above. HRP-conjugated goat anti-rabbit antibody (1:5,000; Southern Biotech) was incubated at 37°C for 1 h followed by three washings with PBST and water. A total of 100 μL of 3,3′,5,5′-tetramethyl benzidine substrate was loaded to each well and incubated at 24°C for 10 to 15 min. The reaction was terminated by the addition of 50 μL of 2 n H2SO4. The plate was read at 450 nm using a plate reader (model 680; Bio-Rad). Western-blot analysis was performed using monoclonal anti-PA primary antibody (1:3,000; Advanced Immunochemical Lab) in PBST and HRP-conjugated goat anti-mouse secondary antibody (1:5,000; American Qualex).
Plastid Isolation
Intact tobacco plastids were isolated according to Yukawa et al. (2007). Fully expanded green leaves (200 g) were collected from 4- to 5-week-old greenhouse-grown tobacco cv TN90. Leaves were homogenized in 50-g batches with 150 mL of MCB1 (50 mm HEPES/KOH, pH 8.0, 0.3 m mannitol, 2 mm EDTA, 5 mm β-mercaptoethanol, 0.1% bovine serum albumin, and 0.6% polyvinylpyrrolidone). Homogenates were filtered through four layers of cheesecloth and then two layers of Miracloth (Calbiochem) and centrifuged for 5 min at 1, 000g in a Sorvall SS-34 fixed-angle rotor at 4°C. Pellets were resuspended by gentle agitation with a soft paintbrush in 30 mL of MCB1, and 5 mL was layered onto 25 mL of Percoll (Sigma) gradients (20%-50%-80% in MCB1). Gradients were centrifuged for 10 min at 10,000g in an L-90K ultracentrifuge SW32-ti rotor at 2°C. The lower dark green band containing intact plastids was harvested and washed in 3 volumes of MCB2 (50 mm HEPES/KOH, pH 8.0, 0.32 m mannitol, 2 mm EDTA, and 5 mm β-mercaptoethanol). Plastids were collected by 1 min of centrifugation at 600g in a Sorvall SS-34 rotor at 4°C. The plastid pellet was resuspended in a minimal volume of EMSA binding buffer (Alexander et al., 1998) with rigorous agitation to disrupt plastid envelopes. Membranes were sedimented by centrifugation in a SS-34 rotor at 27,000g for 15 min. Stromal extracts were adjusted to 15% glycerol, divided into aliquots, and stored at −80°C.
Intact lettuce plastids were isolated according to Gruissem et al. (1986). Fully expanded green leaves (200 g) of 6- to 8-week-old hydroponically grown lettuce cv Longifolia were collected from the greenhouse and homogenized in 50-g batches with 150 mL of 1× GM (50 mm HEPES/KOH, pH 6.8, 1 mm sodium pyrophosphate, 33 mm sorbitol, 2 mm EDTA, 1.25 mm MgCl2, 1.25 mm MnCl2, and 2 mm dithiothreitol) buffer. Homogenates were filtered and centrifuged, and pellets were resuspended as above in 1× GM buffer; 5 mL was layered onto 25 mL of PCBF (100 mL of Percoll, 3% polyethylene glycol 6000, 1% bovine serum albumin, and 1% Ficoll [Sigma]) density gradients. Gradients were centrifuged for 20 min at 8,100g at 2°C. The lower dark green band containing intact plastids was harvested and washed in 2 volumes of 1× GM buffer. Plastids were collected by 3 min of centrifugation at 1,500g at 4°C. Plastid pellet was resuspended in a minimal volume of the above EMSA binding buffer and preserved as for tobacco.
In Vitro Transcription of Radiolabeled Transcripts
Plasmids (pBluescript SK+; Stratagene) containing the psbA 5′ UTR were digested with NdeI (lettuce) or NcoI (tobacco) to generate linearized templates for T7 in vitro transcription using the MAXIscript Kit (Ambion) according to the manufacturer's instructions. For labeled UTR species UTP was replaced with 3.125 μm [α-32P]UTP (Perkin-Elmer). Reaction products were separated by denaturing polyacrylamide and eluted following the manufacturer's instructions. Following ethanol precipitation, supernatants were discarded and RNA pellets were vacuum dried. Pellets were resuspended in 50 μL of nuclease-free water and quantified by spectrophotometry and liquid scintillation counting. Single-use aliquots were prepared and stored at −80°C.
