Abstract
The two subunits of the circadian RNA-binding protein CHLAMY1 from Chlamydomonas reinhardtii are involved in maintaining period (C1 subunit) and phase (C3 subunit) of the circadian clock. C1 coregulates the level of C3. Overexpression of C1 causes a parallel increase in C3. Both subunits can also integrate temperature information, resulting in hyperphosphorylation of C1 and up-regulation of C3 at low temperature. Temperature-dependent up-regulation of C3 is mediated predominantly by an E-box element and only partially by two DREB1A-boxes that are situated within the C3 promoter. The E-box element is also involved in circadian C3 expression. Here, we show that the C3 promoter region drives C3 coregulation by C1. We also found that replacement of the E-box prevents the coregulation of C3 in strains overexpressing C1. In contrast, replacement of any of the two DREB1A-boxes does not influence either the coregulation of C3 by increased levels of C1 or circadian C3 expression. Thus, the E-box has multiple key roles, including temperature-dependent up-regulation of C3, its circadian expression, and its coregulation by C1. Using mobility shift assays and DNA-affinity chromatography along with mass spectrometry, we characterized proteins binding specifically to the E-box region and identified five of them. By immunoblotting, we could further show that C3 that was detected in nuclear extracts can be found in the E-box-binding protein complex. Our data indicate a complex transcriptional mechanism of C3 up-regulation and a positive feedback of C3 on its own promoter region.
Circadian rhythms are endogenous rhythms that persist with a period of about 24 h under constant conditions of light and temperature. They can be entrained by light-dark or temperature cycles. Circadian clocks are temperature compensated (Q10 ∼ 0.8–1.3); thus, their period length is almost unchanged at different temperatures (Rensing and Ruoff, 2002).
While physiological properties of circadian rhythms are well conserved among different species, only certain molecular components are conserved. However, in the different studied model organisms, positive and negative feedback loops of relevant molecular components have been shown to be involved in the oscillatory machinery (for review, see Dunlap et al., 2007; Rosbash et al., 2007; Harmer, 2009). So far, relatively few components that are involved in RNA metabolism have been shown to play a role in the circadian system. For example, the Xenopus and mouse NOCTURNINs that function as deadenylases [poly(A)-specific ribonucleases] likely regulate the mRNA decay and/or translational properties of circadian-regulated transcripts (Garbarino-Pico and Green, 2007). The Drosophila RNA-binding proteins, LARK and FMRP, function together to regulate circadian behavior (Sofola et al., 2008). The Gly-rich RNA-binding proteins of Arabidopsis (Arabidopsis thaliana), AtGRP7, a circadian slave oscillator component, and AtGRP8, are regulated via an interlocked feedback loop coupling splicing to nonsense-mediated decay (Schöning et al., 2008). Recently, the role of microRNAs has emerged (e.g. in the circadian clock of Drosophila [Kadener et al., 2009]).
In the green alga Chlamydomonas reinhardtii, several processes are under control of the circadian clock, such as phototaxis and chemotaxis (for review, see Mittag et al., 2005). The sequencing of the entire genome of C. reinhardtii has opened an avenue for performing homology searches and applying diverse “-omics” methods to study the circadian system of C. reinhardtii (Kucho et al., 2005; Merchant et al., 2007; Rolland et al., 2009; Wagner et al., 2009). Meanwhile, some components that are involved in the oscillatory machinery of C. reinhardtii have been described, including CASEIN KINASE1 (Schmidt et al., 2006) and the two subunits C1 and C3 of the heteromeric RNA-binding protein CHLAMY1 (Iliev et al., 2006). An insertional mutagenesis approach revealed in addition several key components that are relevant for normal circadian rhythmicity of a chloroplast bioluminescence reporter in C. reinhardtii (Matsuo et al., 2008). Also, a homologous gene of the circadian-regulated CONSTANS that is involved in the photoperiodic control of the floral transition in Arabidopsis was found in C. reinhardtii, where it coordinates processes regulated by photoperiod and the circadian clock (Serrano et al., 2009).
Studies in our laboratory focused on the RNA-binding protein CHLAMY1 in recent years. It recognizes specifically (UG)≥7-repeat sequences that are present in the 3′ untranslated regions (UTRs) of several mRNAs of C. reinhardtii, and its binding activity is controlled by the circadian clock (Mittag, 1996; Waltenberger et al., 2001). The insertion of such UG-repeat regions within the 3′ UTR of a reporter gene triggers circadian oscillations (Kiaulehn et al., 2007). CHLAMY1 represents a heteromeric RNA-binding protein that consists of two subunits comprising three RNA recognition motifs (RRM; C3 subunit) and three K homology domains (KH; C1 subunit; Zhao et al., 2004). Their interaction is necessary to recognize the binding motif, the UG-repeat region. While the protein level of both subunits is rather constant (Zhao et al., 2004), cDNA macroarray analysis showed that C3 mRNA fluctuates over a circadian cycle with a peak during night phase. In contrast, C1 mRNA appears to be constantly present (Kucho et al., 2005). The role of CHLAMY1 within the circadian system was characterized by independent up- or down-regulation of its two subunits. Changes in the level of the C3 or C1 subunit resulted in disturbances of the circadian phototaxis rhythm either affecting its acrophase (changes in the C3 level) or causing arrhythmicity (changes in the C1 level). Thus, CHLAMY1 is involved in the maintenance of phase and period of the circadian clock (Iliev et al., 2006). It was also observed that an altered expression of the C1 subunit (overexpression or silencing) changed in parallel the level of the C3 subunit in the same way. Thus, C1 can coregulate the protein level of C3 (Iliev et al., 2006).
Recently, it was shown that both subunits of CHLAMY1 can integrate temperature information (Voytsekh et al., 2008). Temperature entrainment as well as the temperature-dependent regulation of C1 and C3 are altered in the clock mutant per1 that was isolated by Bruce (1972). In the wild type, C1 is hyperphosphorylated at low temperature (18°C), which is still in the physiological range of C. reinhardtii. At the same time, the expression level of the C3 subunit is high at 18°C and low at 28°C. This temperature-dependent regulation of C3 occurs at the transcriptional level and involves predominantly an E-box. Replacement of the E-box results in the loss of C3 up-regulation at low temperature. Replacement of any of two DREB1A-boxes that are also situated in the C3 promoter region and that are known for cold stress integration in higher plants only reduces the amplitude of C3 up-regulation to some extent. Notably, the E-box is also involved in circadian C3 expression. Its replacement results in arrhythmicity of a C3 promoter-driven reporter (Voytsekh et al., 2008).
In Drosophila, E-box regulatory elements, which are situated in the promoters of the clock-relevant genes Period (Per) and Timeless (Tim), are recognized by the basic helix-loop-helix (bHLH) factors CLOCK (CLK) and CYCLE (CYC). The binding of the CLK-CYC complex to the E-boxes promotes the transcription of Per and Tim. Interaction of the PER-TIM heterodimer with the CLK-CYC complex represses the transcriptional activation (Yu et al., 2008, and refs. therein). But E-boxes can also recruit other factors. Two E-box elements in the promoter of the mammalian Ornithine decarboxylase gene are recognized by the bHLH/leucine zipper domain transcription factor complex c-MYC/MAX (Auvinen et al., 2003). Recent experiments suggest that the heterogenous nuclear ribonucleoprotein U (hnRNP U) acts as a coactivator of this transcriptional activator complex (Matsuoka et al., 2009). Also zinc finger E-box homeobox factors interacting with bipartite E-boxes are known (Peinado et al., 2007).
Here, we analyzed if the up-regulation of C3 in strains overexpressing C1 also involves the C3 promoter region and any of the E- or DREB1A-boxes. We show that the C3 promoter region is necessary for the coregulation of C3. Replacement of the E-box prevents the coregulation. In contrast, replacement of any of the two DREB1A-boxes does not influence it. Thus, the E-box has three different functions within the C3 promoter. We further identified five proteins as well as C3 as factors interacting with the E-box region. Our data suggest that C3 feeds back on its own promoter.
RESULTS
The C3 Promoter/5′ UTR But Not the C3 3′ UTR Is Involved in the Up-Regulation of C3 in C1 Overexpression Strains
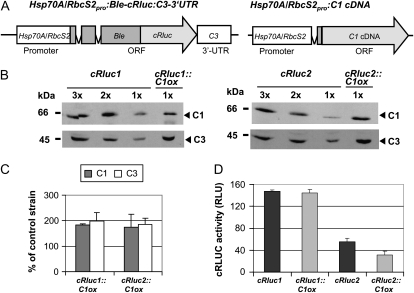
We aimed to analyze by which mechanism C3 is up-regulated in transgenic lines overexpressing C1 (abbreviated as C1ox). At first, we checked if the up-regulation could be mediated at the translational level involving the C3 3′ UTR. Therefore, we took two transgenic lines (cRluc1 and cRluc2; Voytsekh et al., 2008) that bear the C3 3′ UTR next to a reporter gene. As a reporter, we used a fusion consisting of the zeozin resistance gene Ble and the gene encoding Renilla reniformis LUCIFERASE that had been adapted to the nuclear codon usage of C. reinhardtii (cRLUC; Fig. 1A, left panel). The chimeric gene was put under the control of the strong tandem promoter Hsp70A/RbcS2 (Fuhrmann et al., 2004). For overexpression of C1 in cRluc1 and -2, a C1 cDNA construct that was used previously was transformed into these strains (Fig. 1A, right panel; Iliev et al., 2006). Selection was done for paromomycin resistance encoded by the AphVIII gene. Two strains, where C1 was overexpressed about 2-fold, named cRluc1::C1ox and cRluc2::C1ox, were used for further analysis (Fig. 1, B and C). In both strains, C3 was also overexpressed about 2-fold.
