Abstract
Prostacyclin is a potent mediator of vasodilation and anti-platelet aggregation. It is synthesized from prostaglandin H2 by prostacyclin synthase (PGIS), a member of Family 8 in the cytochrome P450 superfamily. Unlike most P450s, which require exogenous reducing equivalents and an oxygen molecule for mono-oxygenation, PGIS catalyzes an isomerization with an initial step of endoperoxide bond cleavage of prostaglandin H2 (PGH2). The low abundance of PGIS in natural tissues necessitates heterologous expression for studies of structure/function relationships and reaction mechanism. We report here a high-yield prokaryotic system for expression of enzymatically active human PGIS. The PGIS cDNA is modified by replacing the hydrophobic amino-terminal sequence with the more hydrophilic amino-terminal sequence from P450 2C5 and by adding a four-histidine tag at the carboxyl terminus. The resulting recombinant PGIS associates with host cell membranes and was purified to electrophoretic homogeneity by nickel affinity, hydroxyapatite and CM Sepharose column chromatography. The recombinant PGIS, with a heme:protein ratio of 0.9:1, catalyzes prostacyclin formation at a Km of 13.3 µM PGH2 and a Vmax of 980 per min. The dithionite-reduced PGIS binds CO with an on-rate of 5.6 × 105 M−1 s−1 and an off-rate of 15 s−1. The ferrous–CO complex of PGIS is very short-lived and decays at a rate of 0.7 s−1. Spectral binding assays showed that imidazole binds weakly to PGIS (Kd ~0.5 mM,) but clotrimazole, a bulky and rigid imidazole derivative, binds strongly (Kd ~1 µM). The transient nature of the CO complex and the weak imidazole binding seem to support an earlier proposal that PGIS active site has a limited space, but the tight binding of clotrimazole argues against this view. It appears that the heme distal pocket of PGIS is fairly adaptable to ligands of various structures. UV-visible absorption, magnetic circular dichroism and electron paramagnetic resonance spectra indicate that PGIS has a typical low-spin heme with a hydrophobic active site. PGIS catalyzes homolytic scission of the peroxide bond of a test substrate, 10-hydroperoxyoctadeca-8,12-dienoic acid, accompanied by formation of a heme intermediate with a Compound II-like optical spectrum.
Keywords: Cytochrome P450, Prostanoid, Heterologous expression, Homolysis of peroxide bond, Active site
1. Introduction
Prostacyclin synthase (PGIS) catalyzes an isomerization of prostaglandin H2 (PGH2) to form prostacyclin (also known as prostaglandin I2, PGI2), which enhances vasodilation and is the most potent endogenous antagonist of platelet aggregation [1]. PGIS activity was first found in aorta [2] and the enzyme was later purified as a 50-kDa hemoprotein with spectroscopic characteristics of a cytochrome P450 [3,4]. It was assigned to the P450 superfamily as CYP8A1 when its cDNA sequence was discovered [5,6]. Unlike most microsomal P450s, which require an oxygen molecule and P450 reductase to transfer external electrons for a mono-oxygenase reaction, PGIS does not need oxygen or reductase. The atypical nature of PGIS catalysis invited detailed structure/function and reaction mechanism studies to gain better understanding of the variety of catalytic strategies used by the P450 superfamily.
Despite low sequence identity among twenty structurally known P450s to date, they share a common overall fold and topology which is unique for P450 proteins [7]. Although the P450 fold is conserved, the precise structural element differs substantially, allowing the binding of substrates of significantly different sizes to different P450s. Some P450s are very rigid and stereo-specific for their substrates, whereas others are flexible, moving upon substrate binding in an induced-fit mechanism [7]. The active site of PGIS has been probed using substrate analogs and heme ligands [8,9]. Surprisingly, most common P450 ligands, even imidazole or pyridine derivatives, do not bind PGIS. Only a few nitrogen-containing compounds bind PGIS with notable affinities. These studies thus led to the hypothesis that the active site of PGIS has a very limited space and is accessible only for nitrogen compounds with side chains extending almost perpendicular to the Fe–N coordination axis [10]. The hypothesis which suggests a rigid active site has not been tested with a wider range of heme ligands. Additionally, a recent report showed that the difference optical spectrum of the carbon monoxide-ferrous PGIS exhibited a peak at 418 nm rather than the characteristic 450 nm [11]. This is in contrast with previous works in which a 440–450 nm species was reported for carbon monoxide-ferrous PGIS [3,4]. Hence, it warrants a revisit to these controversial results.
The prevailing reaction mechanism initially proposed by Hecker and Ullrich [8] involves the key step of an O–O homolytic scission of PGH2 to generate an alkoxyl radical from the substrate intermediate and a [Fe(IV)–O–R] species from the enzyme. The [Fe(IV)–O–R] species is a ferryl–oxo complex that is electronically analogous to Compound II (Cpd II) of peroxidases. In contrast, Compound I (Cpd I) of peroxidases is a ferryl–oxo coupled with porphyrin π-cation radical that results from heterolytic scission of the O–O hydroperoxide bond. Reaction of a chemical model of the P450 active site, using an Fe(III)–porphyrin coordinated by thiolate, with peroxy acid favored heterolytic O–O bond scission over homolytic O–O bond scission [12,13]. Another chemical model proposed homolytic O–O bond scission using Fe(II) [14]. Although some P450s that use hydroperoxides as substrates catalyze homolytic scission to form radicals as transient intermediates [15–17], homolytic scission of the O–O bond has not yet been established for PGIS.
Although the PGIS protein and its cDNA were isolated more than a decade ago, limited information has been obtained about its heme environment and its reaction mechanism. This is mainly due to difficulties in obtaining sufficient enzyme for biophysical studies. PGIS comprises only approximately 0.1% of aorta microsomal proteins, making it difficult to isolate sufficient protein for detailed spectroscopic examination [18]. Recombinant PGIS has been expressed in various eukaryotic cells [11,19,20], but these expression systems produced insufficient PGIS for mechanistic studies. We report here the development of a prokaryotic expression system for generation of milligram levels of active PGIS suitable for biophysical and biochemical studies. With sufficient PGIS, we are able to carry out spectroscopic studies such as stopped-flow absorption, resonance Raman, magnetic circular dichroism (MCD) and electron paramagnetic resonance (EPR) to advance our understanding of this physiologically important P450. Probing the active site environment with the heme ligands, we show that PGIS active site is very flexible to accommodate large and hydrophobic ligands. We also show that the ferrous–CO complex of PGIS is very short-lived. Results from reactions of the recombinant PGIS with a test peroxide substrate demonstrate homolytic cleavage of the O–O bond and formation of an enzyme intermediate resembling Cpd II.
2. Experimental procedures
2.1. Materials
Nickel nitrilotriacetate agarose (Ni-NTA) was obtained from Qiagen. CM Sepharose and hydroxyapatite were from Amersham Pharmacia Biotech and Bio-Rad, respectively. The pCW plasmid, containing two tac promoters and the lacIq gene, was a gift from Dr. Amy Roth (University of Oregon). U44069 (15- hydroxy-9,11-[epoxymethano]prosta-5,13-dienoic acid), U46619 (15-hydroxy-9,11-[methanoepoxy]prosta-5,13-dienoic acid), dihomo-γ-linolenic acid and 6-keto prostaglandin F1α were from Cayman. Minoxidil (6-(1-piperidyl) pyrimidine-2,4-diamine 3-oxide), clotrimazole (1-[o-chloro-α,α-diphenyl-benzyl]imidazole), cholate and Lubrol PX were from Sigma. CHAPS and n-octyl-β-d-glucopyranoside were purchased from Anatrace (Maumee, OH). Sheep antiserum against 6-keto prostaglandin F1α was acquired from Oxford Biochemical Research. 6-Keto[5,8,9,11,12,14,15-3H] prostaglandin F1α (6.88 TBq/mmol) and [1-14C]-linoleic acid (2.04 GBq/mmol) were acquired from Amersham Pharmacia Biotech. PGH1 and PGH2 were synthesized from dihomo-γ-linolenic acid and arachidonic acid, respectively, using detergent-solubilized ovine prostaglandin H synthase-1 and purified by a normal phase silica HPLC column as [21]. Prostaglandin H synthase-1 was reconstituted with heme or mangano protoporphyrin IX [22].
2.2. Construction of PGIS expression vectors
PGIS expression vectors with three different N-terminal modifications were made by PCR using human PGIS cDNA as the template [19]. For all three constructs, the downstream primer was designed to have a BglII site, a stop codon, a 4-histidine sequence and the C-terminal sequence of PGIS. The upstream primers had the N-terminal sequence of P450 2C5, MAKKTSS, with the initiator codon ATG within an NdeI site, followed by the sequence encoding the amino acid sequence AWAALLG (PGIS residues 2–8) or LLSRRRT (PGIS residues 18–24) for the 2C5/PGIS and 2C5/Δ(1–17)PGIS constructs, respectively (Fig. 1A). The third upstream primer was designed to encode the N-terminal sequence of P450 17A, MALLLAVF, followed by sequence LLSRRRT (PGIS residues 18–24), with an NdeI site at the 5′-end, to make the 17A/Δ(1–17)PGIS construct. All PCR products were digested with NdeI and BglII and ligated into the corresponding sites of the pCW plasmid. These constructs were transformed into E. coli BL21(DE3)pLys for protein expression.
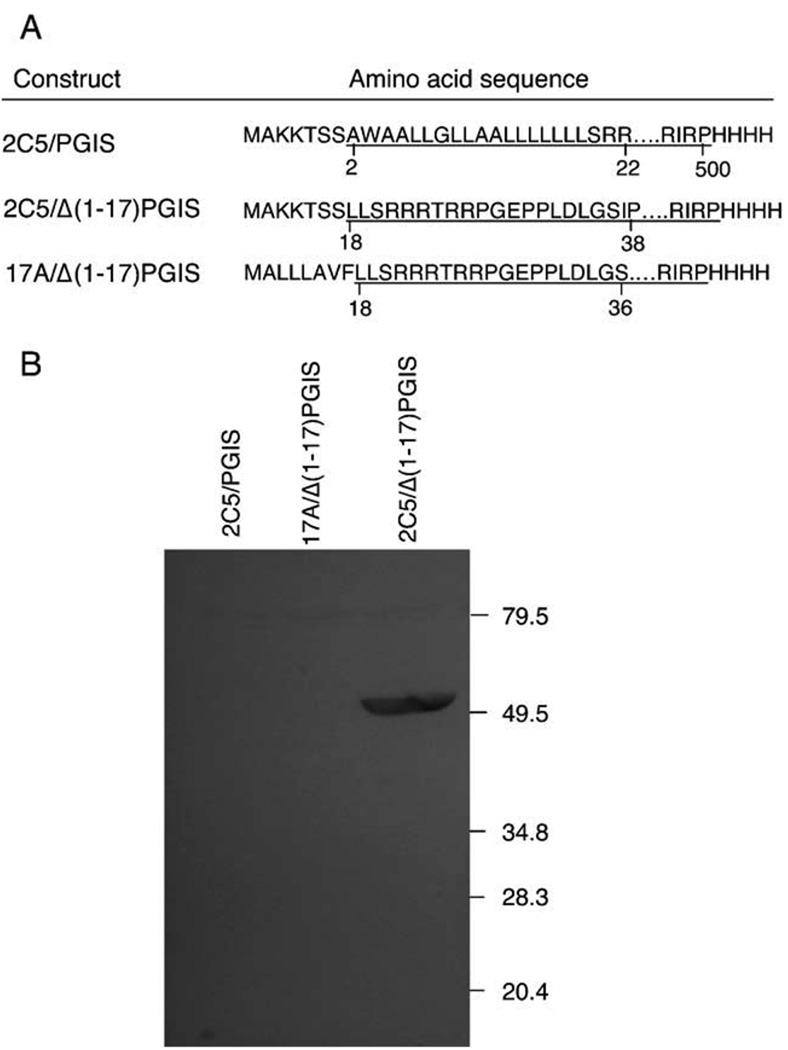
Fig. 1.