EMSA
Competitive RNA EMSA was adapted from Alexander et al. (1998). Stromal extracts were thawed on ice, and total protein content was determined using the Bio-Rad reagent. Reactions were evaluated using increasing quantities of radiolabeled psbA 5′ UTR and stromal proteins to achieve saturation of binding in the absence of competition. Stromal proteins of tobacco (20 μg) or lettuce (40 μg) were incubated with 5 fmol of endogenous radiolabeled psbA 5′ UTR with or without unlabeled competitor psbA 5′ UTR. All reactions were supplemented with 0.5 μg μL−1 yeast tRNA (Ambion) to reduce nonspecific binding, and total volume was adjusted to 20 μL using EMSA binding assay buffer. Control reactions included no competition, competition with 50× molar excess of unlabeled endogenous UTR, and labeled probe only (no protein). Experimental reactions contained 50×, 100×, and 200× molar excess of unlabeled heterologous competitor UTR (i.e. tobacco protein with lettuce UTR as competitor). Competitors were added 5 min prior to the labeled probe. Reactions were allowed to proceed for 15 min at 22°C in the presence of labeled probe. A total of 4 μL of 5× nondenaturing gel loading buffer was added (225 mm Tris-HCl, pH 6.8, 50% [v/v] glycerol, and 0.05% [w/v] bromphenol blue), and reactions were separated through 8% polyacrylamide.
Polyribosome Association Assay and Northern Blotting
The polysome assay was adapted from Barkan (1988). Green leaves from in vitro-grown plants were ground to a fine powder in liquid N2. Approximately 300 mg of each sample was transferred to a 2-mL tube and vortexed with 1 mL of extraction buffer (0.2 m Tris-HCl, pH 9, 0.2 m KCl, 35 mm MgCl2, 25 mm EGTA, pH 8.3, 1% Triton X-100, and 2% polyoxyethylene-10-tridecylether) with 0.5 mg mL−1 heparin, 100 μg mL−1 chloramphenicol, and 25 μg mL−1 cyclohexamide. Homogenates were forced gently through glass wool packed in a 3-mL syringe into a microcentrifuge tube on ice, and extracts were held on ice for 10 min. Extracts were centrifuged at 17,900g for 5 min at 4°C. Supernatants were transferred to new tubes, and 1/20th volume of 10% sodium deoxycholate was added. Control reactions were incubated with puromycin (3 mg mL−1) at 37°C for 10 min. Reactions were held on ice for 5 min prior to centrifugation at 17,900g for 15 min at 4°C. Supernatants (500 μL) were layered onto Suc gradients (15% to 30%–40% to 50%) in 10× salts (0.4 m Tris-HCl, pH 8, 0.2 m KCl, and 0.1 m MgCl2) prepared in Beckman ultraclear 0.5- × 2-inch tubes (13 × 51 mm; 344057). One aliquot of supernatant from each sample was reserved for isolation of total RNA. Gradients were centrifuged at 4°C in a SW55-ti rotor for 65 min at 192,000g. Fractions of approximately 500 μL were collected into microfuge tubes containing 50 μL of 5% SDS and 0.2 m EDTA, pH 8, by puncturing gradient tubes with an 18-gauge needle. One volume of phenol:chloroform:isoamyl (25:24:1) was added to each fraction, vortexed, and then centrifuged at 17,900g for 5 min. The aqueous phase was transferred to a new tube, and 2 volumes of absolute ethanol was added and mixed by inversion. RNA was pelleted by centrifugation, the supernatant was discarded, and pellets were dried under vacuum. Pellets were resuspended in 30 μL of Tris-EDTA. RNA sample buffer (80 mm MOPS, pH 7, 4 mm EDTA, 0.9 m formaldehyde, 20% glycerol, 30.1% formamide, 5 mm sodium acetate, and 0.25% bromphenol blue) was added, and fractions were separated on formaldehyde-agarose gels under denaturing conditions. Gels were washed in RNase-free water, and RNA was transferred to nylon membranes (Nytran SPC; Whatman) by capillary action in 20× SSC. Membranes were rinsed in RNase-free water and fixed by UV light cross-linking. The full-length CTB-Pins coding region was used to generate α-32P-labeled, single-stranded DNA probes according to the procedure described above for Southern blotting. Prehybridization and hybridization steps for polyribosome blots were carried out in Denhardt's buffer. Blots of total RNA extractions were hybridized in QuikHyb (Stratagene). For analysis of total transcripts, RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen) and quantified by spectrophotometry. Total RNA (2 μg, except Nt-Ls-PA and Nt-Nt-PA) was prepared, electrophoresed, and blotted as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Taxa included in genomic analysis.
Supplemental Table S2. Intergenic regions included in analysis.
Supplementary Material
Acknowledgments
We thank Prof. R.K. Jansen and Zhengqui Cai (University of Texas, Austin) for help with genomic analysis and Diane Burberry, Andrew Devine, and Vijay Koya (University of Central Florida) for providing the Ls-Ls-Cp, Nt-Nt-Cp, and Nt-Nt-PA transplastomic lines.
