Abstract
Macroautophagy (hereafter autophagy) is a ubiquitous degradative process in eukaryotic cells.1 Mitochondria autophagy (mitophagy) is a type of specific autophagy that degrades mitochondria selectively.2 Mitophagy is thought to play an important role for maintaining the quality of these organelles by eliminating damaged mitochondria, and it is involved in cellular differentiation, whereas dysfunctional mitophagy is related with neurodegenerative diseases;3-5 however, the mechanism of mitophagy is poorly understood. To facilitate the analysis of mitophagy, we recently established a simple method to monitor mitophagy in yeast, the Om45-GFP processing assay.6 Om45-GFP is a mitochondrial outer membrane protein. Following the uptake of mitochondria into the vacuole, Om45-GFP is degraded, releasing the intact form of GFP, which is detected by immunoblotting. Therefore, the amount of free GFP reflects the level of mitophagy.
Keywords: mitochondria, autophagy, vacuole, Om45, GFP
1. Introduction
Autophagy is originally considered to be the nonspecific degradation of cytoplasmic components.7 Recent studies, however, reveal that there are specific types of autophagy in which particular proteins or organelles are delivered to the vacuole/lysosome for degradation. Such specific autophagy includes the Cvt pathway, pexophagy (selective peroxisome autophagy) and mitophagy.6,8,9 Substantial progress has been made in determining the molecular mechanism of these pathways during the past decade.10-13 Yeast genetic screens for autophagy reveal 33 AuTophaGy-related (ATG) genes at present, and the characterization of each gene product has contributed to the clarification of the molecular mechanism of autophagy. Mitophagy, on the other hand, is poorly understood, presumably due to the difficulty of monitoring this process. Here we describe a useful assay for monitoring yeast mitophagy, Om45-GFP processing (see Note 1).
The Om45 protein is a mitochondrial outer membrane protein of unknown function.14 The Om45 protein tagged with the green fluorescent protein (Om45-GFP) also localizes on the mitochondrial outer membrane, and accumulates in the vacuole when mitophagy is induced. Om45-GFP delivered into the vacuole is degraded; however, the GFP is relatively stable within the vacuole and is often released as an intact protein. The processed GFP can be detected by immunoblotting as semi-quantitative evidence for mitophagy (Fig. 1). Mitophagy can also be monitored as an accumulation of green fluorescence in the vacuole lumen observed by fluorescence microscopy (Fig. 2).
Figure 1.
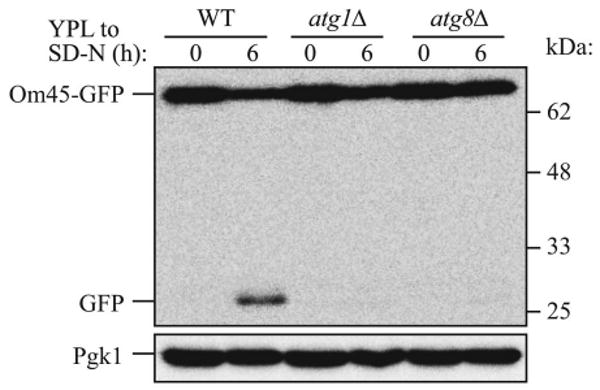
Monitoring mitophagy by western blot using the mitochondrial protein Om45-GFP during starvation. Wild-type (WT), atg1Δ or atg8Δ strains expressing Om45-GFP were cultured in YPL medium to the mid-log growth phase, then shifted to SD-N medium for 0 and 6 h. GFP processing was monitored by immunoblotting with anti-YFP and anti-Pgk1 (loading control) antibody. The positions of molecular mass markers are indicated on the right.
Figure 2.