PGIS constructs and their expression levels. (A) The constructs, each with a distinctive amino terminus, were made by PCR and subcloned into the pCW vector. The construct names and their amino acid sequences are shown. The sequence derived from PGIS is underlined and the numbering of the amino acid residues in the wild type PGIS sequence is indicated. (B) Cells expressing each construct were sonicated, solubilized with n-octyl-β-d-glucopyranoside and subjected to ultracentrifugation. Aliquots of solubilized material, containing approximately 20 µg total protein, were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with antibody against anti-GST-PGIS fusion protein [19]. The positions of molecular weight markers (in kDa) are indicated.
2.3. Expression and purification of recombinant PGIS
Bacteria transformed with PGIS expression vector were grown overnight in LB media containing 35 µg/ml chloramphenicol and 100 µg/ml ampicillin. An overnight culture was inoculated at a 1:50 ratio into 2YT medium containing 100 µg/ml ampicillin. Bacteria were grown at 37 °C in a shaker at 250 rpm until the A600 was between 0.4 and 0.6 and δ-aminolevulinic acid was added to a final concentration of 0.25 mM. When the absorbance reached 0.8, expression of PGIS was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside, and the culture was continued for 18–22 h at 200 rpm and 30 °C before cells were harvested by centrifugation.
Frozen cell pellets from 4 l of cultured medium (~ 32 g) were thawed and resuspended in 120 ml of 50 mM NaPi, pH 7.5 containing 10% glycerol, 0.1 M NaCl, 50 µg/ml DNase and 4 mM MgCl2. The cells were lysed by a Fisher Sonic Model 500 Dismembrator with twenty cycles of 10-s burst/10-s cooling. Phenylmethylsulfonyl fluoride (1 mM) was added immediately after sonication. All the subsequent steps were performed at 4 °C. To solubilize PGIS, n-octyl-β-d-glucopyranoside was added to a final concentration of 30 mM and the homogenate was stirred overnight. Cell debris was first removed by centrifugation (8,000×g for 20 min), and solubilized recombinant PGIS was recovered in the supernatant following further centrifugation at 100,000×g for 1 h. The crude solubilized enzyme was applied to a Ni-NTA column (8-ml bed volume) pre-equilibrated with 20 mM NaPi, pH 7.5, containing 10% glycerol and 0.5 M NaCl. All the following buffers contained 0.2% cholate, 0.05% Lubrol PX and 10% glycerol. The column was first washed with 20 mM NaPi, pH 7.5, and 0.5 M NaCl until the A280 was less than 0.1. The column was then washed with 200 ml of 20 mM NaPi, pH 7.5 containing 2 mM histidine and 20 ml of the same buffer containing 4 mM of histidine. PGIS was finally eluted with 20 mM NaPi, pH 7.5 containing 100 mM histidine, diluted 3-fold with 10 mM NaPi, pH 7.2, and loaded on a hydroxyapatite column (5-ml bed volume). The column was first washed with 10 mM NaPi, pH 7.2 until the A280 was less than 0.05, and then washed with 20 mM NaPi, pH 7.2. PGIS was eluted with 100 mM NaPi, pH 7.2, diluted 5-fold with PGIS 10 mM NaPi, pH 7.2 and loaded on a CM sepharose column (5-ml bed volume). The column was washed with 20 mM NaPi, pH 7.2, containing 20 mM NaCl, and PGIS was eluted with 20 mM NaPi, pH 7.2, containing 150 mM NaCl.
Protein concentrations were determined by bicinchoninic acid assay using bovine serum albumin as a standard [23]. PGIS heme content was determined by formation of pyridine hemochromogen [24]. PGIS was also quantified from the tryptophan content by MCD using l-tryptophan as a standard [25].
2.4. Enzyme assay
For routine assay, we utilized the ability of PGIS to cleave PGH1 into 12- hydroxy-5,8,10-heptadecadienoic acid and malondialdehyde (MDA) [8], with formation of MDA monitored increase in A268 (ε = 31.5 mM−1 cm−1). The assay was carried out at 23 °C in a cuvette containing 400 µl of 20 mM NaPi, pH 7.4, 0.2% Lubrol PX and appropriate amounts of PGIS. The reaction was started with addition of 40 µM PGH1 and absorbance changes within the first 30 s were recorded for activity analysis.
To determine steady-state kinetic parameters using PGH2 as the substrate, PGIS (10 pmol) was added to 200 µl of 20 mM NaPi, pH 7.4, and 0.2% Lubrol PX at 23 °C. The reaction was initiated by adding 2.5–50 µM PGH2 and terminated after 20 s with 20 µl of 2 M citric acid. Formation of 6-keto prostaglandin F1α, the stable hydration product of PGI2, was measured by radio-immunoassay [26]. Non-linear regression analysis of the relationship between PGIS activity and PGH2 concentration was used to estimate Km and Vmax.
2.5. UV-Vis, MCD and EPR spectroscopy
UV-Vis absorbance spectra were recorded with a Shimadzu UV-2501PC spectrophotometer. Dissociation constants (Kd) of heme ligands with PGIS were determined by titration of PGIS with a concentrated stock solution of ligands. Perturbation spectra were recorded after each addition, and difference spectra were generated by subtraction of the spectrum of PGIS from each ligand-bound spectrum. Kd were calculated by fitting the peak minus trough amplitudes obtained from the difference spectra and the corresponding ligand concentrations to a one-site binding model.
For kinetic studies of carbon monoxide complex formation, PGIS was first reduced with dithionite under anaerobic conditions. A titration vessel containing PGIS in 20 mM NaPi, pH 7.4, and 0.2% Lubrol PX was made anaerobic by five cycles of alternating vacuum (30 s) and argon replacement (5 min). Dithionite was then added stepwise through a gastight Hamilton syringe attached to the titration vessel with absorption spectra recorded after each addition of dithionite to ensure a complete conversion of ferric PGIS to the ferrous form. A CO-saturated solution (0.94 mM at 23 °C) was prepared by bubbling of the buffer with CO for more than 5 min; dilution were made as needed. The reduced PGIS and CO-containing buffer were then loaded on a Bio-Sequential DX-18MV stopped-flow apparatus (Applied Photophysics, Leatherhead, UK) for equal volume mixing reaction at 23 °C. PGIS heme spectral changes were monitored in either photodiode array detection mode or single wavelength mode. For single wavelength kinetic data, the built-in software was used for rate analysis, and the rapid-scan data were analyzed with the Pro-K software package (Applied Photophysics).
Magnetic circular dichroism spectra were recorded at room temperature using a Jasco Model J-500C spectropolarimeter with a 1.4-T electromagnet. EPR spectra were recorded at 10K on a Bruker EMX using a GFS600 transfer line, an ITC503 temperature controller, and an Oxford ESR900 cryostat.
2.6. Preparation of [1-14C]-10-hydroperoxyoctadeca-8,12-dienoic acid ([1-14C]-10-OOH-18:2) and its reaction with PGIS
[1-14C]-10-OOH-18:2 was prepared by photooxygenation of [1-14C]-linoleic acid as described by Labeque and Marnett [27]. Briefly, linoleic acid (45 mg, 45 µCi) in 40 ml of methanol containing 3 mg of methylene blue and 5 mg of butylated hydroxytoluene was irradiated with a 150 W flood lamp for 67 h at 23 °C. The 10-OOH-18:2 was first separated from unreacted linoleic acid by silicic acid chromatography and subsequently from other hydroperoxides by HPLC using a semi-preparative silica column (Waters Spherisorb S10W, 10 µm, 10 × 250 mm). The authenticity of 10-OOH-18:2, synthesized from the non-radiolabelled linoleic acid, was confirmed by electrospray ionization mass spectroscopy: molecular ions of the compound complexed with Ag+ gave m/z of 419 and 421.
PGIS (2 µM) was reacted with [1-14C]-10-OOH-18:2 in 0.8 ml of 20 mM NaPi, pH 7.5, 10% glycerol and 0.2% Lubrol PX for 0.5 min. The reaction was stopped by acidification with 6 N HCl, followed by immediate extraction 3 times with 3 volumes of ether. The ether layers were dried over MgSO4, and the solvent was evaporated under nitrogen. The dried products were dissolved in a minimal volume of hexane and were separated on a Waters HPLC system equipped with a Spherisorb S5W silica column (5 µm, 4.6 × 250 mm). Products were eluted isocratically in hexane/isopropanol/acetic acid (985/15/1) at a flow rate of 1 ml/min. Radioactivity eluting from the column was quantitated by on-line liquid scintillation with a β-RAM Model 2 β-detector (IN/US system).
3. Results
3.1. Expression of PGIS constructs
In contrast to bacterial P450s, eukaryotic P450s are almost invariably membranous, anchoring in mitochondria or the endoplasmic reticulum. Extensive protein engineering studies over the past decade suggested a general approach for overexpression of eukaryotic P450s in E. coli, replacing the putative membrane-binding domain at the amino-terminus with another P450 amino-terminal sequence favoring prokaryotic expression [28]. The most widely used sequence is MALLLAVFL, the N-terminal sequence of bovine P450 17A [29]. However, this leader sequence failed to produce significant amounts of recombinant PGIS. Another leader sequence, MAKKTSS from human P450 2C5, has been utilized to enhance heterologous P450 expression levels [30]. We made two constructs that inserted this segment into the PGIS N-terminus (2C5/PGIS) or replaced the PGIS N-terminus with this segment (2C5/Δ(1–17)PGIS) (Fig. 1A). For comparison, we also replaced the N-terminal sequence in 2C5/Δ(1–17)PGIS with MALLLAVF (17A/Δ(1–17)PGIS). Four histidine residues were added to the carboxyl-termini of all these constructs. The recombinant proteins were solubilized with n-octyl-β-d-glucopyranoside and analyzed by immunoblotting. As seen in Fig. 1B, 2C5/Δ(1–17)PGIS was the only construct that gave a substantial level of recombinant PGIS expression. The 2C5/Δ(1–17)PGIS construct was also transformed in other E. coli strains, such as JM109 and Rosetta, but there was little change in PGIS expression levels; neither was expression enhanced when the modified PGIS cDNA was controlled by the T7 polymerase promoter (data not shown). For simplicity, we refer below the recombinant protein expressed from the 2C5/Δ(1–17)PGIS construct as PGIS.