References
- Alexander C, Faber N, Klaff P. (1998) Characterization of protein-binding to the spinach chloroplast psbA mRNA 5′ untranslated region. Nucleic Acids Res 26: 2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Bally J, Nadai M, Vitel M, Rolland A, Dumain R, Dubald M. (2009) Plant physiological adaptations to the massive foreign protein synthesis occurring in recombinant chloroplasts. Plant Physiol 150: 1474–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. (2007) Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol 18: 100–106 [DOI] [PubMed] [Google Scholar]
- Cerutti H, Ibrahim HZ, Jagendorf AT. (1993) Treatment of pea (Pisum sativum L.) protoplasts with DNA-damaging agents induces a 39-kilodalton chloroplast protein immunologically related to Escherichia coli RecA. Plant Physiol 102: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Osman M, Grandoni P, Jagendorf AT. (1992) A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA 89: 8068–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK. (2006) Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet 112: 1503–1518 [DOI] [PubMed] [Google Scholar]
- Daniell H, Ruiz G, Denes B, Sandberg L, Langridge L. (2009a) Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function. BMC Biotechnol 9: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Ruiz ON, Dhingra A. (2005) Chloroplast genetic engineering to improve agronomic traits. Methods Mol Biol 286: 111–138 [DOI] [PubMed] [Google Scholar]
- Daniell H, Singh ND, Mason H, Streatfield SJ. (2009b) Plant-made vaccine antigens and biopharmaceuticals. Trends Plant Sci 14: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi-Semiromi A, Samson N, Daniell H. (2009) The green vaccine: a global strategy to combat infectious and autoimmune diseases. Hum Vaccin 5: 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127: 852–862 [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Jr, Daniell H. (2004) Enhanced translation of a chloroplast-expressed rbcS gene restores small subunit levels and photosynthesis in nuclear rbcS antisense plants. Proc Natl Acad Sci USA 101: 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, Garcon F, Muhr C, Jansens S, Pelissier B, Peltier G, Dubald M. (2005) Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol 58: 659–668 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop HU. (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19: 333–345 [DOI] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingo-Castel A, Miller M, Daniell H. (2003) A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J 1: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EF, Hall MA, Klerk GJK, (2008) Plant Propagation by Tissue Culture. Springer, Basingstoke, UK [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. (2008) The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W, Greenberg G, Zurawski G, Hallick RB. (1986) Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol 118: 253–270 [DOI] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H. (2000) Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Rep 19: 257–265 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Shiina T, Ishii N, Iwai K, Ishizaki Y, Morikawa K, Toyoshima Y. (2003) A role of the −35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Hess WR, Borner T. (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190: 1–59 [DOI] [PubMed] [Google Scholar]
- Hirose T, Sugiura M. (1996) Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J 15: 1687–1695 [PMC free article] [PubMed] [Google Scholar]
- Inouye T, Odahara M, Fujita T, Hasebe M, Sekine Y. (2008) Expression and complementation analyses of a chloroplast-localized homolog of bacterial RecA in the moss Physcomitrella patens. Biosci Biotechnol Biochem 72: 1340–1347 [DOI] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa K. (2006) Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res 15: 205–217 [DOI] [PubMed] [Google Scholar]
- Kavanagh TA, Thanh ND, Lao NT, McGrath N, Peter SO, Horvath EM, Dix PJ, Medgyesy P. (1999) Homeologous plastid DNA transformation in tobacco is mediated by multiple recombination events. Genetics 152: 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaff P, Mundt SM, Steger G. (1997) Complex formation of the spinach chloroplast psbA mRNA 5′ untranslated region with proteins is dependent on the RNA structure. RNA 3: 1468–1479 [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Mason HS, Mullet JE. (1988) Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol 106: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya V, Moayeri M, Leppla SH, Daniell H. (2005) Plant-based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immun 73: 8266–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Daniell H. (2004) Engineering the chloroplast genome for hyperexpression of human therapeutic proteins and vaccine antigens. Methods Mol Biol 267: 365–383 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2001) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelavathi S, Reddy VS. (2003) Chloroplast expression of His-tagged GUS-fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed 11: 49–58 [Google Scholar]
- Lelivelt CL, McCabe MS, Newell CA, deSnoo CB, van Dun KM, Birch-Machin I, Gray JC, Mills KH, Nugent JM. (2005) Stable plastid transformation in lettuce (Lactuca sativa L.). Plant Mol Biol 58: 763–774 [DOI] [PubMed] [Google Scholar]