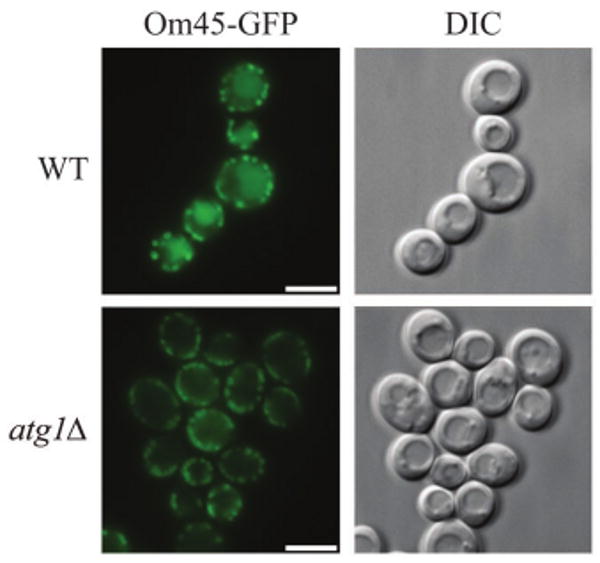
Monitoring mitophagy by microscopy using the mitochondrial protein Om45-GFP at the post-log phase. Wild-type (WT) and atg1Δ strains expressing Om45-GFP were cultured in YPL medium for 48 h. The localization of GFP was visualized by fluorescence microscopy. DIC, differential interference contrast. Scale bars, 5 μm.
2. Materials and Methods
2.1. Culture medium
Growth medium (YPD; 1% yeast extract, 2% peptone, 2% glucose), lactate medium (YPL; 1% yeast extract, 2% peptone, 2% lactic acid, pH 5.5), synthetic minimal medium with glucose (SMD; 0.67% yeast nitrogen base, 2% glucose, auxotrophic amino acids and nucleoside, and vitamins).
Synthetic minimal medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose).
2.2. Reagents
-
Anti-Pgk1 antibody,
Nordic Immunology, NE130/7S
-
Anti-YFP antibody,
JL-8 Clontech, 632380 (see Note 2)
-
Bromophenol blue,
Sigma, 32712
-
ECL detection kit,
Pierce, 32106
-
Glass beads,
Thomas Scientific, 5663R50
-
Glycerol,
Sigma, G5516
-
HRP-conjugated anti-rabbit IgG,
Fisher Scientific, ICN55676
-
Lactic acid,
Sigma, L6661
-
Polyethylene glycol 3350,
Sigma, P3640
-
PVDF membrane,
Millipore, IPVH00010
-
Salmon sperm DNA,
Sigma, D1626
-
SDS,
Sigma, L4509
-
TCA,
Sigma, T0699
2.3. Strain construction
The chromosomal GFP tagging on the C terminus of OM45 is done by the method shown by Longtine et al.15 The individual steps are described below.
A DNA fragment encoding GFP with a selective marker is PCR-amplified using pFA6a-GFP(S65T)-HIS3MX6,15 as a template plasmid (see Note 3) and primers (5′-TGA TAA GGG TGA TGG TAA ATT CTG GAG CTC GAA AAA GGA CCG GAT CCC CGG GTT AAT TAA and 5′-GAG AAA CAT GTG AAT ATG TAT ATA TGT TAT GCG GGA ACC AGA ATT CGA GCT CGT TTA AAC).
A wild-type and ATG knockout strain should be used as a positive and negative control, respectively (see Note 4). For transformation, strains are grown in YPD to mid-log phase (A600 = 0.5 to 1.0). Cells equivalent to 2.0 A600 units (a unit of cells is defined as the amount of cells that give an A600 = 1.0 in one ml) are collected by centrifugation (1,500 ×g, 2 min; 3,000 rpm in ST-720 rotor (Kubota) or equivalent) and the supernatant fraction is discarded by decanting. The cell pellets are resuspended in 1 ml of sterilized water, transferred to microcentrifuge tubes and recentrifuged (750 ×g, 1 min; 3,000 rpm in T15A22 rotor (Hitachi Koki) or equivalent); the supernatant fraction is again discarded. Cells are resuspended in 25 μl of sterile TE-lithium acetate (10 mM Tris-HCl, pH 7.5, 1 mM EDTA and 100 mM lithium acetate (pH 7.5 adjusted by acetic acid)). This solution is kept at room temperature and is stable indefinitely. Next, add 2.5 μl of single-stranded salmon sperm DNA (10 mg/ml) (see Note 5), 150 μl of 40% PEG solution (40% polyethylene glycol 3350 in TE-lithium acetate) (see Note 6) and 20 μl of the PCR product. Cells are incubated at 30°C with agitation for 30 min, then incubated at 42°C for 15 min. Cells are centrifuged (750 ×g, 1 min) and the supernatant fraction is discarded. The cells are resuspended in 50 μl of sterilized water and spread on agar plates containing selective medium (SMD without histidine; see Note 3).