Figure 1.
The C3 3′ UTR is not involved in the coregulation of C3 by C1. A, The chimeric construct Hsp70A/RbcS2pro:Ble-cRluc:C3-3′UTR of pSK27 (Voytsekh et al., 2008) that was used for transformation resulting in transgenic lines cRluc1 and -2 is schematically shown as well as the construct used for C1 overexpression (C1ox; Iliev et al., 2006). ORF, Open reading frame. B, Different amounts of proteins from a crude extract (90, 60, and 30 μg per lane) labeled as 3x, 2x, and 1x, respectively, of cRluc1 and -2 cells that had been transformed in some cases with the C1ox vector (cRluc1::C1ox and cRluc2::C1ox) were separated by SDS-PAGE. They were used for immunodetection with anti-C1 and anti-C3 antibodies, respectively. C, Quantification of the expression levels of C1 and C3 via ImageMaster 2D Elite version 4.01 (GE Healthcare) according to “Materials and Methods” in C1ox strains in percentage in comparison with the appropriate control strain (n = 2). D, Measurements of luciferase activities (cRLUC) in the cRluc1 and -2 strains in comparison with the cRluc1::C1ox and cRluc2::C1ox strains in relative light units (RLU; n = 3). Error bars represent the se of technical replicates. Cells were grown at 23°C and used at LD2 to measure the cRLUC activities.
Then, we analyzed cRLUC activities in the original reporter containing strains in comparison with the strains overexpressing in addition C1. In the cRluc1::C1ox and cRluc2::C1ox strains overexpressing C1, no increase in cRLUC activity was visible in comparison with the cRluc1 and cRluc2 strains (Fig. 1D). Thus, the C3 3′ UTR is not able to mediate the up-regulation of C3 in C1-overexpressing strains.
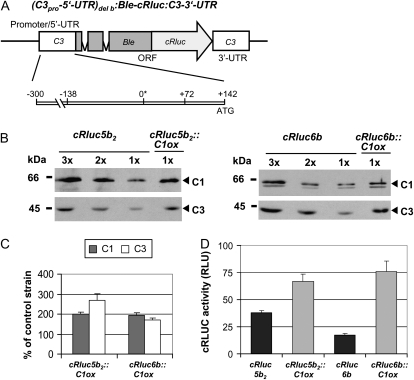
In a next step, we examined the C3 promoter region, including its 5′ UTR (−300 to +142). This region of the C3 promoter is sufficient for the up-regulation of C3 at low temperature and for its circadian regulation (Voytsekh et al., 2008). Strains expressing the chimeric cRluc reporter construct with the C3 3′ UTR that is under control of the above-mentioned C3 promoter region (Fig. 2A) had been generated before (cRluc6b) or were newly generated (cRluc5b2).
Figure 2.
The −300 to +142 region of the C3 promoter is sufficient to mediate the coregulation of C3 by C1. A, The chimeric construct (C3pro-5′UTR)del b:Ble-cRluc:C3-3′UTR of pDI23 having a shortened C3 promoter (−300 to +142; Voytsekh et al., 2008) that was used for transformation resulting in transgenic lines cRluc5b2 and -6b is schematically shown. ORF, Open reading frame. B, Different amounts of proteins from a crude extract (90, 60, and 30 μg per lane) labeled as 3x, 2x, and 1x, respectively, of cRluc5b2 and -6b cells that had been transformed in some cases with the C1ox vector (cRluc5b2::C1ox and cRluc6b::C1ox) were separated by SDS-PAGE. They were used for immunodetection with anti-C1 and anti-C3 antibodies, respectively. C, Quantification of the expression levels of C1 and C3 via ImageMaster 2D Elite version 4.01 (GE Healthcare) according to “Materials and Methods” in C1ox strains in percentage in comparison with the appropriate control strain (n = 2). D, Measurements of luciferase activities (cRLUC) in the cRluc5b2 and -6b strains in comparison with the cRluc5b2::C1ox and cRluc6b::C1ox strains in relative light units (RLU; n = 3). Error bars represent the se of technical replicates. Cells were grown at 23°C and used at LD2 to measure the cRLUC activities.
The C1ox construct was transformed into both cRluc5b2 and -6b. Paromomycin-resistant strains were again analyzed for overexpression of C1. An overexpression of about 2-fold was found in two strains (Fig. 2, B and C). C3 was coregulated and overexpressed about 2- to 3-fold in the transgenic lines. Measurement of cRLUC activities in the cRluc5b2 and -6b lines and in the strains transformed in addition with the C1ox constructs (cRluc5b2::C1ox and cRluc6b::C1ox) showed that cRLUC activities were significantly increased in the C1 overexpression lines (Fig. 2D). Thus, overexpression of C1 controls C3 up-regulation via the (−300 to +142) promoter region of C3.
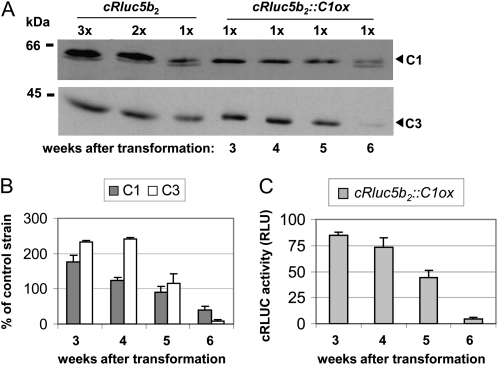
In the past, we had found that strains where the C1 subunit is silenced or overexpressed are not stable with regard to their expression level over time (Iliev et al., 2006). This was observed again when transforming the different cRluc strains with the C1ox construct. Overexpression of C1, which was only found with amplitude of about 2-fold as seen before (Iliev et al., 2006), returned back to wild-type levels after several weeks. We checked with one strain the reversion of C1 over time and analyzed if the coregulation of C3 would also be concerned (Fig. 3). Five weeks after transformation, the level of C1 was reversed back to the wild-type level, and the same was observed with the level of C3 (Fig. 3, A and B). After 6 weeks, the level of C1 was even below the wild-type level as well as the C3 level. These data are also in accordance with the reduction of the C3 promoter-triggered cRLUC activities within this time frame (Fig. 3C). Thus, the reversion of C1 back to the wild type includes a tight coregulation of the level of C3 back to the wild type. The reporter assays show that this reversion is mediated via the C3 promoter region.
Figure 3.
Reversion of C1 in the C1-overexpressing strain cRluc5b2 includes reversion of the C3 level. A, Different amounts of proteins from a crude extract (90, 60, and 30 μg per lane) labeled as 3x, 2x, and 1x, respectively, of cRluc5b2 cells that had been transformed in some cases with the C1ox vector (cRluc5b2::C1ox) were separated by 9% SDS-PAGE. They were used for immunodetection with anti-C1 and anti-C3 antibodies, respectively. cRluc5b2::C1ox cells were grown and harvested at different time points after transformation, as indicated. B, Quantification of the expression levels of C1 and C3 via ImageMaster 2D Elite version 4.01 (GE Healthcare) according to “Materials and Methods” in C1ox strains in percentage in comparison with the appropriate control strain (n = 2). C, Measurements of cRLUC activities in the cRluc5b2::C1ox strain at different time points after transformation in relative light units (RLU; n = 3). Error bars represent the se of technical replicates. Cells were grown at 23°C and used at LD2 to measure the cRLUC activities.
Replacement of Any of the Two DREB1A-Boxes Does Not Influence Circadian C3 Expression and the Coregulation of C3 by C1
As already mentioned, temperature-dependent up-regulation of C3 was reduced in amplitude when either of the two DREB1A-boxes had been replaced (Voytsekh et al., 2008). A possible function of these two boxes for circadian regulation had not been analyzed at this time. We now examined if any of the two DREB1A-boxes influence circadian expression of C3 and/or the coregulation of C3 in C1-overexpressing lines. Therefore, we used different cRluc transgenic lines that bear the already described chimeric cRluc construct including the C3 3′ UTR that is under the control of the (−300 to +142) C3 promoter in different variant forms (Voytsekh et al., 2008). In the cRluc strains ΔD(+72)1 and ΔD(+72)2, the DREB1A-box at position +72 that is situated in the C3 5′ UTR had been replaced by other nucleotides representing a HindIII restriction site (Supplemental Fig. S1A). In the cRluc strains ΔD(−130)1 and ΔD(−130)2, the DREB1A-box at position −130 had been replaced by other nucleotides representing a HindIII site (Supplemental Fig. S2A). Measurements of cRLUC activities in two transgenic cRluc ΔD(+72) lines and in two ΔD(−130) lines were carried out over a circadian cycle. Exemplary cRLUC activities of the transgenic cRluc lines ΔD(+72)1 (Supplemental Fig. S1B) and ΔD(−130)2 (Supplemental Fig. S2B) are shown. In both cases, a rhythm of cRLUC activity was observed with a peak at the end of subjective night similar to that observed with the wild-type promoter (Voytsekh et al., 2008). These data suggest that the two DREB1A-boxes are not relevant for circadian expression of C3.