3.2. Purification of recombinant PGIS
To improve yield, several detergents were assessed for solubilization of recombinant PGIS from E. coli membrane. We tested n-octyl-β-d-glucopyranoside (20 mM), Emulgen 913 (1%), CHAPS (16 mM), Renex (1%) and a mixture of cholate (0.2%) and Lubrol PX (0.1%) and found that n-octyl-β-d-glucopyranoside gave the highest yield (data not shown). We thus used n-octyl-β-d-glucopyranoside for solubilization, but switched to the less expensive cholate/Lubrol mixture to preserve PGIS solubility for the rest of purification procedures. The cholate/Lubrol mixture was also used to purify endogenous bovine PGIS [8].
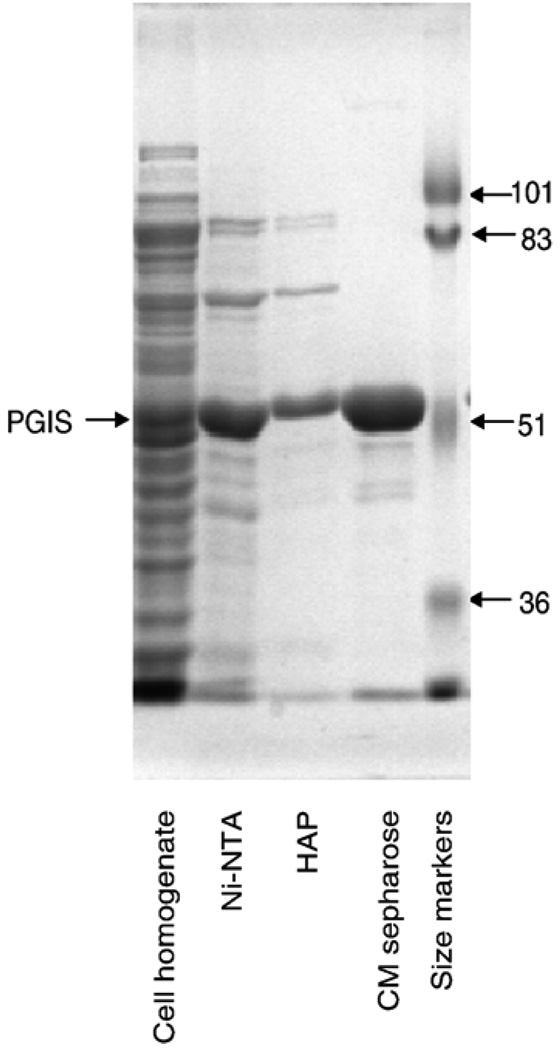
PGIS was solubilized directly from the cell homogenate without isolation of the membrane fraction. A protocol that includes chromatography on Ni-NTA, hydroxyapatite and CM Sepharose was developed for purification of recombinant PGIS (Table 1). The Ni-NTA affinity column chromatography step provided the most effective purification, increasing the PGIS specific activity by 25-fold. After hydroxyapatite and CM Sepharose column chromatography, the PGIS was more than 95% pure by electrophoretic analysis (Fig. 2). The authenticity of PGIS was confirmed by immunoblot analysis. Overall, the recombinant PGIS was purified about 45-fold with a yield of near 40% (Table 1). Purified PGIS is stable at room temperature for at least ten days without significant loss of catalytic activity.
Table 1.
Purification of recombinant PGIS expressed in E. coli
| Step | Total protein (mg) |
Specific activity * (µmol MDA/min/mg protein) |
Yield (%) |
|---|---|---|---|
| Homogenate | 1740 | 1.1 | 100 |
| Ni-NTA column | 63 | 25 | 82 |
| HTP column | 43 | 32 | 72 |
| CM column | 16 | 46 | 39 |
Results shown are for a typical run with cells from 2 l culture of E. coli BL21 (DE3)pLys transformed with the 2C5/Δ(1–17)PGIS construct.
Activity was assayed at 23 °C using 30 µM PGH1 as the substrate. Absorbance changes at 268 nm were monitored for MDA formation as described in Experimental procedures.
Fig. 2.
SDS-PAGE of PGIS at various stages of purification. Aliquots taken at each purification step, as indicated at the bottom, were separated by electrophoresis on a 10% polyacrylamide gel and the proteins stained by Coomassie Blue. The positions of PGIS and size markers are indicated.
3.3. Steady-state kinetic analysis of PGIS
A fixed amount of PGIS was incubated with various concentrations of PGH2 for 20 s at 23 °C. Quantitation of 6-keto PGF1α, the hydrolyzed product of PGI2, was carried out by radio-immunoassay. Analysis of the initial rates versus PGH2 concentrations indicated a Km value of 13.3±1.4 µM and a Vmax value of 980±42 mole of PGI2/min/mol of protein. These values are comparable to those reported for endogenous bovine PGIS: Km, 9.0±5 µM; Vmax, 147±45 mole of PGI2/min/mole [5]. Using PGH1 as the substrate, PGIS exhibited a Km value of 6.2±1.2 µM and a Vmax value of 2400±200 mole of MDA/min/mole.
3.4. UV-Vis spectra of PGIS
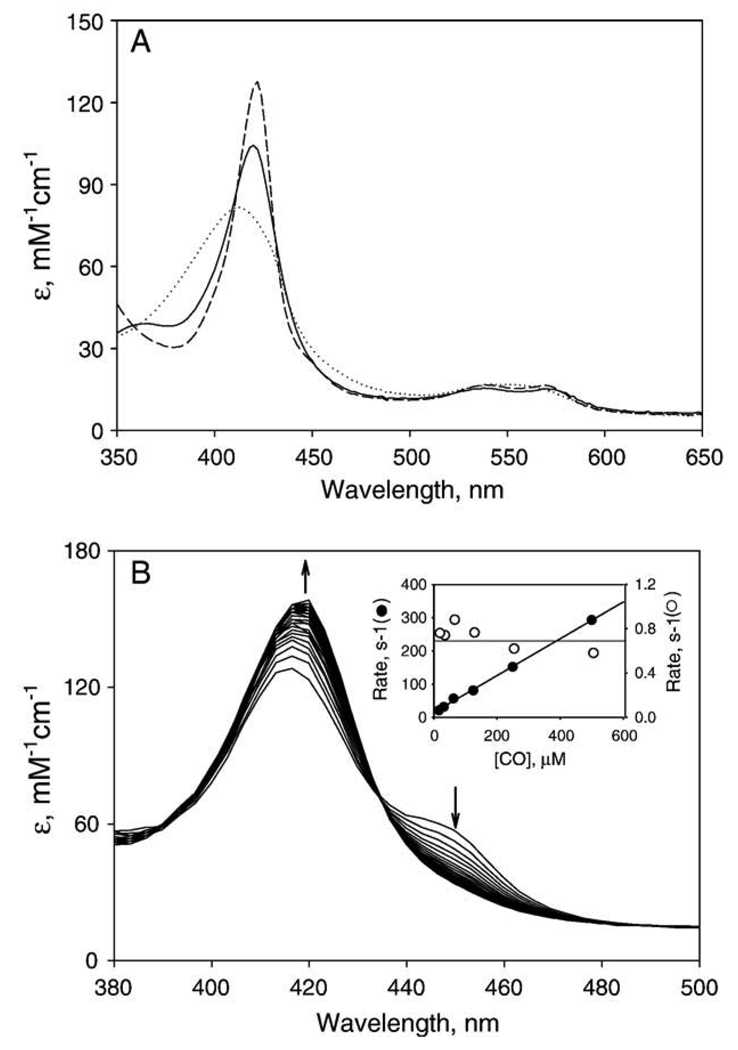
Resting PGIS exhibited an absorption spectrum consistent with a low-spin ferric P450, with a Soret peak at 418 nm and α and β bands at 570 and 537 nm, respectively (Fig. 3A and Table 2). The heme content determined by pyridine-hemochromogen analysis was 0.91±0.02 mol of heme/mol of protein. Our recombinant PGIS which has eight tryptophan residues was compared with l-tryptophan standard by MCD spectra to determine the extinction coefficients of given wavelengths [25]. The extinction coefficients at 280 nm and 418 nm thus determined were 100 and 103 mM−1 cm−1, respectively.
Fig. 3.
Absorption spectra of PGIS. (A) Resting PGIS (5 µM) in 20 mM NaPi, pH 7.4, 10% glycerol and 0.2% Lubrol PX (solid line) was reduced by dithionite under anaerobic conditions (dotted line) before adding CO-saturated buffer to form the ferrous–CO complex (dashed line). (B) Absorption spectrum changes during reaction with ferrous PGIS. Stopped-flow time-resolved absorption spectra were taken by mixing 7 µM dithionite-treated PGIS with 125 µM CO in 20 mM NaPi, pH 7.4, containing 10% glycerol and 0.2% Lubrol PX. The first trace was recorded at 0.02 s, the others followed at 0.2 s intervals, for a total of 8.2 s. Inset, CO concentrations vs. the observed rate constants of the first phase (filled circles) and the second phase (open circles). The pseudo first-order rate constant for the increases in A450 (first phase) is dependent on CO concentration, whereas the rate constant for the decreases in A450 (second phase) is independent. The solid lines are the results from linear regression analyses.
Table 2.
Absorption and MCD parameters of PGIS and its ligand complexes
| Ligand | Absorption |
MCD |
Kd (µM) | ||
|---|---|---|---|---|---|
| λmax (nm) |
Am·10−3 (cm−1 M−1) |
λ (nm) | ΔAm/ H (cm−1 M−1 T−1) |
||
| None | 418 | 103.5 | 409 | 49.3 | |
| 419 | 0 | ||||
| 429 | −49.2 | ||||
| Sodium | 427 | 71.0 | 421 | 60.5 | 15,800±4900 |
| Cyanide | 436 | 0 | |||
| 448 | −42.7 | ||||
| Minoxidil | 424 | 68.6 | 416 | 46.2 | 5.0±0.2 |
| 430 | 0 | ||||
| 440 | −32.9 | ||||
| Clotrimazole | 424 | 87.0 | 416 | 67.4 | 0.94±0.02 |
| 427 | 0 | ||||
| 436 | −51.8 | ||||
| Pyridine | 423 | 82.0 | 414 | 58.4 | 2400±200 |
| 425 | 0 | ||||
| 434 | −48.4 | ||||
| U44069 | 416 | ND* | 414 | 49.8 | >190 |
| 425 | 0 | ||||
| 434 | −40.9 | ||||
| U46619 | 410 | 95.0 | 405 | 46.6 | 36±2 |
| 414 | 0 | ||||
| 423 | −47.9 | ||||
Not determined; little changed from control PGIS.