Colonies grown up after two to three days are inoculated in SMD medium without histidine. If GFP tagging at the 3′ end of OM45 was correct, mitochondrial GFP fluorescence can be observed by fluorescence microscopy (for example, Fig. 2, atg1Δ). Correct integration can be determined by PCR analysis of genomic DNA.
2.4. Induction of mitophagy
2.4.1. Induction of mitophagy by amino acid starvation
Cells expressing Om45-GFP are cultured in YPD to mid-log phase. The cells are centrifuged (1,500 ×g, 2 min), and the supernatant fraction is discarded. The cell pellet is resuspended in 3 ml of YPL (A600 = 0.2), then cultured for 14 to 16 h to allow the cells to grow to log phase (A600 = 0.8 to 1.5).
When the cells are grown to log phase, cell aliquots equivalent to 1.0 A600 are placed in 1.7 ml microcentrifuge tubes to prepare the t = 0 h (before starvation) samples for SDS-PAGE (immediately follow with step 2.5.1).
The remaining cells are washed with 5 ml of sterilized water two times (centrifuge (1,500 ×g, 2 min), discard the supernatant fraction, add sterilized water). After the second washing step, the cells are resuspended in SD-N and cultured for 6 h (see Note 7). After starvation, cell aliquots equivalent to 1.0 A600 unit are placed in 1.7 ml microcentrifuge tubes to prepare samples for SDS-PAGE (immediately follow with step 2.5.1).
2.4.2. Induction of mitophagy at post-log phase
Cells expressing Om45-GFP are cultured in 5 ml of YPD to mid-log phase. Cells are centrifuged (750 ×g, 2 min), and the supernatant fraction discarded. The cells are resuspended in 5 ml of YPL, and inoculated into a flask containing YPL (A600 = 0.2).
After culturing for 12, 36 and 48 h in YPL, cell aliquots equivalent to 1.0 A600 are placed in 1.7 ml microcentrifuge tubes to prepare samples for SDS-PAGE (immediately follow with step 2.5.1).
2.5. Detection of mitophagy
2.5.1. Detection of mitophagy by immunoblotting
Trichloroacetic acid (TCA; 10% final concentration) is added to the samples prepared above and the samples are incubated for 10 min on ice. The proteins are pelleted by centrifugation at 21,000 ×g (15,000 rpm in T15A22 rotor (Hitachi Koki) or equivalent) for 10 min. After washing the pellet twice with 1 ml of ice-cold acetone, the pellet is air-dried (see Note 8).
The air-dried cell pellet is resuspended in 50 μl of sample buffer (150 mM Tris-HCl, pH 8.8, 6% SDS, 25% glycerol, 6 mM EDTA, 0.5% 2-mercaptoethanol, and 0.05% bromophenol blue) and disrupted by vortex with an equal volume of acid-washed glass beads (see Note 9) for 3 min. The samples are incubated at 100°C for 3 min.
10 μl of samples are loaded on a 12% polyacrylamide gel and resolved. A standard semidry western blot transfer procedure is performed using PVDF membrane. After blotting, the membranes are probed with anti-YFP antibody (1:10,000 dilution) by incubation overnight at 4°C. After washing the membranes for 10 min, 3 times with TTBS (50 mM Tris-HCl, pH 7.6, 0.9% NaCl, 0.1% Tween 20), a secondary incubation is performed with HRP-conjugated anti-rabbit IgG (1:10,000 dilution) for 1 h at room temperature. After washing the membrane 10 min, 3 times with TTBS, the signal of GFP is detected using the ECL kit. Om45-GFP and processed GFP can be detected as bands that migrate at approximately 72 and 28 kDa molecular mass, respectively (Fig. 1).
2.5.2. Detection of mitophagy by fluorescence microscopy
Cells expressing Om45-GFP are cultured in YPL for 48 h or in SD-N for 6 h to induce mitophagy as described above in section 2.4.