For analyzing their potential role in the coregulation of C3 by C1, the C1 overexpression construct was transformed in all of these transgenic lines. Paromomycin-resistant strains were used for crude extract preparations. Again, it was examined by immunoblotting if C1 is overexpressed and if C3 would be coregulated (Supplemental Figs. S1 and S2, C and D). Overexpression of C1 ranged from about 1.5- to 2.5-fold depending on the strain, and coregulation of C3 occurred in the range of about 2- to 4-fold depending on the strain.
In all transgenic cRluc ΔD lines as well as in those overexpressing in addition C1 (ΔD::C1ox), cRLUC activities were then measured (Supplemental Figs. S1 and S2E). Replacement of the DREB1A-boxes at either +72 or −130 did not influence the coregulation of C3. cRLUC activities were significantly higher in the ΔD::C1ox strains compared with the corresponding ΔD strains (Supplemental Figs. S1 and S2E). Thus, the absence of any of the two DREB1A-boxes does not influence the up-regulation of C3 by C1.
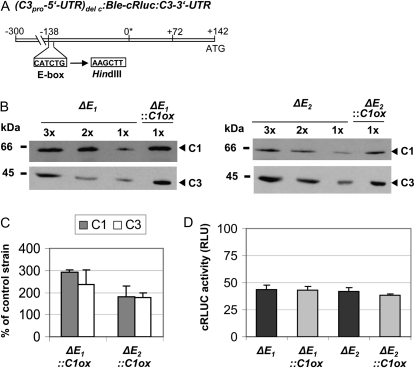
The E-Box Also Mediates C3 Coregulation in C1-Overexpressing Lines
In the past, we had shown that replacement of the E-box prevents temperature-dependent regulation of C3 as well as circadian C3 expression (Voytsekh et al., 2008). To examine its potential involvement in the coregulation of C3 in C1-overexpressing lines, we used the chimeric cRluc strains ΔE1 and ΔE2. There, the cRluc reporter is under control of the C3 promoter region (−300 to +142), but the E-box at position −138 had been replaced by other nucleotides representing again a HindIII site (Fig. 4A). The C1 overexpression construct was transformed in the two transgenic ΔE lines. Paromomycin-resistant strains were used for crude extract preparations. Again, it was examined by immunoblotting if C1 is overexpressed and if C3 would be coregulated (Fig. 4, B and C). Overexpression of C1 ranged from about 2- to 3-fold depending on the strain, and coregulation of C3 occurred in the same range. Measurements of cRLUC activities in the cRluc strains ΔE1::C1ox and ΔE2::C1ox in comparison with the cRluc lines ΔE1 and ΔE2 revealed rather equal activities in the C1ox strains (Fig. 4D). Thus, replacement of the E-box within the C3 promoter disrupts C3 coregulation in the ΔE::C1ox strains. These data suggest that the E-box is also the key element for the coregulation of C3 and underlines that it has multiple roles.
Figure 4.
The replacement of an E-box within the C3 promoter region prevents the coregulation of C3 by C1. A, The chimeric construct (C3pro-5′UTR)del c:Ble-cRluc:C3-3′UTR of pDI26 having a shortened C3 promoter and a replaced E-box at −138 (Voytsekh et al., 2008) that was used for transformation resulting in transgenic cRluc lines ΔE1 and ΔE2 is schematically shown. B, Different amounts of proteins from a crude extract (90, 60, and 30 μg per lane) labeled as 3x, 2x, and 1x, respectively, of the cRluc strains ΔE1 and ΔE2 that had been transformed in some cases with the C1ox vector (ΔE1::C1ox and ΔE2::C1ox) were separated by SDS-PAGE. They were used for immunodetection with anti-C1 and anti-C3 antibodies, respectively. C, Quantification of the expression levels of C1 and C3 via ImageMaster 2D Elite version 4.01 (GE Healthcare) according to “Materials and Methods” in C1ox strains in percentage in comparison with the appropriate control strain (n = 2). D, Measurements of cRLUC activities in the cRluc strains ΔE1 and ΔE2 in comparison with the cRluc strains ΔE1::C1ox and ΔE2::C1ox in relative light units (RLU; n = 3). Error bars represent the se of technical replicates. Cells were grown at 23°C and used at LD2 to measure the cRLUC activities.
Proteins Interacting with the E-Box Region of C3
The different roles of the E-box could be mediated by different factors or different modified forms of the same factor(s) that are activated (1) at low temperature, (2) in a circadian way, or (3) upon C3 coregulation by C1. We have decided to investigate first proteins that are involved in the up-regulation of C3 at 18°C; therefore, we grew the cells at this temperature for the following experiments. Studies on C3 coregulation would have been only possible with transgenic C1ox lines that are not stable over time with regard to the expression level, as mentioned before.
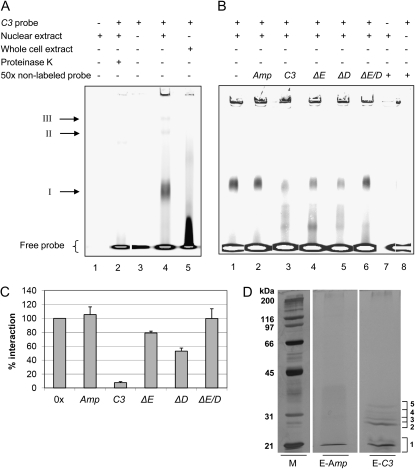
For identification of the factor(s) binding to the E-box region, we started to perform fluorescence-based electromobility shift assays (EMSA; see “Materials and Methods”; Steiner and Pfannschmidt, 2009). Fluorescent dyes were shown to have a similar sensitivity as radioactivity (Ying et al., 2007). Since the binding of a factor to the E-box may be also influenced by its surrounding nucleotides, we used a DY781 dye-labeled 45-bp DNA fragment of the C3 promoter (positions −154 to −109; the C3 probe) that contains the E-box in its center. It should be noted that this probe contains also one of the DREB1A-boxes that is situated only two nucleotides away from the E-box. Cells were harvested at LD2, and crude extracts as well as nuclear extracts were prepared. At LD2 and 18°C, up-regulation of C3 involving the E-box was studied before (Voytsekh et al., 2008). Proteins from crude extracts of C. reinhardtii (Fig. 5A, lane 5) as well as from nuclear preparations (lane 4) were incubated in the presence of a nonspecific competitor [poly(dIdC)] with the dye-labeled DNA fragment, and protein-DNA complexes were separated by native PAGE. As a control, nuclear proteins alone (lane 1) or the labeled DNA fragment alone (lane 3) was loaded. With total proteins from a crude extract, the DNA was only shifted to some extent. In the case of the nuclear proteins, a main shift (I) and two minor shifts (II and III) of the labeled DNA were visible. Incubation of the DNA-protein mixture with proteinase K (lane 2) revealed their disappearance, showing that they are based on DNA-protein interactions. It should be noted that the two minor shifts II and III were not always observed in different mobility shift assays, while shift I was consistently observed in all assays.
Figure 5.
Identification of proteins from a nuclear extract that bind specifically to the E-box region of the C3 promoter. A to C, Fluorescence-based EMSAs along with a 45-bp DNA fragment (named the C3 probe) representing the E-box-containing region of the C3 promoter (−154 to −109) with whole cell or nuclear extracts from cells that were grown at 18°C and harvested at LD2. The 5′ DY781 fluorescence-labeled C3 probe was incubated with 20 μg of proteins from nuclear or whole cell extracts, which had been preincubated with 2 μg of poly(dIdC) for 10 min (see “Materials and Methods”). Samples were analyzed on a 6% native polyacrylamide gel and visualized using the LI-COR Odyssey Infrared Imaging System. A, Proteins from the nuclear extract alone (lane 1), labeled probe in the absence of proteins (lane 3) or in the presence of proteins from a nuclear extract (lane 4) or whole cell extract (lane 5), and after treatment of nuclear extract with proteinase K (lane 2). B and C, EMSA experiments in the absence (lane 1) or presence of 50-fold excess of nonlabeled probes (lanes 2–6) along with proteins from nuclear extracts solely with nonlabeled probe (lane 7) or along with labeled C3 probe and no proteins (lane 8). Nonlabeled probes representing Amp DNA, a random fragment of the E. coli Bla gene (lane 2), C3 probe (lane 3), C3 probe containing either a replacement of the E-box (ΔE; lane 4), of the DREB1A-box (ΔD; lane 5), or of both boxes (ΔE/D; lane 6). C, Fifty-fold molar excess of nonlabeled probes was used. The degree of DNA-protein interaction in shift I was quantified via ImageMaster 2D Elite version 4.01 (GE Healthcare) evaluating independent competition experiments (n = 4). Error bars represent the se. D, Proteins from nuclear extracts of cells that were grown at 18°C and harvested at LD2 were used for an affinity purification by incubating them with biotinylated C3 probe, which was captured by paramagnetic streptavidin particles (see “Materials and Methods”). Proteins were eluted with a 50-fold molar excess of nonbiotinylated C3 probe (E-C3) or unspecific Amp DNA (E-Amp). Eluted proteins were separated by 10% piperazine diacrylamide-SDS-PAGE along with molecular mass standards and were silver stained. The numbered brackets indicate areas that were cut and analyzed by nano LC-ESI-MS/MS in both lanes (E-Amp and E-C3; see “Materials and Methods”). M, Broad range protein standard (BioRad).