One hallmark of P450 proteins is the 450-nm absorption maximum exhibited by their ferrous–CO complexes. When ferric PGIS heme was reduced with dithionite, the Soret peak was blue-shifted to 412 nm and decreased in intensity (Fig. 3A). However, subsequent bubbling with CO did not produce a noticeable 450-nm peak (Fig. 3A). The 450-nm peak was not observed when U44069, a Type I ligand [8], was included in the ferrous–CO complex, or when PGIS was exposed to CO prior to the addition of dithionite. We then examined the possibility that the 450-nm peak was only transiently formed in our recombinant PGIS. Rapid-scan stopped-flow spectroscopy was carried out by reaction of CO-containing buffer with PGIS, reduced by prior anaerobic titration with dithionite. A shoulder at 450 nm was fully developed at the first time point, 20 ms (Fig. 3B). The 450 nm feature subsequently decayed as the absorbance at 422 nm increased. More detailed kinetic observations at 450 nm were conducted for reaction of PGIS with 16–500 µM CO solution. The absorbance changes at 450 nm were bimodal, with an initial increase and later decline back to near the starting level. The first phase followed single-exponential kinetics and the rate constant was linearly dependent on the CO concentrations (Fig. 3B, inset). A second-order rate constant for the formation of the CO complex of 5.6 × 105 M−1 s−1 was calculated from the slope of the secondary plot, and a dissociation rate constant of 15 s−1 was calculated from the ordinate intercept of the plot. The ratio of these two constants gives an estimate for the Kd of reduced PGIS–CO complex of 27 µM. The second phase of the A450 changes also followed single-exponential kinetics, but were independent of CO concentrations; the average rate constant was ~0.7 s−1 (Fig. 3B, inset). Because the disappearance of the 450 nm species was associated with the appearance of the 422 nm feature, stopped flow experiments were also performed at 422 nm. The rate constants thus obtained were essentially the same as those of the second phase at 450 nm (data not shown). Taken together with the observation of an isosbestic point at 434 nm (Fig. 3B), these results indicate that the ferrous–CO complex converted directly to the 422-nm species, without any intermediates. A 420-nm species is commonly found as an inactive form in P450 superfamily proteins. Initial experiments using the 422-nm species for activity assays found no detectable PGI2 formation. We then re-oxidized the 422-nm species by gently pipetting the solution up and down along the cuvette wall till the Soret peak shifted to 416.5 nm and assayed the catalytic activity of the 416.5-nm species. The rate of PGI2 formation of 416.5-nm species is about 50% of the un-treated PGIS, indicating that the ferrous–CO complex of PGIS is unstable and decays partially to an inactive form through an irreversible process.
3.5. Comparison of PGIS interactions with various heme ligands
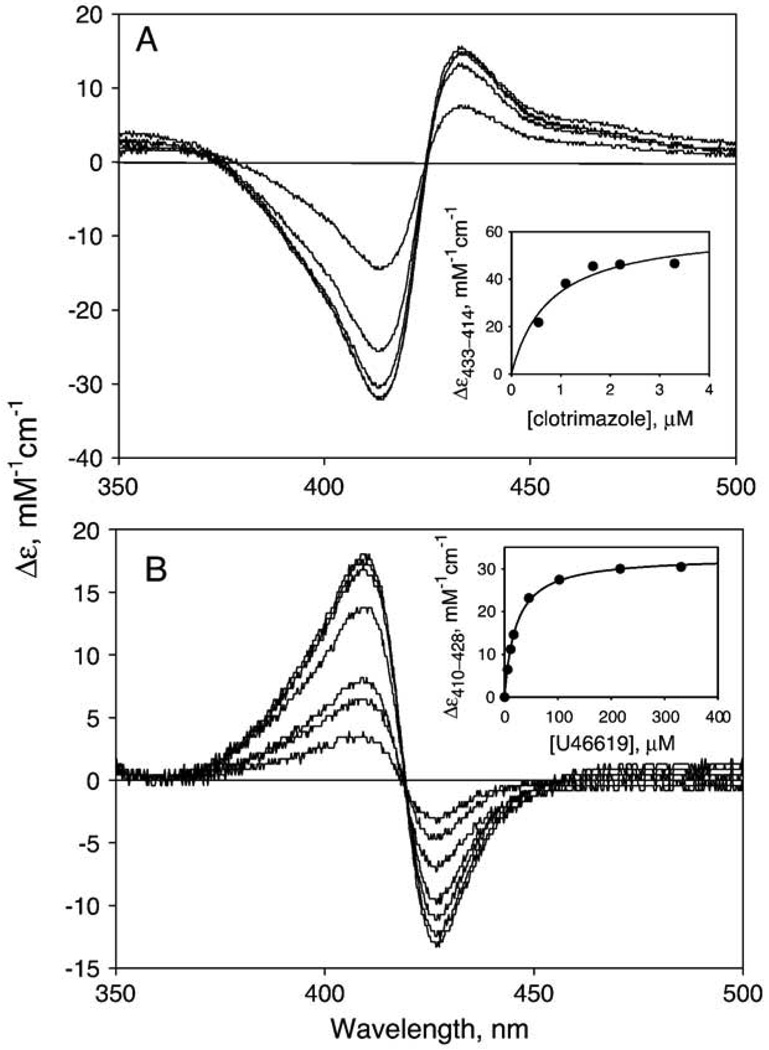
P450s bind a variety of imidazole- and pyridine-derivatives at their distal heme sites and form a six-coordinate nitrogen-based complex, causing the Soret peak shift to 420–430 nm. In contrast, PGIS was reported to have only a few known heme ligands and was unable to bind imidazole [8,9]. We first examined whether the recombinant PGIS bound imidazole. Spectral perturbation titration was carried out to examine the interaction of PGIS with imidazole (see Materials and methods). PGIS did indeed bind imidazole, albeit very weakly, as the difference spectra showed a peak at 433 nm and a trough at 412 nm (data not shown). The dissociation constant was estimated to be 0.51±0.02 mM. PGIS also bound imidazole derivatives, such as 4-phenylimidazole (Kd = 1.19±0.12 mM), 1-phenylimidazole (Kd = 0.12 ± 0.02 mM) and 1-benzylimidazole (Kd = 58±4 µM). Surprisingly, PGIS bound strongly to a large and structurally rigid imidazole derivative, clotrimazole, with a Kd of 0.94±0.02 µM (Fig. 4A). We also determined the dissociation constant for minoxidil, a known N-based PGIS ligand. A Kd of 5.0±0.2 µM was obtained, consistent with previous report of 2.4 µM for PGIS isolated from bovine tissue [31]. We also measured the dissociation constants of two stable substrate analogs, U46619 and U44069. U44069 has a C-11 carbon replacing an oxygen atom of PGH2, whereas in U46619 the C-9 carbon replaces an oxygen atom. Because the endoperoxide O–O bond is replaced by C–O bond, neither analog can be converted to prostacyclin. PGIS was proposed to form a six-coordinate complex with U46619 (Kd = 30 µM) involving the C-11 oxygen but a five-coordinate complex with U44069 (Kd>180 µM) [8]. Addition of U46619 to PGIS resulted in a blue shift of the Soret peak, with the difference spectrum showing a peak at 410 nm and a trough at 428 nm (Fig. 4B); analysis indicated a Kd value of 36±2 µM for U46619 (Fig. 4B, inset). The interaction between PGIS and U44069 was much weaker (Kd>190 µM), with the difference spectrum showing a peak at 393 nm and a trough at 432 nm. The binding results for the substrate analogs and PGIS thus are consistent with those from endogenous bovine PGIS. Other potential heme ligands were also tested for their interactions with recombinant PGIS. Similar to other P450s, PGIS was found to have weak affinities for NaCN and pyridine, with Kd values of 15.8±4.9 mM and 2.4±0.2 mM, respectively.
Fig. 4.
Absorption spectra of PGIS during titration with (A) clotrimazole and (B) U46619. The difference spectra shown were obtained with 1.4 µM PGIS (in 20 mM NaPi, pH 7.4, 10% glycerol and 0.2% Lubrol PX) and 0.55, 1.1, 1.65, 2.2, and 3.3 µM clotrimazole and 6, 11, 17, 46, 103, 216 and 330 µM U46619. The insets show the titration curves fitted to a one-site binding model to obtain the dissociation constants of 0.94±0.02 µM clotrimazole and 36±2 µM U46619. The lower signal/noise ratio in panel B is due to the use of a faster scan mode for spectra acquisition.
MCD spectroscopy was applied to characterize resting PGIS and its complexes with a C-based ligand (sodium cyanide), N-based ligands (minoxidil, clotrimazole and pyridine), and O-based ligands (U44069 and U46619). As shown in Fig. 5A, the MCD spectrum of PGIS is typical both in line shape and in amplitude for a low-spin P450 hemeprotein. There is no sign of high-spin heme charge-transfer signal present in the region of 600–700 nm. The Soret crossovers of the sodium cyanide, minoxidil, clotrimazole, U44069 and pyridine-ligated PGIS are red-shifted 6–17 nm relative to resting PGIS (Table 2). These red-shifts of the Soret crossover are also accompanied with an increase of the trough at 350 nm region and a decrease in the 500–600 nm spectral features (data not shown). These changes are typical for the “normal” to “hyper” porphyrin spectral transitions found for other thiolate-ligated heme complexes [32]. This axial ligand-induced spectral transition was interpreted as a mixing of the sulfur P orbital→porphyrin π* charge-transfer to the porphyrin π→π* transitions that causes the splitting of the single Soret band. The Soret crossovers of resting PGIS and its complexes with imidazole derivatives, pyridine, and U46619 are close to the positions of their Soret peaks in the electronic absorption spectra (Table 2), suggesting that this feature has a pure MCD origin, but whether it is an “A” or “C” term requires a temperature dependence study. The only inconsistency of ligand-induced MCD spectral shift is with U44069, which caused a blue-shift in the electronic absorption spectrum but a red-shift in the MCD spectrum (data not shown). This result indicates that there are multiple terms present in the MCD of the U44069–PGIS complex, so that the sum of these terms gives an opposite shift of the MCD crossover relative to the shift in the optical peak.
Fig. 5.