- Majewski J, Cohan FM. (1998) The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics 148: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder J, Chapman D, Telfer A, Nixon P, Barber J. (1987) Identification of psbA and psbD gene products, D1 and D2, as reaction centre proteins of photosystem 2. Plant Mol Biol 9: 325–333 [DOI] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhige PM, Both-Kim D, Robida MD, Hollingsworth MJ. (2005) RNA-protein complexes that form in the spinach chloroplast atpI 5′ untranslated region can be divided into two subcomplexes, each comprised of unique cis-elements and trans-factors. Curr Genet 48: 256–264 [DOI] [PubMed] [Google Scholar]
- Minami E, Shinohara K, Kawakami N, Watanabe A. (1988) Localization and properties of transcripts of psbA and rbcL genes in the stroma of spinach chloroplast. Plant Cell Physiol 29: 1303–1309 [Google Scholar]
- Murashige T, Skoog A. (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamura T, Ohta M, Sugiura M, Sugita M. (1999) Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett 460: 437–441 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Ohta M, Sugiura M, Sugita M. (2001) Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J Biol Chem 276: 147–152 [DOI] [PubMed] [Google Scholar]
- Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH. (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Harris EH, Johnson AM, Boynton JE, Gillham NW. (1992) Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: evidence for a hotspot and an adjacent cold region. Genetics 132: 413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J. (2003) Chloroplast RNA-binding proteins. Curr Genet 43: 392–399 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445 [DOI] [PubMed] [Google Scholar]
- Ovecka M, Bobak M, Samaj J. (2000) A comparative structural analysis of direct and indirect shoot regeneration of Papaver somniferum L. in vitro. J Plant Physiol 157: 281–289 [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19: 870–875 [DOI] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. (2007) Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts: oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J 5: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. (2007) Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet 115: 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Barkan A. (2007) RNA splicing and RNA editing in chloroplasts. Bock R, , Cell and Molecular Biology of Plastids. Springer, Berlin, 213–248 [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Huang HV. (1986) Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112: 441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Danon A, Christopher DA. (2001) RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5′ untranslated region in a redox-dependent manner. Plant Cell Physiol 42: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS. (1999) Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19: 209–216 [DOI] [PubMed] [Google Scholar]
- Singh ND, Ding Y, Daniell H. (2009) Chloroplast-derived vaccine antigens and biopharmaceuticals: protocols for expression, purification, or oral delivery and functional evaluation. Methods Mol Biol 483: 163–192 [DOI] [PubMed] [Google Scholar]
- Singh ND, Li M, Lee SB, Schnell D, Daniell H. (2008) Arabidopsis Tic40 expression in tobacco chloroplasts results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell 20: 3405–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman P, Silhavy D, Maliga P. (1998) Transcription from heterologous rRNA operon promoters in chloroplasts reveals requirement for specific activating factors. Plant Physiol 117: 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al. (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18: 333–338 [DOI] [PubMed] [Google Scholar]
- Staub JM, Maliga P. (1994) Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J 6: 547–553 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Ingersoll J, Stern DB, Kindle KL. (1997) Generation and maintenance of tandemly repeated extrachromosomal plasmid DNA in Chlamydomonas chloroplasts. Plant J 11: 635–648 [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Sriraman P, Svab Z, Maliga P. (2003) Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Kuehl JV, Boore JL, Jansen RK. (2007) A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot 94: 302–312 [DOI] [PubMed] [Google Scholar]
- Tregoning JS, Nixon P, Kuroda H, Svab Z, Clare S, Bowe F, Fairweather N, Ytterberg J, van Wijk KJ, Dougan G, et al. (2003) Expression of tetanus toxin fragment C in tobacco chloroplasts. Nucleic Acids Res 31: 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Daniell H. (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Samson NP, Koya V, Daniell H. (2008) A protocol for expression of foreign genes in chloroplasts. Nat Protoc 3: 739–758 [DOI] [PubMed] [Google Scholar]
- Xinrun Z, Conner AJ. (1992) Genotype effects on tissue culture response of lettuce cotyledons. J Genet Breed 46: 287–290 [Google Scholar]
- Yamamoto Y. (2001) Quality control of photosystem II. Plant Cell Physiol 42: 121–128 [DOI] [PubMed] [Google Scholar]
- Yang J, Usack L, Monde RA, Stern DB. (1995) The 41 kDa protein component of the spinach chloroplast petD mRNA 3′ stem-loop:protein complex is a nuclear encoded chloroplast RNA-binding protein. Nucleic Acids Symp Ser 33: 237–239 [PubMed] [Google Scholar]
- Ye GN, Hajdukiewicz PTJ, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25: 261–270 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Kuroda H, Sugiura M. (2007) A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. Plant J 49: 337–376 [DOI] [PubMed] [Google Scholar]
- Zou Z, Eibl C, Koop HU. (2003) The stem-loop region of the tobacco psbA 5′UTR is an important determinant of mRNA stability and translation efficiency. Mol Genet Genomics 269: 340–349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.