In a next step, we examined the specificity of binding using in addition to poly(dIdC) (Fig. 5B, lane 1) a 50-fold excess of nonlabeled oligonucleotides for the assay, representing (1) the C3 probe as a control (lane 3), (2) a nonrelated fragment from the Ampicillin resistance gene, named Amp (lane 2), (3) the C3 probe with both replaced E- and DREB1A-boxes (ΔE/D; lane 6), (4) the C3 probe with only the replaced E-box (ΔE; lane 4), or (5) only the replaced DREB1A-box (ΔD; lane 5). Shift I was fully maintained in the case of the nonspecific Amp probe, where it was even slightly strengthened, as well as with the ΔE/D probe (Fig. 5, B and C). Addition of the unlabeled ΔE probe led to reduction of shift I down to approximately 80%, and addition of ΔD probe reduced shift I down to approximately 50% (Fig. 5, B and C). Thus, a protein or protein complex interacting specifically with the E-box is present in shift I, but the only two nucleotides distinct DREB1A-box also recruits protein(s) within this complex to some extent.
To identify the factor(s) interacting with the E-box region, we used a biochemical approach. The 45-bp C3 probe was biotinylated and used as bait along with proteins from nuclear extracts in the presence of poly(dIdC) (for details, see “Materials and Methods”). At first, the biotinylated DNA was captured with streptavidin paramagnetic beads, and then the proteins from the nuclear extract were incubated with the beads. Nonbound proteins were removed in three washing steps. The release of the bound proteins was done by adding 50-fold excess of nonlabeled C3 probe. As a control, the same procedure was carried out and an excess of 50-fold Amp probe was used.
Proteins from eluates with either the C3 probe or Amp were then separated on SDS-PAGE and silver stained (Fig. 6D). Stained protein bands deriving from the eluate of the C3 probe were cut out of the gel and in parallel the same regions from the eluate of the Amp probe (control) were cut out, even when no protein band was visible in the latter. Proteins were in-gel tryptic digested and used for nano liquid chromatography-electrospray ionization-tandem mass spectrometry (nano LC-ESI-MS/MS) analysis (for details, see “Materials and Methods”). Evaluation was done with a false discovery rate of not more than 1%. Only proteins that were found solely in the C3 and not in the Amp eluate were considered as interaction partners of the E-box region. These are listed in Table I if they were identified in two independent experiments with at least one peptide or in only one experiment (labeled with an asterisk) with more than one peptide. The stained band 2 that is most prominent in the C3 probe eluate contained a protein that has been described as G-strand telomere-binding protein 1 (GBP1; Petracek et al., 1994; Johnston et al., 1999). In the weaker stained bands 3 to 5, HLA8, annotated as a nuclease that bears a conserved nuclease domain as well as a transcription factor TFIIB family domain (predicted by InterPro; IPR000812) was found, as were two proteins with conserved short-chain dehydrogenase/reductase (SDR) domains. In band 1, a HU-like protein (HLP1) that is known to bend DNA was present. The potential functions of these factors for transcriptional activation of C3 will be discussed later.
Figure 6.
C3 can be immunodetected in the nucleus along with GBP1, and both are present in the C3 probe eluate and in shift I. A, C3 probe eluates after affinity chromatography from nuclear extracts were prepared from cells harvested at LD2 and LD22 (as described in Fig. 5D; 100 μL). Proteins were separated by 9% SDS-PAGE and immunoblotted with anti-GBP1 antibodies (Johnston et al., 1999). B, Cells were harvested at LD2 and LD22. Crude (CE) and nuclear (NE) extracts were prepared, and proteins (25 μg per lane) were separated by 9%, and for GBP1 detection by 14%, SDS-PAGE and immunoblotted with anti-C1, anti-C3, and anti-GBP1 antibodies, respectively. C, The same procedure as described in A was carried out, but immunoblotting was done with anti-C3 antibodies. D, Supershift experiments with anti-C3 and anti-GBP1 antibodies. EMSAs were basically performed as described above using nuclear extracts of cells grown at 18°C and harvested at LD2 (lanes 1–3) or LD22 (lanes 4–6). For supershifts, 0.5 μL of anti-C3 (lanes 2 and 5) or 0.25 μL of anti-GBP1 antibody (lanes 3 and 6) was added (see “Materials and Methods”). 5′ DY781 fluorescence-labeled C3 probe alone (lane 7) was used as a control. The position of the protein-DNA complex I is indicated by the arrow. E, Shift I was cut out of the native gel, and the slice was loaded on a standard 12% SDS-PAGE apparatus. Proteins present on the gel slice as well as proteins from a nuclear extract and a molecular mass standard were separated by SDS-PAGE and immunoblotted with anti-C3 and anti-GBP1 antibodies, respectively.
Table I. Nano LC-ESI-MS/MS analysis of proteins binding to the E-box region.
The name/conserved domains (CD) of depicted proteins are given as determined by National Center for Biotechnology Information BLAST and their e-values. Xcorr, Cross-correlation factor; z, charge.
| Protein Identifier |
Name/Conserved Domains | Identified Peptides | z | Xcorr | |
| Chlre2.0 | Chlre4.0 | ||||
| 154362 | 146567 | G-strand telomere-binding protein 1; CDs: 2× RRM; Expect = 1e-19, 3e-14 | DMFAEVGGVDR | 2 | 3.18 |
| QADGAPGAPAER | 2 | 2.93 | |||
| RLAVFIDR | 2 | 3.03 | |||
| ADVVTGYDGR | 2 | 3.17 | |||
| 158312 | 129012 | High-light-induced nuclease; CDs: staphylococcal nuclease homolog domain; Expect = 8e-21; InterPro: transcription factor TFIIB-related domain | FFCTESEATSAGWR | 2 | 3.96 |
| GIWAGEFQVPSEWR | 2 | 4.15 | |||
| 164634 | 137062 | Predicted protein; CD: short-chain dehydrogenase/reductase; Expect = 9e-46 | GLAAELGPEGIRa | 2 | 3.29 |
| 152972 | 59969 | Predicted proteinb; CD: short-chain dehydrogenase/reductase; Expect = 2e-64 | AEGVTAMGLQGDVR | 2 | 4.26 |
| ELVQSTIPLR | 2 | 2.71 | |||
| SAVDSLTR | 1 | 2.08 | |||
| SLALEWGEFNVR | 2 | 4.17 | |||
| VALVTGGSSGIGFEIAR | 2 | 3.71 | |||
| VNGVAPGPIDGTAGMTK | 2 | 3.06 | |||
| 162329 | 307591 | Predicted proteinb; CD: HU_IHF, a member of the DNABII protein family that binds and bends DNA, functioning as architectural factors in many cellular processes, including transcription, site-specific recombination, and higher order nucleoprotein complex assembly; Expect = 2e-23 | AFDSLIGGIEDALINGDR | 2 | 2.65 |
| LVEAIATEVGLTK | 2 | 4.59 | |||
| NPSTGAVLQIAASK | 2 | 4.01 | |||
The spectra of the peptide were manually evaluated.
The protein was only identified in one of two independent large-scale experiments.
GBP1 and C3 Interact with the E-Box Region of C3 during Early Day and Late Night
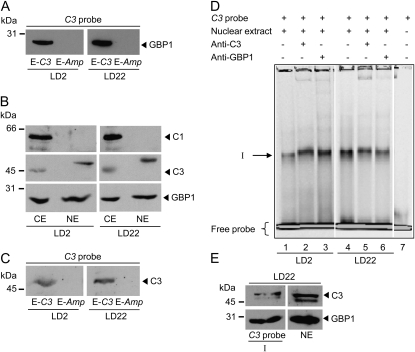
As already mentioned, the E-box is involved in circadian expression of C3. Its mRNA peaks at the end of the night (LD22) within a 12-h-light/12-h-dark cycle. All so far described experiments for DNA-protein interactions were conducted with cells that had been grown at 18°C and harvested at LD2. We were also interested to see if any of the interacting factors would be present in the C3 probe-protein complex at the end of the night (LD22) when C3 mRNA is increased (Kucho et al., 2005; Voytsekh et al., 2008). In the case of GBP1 from C. reinhardtii, an anti-GBP1 antibody was available that allowed immunological detection (Petracek et al., 1994).
In a next step, we performed the affinity purification with the biotinylated C3 probe along with nuclear extracts from cells grown at 18°C and harvested at LD2 and LD22, respectively. Proteins were again eluted with nonbiotinylated C3 or Amp probe. The presence of GBP1 in the eluates was then verified by immunoblotting using the anti-GBP1 antibodies. At both time points, a strong GBP1 signal was visible in the C3 probe eluate but not in the Amp eluate (Fig. 6A). The signal appeared even stronger in the LD22 nuclear extracts, which is in accordance with an increased C3 expression at LD22. However, comparative quantification with different cellular subfractions and after affinity purification may be not fully reliable for quantitative analysis.