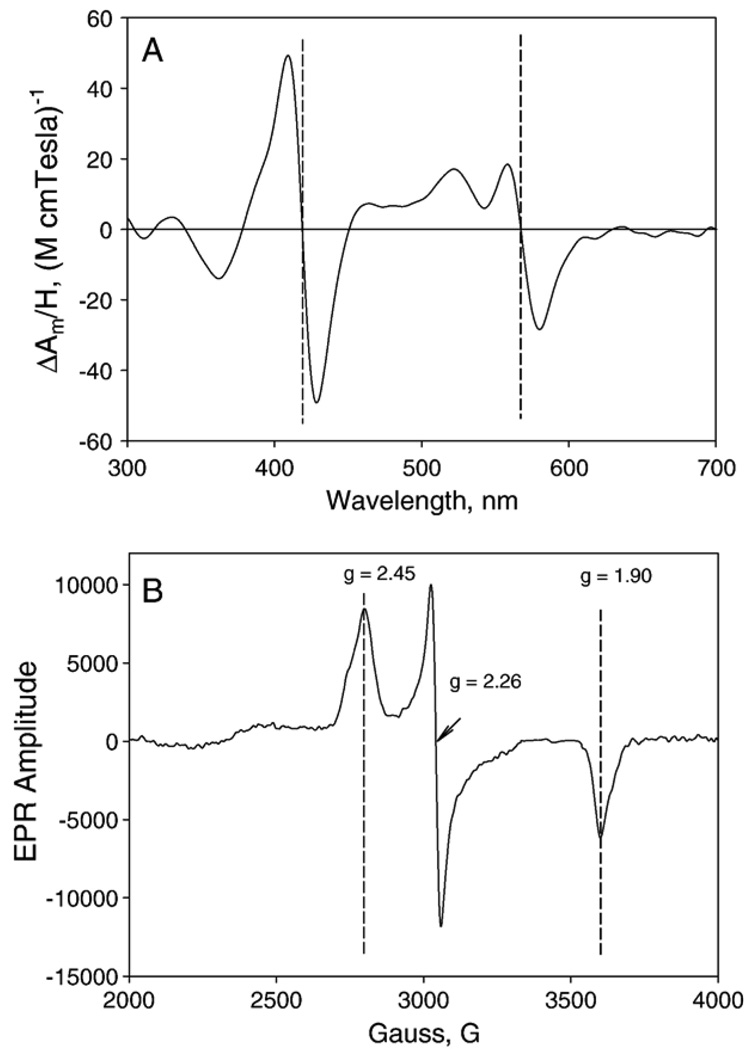
MCD and EPR spectra of PGIS. Spectra were recorded for PGIS (10 µM in 20 mM NaPi, pH 7.4, 10% glycerol and 0.2% Lubrol PX). (A) MCD spectrum was taken at 2 nm spectral band width and 1 millidegree sensitivity at 25 °C. Left and right vertical broken lines indicate the zero-crossovers at Soret and α bands, respectively. (B) EPR spectrum was recorded at 10K. Microwave frequency, 9.60 GHz; power, 4 mW; modulation amplitude, 10 G, and 0.3 s time constant. The vertical broken lines indicate g values of 2.45, 2.26 and 1.90.
3.6. Characterization of PGIS by EPR spectroscopy
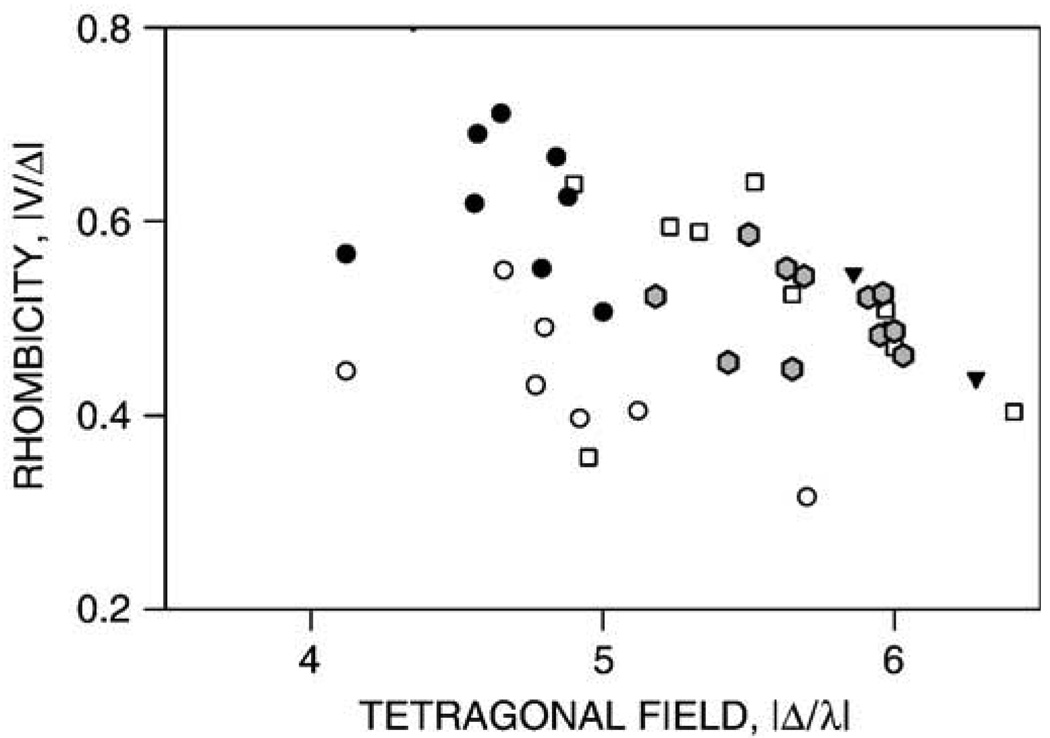
EPR spectra were obtained for resting PGIS and PGIS complexes with substrate analogs or heme ligands of various sizes. The EPR spectrum of the resting enzyme shows a typical low-spin P450 feature, with anisotropic g values of 2.45 (2.50 shoulder), 2.26 and 1.90 (Fig. 5B and Table 3). There is no additional EPR signal at low-field side of the gmax of the low-spin heme. The low-spin heme features showed a microwave power relaxation similar to other P450 heme centers, with a P1/2 of 0.43 mW and an inhomogeneous broadening of the EPR signal with increasing power (data not shown). This power saturation behavior is fairly typical for a low-spin P450 heme [33]. U44069 and U46619 had little effects on the EPR spectrum, whereas minoxidil only slightly increased the heme rhombicity, as indicated by a slight shift of the gmax toward the low-field region (gmax = 2.46). Histidine also caused a slight shift of gmax (gmax = 2.47). In contrast, imidazole caused a shift of the gmin to 1.89 with little shift of the other g components. Clotrimazol, pyridine and cyanide caused much larger changes in heme rhombicity, with g values of 2.52/2.27/1.86, 2.48/2.27/1.88, and 2.50/2.25/1.88, respectively. The g parameters of the resting PGIS and its complexes with various ligands are listed in Table 3. The rhombic and axial ligand field components (V and D, respectively) and the rhombicity of the heme center (in %) were calculated using these g parameters [33] (Table 3). The “Truth Diagram” that correlates the rhombicity and tetragonal ligand field strength for hemoproteins with a thiolate proximal ligand is shown in Fig. 6. Regardless of the perturbation of the heme rhombicity in PGIS by various heme ligands, the PGIS ligand complexes fall in the “P” zone and overlap nicely with those P450s having a hydrophobic distal heme domain, such as P450cam and thromboxane synthase [34]. In contrast, the heme ligand complexes of chloroperoxidase or nitric oxide synthase, which have a relatively polar distal heme pocket, are located in the region with lower tetragonal field strength (Fig. 6). This is a strong indication that the heme distal pocket of PGIS is rather hydrophobic, in contrast to chloroperoxidase or nitric oxide synthase, where a hydrophilic environment is needed to accommodate polar substrates. Taken together, the present spectroscopic data indicates that PGIS active site is hydrophobic and constrained, but adaptable to accommodate ligands of various sizes.
Table 3.
EPR parameters of PGIS and its ligand complexes
| Ligand | gmax | gmid | gmin | Δa | Va | Rhombicity (%) b |
|---|---|---|---|---|---|---|
| None | 2.45 | 2.26 | 1.90 | 5.95 | 2.87 | 48.2 |
| Sodium Cyanide | 2.50 | 2.25 | 1.88 | 5.63 | 3.10 | 55.1 |
| Minoxidil | 2.46 | 2.25 | 1.90 | 5.96 | 3.13 | 52.5 |
| Clotrimazole | 2.52 | 2.27 | 1.86 | 5.18 | 2.71 | 52.2 |
| Pyridine | 2.48 | 2.27 | 1.88 | 5.43 | 2.47 | 45.4 |
| U44069 | 2.44 | 2.26 | 1.91 | 6.03 | 2.78 | 46.1 |
| U46619 | 2.45 | 2.25 | 1.90 | 6.00 | 2.92 | 48.6 |
| 2.52 | 2.25 | 1.88 | 5.50 | 3.22 | 58.6 | |
| Imidazole | 2.46 | 2.26 | 1.89 | 5.65 | 2.53 | 44.7 |
| Histidine | 2.47 | 2.25 | 1.90 | 5.91 | 3.08 | 52.1 |
| 2.49 | 1.89 | 5.69 | 3.09 | 54.3 |
Rhombicity (%) calculated as 100×V/Δ.
Fig. 6.
Truth Diagram analysis. Correlations between tetragonal field strength (Δ) and rhombicity (%) of low-spin PGIS complexes (filled hexagons) are compared with those for endothelial nitric oxide synthase (open circles), chloroperoxidase (filled circles), P450cam (squares), and thromboxane synthase (filled triangles). The PGIS complexes were prepared using the ligands listed in Table 3. For clarity, only the “P zone” of the Truth Diagram is shown.