An aliquot of cells (100 μl) is collected by centrifugation (750 ×g, 1 min), the supernatant fraction is discarded and the cells are resuspended in 50 μl of phosphate buffered saline (3.2 mM Na2HPO4, 0.5 mM KH2PO4, pH 7.4, 1.3 mM KCl, 135 mM NaCl). Spot 2 μl of the cell suspension on a microscope slide and immediately cover the slide with a cover slip.
GFP is observed by fluorescence microscopy with excitation 510–550 nm and emission 470–490 nm. By comparing with the DIC (differential interference contrast) image, the accumulation of the GFP signal in the vacuole becomes clear, when mitophagy occurs (Fig. 2; see Note 10).
Acknowledgments
This work was supported by National Institutes of Health Grant GM53396 (to D.J.K.) and Grants-in-Aid for Scientific Research #19209019 from the Ministry of Education, Science, Technology, Sports and Culture of Japan (to D.K.).
Notes
Although we focus on Om45-GFP in this protocol, isocitrate dehydrogenase (Idh1)-GFP and mito-DHFR-GFP are alternative constructs that have been used successfully.6,16 In theory, most mitochondrial proteins can be tagged with GFP for this purpose. We tagged the C terminus of several different mitochondrial proteins with GFP and monitored mitophagy, and we found that Om45-GFP and Idh1-GFP showed the most sensitive results for this assay. Idh1 is a mitochondrial matrix protein. To construct an Idh-GFP expressing strain, use the primers 5′-TGA CTT CAC GAA TGA AAT CAT CAA CAA ATT ATC TAC CAT GCG GAT CCC CGG GTT AAT TAA and 5′-AAT TAT AGA TAA GAA TTT GAA CAC ACT TAA GTT GCA GAA CGA ATT CGA GCT CGT TTA AAC. The method for induction and detection of mitophagy are the same as for Om45-GFP.
One important point is that the chimera should not be overexpressed, which can lead to mislocalization and degradation that is independent of mitophagy. Thus, it is recommended to use the endogenous promoter, or a heterologous mitochondrial gene promoter.
This antibody in our hands is very specific, but other antibodies should also work.
Other plasmids with different selection markers (pFA6a-GFP(S65T)-kanMX6 or pFA6a-GFP(S65T)-TRP1) are available. In that case, different selective medium is required to select the positive strains (YPD with G418 (250 μg/ml) or SMD lacking tryptophan, respectively).
Most of the ATG deleted strains block mitophagy; we have confirmed that ATG1, 2, 4, 6–16, 18 and 32 deleted strains completely block mitophagy.16,17
The salmon sperm DNA is prepared in sterile water or TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA) and boiled for 5 min followed by quickly cooling on ice. It is not necessary to subsequently boil the carrier DNA. The salmon sperm DNA should be aliquoted and stored at -20°C. Other types of carrier can be used including tRNA.
The PEG solution should be less than three weeks old and filter sterilized.
A six-hour time point provides a clear signal for mitophagy-dependent processing of Om45-GFP under the conditions described here; however, there may be strain-dependent differences, and a time course may be used to monitor the kinetics of processing.
The air-dried pellet is stable at -20°C for several months. The samples can also be kept in TCA or acetone if necessary, and processed when convenient.
Acid washing for glass beads:
-
(9.1)Glass beads (1 kg of 0.4–0.6 mm size) are soaked in 1 liter of 1 N HCl overnight in a glass beaker for etching, which improves the efficiency of yeast breakage. The beaker should be covered and placed in a fume hood for safety.
-
(9.2)The beads are rinsed 10 to 15 times with the same volume (1 liter) of water.
-
(9.3)The beads are rinsed at least five times with deionized water. Mix the beads with a spatula during the washing to break up any clumps. Rinse again with deionized distilled water at least five times.
-
(9.4)After rinsing, the beads are dried in a drying oven overnight. We recommend that the container for storing the beads is used for this step because the dry beads are difficult to transfer.
The vacuole membrane can also be stained with the red dye FM 4-64 (Molecular Probes/Invitrogen, T-3166).
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 3.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–93. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–8. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 9.Dunn WA, Jr, Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 10.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 14.Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–6. [PubMed] [Google Scholar]
- 15.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