In other organisms, components of the oscillatory machinery like PER in Drosophila or FREQUENCY in Neurospora feed back on their own transcription, as mentioned before. In the purification of the factors interacting with the E-box region, we did not observe a stained band in the region of about 45 or 65 kD, where C3 and C1 are migrating. However, if any of the two proteins are interacting indirectly with GBP1 or any of the other factors, their amount might be low depending on the stability of the protein-protein interaction. Thus, they may not be visible by silver staining. We first analyzed at both LD2 and LD22 if C1 and C3, which we considered as cytosolic RNA-binding proteins, can be found in the nuclear fraction. For these preparations, cells were grown at 23°C. Immunoblotting was done with the same amounts of proteins from crude extracts and nuclear fractions along with anti-C1, anti-C3, and anti-GBP1 antibodies (Fig. 6B). As expected, GBP1 was found in the nuclear fractions at both LD2 and LD22. C1 was only detected in the crude extract but not in the nuclear fraction. However, C3 was present in the nuclear fraction (LD2 and LD22). It migrates slightly higher in comparison with its migration behavior in the crude extract, indicating that it may be present in some posttranslationally modified form within the nucleus. We then analyzed in the C3 probe and Amp eluates if C3 can be detected by immunoblotting. At both LD2 and LD22, C3 was found in the C3 probe eluate and was missing in the Amp eluate (Fig. 6C). These data suggest that C3 can interact with its own promoter, most likely in an indirect way via protein-protein interaction, as will be discussed later.
To further characterize the binding of GBP1 and C3 to the E-box-containing region, we tried supershift experiments with anti-C3 and anti-GBP1 antibodies (Fig. 6D). However, the results were not clear. Shift I was only very slightly if at all retarded (lanes 2/3 and 5/6) in comparison with a control without antibodies (lanes 1 and 4) using both nuclear extracts prepared from cells harvested at LD2 and LD22. It could be that the applied conditions are not well suited for the protein-antibody interactions or that the protein complex interacting with the E-box region prevents efficient access of the antibodies. Thus, we tried another approach. The region of shift I was cut out of the native gel, and the slice was loaded on a SDS-PAGE apparatus and immunoblotted with anti-GBP1 and anti-C3 antibodies (Fig. 6E). As a positive control, proteins from nuclear preparations were loaded. Both GBP1 and C3 were immunodetected in shift I. Thus, both proteins are present in the DNA-protein complex of shift I.
The E-Box Alone Is Sufficient for the Binding of GBP1 and C3 at Both Low and High Temperatures
Since the binding of a factor to the E-box may be influenced also by its surrounding nucleotides, we had chosen a 45-bp fragment where the E-box is situated in the center for all assays. However, it also contains one of the two DREB1A-boxes that is only two nucleotides away from the E-box and has some influence in the amplitude of the temperature-dependent up-regulation of C3 (Voytsekh et al., 2008) but not for the coregulation of C3 in C1ox strains (Supplemental Fig. S2E) and for its circadian expression (Supplemental Fig. S2B). The assays with unlabeled probe (Fig. 5, B and C) had shown that replacement of the E-box still recruits about 80% interaction within shift I, but not 100%. This was only achieved when both E- and DREB1A-boxes had been replaced.
We wanted to know if the E-box alone is sufficient for the binding of GBP1 and C3, resulting in shift I. Therefore, we created an E-box construct that has the six-nucleotide-long E-box as well as two nucleotides upstream and downstream of it five times in a row and used this for a mobility shift assay. As seen with the C3 probe, three shifts were visible that we labeled again as I, II, and III, according to their similar migration pattern as observed with the C3 probe containing the E-box region (Fig. 7A, lane 2). All shifts disappeared after treatment with proteinase K (lane 4). We cut out three regions of the native gel corresponding to the positions of the shifted bands and separated the protein constituents by SDS-PAGE. GBP1 and C3 were detected by immunoblotting. As a control, nuclear extracts alone were used. In shifts II and III, there was no reaction detectable with any of the two antibodies, while shift I resulted in a strong signal with anti-GBP1 antibodies and a weaker signal with anti-C3 antibodies (Fig. 7B). When proteins from the nuclear extract alone without incubation of labeled DNA were separated on a native gel and the region of shift I was excised and treated as described above, it was found that neither GBP1 nor C3 alone migrates on native gels at the position of shift I (Fig. 7C). Thus, GBP1 and C3 can interact directly or indirectly (via a protein that is bound to the E-box) with the E-box alone and do not require the extended other surrounding nucleotides, including the DREB1A-box, that are present in the 45-bp C3 probe also named the E-box region.
Figure 7.
C3 and GBP1 are present in shift I along with an E-box probe at both low and high temperatures. A, EMSAs were performed as described before, but a 5′ DY781 fluorescence-labeled five-times repeat of the E-box including two surrounding nucleotides at each site (5×E-box probe) instead of the C3 probe was used. Nuclear extracts were prepared from cells grown at 18°C and harvested at LD22. Nuclear extract alone (lane 3), labeled probe in the presence of nuclear extract (lane 2) or in its absence (lane 1), as well as nuclear extract treated with proteinase K (lane 4) are shown. The positions of protein-DNA complexes I, II, and III are indicated by arrows. B, Shifts I, II, and III were cut out of the gel, and proteins on the gel slices were subjected to SDS-PAGE (as described before) along with proteins from nuclear extracts (NE). Immunoblotting was done using anti-C3 and anti-GBP1 antibodies, respectively. C, The same procedure as described for B was carried out, but the region of shift I from a sample lane, which contained proteins from nuclear extracts that had not been incubated with labeled DNA probe, was cut out. D, The same procedure as described for B was carried out, but nuclear extracts were prepared from cells grown at 28°C and 18°C and harvested at LD2.
We also carried out EMSAs along with the 5×E-box probe and proteins from nuclear extracts of cells grown at either 18°C or 28°C and harvested at LD2 to check the influence of temperature (data not shown). Shifts I, II, and III were again excised from the gel and treated as described above. Both C3 and GBP1 were identified at both temperatures (Fig. 7D).
DISCUSSION
The Mechanism of Coregulation of C3 and the Different Roles of the E-Box
In the green alga C. reinhardtii, the duo of subunits of the RNA-binding protein CHLAMY1 was shown to maintain period (C1 subunit) and phase (C3 subunit) of the circadian clock (Iliev et al., 2006). It was also found that overexpression and silencing of the C1 subunit caused a parallel increase (in the case of overexpression) or decrease (by silencing) of the C3 subunit. In contrast, changes in the level of the C3 subunit did not influence the level of C1 significantly. In this study, we aimed to analyze the mechanism of the coregulation of C3 by C1. In some cases, like with the assembly-governed regulation of the biogenesis of subunits from the photosystems (PSI and PSII), translation is involved in such a process, including the 5′ UTR (Minai et al., 2006). Here, we found that an E-box element that is situated in the C3 promoter mediates the coregulation of C3 in C1 overexpression lines. Two DREB1A-boxes are also situated in the C3 promoter region. They play a limited role in the temperature-dependent regulation of C3 that is up-regulated at 18°C, a temperature being still in the physiological range of C. reinhardtii. DREB1A-boxes are recognized by cold stress (usually 4°C or below)-induced transcription factors in higher plants (Agarwal et al., 2006). Replacement of any of the two DREB1A-boxes did not influence C3 circadian expression and the coregulation of C3 by C1. Thus, the E-box has three different roles. It is the key element for (1) the up-regulation of C3 at low temperature, (2) circadian C3 expression (Voytsekh et al., 2008), and (3) the up-regulation of C3 by increased levels of C1. For the control of these regulatory processes, it may be recognized by different factors depending on its role. But it is also possible that posttranslational modifications such as reversible phosphorylation or small ubiquitin-like modifiers of a common factor or a set of factors are involved.
The increase in C1 results in tight coregulation of C3 via the E-box. Thus, C1 could either directly activate C3 expression or induce a yet unknown factor that activates C3. KH domain-containing proteins such as C1 are able to recognize RNA and DNA sequences, as was found in the case of poly(C)-binding proteins (Du et al., 2007). Also, it was shown that DNA-protein-binding complexes such as nuclear factor κB can contain KH domain subunits. Ribosomal protein S3 was found as a subunit of this complex (Wan et al., 2007). But within the frame of these studies, we also examined the presence of C1 in nuclear extracts, a prerequisite for acting directly or within a protein complex on the C3 promoter. But C1 was not found by immunodetection in nuclear extracts from cells harvested at LD2 and LD22, in contrast to the C3 subunit. Thus, it seems unlikely that C1 is directly coregulating C3.
Factors Interacting with the E-Box Region
Another aim of our work was to characterize and identify binding factors that interact with the E-box region within the C3 promoter. Therefore, we grew the cells at 18°C when C3 was shown to be up-regulated (Voytsekh et al., 2008). In EMSAs, three mobility shifts were found with proteins from nuclear extracts, and the main shift I is based on a specific interaction of protein(s) with the E-box region. EMSAs along with different unlabeled probes showed that the E-box along with the two nucleotide distinct DREB1A-box recruit proteins present in shift I. Replacement of the E-box showed 80% interaction of the proteins, while replacement of both boxes kept the 100% level. These data suggest that these motifs act partially in concert as binding sites for a protein complex. To identify the proteins interacting with the E-box region, we applied an affinity approach that is based on a biotinylated C3 probe that was captured by paramagnetic streptavidin beads. Elution of proteins from a nuclear extract was done with 50-fold excess of unlabeled C3 probe and the same amount of Amp probe as a control.