3.7. Homolytic and heterolytic O–O bond scission of 10-OOH-18:2 by PGIS
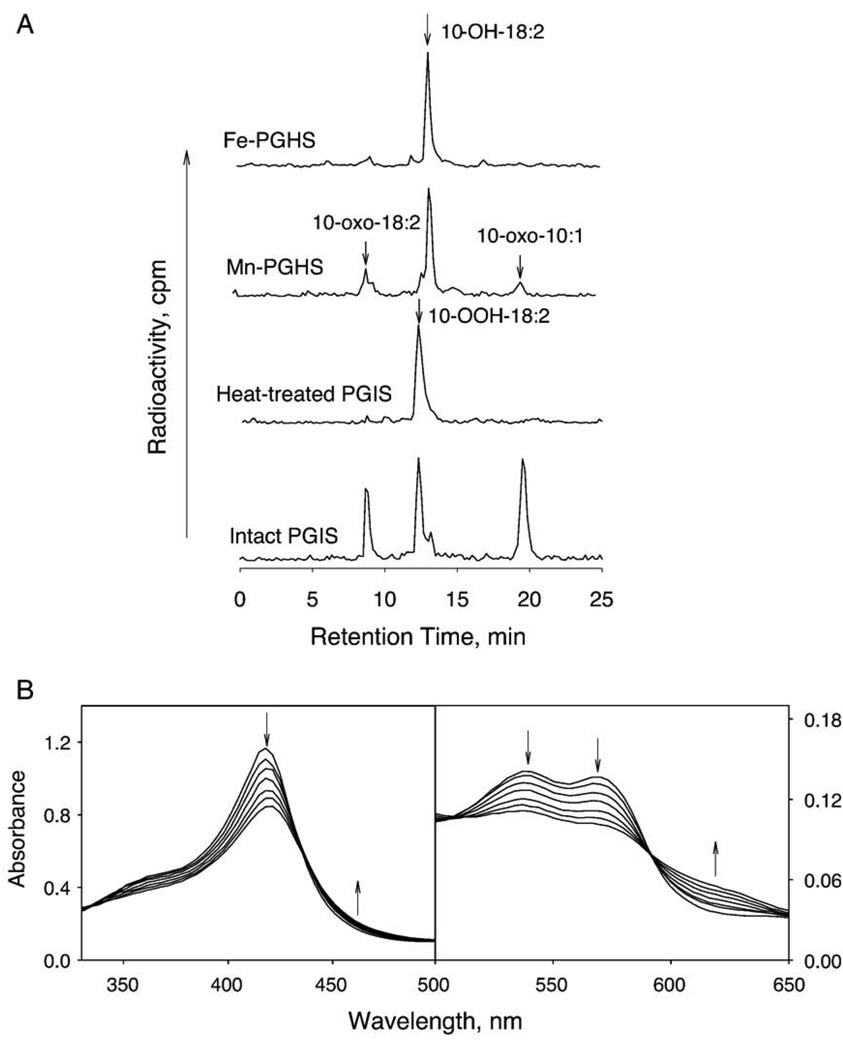
Hecker and Ullrich proposed an O–O homolytic scission of PGH2 to generate an alkoxyl radical in the PGIS-catalyzed reaction [8]. To test this hypothesis, we used 10-OOH-18:2 as a substrate. This lipid hydroperoxide has been extensively used to differentiate heterolytic and homolytic scission of the hydroperoxide bond [27,35–37]. With heterolytic scission of the O–O bond, 10-hydroxyoctadeca-8,12-dienoic acid (10-OH-18:2) is formed. If homolytic scission occurs, the resulting alkoxyl radical either further oxidizes to 10-oxooctadeca-8,12-dienoic acid (10-oxo-18:2) or undergoes β-scission to form 10-oxodec-8-enoic acid (10-oxo-10:1) and an allylic radical. Formation of 10-oxo-10:1 is a strong indication that homolytic scission takes place as β-scission can only occur via 10-alkoxyl radical intermediate [27,35,36]. Recombinant PGIS (2 µM) was reacted with [1-14C]-10-OOH-18:2 (80 µM) at 23 °C for 30 s and the products were analyzed by HPLC. Three product peaks, in addition to the un-consumed substrate, were observed in the chromatogram (Fig. 7A). The position of 10-OH-18:2 was independently identified by reaction of 10-OOH-18:2 with prostaglandin H synthase-1, which produces primarily the heterolytic scission product [27], whereas the peaks of two homolytic scission products were identified from the products of 10-OOH-18:2 reaction with mangano-prostaglandin H synthase-1 [37]. In PGIS-catalyzed reaction, the homolytic scission products, 10-oxo-18:2 and 10-oxo-10:1, constituted the major portion (32% and 59% of the total products, respectively), whereas the heterolytic scission product made up only 9% of total product. Comparable amounts of homolytic products were obtained even when a peroxidase co-substrate, such as guaiacol, was present in the reaction mixture (data not shown), suggesting that the oxidized heme intermediate acquires an electron from the alkoxyl radical to re-establish the resting state and does not interact with exogenous reducing agents. Shortening the reaction time to 10 s did not alter the ratio of three products generated, although, as expected, less 10-OOH-18:2 was consumed (35% for 10-s incubation vs. 65% for 30-s incubation). A longer incubation (60 s) also had a similar product ratio, with 75% of substrate consumed. Only trace amounts of products were detected from a control incubation of 10-OOH-18:2 with heat-treated PGIS (Fig. 7A). Collectively, these results establish that the PGIS very much favors homolytic cleavage of the hydroperoxide.
Fig. 7.
Reaction of 10-OOH-18:2 with PGIS. A. HPLC profile of the products from the reactions of wild type prostaglandin H synthase-1 (Fe-PGHS; 0.5 µM), mangano-prostaglandin H synthase-1 (Mn-PGHS; 0.5 µM), heat-treated and intact PGIS (2 µM) with [1-14C]-10-OOH-18:2 (80 µM) at 23 °C for 30 s in 20 mM NaPi, pH 7.4, containing 10% glycerol and 0.2% Lubrol PX. Guaiacol at a final concentration of 0.55 mM was included in the reaction of prostaglandin H synthase-1. Reaction products were analyzed as described in Materials and methods Procedures“. The positions of the substrate and products were indicated. B. Absorption spectrum changes upon reaction of PGIS (15 µM) with 10-OOH-18:2 (150 µM) in 20 mM NaPi, pH 7.4, containing 10% glycerol and 0.2% Lubrol PX at 23 °C. Spectra were recorded at 0.04, 0.41, 0.82, 1.64, 3.36, 4.91, 6.55 and 16.4 s. Arrows show the directions of spectral changes with increasing time.
Homolytic scission of O–O bond is expected to produce a ferryl–oxo complex (Cpd II). We carried out rapid-scan stopped-flow absorption spectroscopy to determine whether Cpd II is actually formed in the PGIS-catalyzed reaction. The reaction of 15 µM of PGIS with 150 µM of 10-OOH-18:2 revealed the formation of an intermediate with a Soret peak at ~422 nm with an apparent rate of 0.67 s−1 (Fig. 7B). The presence of four isosbestic points, at 334, 438, 507 and 592 nm, suggest that a single chemical reaction of PGIS proceeds to the initial intermediate within 16 s. This initial intermediate which has a red-shifted Soret peak is possibly the Cpd II. The position of Soret peak seems to be a reasonable gauge in distinguishing Cpd I from Cpd II of P450s. The Cpd I of the P450 enzyme chloroperoxidase has its Soret peak at 367 nm whereas the Cpd II has a predominant band at 436 nm [38]. The Soret peak of Cpd I of P450 1A2 is at 389 nm, whereas the Cpd II is at 423 nm [39]. Furthermore, the Cpd I of P450 101 and P450 109 also has a Soret peak at 367 and 370 nm, respectively [40,41]. In addition to the red-shift of the PGIS Soret peak, the spectral features of the transient α band (570 nm) and β band (537 nm) are also similar to those observed for P450 Cpd II. The absorption spectra of Cpd II of chloroperoxidase and P450 1A2 have slightly lower intensities in α bands (572 and 567 nm, respectively) than β band (542 and 535 nm, respectively) [38,39]. In contrast, the Cpd I of chloroperoxidase completely loses its α and β bands, rather, a prominent band is seen at 687 nm [38]. These results suggest that one-electron reduction of the peroxide (i.e., homolytic scission) resulted in formation of a Cpd II form of PGIS. To determine the rate constant of Cpd II formation, we carried out stopped-flow spectroscopy at 418 nm for reaction of 2.75 µM of PGIS with a 12 to 56-fold excess of 10-OOH-18:2. Biphasic exponential kinetics was observed for the first 50 s. The rate constants of the first phase were between 0.5 and 0.6 s−1 and were independent of the 10-OOH-18:2 concentration, suggesting that the absorption spectrum observed in the rapid-scan stopped flow experiments actually was preceded by a faster step for binding of PGIS and 10-OOH-18:2. The rate constant for the second phase was ~0.06 s−1 and was independent of the substrate concentration. Formation of the initial optical intermediates in the first phase seems to coincide with accumulation of the homolytic cleavage products revealed by HPLC analyses. The second phase appears to reflect a slower heme-bleaching event, with significant decrease in the Soret peak and disappearance of the isosbestic points (data not shown). Heme bleaching caused by hydroperoxides is a common phenomenon in many P450s. Taken together, the results indicated that PGIS catalyzed predominantly a homolytic scission of 10-OOH-18:2 with concomitant formation of Cpd II.
4. Discussion
It has been a challenge to express recombinant PGIS in quantities sufficient for biophysical studies. Wada et al [11] used a baculovirus expression system and obtained the human PGIS at sub-milligram levels. It was speculated that a tight secondary structure at the PGIS active site might hinder heme incorporation in expression systems [42]. Earlier efforts in our laboratory to express PGIS, with or without N-terminal modifications, in insect cells or yeast cells produced predominantly inactive protein or meager amounts of active enzyme. We also experienced low levels of expression in E. coli using various strategies including introduction of modified N-terminal sequences (e.g., OmpA), co-expression with chaperones or thioredoxin [43], adoption of other expression vectors and host strains. It was only the new approach of substituting the peptide MAKKTSS for the first 17 amino acid residues that gave significantly improved expression of active PGIS in E. coli. Retaining the amino-terminal sequence or replacing the first 17 amino acid sequence with other leader sequences did not yield a high level of expression. However, we did not exhaustively explore other modifications of the amino terminus and further improvements in expression may be present.
The amino-terminal segments of microsomal P450s are hydrophobic and are thought to comprise a membrane-anchoring domain [44]. However, removal of the amino-terminal segment does not always lead to a soluble protein because the F–G loop is thought to be another membrane-associated domain [45]. For some microsomal P450s lacking a hydrophobic amino-terminal segment, the membrane-association is weak and can be disrupted by high concentration of salt [45,46]. This does not appear to be the case with PGIS because recombinant PGIS remains associated with membrane fractions when the E. coli lysate is separated by sucrose gradient ultracentrifugation [45] regardless of the salt treatment (data not shown). Membrane-association via the F–G loop in PGIS is also suggested by the very low yield of the recombinant protein if the detergent extraction step is omitted (data not shown).
The PGIS amino-terminal segment, as in most microsomal P450s, is not required for catalytic activity. The specific activity of the recombinant PGIS is actually higher than that of endogenous bovine PGIS and comparable to that of wild type recombinant PGIS expressed in insect cells, which has a specific activity of 15 µmole/min/mg protein [11]. The recombinant PGIS can also catalyze cleavage of PGH1 to form 12-hydroxy-5,8,10-heptadecadienoic acid and MDA as efficiently as does wild type PGIS. Recombinant PGIS also has similar affinities as wild type PGIS for the substrate analogs. UV-vis absorption, MCD and EPR spectra indicated that the heme-iron of the resting enzyme is most likely ligated to an O-based ligand, presumably a water molecule. When N- or C-based ligand, such as imidazole or cyanide, is added to the recombinant PGIS, the optical absorption and MCD spectra are changed in a manner similar to those observed for other P450s, i.e. gradual red-shifts of the Soret peak (optical) and crossover (MCD). Overall, the MCD and EPR spectral changes induced by various heme ligands are modest in comparison with those hemoproteins using the less field-strength ligands, such as imidazole from histidine residue, as the proximal ligand. This indicates that the strong thiolate ligand in PGIS dominates the axial ligand effect, thus prohibiting a large spectral shift caused by various distal ligands. This is a common property of P450 enzymes [47]. Although both are N-based ligands, imidazole binds weakly to PGIS but clotrimazole which is a bulky, rigid and hydrophobic imidazole derivative binds strongly. These results may suggest that the active site of PGIS is very flexible to accommodate large and hydrophobic ligands and is therefore capable of binding the ligand in an induced-fit mechanism. Alternatively, in contrast to the previous view that limited space is present in the PGIS active site, the active site may be ample to accommodate ligands with varying affinities depending on the interaction of amino acid residues of the active site with the hydrophobic ligand.