Three proteins were identified by nano LC-ESI-MS/MS within the C3 probe eluate in two independent experiments and two others solely in one of the two experiments (but with more than one peptide). None of them was found in the Amp eluate (control). The most prominent stained band 2 included GBP1, a protein that was characterized before in C. reinhardtii (Petracek et al., 1994). It has two RRM domains and was shown to recognize the G-strand telomere sequence (TTTTAGGG)n. Moreover, it was found that monomeric GBP1 associates with either single-stranded (ss)DNA or RNA and that dimeric GBP1 has a strong preference for binding ssDNA (Johnston et al., 1999). In these studies, proteins from a crude extract (1 μg of total protein per binding assay) were used for the detection of the mobility shift of GBP1 with the telomere sequence. When we used proteins from a protein crude extract (20 μg of total protein per binding assay) along with the C3 probe, shift I that contains GBP1 was not detected. Only when proteins from nuclear extracts were used was it observed. We assume that either some posttranslationally modified form of GBP1 or its interaction with other proteins that were also found in shift I leads to the specific binding of GBP1 to the E-box region. It is also possible that any of the other factors is involved in the direct interaction with the DNA and that GBP1 is recruited by protein-protein interaction with the E-box region. In any case, our data indicate multiple functions for GBP1. Multiple functions of proteins with RRM domains are not unusual. For example, TAR DNA-binding protein 43 (TDP-43), which is a dimeric protein with two RRM domains, is involved in DNA and RNA binding (Kuo et al., 2009). TDP-43 was originally identified as a transcriptional factor, repressing the transcription of the Hiv1 gene (Ou et al., 1995), but it is also involved in the transcriptional regulation of mouse Sp-10 gene by interaction with the Sp-10 insulator (Abhyankar et al., 2007). But TDP-43 is also acting as a splicing factor (Buratti and Baralle, 2001; Ayala et al., 2008), and it was suggested that it is in addition involved in mRNA stability, transport, and local translation in neurons (Strong et al., 2007; Wang et al., 2008).
The availability of anti-GBP1 antibodies allowed us to confirm by immunodetection its presence in the C3 probe eluate after the affinity approach conducted with cells harvested at LD2 and LD22. We could also reveal its presence in shift I using the E-box alone as probe at both l8°C and 28°C. Thus, GBP1 seems to be involved in both temperature-dependent regulation of C3 and its circadian expression. An RNA interference approach of GBP1 will be necessary in the future to study its specific roles in these two processes. It will also be of interest to examine if it might also be involved in the coregulation of C3 by C1. We cannot rule out that the proteins present in shift I may include in addition to C3 and GBP1 different partners or posttranslational modifications of identified partners when cells are grown at different temperatures or harvested at different times of the day/night cycle.
A second identified factor, HLA8, is annotated as nuclease. In the past, it was shown that the C. reinhardtii Hla8 gene is induced under high-light conditions (Im and Grossman, 2001). HLA8 has a conserved nuclease domain, and InterPro additionally predicts a transcription factor TFIIB-related domain (IPR000812). The presence of this factor along with the RRM domain bearing GBP1 and C3 might indicate some kind of similar mechanism of transcriptional control as was found in c-Myc transcriptional control (Crichlow et al., 2008, and refs. therein). Transcription of this oncogene involves FUSE-binding protein (FBP), which binds to the ssDNA sequence upstream of the c-Myc promoter, known as far upstream element (FUSE). FUSE becomes single stranded due to forces generated during c-Myc transcription, and transcription stalls. FBP is a stimulator of the p89 helicase subunit of the transcription factor TFIIH, allowing c-Myc transcription to proceed. The FBP action is counterbalanced by FBP-interacting repressor (FIR), which binds the FUSE ssDNA, FBP, and TFIIH, thus reducing c-Myc transcription back to basal levels. Both FBP and FIR are nucleic acid-binding proteins with RNA-binding domains, bearing either four KH domains (FBP) or two RRM domains (FIR).
Two other factors that were found (one of them only in one of the two experiments) have conserved SDR domains. SDRs constitute a large family of NAD(P) (H)-dependent oxidoreductases. They have critical roles in lipid, amino acid, carbohydrate, and other metabolic reactions as well as in redox sensor mechanisms (for review, see Kavanagh et al., 2008). Their NAD(P)(H) binding can include nonenzymatic functions (redox sensoring). For example, the SDR-fold fungal transcriptional regulator NmrA differentially binds oxidized nucleotide cofactors, thus linking redox status to interactions with transcription factors (Lamb et al., 2003). The SDR proteins within the C3-binding complex may also exert such a function.
Finally, a HU-like protein was identified in the C3 probe eluate that constitutes a member of the DNABII protein family, binding and bending DNA. This protein was only found in one of two experiments, but with more than one peptide. Integration host factor (IHF) and HU proteins belong to a family of proteins that introduce sharp bends into DNA and are commonly found in prokaryotes (Rice, 1997).
The just described factors that were identified in the C3 probe eluate implicate a novel mechanism of transcriptional control via the E-box region, which has to be further investigated in the future. At the moment, we cannot exclude that so far known transcription factors interacting with E-box elements that have been described in the introduction have been missed by our approach. This could be due, for example, to very low abundance of such factors. Thus, the identified factors here may act as coregulators, as was shown with hnRNP U and the c-MYC-MAX complex recently (Matsuoka et al., 2009). Homologues of the CLK/CYC duo that bind to the E-box of the Per gene in Drosophila have not been found in the genome of C. reinhardtii (Mittag et al., 2005). bHLH factors in C. reinhardtii are rare. Only four have been predicted, while in Arabidopsis, predictions for 160 were made (Riaño-Pachón et al., 2008). Stepwise functional characterization of the identified factors, binding directly or indirectly to the E-box region (e.g. by RNA interference) will be necessary to depict their specific roles.
The Feedback of C3
As mentioned before, positive and negative feedback loops have been observed in the oscillatory machinery of different model organisms. Since C3 was immunologically detected in nuclear extracts, we wanted to examine if C3 could feed back on the C3 promoter region, most likely by protein-protein interaction with any of the identified factors. C3 is a member of the CELF/Bruno-like RNA-binding proteins (Zhao et al., 2004) including CUG-binding proteins (CUG-BP) that have three RRM domains and are implicated in major posttranscriptional regulatory processes, including alternative splicing, the control of translation, and the stability of target mRNAs (Barreau et al., 2006). Recently, it was shown that RRM3 is the structural basis for the sequence-specific RNA binding (Tsuda et al., 2009). To our knowledge, there are no indications so far that members of this family can bind to DNA. However, protein-protein interactions are known, such as the interaction of C3 with C1 (Zhao et al., 2004) or of CUG-BP1 with the eIF2 complex (Salisbury et al., 2008).
In the C3 probe eluate, there was no evidence for the presence of an approximately 45-kD protein after separating the proteins by SDS-PAGE and silver staining them. Thus, if C3 were part of the protein complex interacting with the E-box, it could only be present in very low amounts there. Immunological detection with anti-C3 antibody showed a signal in the C3 probe eluate as well as in shift I caused by the E-box region and the E-box alone. Therefore, we conclude that C3 can feed back most likely by protein-protein interaction to any of the identified factors to its own promoter. This was observed when cells were grown at different temperatures or harvested at both LD2 and LD22. These data suggest that C3 is part of a positive feedback loop influencing its temperature-dependent and circadian regulation.
MATERIALS AND METHODS
Standard molecular biology methods were followed according to Sambrook and Russell (2001).
Cell Culture
Chlamydomonas reinhardtii cells (wild-type strain SAG73.72 and the cell wall-less cw15 mt+ strain SAG83.81 that was used for the nuclear preparations) were grown in Tris-acetate phosphate medium (Harris, 1989) under a 12-h-light/12-h-dark cycle (LD12:12) with a light intensity of 71 μE m−2 s−1 (1 E = 1 mol of photons) at 18°C, 23°C, or 28°C, as indicated for each experiment. The beginning of the light period is defined as time zero (LD0) and the beginning of the dark period as LD12. Cells were harvested either at LD2 or LD22, as indicated for each experiment. In some cases, cells were released after light/dark entrainment into constant conditions of dim light (20 μE m−2 s−1) and temperature (23°C).