The most striking difference between the recombinant human PGIS and most other P450s is the transient character of its ferrous–CO complex. The rate constant for formation of the CO complex with PGIS was 5.6 × 105 M−1 s−1, much slower than the values of 106–107 M−1 s−1 reported for other P450s [48–53]. Further, the rate constants for dissociation of the ferrous–CO complex is one to two orders of magnitude larger than for most P450s, indicating a less stable PGIS ferrous–CO complex. These results suggested that the PGIS distal site is more sterically constrained and less favorable for CO binding than in other P450s. In addition, the ferrous–CO complex in PGIS readily converted to a 422-nm species with a half-life of approximately 1 s. This is in contrast with endogenous bovine PGIS where a distinct 450-nm peak was stable enough to be seen in a static spectrum [3]. It is unlikely that the amino-terminal modifications in the recombinant PGIS are responsible for the instability of ferrous–CO complex because wild type human PGIS expressed in insect cells also exhibited the Soret peak of ferrous–CO difference spectrum at 418 nm [11]. The chemical nature of the 422-nm species of PGIS is unclear at present. One possibility is that the proximal thiolate is protonated to become a thiol or is replaced by another ligand, such as histidine; both thiol- and imidazole-ligated ferrous–CO complexes have Soret peaks at 420 nm [54–58]. The labile nature of the ferrous–CO complex of recombinant PGIS suggests the structure of the PGIS heme environment is rather malleable, allowing proximal ligand exchange, or protonation of the thiolate ligand.
The PGIS reaction mechanism remains an intriguing topic in the P450 field. Although most P450s are able to bypass the requirement for NAD(P)H and catalyze hydroperoxide-dependent oxidations in vitro (also known as the “peroxide shunt”) analogous to peroxidase reactions [59], PGIS is obligatory for peroxides as the substrates. In this regard, PGIS is more like a peroxidase than ordinary P450s. PGIS catalysis is proposed to involve attack on the O–O bond in PGH2 [8]. It is thus relevant to use hydroperoxides as model substrates for exploring this aspect of the reaction mechanism. Peroxidases generally catalyze heterolysis of hydroperoxides forming Cpd I, whereas P450s can support both homolytic and heterolytic cleavage of the O–O bond, depending on the individual P450 and the substrate structure [16,17,39,60]. The products of PGIS reacting with the diagnostic substrate 10-OOH-18:2 clearly indicate that homolysis is the predominant pathway. As expected with homolytic cleavage of the O–O bond, one-electron oxidation of PGIS ferric heme appears to generate a ferryl–oxo species (Cpd II) which is preliminarily characterized by the red-shift of the Soret peak. Certainly, further spectroscopic studies such as MCD and resonance Raman are required to confirm the identity of the PGIS Cpd II. Nevertheless, our results hitherto supported the reaction mechanism of homolytic scission of O–O endoperoxide of PGH2 [8]. Characterization of the PGIS Cpd II reacting with the diagnostic substrate will thus pave the way to understand the reaction mechanism of PGIS with its physiological substrate.
In summary, we reported an over-expression system and a purification scheme for human PGIS. This protocol provides sufficient amounts of active enzyme for biophysical characterization, drug design, structure/function and reaction mechanism studies. Spectroscopic characterization of the recombinant PGIS indicates that it has a hydrophobic active site with a flexible heme environment for an induced-fit substrate binding. Formation of CO-ferrous PGIS complex is transient. The active site favors a homolytic cleavage of the O–O bond to form an alkoxyl radical and Cpd II.
Acknowledgements
This work is supported by Grants HL60625 (to L.-H.W.) and GM44911 (to A.-L. T.) from the National Institutes of Health. We thank Drs. Phil Kingsley and Lawrence Marnett at Vanderbilt University for characterization of 10-OOH-18:2 by electrospray ionization mass spectroscopy and Dr. Graham Palmer at Rice University for use of the MCD spectrophotometer. We also thank Dr. Richard J. Kulmacz for valuable suggestions.
Abbreviations
- PGIS
prostacyclin synthase
- PGI2
prostacyclin or prostaglandin I2
- Ni-NTA
Nickel nitrilotriacetate agarose
- MCD
magnetic circular dichroism
- EPR
electron paramagnetic resonance
- MDA
malondialdehyde
- 10-OOH-18:2
10-hydroperoxyocatdeca-8,12-dienoic acid
References
- 1.Vane JR, Botting RM. Pharmacodynamic profile of prostacyclin. Am. J. Cardiol. 1995;75:3A–10A. doi: 10.1016/s0002-9149(99)80377-4. [DOI] [PubMed] [Google Scholar]
- 2.Moncada S, Gryglewski R, Bunting S, Vane JR. An enzyme isolated from arteries transforms prostaglandin endoperoxides to an unstable substance that inhibits platelet aggregation. Nature. 1976;263:663–665. doi: 10.1038/263663a0. [DOI] [PubMed] [Google Scholar]
- 3.Ullrich V, Castle L, Weber P. Spectral evidence for the cytochrome P450 nature of prostacyclin synthetase. Biochem. Pharmacol. 1981;30:2033–2036. doi: 10.1016/0006-2952(81)90218-5. [DOI] [PubMed] [Google Scholar]
- 4.DeWitt DL, Smith WL. Purification of prostacyclin synthase from bovine aorta by immunoaffinity chromatography. Evidence that the enzyme is a hemoprotein. J. Biol. Chem. 1983;258:3285–3293. [PubMed] [Google Scholar]
- 5.Hara S, Miyata A, Yokoyama C, Inoue H, Brugger R, Lottspeich F, Ullrich V, Tanabe T. Isolation and molecular cloning of prostacyclin synthase from bovine endothelial cells. J. Biol. Chem. 1994;269:19897–19903. [PubMed] [Google Scholar]
- 6.Pereira B, Wu KK, Wang LH. Molecular cloning and characterization of bovine prostacyclin synthase. Biochem. Biophys. Res. Commun. 1994;203:59–66. doi: 10.1006/bbrc.1994.2148. [DOI] [PubMed] [Google Scholar]
- 7.Poulos TL, Johnson EF. Structures of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. New York: Plenum Publishers; 2005. pp. 87–114. [Google Scholar]
- 8.Hecker M, Ullrich V. On the mechanism of prostacyclin and thromboxane A2 biosynthesis. J. Biol. Chem. 1989;264:141–150. [PubMed] [Google Scholar]
- 9.Hecker M, Haurand M, Ullrich V, Terao S. Spectral studies on structure–activity relationships of thromboxane synthase inhibitors. Eur. J. Biochem. 1986;157:217–223. doi: 10.1111/j.1432-1033.1986.tb09659.x. [DOI] [PubMed] [Google Scholar]
- 10.Ullrich V, Brugger R. Prostacyclin and thromboxane synthase: new aspects of hemethiolate catalysis. Angew. Chem., Int. Ed. Engl. 1994;33:1911–1919. [Google Scholar]
- 11.Wada M, Yokoyama C, Hatae T, Shimonishi M, Nakamura M, Imai Y, Ullrich V, Tanabe T. Purification and characterization of recombinant human prostacyclin synthase. J. Biochem. (Tokyo) 2004;135:455–463. doi: 10.1093/jb/mvh059. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi K, Watanabe Y, Morishima I. Push effect on the heterolytic O–O bond cleavage of peroxoiron (III) porphyrin adducts. Inorg. Chem. 1992;31:156–157. [Google Scholar]
- 13.Higuchi T, Shimada K, Maruyama N, Hirobe M. Heterolytic oxygen–oxygen bond cleavage of peroxy acid and effective alkane hydroxylation in hydrophobic solvent mediated by an iron porphyrin coordinated by thiolate anion as a model for cytochrome P450. J. Am. Chem. Soc. 1993;115:7551–7552. [Google Scholar]
- 14.Turner JA, Herz W. Fe(II)-induced decomposition of epidioxides. A chemical model for prostaglandin E, prostacyclin and thromboxane biosynthesis. Experientia. 1977;33:1133–1134. doi: 10.1007/BF01922283. [DOI] [PubMed] [Google Scholar]
- 15.Chamulitrat W, Takahashi N, Mason RP. Peroxyl, alkoxyl, and carbon-centered radical formation from organic hydroperoxides by chloroperoxidase. J. Biol. Chem. 1989;264:7889–7899. [PubMed] [Google Scholar]
- 16.Barr DP, Martin MV, Guengerich FP, Mason RP. Reaction of cytochrome P450 with cumene hydroperoxide: ESR spin-trapping evidence for the homolytic scission of the peroxide O–O bond by ferric cytochrome P450 1A2. Chem. Res. Toxicol. 1996;9:318–325. doi: 10.1021/tx9501501. [DOI] [PubMed] [Google Scholar]
- 17.Correia MA, Yao K, Allentoff AJ, Wrighton SA, Thompson JA. Interactions of peroxyquinols with cytochromes P450 2B1, 3A1, and 3A5: influence of the apoprotein on heterolytic versus homolytic O–O bond cleavage. Arch. Biochem. Biophys. 1995;317:471–478. doi: 10.1006/abbi.1995.1190. [DOI] [PubMed] [Google Scholar]
- 18.Siegle I, Nusing R, Brugger R, Sprenger R, Zecher R, Ullrich V. Characterization of monoclonal antibodies generated against bovine and porcine prostacyclin synthase and quantitation of bovine prostacyclin synthase. FEBS Lett. 1994;347:221–225. doi: 10.1016/0014-5793(94)00504-4. [DOI] [PubMed] [Google Scholar]
- 19.Shyue SK, Ruan KH, Wang LH, Wu KK. Prostacyclin synthase active sites. Identification by molecular modeling-guided site-directed mutagenesis. J. Biol. Chem. 1997;272:3657–3662. doi: 10.1074/jbc.272.6.3657. [DOI] [PubMed] [Google Scholar]
- 20.Shyue SK, Tsai MJ, Liou JY, Willerson JT, Wu KK. Selective augmentation of prostacyclin production by combined prostacyclin synthase and cyclooxygenase-1 gene transfer. Circulation. 2001;103:2090–2095. doi: 10.1161/01.cir.103.16.2090. [DOI] [PubMed] [Google Scholar]