Preparation of Plasmid Constructs, Characterization of Transgenic Algae, and Immunoblots
The construction of the reporter luciferase vectors Hsp70A/RbcS2pro:Ble-cRLUC:C3-3′UTR (pSK27), (C3pro-5′UTR)del b:Ble-cRLUC:C3-3′UTR (pDI23), (C3pro-5′UTR)del c:Ble-cRLUC:C3-3′UTR (pDI26), (C3pro-5′UTR)del d:Ble-cRLUC:C3-3′UTR (pDI27), and (C3pro-5′UTR)del e:Ble-cRLUC:C3-3′UTR (pDI28) were described by Voytsekh et al. (2008). For ease of understanding, we renamed some of the cRluc strains described by Voytsekh et al. (2008): ΔD(+72)1 (formerly crluc6e), ΔD(+72)2 (formerly crluc5e2), ΔD(−130)1 (formerly crluc6d2), ΔD(−130)2 (formerly crluc5d), ΔE1 (formerly crluc5c), and ΔE2 (formerly crluc6c2). The cRLUC-expressing strains were transformed with the C1ox vector (pDI6), which was described by Iliev et al. (2006). The transformation procedure and subsequent characterization of the transgenic strains by immunoblots were done as described before along with chemiluminescence detection (Zhao et al., 2004; Iliev et al., 2006). Densitometry analysis was done as described previously (Voytsekh et al., 2008). Thereby, signals of control strains (cRluc) deriving from 30-, 60-, and 90-μg protein amounts were taken as 100%, 200%, and 300% and used for the creation of standard curves. Using these standard curves, the percentages of the signals (based on the 30-μg protein amount) of the cRluc::C1ox strains were calculated. cRLUC activity measurements were carried out as described previously (Kiaulehn et al., 2007).
Nuclei Isolation and Preparation of Nuclear Extracts
Nuclei from C. reinhardtii SAG83.81 were isolated with the CelLytic PN plant nuclei isolation/extraction kit (Sigma-Aldrich; Del Viso et al., 2007) according to the manufacturer's manual with some modifications. Briefly, cells were harvested in the middle log phase (6 × 106 cells mL−1) by centrifugation (2 min, 3,000g, 4°C). The pellet was suspended and washed in 0.005× volume of 1× NIB (CelLytic PN kit [Sigma-Aldrich], 0.01% [w/v] protease inhibitor cocktail [Roche], and 0.1 mm dithiothreitol [DTT]). Subsequently, the cell suspension was homogenized by grinding it in liquid N2 to a fine powder. 1× NIB buffer (3 mL g−1 cells) was added, and the suspension was passed through a nylon mesh (Sigma-Aldrich). After centrifugation (10 min, 1,260g, 4°C), the supernatant was decanted and the pellet was resuspended completely in 2 mL of 1× NIBA (1× NIB buffer with 1% [v/v] Triton X-100). This step was repeated four times, although with different centrifugation conditions: two times for 30 min at 1,000g and 4°C, and two times for 10 min at 600g and 4°C. Each time, the supernatant was decanted and the pellet was suspended completely in 2 mL of 1× NIBA. Finally, the residual white/grayish pellet was suspended completely in 2 mL of 1× NIB. Then, the buffer was removed by centrifugation (10 min, 12,000g, 4°C). The quality of the nuclear preparations was positively verified by fluorescence microscopy using a DNA stain with mithramycin (Supplemental Fig. S3; Hill and Whatley, 1975). The preparation of nuclear extracts was done according to the kit's manual by gentle agitation (30 min, 4°C) in the provided nuclear extraction buffer that is described as suitable for the detection of DNA-protein interactions (CelLytic PN kit manual; Sigma-Aldrich; Del Viso et al., 2007). Protein concentration was measured with the BioRad Protein Assay according to its manual with bovine serum albumin as a standard.
Fluorescence-Based EMSA, including Competitor and Supershift Experiments
The binding reaction and the EMSA were performed according to Steiner and Pfannschmidt (2009) with some modifications. Briefly, 20 μg of protein in extraction buffer was preincubated in the presence of 2 μg of poly(dIdC) (Sigma-Aldrich) for 10 min at room temperature in 29.4-μL volume (filled up with 10 mm Tris-HCl, 1.67 mm EDTA, pH 7.0, 1.67 mm DTT, and 10 mm MgCl2) before 0.6 μL (0.4 ng μL−1) of double-stranded DY781-labeled probe (for preparation, see below) was added. Some experiments include unlabeled, double-stranded DNA probes in 50-fold molar excess or proteinase K (3 μg μg−1 nuclear protein; Sigma-Aldrich). It was added to the binding reaction, allowing an additional incubation time of 10 min at room temperature, according to Lee et al. (2000). The binding reaction was carried out for 30 min at room temperature before the sample was loaded to a 6% native polyacrylamide gel (Steiner and Pfannschmidt, 2009). Gel electrophoresis and scanning using a LI-COR Odyssey fluorescence imager were done according to Steiner and Pfannschmidt (2009). Densitometry analysis was performed as described before (Voytsekh et al., 2008). For the supershift experiments (according to Zhao et al., 2004), 0.5 μL of anti-C3 or 0.25 μL of anti-GBP1 antibody was added to the binding reaction, allowing an additional 20-min incubation time prior to loading to the EMSA gel.
Purification of DNA-Binding Proteins Using a Biotin/Streptavidin Affinity System
The double-stranded biotinylated DNA (for preparation, see below) was immobilized on 1 mg of equilibrated paramagnetic streptavidin beads (Promega) according to the manufacturer's manual. Nuclear extracts were obtained from nuclei isolated from 0.5 L of cell culture (about 1.6 mg of protein). Proteins from nuclear extracts had been preincubated according to the binding reaction described above with poly(dIdC). They were added to the biotinylated DNA-streptavidin beads, and incubation was continued for 30 min at 4°C with gentle agitation. Beads were washed three times with 600 μL of 10 mm Tris-HCl, 1.67 mm EDTA, pH 7.0, 1.67 mm DTT, and 10 mm MgCl2. Elution of the bound proteins was obtained by incubation with 50-fold molar excess of nonbiotinylated DNA probe for 30 min at 4°C and gentle agitation.
Sequences and Annealing Procedure of Oligonucleotides
To obtain the DY781- or biotin-labeled probes and the unlabeled competitor DNA, the appropriate sequences were selected and obtained as sense (with or without labeling) or antisense (always nonlabeled) oligonucleotides from Biomers.net. Oligonucleotide OMM494 represents the C3 probe sense sequence (5′-TTATTTGTGTGTGAAACATCTGTGGTCGGCGTCGAGCGCGAGTGC-3′) with 5′-DY781 fluorescence label, OMM552 with 5′-biotinylation, and OMM516 is the unlabeled form. OMM517 represents the unlabeled antisense C3 probe sequence (5′-GCACTCGCGCTCGACGCCGACCACAGATGTTTCACACACAAATAA-3′). Oligonucleotide OMM528 represents the C3 probe sense sequence with a random replacement of the E-box (5′-TTATTTGTGTGTGAAAAAGCTTTGGTCGGCGTCGAGCGCGAGTGC-3‘) and OMM520 the unlabeled antisense probe (5′-GCACTCGCGCTCGACGCCGACCAAAGCTTTTTCACACACAAATAA-3′). Oligonucleotide OMM529 has the C3 probe sense sequence with a random replacement of the DREB1A-box (5′-TTATTTGTGTGTGAAACATCTGTGAACGAAGTCGAGCGCGAGTGC-3′) and OMM522 is the unlabeled antisense probe (5′-GCACTCGCGCTCGACTTCGTTCACAGATGTTTCACACACAAATAA-3′). Oligonucleotide OMM541 represents the C3 probe sense sequence with a random replacement of both E- and DREB1A-boxes (5′-TTATTTGTGTGTGAAAGAGCGCAGAAGTGGGTCGAGCGCGAGTGC-3′) and OMM522 the unlabeled antisense probe (5′-GCACTCGCGCTCGACCCACTTCTGCGCTCTTTCACACACAAATAA-3′). The 5×E-box probe sense sequence (5′-AACATCTGTGAACATCTGTGAACATCTGTGAACATCTGTGAACATCTGTG-3′) is present in OMM691 (with 5′-DY781 fluorescence label) and OMM684 (unlabeled). Its unlabeled antisense sequence (5′-CACAGATGTTCACAGATGTTCACAGATGTTCACAGATGTTCACAGATGTT-3′) is present in OMM685. The Amp probe sense sequence, being a random fragment of the Escherichia coli Bla gene (5′-AGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAAC-3′), is contained in OMM535 and the appropriate antisense sequence (5′-GTTGCAGGACCACTTCTGCGCTCGGCCCTTCCGGCT-3′) in OMM536. Equimolar amounts of the complementary oligonucleotides that had been solved in 10 mm Tris, pH 7.0, were heated to 95°C and allowed to cool slowly to 4°C in a Thermocycler.
Peptide Identification by Nano LC-ESI-MS/MS and Data Analysis
Proteins in the eluates from the affinity approach were prepared for nano LC-ESI-MS/MS analysis by separating them by 10% SDS-PAGE that contains 0.3% piperazine diacrylamide as a cross-linker (Wagner et al., 2004). Subsequent to the gel electrophoresis, the gel was silver stained (Heukeshoven and Dernick, 1986), and appearing bands were sliced out. The slices were destained with freshly prepared destaining solution (15 mm K3[Fe(CN)6] and 50 mm NaS2O3) for 8 min at room temperature and washed four times with MS-grade water. Proteins in the gel slices were in-gel tryptic digested, and peptides were subjected to nano LC-ESI-MS/MS using an UltiMate 3000 nano HPLC apparatus (Dionex) coupled online with a linear ion trap nano ESI mass spectrometer (Finnigian LTQ; Thermo Electron) as described previously (Schmidt et al., 2006). Data analysis was done using the Proteome Discoverer software (version 1.0) from Thermo Electron including the SEQUEST algorithm (Link et al., 1999). The parameters for all database searches were set to achieve a false discovery rate of not more than 1% for each individual analysis according to Boesger et al. (2009). Data were searched against the Joint Genome Institute C. reinhardtii database (versions 2.0 and 4.0).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The replacement of a DREB1A element within the C3 5′ UTR does not influence circadian C3 expression and coregulation of C3 by C1.
Supplemental Figure S2. The replacement of a DREB1A element at −130 of the C3 promoter does not influence circadian C3 expression and coregulation of C3 by C1.
Supplemental Figure S3. Fluorescence and phase-contrast microscopy of C. reinhardtii cells and isolated nuclei.
Supplementary Material
Acknowledgments
We thank C. Milkowski, K.M. Mohan, and O. Voytsekh for helpful comments on the manuscript, J. Berman for giving us anti-GBP1 antibody, F.V. Winck for helpful comments on the nuclear isolation protocol, N. Alekseeva and E.M. Schmidt for initial experiments on the nuclear localization of C1 and C3, the laboratory of T. Pfannschmidt for letting us use its LI-COR system, as well as C. Schimek and J. Wöstemeyer for letting us use their fluorescence microscope. We also appreciate very much the free delivery of the EST and genome sequences from the genome projects in the United States (Joint Genome Institute) and Japan.
References
- Abhyankar MM, Urekar C, Reddi PP. (2007) A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem 282: 36143–36154 [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Auvinen M, Järvinen K, Hotti A, Okkeri J, Laitinen J, Jänne OA, Coffino P, Bergman M, Andersson LC, Alitalo K, et al. (2003) Transcriptional regulation of the ornithine decarboxylase gene by c-Myc/Max/Mad network and retinoblastoma protein interacting with c-Myc. Int J Biochem Cell Biol 35: 496–521 [DOI] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE. (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA 105: 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Méreau A, Osborne HB. (2006) Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie 88: 515–525 [DOI] [PubMed] [Google Scholar]
- Boesger J, Wagner V, Weisheit W, Mittag M. (2009) Analysis of flagellar phosphoproteins from Chlamydomonas reinhardtii. Eukaryot Cell 8: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG. (1972) Mutants of the biological clock in Chlamydomonas reinhardtii. Genetics 70: 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Crichlow GV, Zhou H, Hsiao HH, Frederick KB, Debrosse M, Yang Y, Folta-Stogniew EJ, Chung HJ, Fan C, De la Cruz EM, et al. (2008) Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J 27: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Viso F, Casaretto JA, Quatrano RS. (2007) 14-3-3 proteins are components of the transcription complex of the ATEM1 promoter in Arabidopsis. Planta 227: 167–175 [DOI] [PubMed] [Google Scholar]
- Du Z, Lee JK, Fenn S, Tjhen R, Stroud RM, James TL. (2007) X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains poly(C)-binding protein-2. RNA 13: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, Colot HV, Mehra A, Belden WJ, Shi M, Hong CI, Larrondo LF, Baker CL, Chen CH, et al. (2007) A circadian clock in Neurospora: how genes and proteins cooperate to produce a sustained, entrainable and compensated biological oscillator with a period of about a day. Cold Spring Harb Symp Quant Biol 72: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Hausherr A, Ferbitz L, Schödl T, Heitzer M, Hegemann P. (2004) Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol 55: 869–881 [DOI] [PubMed] [Google Scholar]
- Garbarino-Pico E, Green CB. (2007) Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol 72: 145–156 [DOI] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, New York [Google Scholar]
- Heukeshoven J, Dernick R. (1986) Improved silver staining for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9: 28–32 [DOI] [PubMed] [Google Scholar]
- Hill BT, Whatley S. (1975) A simple rapid microassay for DNA. FEBS Lett 56: 20–23 [DOI] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt E-M, Fiedler M, Nykytenko A, Mittag M. (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Grossman A. (2001) Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J 30: 301–313 [DOI] [PubMed] [Google Scholar]
- Johnston SD, Lew JE, Berman J. (1999) Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol Cell Biol 19: 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. (2009) A role for microRNAs in the Drosophila circadian clock. Genes Dev 23: 2179–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh KL, Jörnvall H, Persson B, Oppermann U. (2008) The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65: 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiaulehn S, Voytsekh O, Fuhrmann M, Mittag M. (2007) The presence of UG-repeat sequences in the 3′ UTRs of reporter luciferase mRNAs mediates circadian expression and determines acrophase in Chlamydomonas reinhardtii. J Biol Rhythms 22: 275–277 [DOI] [PubMed] [Google Scholar]
- Kucho K, Okamoto K, Tabata S, Fukuzawa H, Ishiura M. (2005) Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol Biol 57: 889–906 [DOI] [PubMed] [Google Scholar]
- Kuo P-H, Doudeva LG, Wang Y-T, Shen C-KJ, Yuan HS. (2009) Structural insights into TDP-43 in nucleic acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb HK, Leslie K, Dodds AL, Nutley M, Cooper A, Johnson C, Thompson P, Stammers DK, Hawkins AR. (2003) The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J Biol Chem 278: 32107–32114 [DOI] [PubMed] [Google Scholar]
- Lee M, Yu S, Park J-S. (2000) Biochemical characterization of a nuclear factor that binds to NF1-like elements in the rat p53 promoter. J Cell Biochem 78: 1–7 [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III (1999) Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17: 676–682 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Uehara N, Tsubura A. (2009) hnRNP U interacts with the c-Myc-Max complex on the E-box promoter region inducing the ornithine decarboxylase gene. Oncol Rep 22: 249–255 [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The evolution of key animal and plant functions is revealed by analysis of the Chlamydomonas genome. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman FA, Choquet Y. (2006) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M. (1996) Conserved circadian elements in phylogenetically diverse algae. Proc Natl Acad Sci USA 93: 14401–14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH. (2005) The circadian clock in Chlamydomonas reinhardtii. What is for? What is it similar to? Plant Physiol 137: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S-HI, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol 69: 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. (2007) Snail, ZEB and bHLH factors in tumor progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Konkel LMC, Kable ML, Berman J. (1994) A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J 13: 3648–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing L, Ruoff P. (2002) Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int 19: 807–864 [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Corrêa LGG, Trejos-Espinos R, Mueller-Roeber B. (2008) Green transcription factors: a Chlamydomonas overview. Genetics 179: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA. (1997) Making DNA do a U-turn: IHF and related proteins. Curr Opin Struct Biol 7: 86–93 [DOI] [PubMed] [Google Scholar]
- Rolland N, Atteia A, Decottignies P, Garin J, Hippler M, Kreimer G, Lemaire SD, Mittag M, Wagner V. (2009) Chlamydomonas proteomics. Curr Opin Microbiol 12: 285–291 [DOI] [PubMed] [Google Scholar]
- Rosbash M, Bradley S, Kadener S, Li Y, Luo W, Menet JS, Nagoshi E, Palm K, Schoer R, Shang Y, et al. (2007) Transcriptional feedback and definition of the circadian pacemaker in Drosophila and animals. Cold Spring Harb Symp Quant Biol 72: 75–84 [DOI] [PubMed] [Google Scholar]
- Salisbury E, Sakai K, Schoser B, Huichalaf C, Schneider-Gold C, Nguyen H, Wang GL, Albrecht JH, Timchenko LT. (2008) Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp Cell Res 314: 2266–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al. (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18: 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL. (2008) The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci 28: 10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Pfannschmidt T. (2009) Fluorescence-based electrophoretic mobility shift assay in the analysis of DNA-binding proteins. Methods Mol Biol 479: 273–289 [DOI] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci 35: 320–327 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. (2009) Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res 37: 5151–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytsekh O, Seitz SB, Iliev D, Mittag M. (2008) Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol 147: 2179–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V, Boesger J, Mittag M. (2009) Sub-proteome analysis in the green flagellate alga Chlamydomonas reinhardtii. J Basic Microbiol 49: 32–41 [DOI] [PubMed] [Google Scholar]
- Wagner V, Fiedler M, Markert C, Hippler M, Mittag M. (2004) Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett 559: 129–135 [DOI] [PubMed] [Google Scholar]
- Waltenberger H, Schneid C, Grosch JO, Bareiß A, Mittag M. (2001) Identification of target mRNAs from C. reinhardtii for the clock-controlled RNA-binding protein Chlamy1. Mol Genet Genomics 265: 180–188 [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. (2007) Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131: 927–939 [DOI] [PubMed] [Google Scholar]
- Wang I-F, Wu L-S, Chang H-Y, Shen C-KJ. (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 105: 797–806 [DOI] [PubMed] [Google Scholar]
- Ying B-W, Fourmy D, Yoshizawa S. (2007) Substitution of the use of radioactivity by fluorescence for biochemical studies of RNA. RNA 13: 2042–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. (2008) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt E-M, Wagner V, Wollnik F, Mittag M. (2004) The circadian RNA-binding protein CHLAMY1 represents a novel type heteromer of RNA recognition motif and lysine homology-containing subunits. Eukaryot Cell 3: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.