- 21.Wang LH, Tsai AL. P.Y. Hsu, Substrate binding is the rate-limiting step in thromboxane synthase catalysis. J. Biol. Chem. 2001;276:14737–14743. doi: 10.1074/jbc.M009177200. [DOI] [PubMed] [Google Scholar]
- 22.Kulmacz RJ, Palmer G, Wei C, Tsai AL. Reaction and free radical kinetics of prostaglandin H synthase with manganese protoporphyrin IX as the prosthetic group. Biochemistry. 1994;33:5428–5439. doi: 10.1021/bi00184a011. [DOI] [PubMed] [Google Scholar]
- 23.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 24.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 25.Holmquist B, Vallee BL. Tryptophan quantitation by magnetic circular dichroism in native and modified proteins. Biochemistry. 1973;12:4409–4417. doi: 10.1021/bi00746a018. [DOI] [PubMed] [Google Scholar]
- 26.Tai CL, Tai HH. Radioimmunological assay of prostacyclin synthetase activity. Prostaglandins. Med. 1980;4:399–408. doi: 10.1016/0161-4630(80)90048-8. [DOI] [PubMed] [Google Scholar]
- 27.Labeque R, Marnett LJ. 10-Hydroperoxy-8,12-0ctadecadienoic acid: a diagnostic probe of alkoxyl radical generation in metal-hydroperoxide reactions. J. Am. Chem. Soc. 1987;109:2828–2829. [Google Scholar]
- 28.Barnes HJ. Maximizing expression of eukaryotic cytochrome P450s in Escherichia coli. Methods Enzymol. 1996;272:3–14. doi: 10.1016/s0076-6879(96)72003-7. [DOI] [PubMed] [Google Scholar]
- 29.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wachenfeldt C, Richardson TH, Cosme J, Johnson EF. Microsomal P450 2C3 is expressed as a soluble dimer in Escherichia coli following modification of its N-terminus. Arch. Biochem. Biophys. 1997;339:107–114. doi: 10.1006/abbi.1996.9859. [DOI] [PubMed] [Google Scholar]
- 31.Hecker M, Ullrich V. Studies on the interaction of minoxidil with prostacyclin synthase in vitro. Biochem. Pharmacol. 1988;37:3363–3365. doi: 10.1016/0006-2952(88)90652-1. [DOI] [PubMed] [Google Scholar]
- 32.Gouterman M. Optical spectra and electronic structure of porphyrins and related rings. In: Dolphin D, editor. The Porphyrins. New York: Academic Press; 1978. pp. 1–165. [Google Scholar]
- 33.Tsai AL, Berka V, Chen PF, Palmer G. Characterization of endothelial nitric-oxide synthase and its reaction with ligand by electron paramagnetic resonance spectroscopy. J. Biol. Chem. 1996;271:32563–32571. doi: 10.1074/jbc.271.51.32563. [DOI] [PubMed] [Google Scholar]
- 34.Hsu PY, Tsai AL, Kulmacz RJ, Wang LH. Expression, purification, and spectroscopic characterization of human thromboxane synthase. J. Biol. Chem. 1999;274:762–769. doi: 10.1074/jbc.274.2.762. [DOI] [PubMed] [Google Scholar]
- 35.Labeque R, Marnett LJ. Reaction of hematin with allylic fatty acid hydroperoxides: identification of products and implications for pathways of hydroperoxide-dependent epoxidation of 7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene. Biochemistry. 1988;27:7060–7070. doi: 10.1021/bi00418a059. [DOI] [PubMed] [Google Scholar]
- 36.Labeque R, Marnett LJ. Homolytic and heterolytic scission of organic hydroperoxides by (meso-tetraphenylporphinato)iron(III) and its relation to olefin epoxidation. J. Am. Chem. Soc. 1989;111:6621–6621. [Google Scholar]
- 37.Landino LM, Marnett LJ. Mechanism of hydroperoxide reduction by mangano-prostaglandin endoperoxide synthase. Biochemistry. 1996;35:2637–2643. doi: 10.1021/bi952546h. [DOI] [PubMed] [Google Scholar]
- 38.Egawa T, Proshlyakov DA, Miki H, Makino R, Ogura T, Kitagawa T, Ishimura Y. Effects of a thiolate axial ligand on the pi→pi* electronic states of oxoferryl porphyrins: a study of the optical and resonance Raman spectra of compounds I and II of chloroperoxidase. J. Biol. Inorg. Chem. 2001;6:46–54. doi: 10.1007/s007750000181. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu T, Murakami Y, Hatano M. Glu318 and Thr319 mutations of cytochrome P450 1A2 remarkably enhance homolytic O–O cleavage of alkyl hydroperoxides. An optical absorption spectral study. J. Biol. Chem. 1994;269:13296–13304. [PubMed] [Google Scholar]
- 40.Egawa T, Shimada H, Ishimura Y. Evidence for compound I formation in the reaction of cytochrome P450cam with m-chloroperbenzoic acid. Biochem. Biophys. Res. Commun. 1994;201:1464–1469. doi: 10.1006/bbrc.1994.1868. [DOI] [PubMed] [Google Scholar]
- 41.Kellner DG, Hung SC, Weiss KE, Sligar SG. Kinetic characterization of compound I formation in the thermostable cytochrome P450 CYP119. J. Biol. Chem. 2002;277:9641–9644. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt P, Youhnovski N, Daiber A, Balan A, Arsic M, Bachschmid M, Przybylski M, Ullrich V. Specific nitration at tyrosine 430 revealed by high resolution mass spectrometry as basis for redox regulation of bovine prostacyclin synthase. J. Biol. Chem. 2003;278:12813–12819. doi: 10.1074/jbc.M208080200. [DOI] [PubMed] [Google Scholar]
- 43.Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- 44.Black SD. Membrane topology of the mammalian P450 cytochromes. FASEB J. 1992;6:680–685. doi: 10.1096/fasebj.6.2.1537456. [DOI] [PubMed] [Google Scholar]
- 45.Cosme J, Johnson EF. Engineering microsomal cytochrome P450 2C5 to be a soluble, monomeric enzyme. Mutations that alter aggregation, phospholipid dependence of catalysis, and membrane binding. J. Biol. Chem. 2000;275:2545–2553. doi: 10.1074/jbc.275.4.2545. [DOI] [PubMed] [Google Scholar]
- 46.Hsu PY, Wang LH. Protein engineering of thromboxane synthase: conversion of membrane-bound to soluble form. Arch. Biochem. Biophys. 2003;416:38–46. doi: 10.1016/s0003-9861(03)00287-x. [DOI] [PubMed] [Google Scholar]
- 47.Sono M, Dawson JH. Formation of low spin complexes of ferric cytochrome P-450-CAM with anionic ligands. Spin state and ligand affinity comparison to myoglobin. J. Biol. Chem. 1982;257:5496–5502. [PubMed] [Google Scholar]
- 48.Tetreau C, Di Primo C, Lange R, Tourbez H, Lavalette D. Dynamics of carbon monoxide binding with cytochromes P-450. Biochemistry. 1997;36:10262–10275. doi: 10.1021/bi970619m. [DOI] [PubMed] [Google Scholar]
- 49.Sevrioukova IF, Peterson JA. Reaction of carbon monoxide and molecular oxygen with P450terp (CYP108) and P450BM-3 (CYP102) Arch. Biochem. Biophys. 1995;317:397–404. doi: 10.1006/abbi.1995.1180. [DOI] [PubMed] [Google Scholar]
- 50.Mitani F, Iizuka T, Shimada H, Ueno R, Ishimura Y. Flash photolysis studies on the CO complexes of ferrous cytochrome P-450scc and cytochrome P-45011 beta. Effects of steroid binding on the photochemical and ligand binding properties. J. Biol. Chem. 1985;260:12042–12048. [PubMed] [Google Scholar]
- 51.Unno M, Ishimori K, Ishimura Y, Morishima I. High-pressure flash photolysis study of hemoprotein: effects of substrate analogues on the recombination of carbon monoxide to cytochrome P450CAM. Biochemistry. 1994;33:9762–9768. doi: 10.1021/bi00198a047. [DOI] [PubMed] [Google Scholar]
- 52.Oertle M, Richter C, Winterhalter KH, Di Iorio EE. Kinetics of carbon monoxide binding to phenobarbital-induced cytochrome P-450 from rat liver microsomes: a simple bimolecular process. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4900–4904. doi: 10.1073/pnas.82.15.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray RD. Kinetics and mechanism of CO binding to cytochromes P- 450LM2 and P-450LM4. Effect of phospholipid, nonionic detergent, and substrate binding. J. Biol. Chem. 1983;258:3764–3768. [PubMed] [Google Scholar]
- 54.Perera R, Sono M, Sigman JA, Pfister TD, Lu Y, Dawson JH. Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinis SA, Blanke SR, Hager LP, Sligar SG, Hoa GH, Rux JJ, Dawson JH. Probing the heme iron coordination structure of pressure-induced cytochrome P420cam. Biochemistry. 1996;35:14530–14536. doi: 10.1021/bi961511u. [DOI] [PubMed] [Google Scholar]
- 56.Wells AV, Li P, Champion PM, Martinis SA, Sligar SG. Resonance Raman investigations of Escherichia coli-expressed Pseudomonas putida cytochrome P450 and P420. Biochemistry. 1992;31:4384–4393. doi: 10.1021/bi00133a002. [DOI] [PubMed] [Google Scholar]
- 57.Collman JP, Sorrell TN, Dawson JH, Trudell JR, Bunnenberg E, Djerassi C. Magnetic circular dichroism of ferrous carbonyl adducts of cytochromes P-450 and P-420 and their synthetic models: further evidence for mercaptide as the fifth ligand to iron. Proc. Natl. Acad. Sci. U. S. A. 1976;73:6–10. doi: 10.1073/pnas.73.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharonov Yu A, Pismensky VF, Greschner S, Ruckpaul K. Low- and ultralow-temperature magnetic circular dichroism studies of reduced cytochromes P-450-LM2 and P-420-LM2 and of photo-products of their co-complexes. The spin-state and axial ligation of heme iron. Biochem. Biophys. Res. Commun. 1987;146:165–172. doi: 10.1016/0006-291x(87)90706-6. [DOI] [PubMed] [Google Scholar]
- 59.Marnett LJ, Kennedy TA. Comparison of the peroxidase activity of hemoproteins and cytochrome P450. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. New York: Plenum Press; 1995. pp. 49–80. [Google Scholar]
- 60.White RE, Sligar SG, Coon MJ. Evidence for a homolytic mechanism of peroxide oxygen–oxygen bond cleavage during substrate hydroxylation by cytochrome P-450. J. Biol. Chem. 1980;255:11108–11111. [PubMed] [Google Scholar]
- 61.Taylor CP. The EPR of low spin heme complexes. Relation of the t2g hole model to the directional properties of the g tensor, and a new method for calculating the ligand field parameters. Biochim. Biophys. Acta. 1977;491:137–148. doi: 10.1016/0005-2795(77)90049-6. [DOI] [PubMed] [Google Scholar]