Abstract
Drug Induced Liver Injury (DILI) is one of the main causes of drug attrition. The ability to predict the liver effects of drug candidates from their chemical structure is critical to help guiding experimental drug discovery projects towards safer medicines. In this study, we have compiled a dataset of 951 compounds reported to produce a wide range of effects in the liver in different species, comprising humans, rodents, and non-rodents. The liver effects for this dataset were obtained as assertional meta-data, generated from MEDLINE abstracts using a unique combination of lexical and linguistic methods and ontological rules. We have analyzed this dataset using conventional cheminformatics approaches and addressed several questions pertaining to cross-species concordance of liver effects, chemical determinants of liver effects in humans, and the prediction of whether a given compound is likely to cause a liver effect in humans. We found that the concordance of liver effects was relatively low (ca. 39–44%) between different species raising the possibility that species specificity could depend on specific features of chemical structure. Compounds were clustered by their chemical similarity, and similar compounds were examined for the expected similarity of their species-dependent liver effect profiles. In most cases, similar profiles were observed for members of the same cluster, but some compounds appeared as outliers. The outliers were the subject of focused assertion re-generation from MEDLINE, as well as other data sources. In some cases, additional biological assertions were identified which were in line with expectations based on compounds' chemical similarity. The assertions were further converted to binary annotations of underlying chemicals (i.e., liver effect vs. no liver effect), and binary QSAR models were generated to predict whether a compound would be expected to produce liver effects in humans. Despite the apparent heterogeneity of data, models have shown good predictive power assessed by external five-fold cross validation procedures. The external predictive power of binary QSAR models was further confirmed by their application to compounds that were retrieved or studied after the model was developed. To the best of our knowledge, this is the first study for chemical toxicity prediction that applied QSAR modeling and other cheminformatics techniques to observational data generated by the means of automated text mining with limited manual curation, opening up new opportunities for generating and modeling chemical toxicology data.
1. Introduction
Drug Induced Liver Injury (DILI) is widely regarded as a leading cause of drug attrition both during clinical development and post-approval (1) and therefore it constitutes a major safety concern for drug development (2–6). Elimination of drug candidates likely to cause hepatotoxicity at early stages of drug discovery could significantly decrease the rate of attrition and cut the cost of drug development. There is a great deal of interest both in the US (cf. the ToxCast program, http://www.epa.gov/ncct/toxcast/) and Europe (cf. the REACH program, http://ec.europa.eu/environment/chemicals/reach.htm) in developing fast and accurate experimental and computational approaches to predicting toxic effects of chemicals including hepatotoxicity. Experimental approaches have focused on the development of various in vitro assays (4;7–9) that can be used to assess the in vivo effects. Farkas and Tannenbaum (8) as well as Sutter (7) published very detailed reviews about different in vitro hepatotoxicity assessing techniques. O'Brien et al. (4) demonstrated that most conventional assays that measure cytotoxicity have a poor concordance with human toxicity. However, they still point out the great predictive accuracy of certain assays that evaluate oxidative stress, mitochondrial reductive activity and cell proliferation. In addition, O'Brien et al. suggested a novel promising strategy (involving the High Content Screening (HCS) technique) to monitor cytotoxicity biomarkers in human hepatocytes exposed to drugs and demonstrated good concordance of such in vitro results with drug-induced human hepatotoxicity. In another recent study, Xu et al. (10) reported testing of ca. 300 drugs and chemicals on human hepatocytes for their induced effects (mitochondrial damage, oxidative stress and intracellular glutathione, all measured by HCS imaging assay technology). Xu et al. obtained a true positive rate of 50–60% with very few false-positives. Recent studies, e.g., Blomme et al. (11) or Elferink et al. (12), showed promising results concerning the prediction of in vivo hepatotoxicity from microarray analysis of gene expression profiles (extracted from rat livers treated with a given drug).
Computational predictors of hepatotoxicity have been developed as well. For instance, a classification recursive partitioning model was developed based on 1D and 2D molecular descriptors that was trained using an ensemble of 143 compounds inducing liver injuries and 233 non-toxic molecules (13). A COMFA based approach was utilized (14) for the classification of 654 drugs, which have been experimentally tested using different in vitro assays to characterize their biological effects on liver. The MCASE program (15) was used to analyze liver toxicity and identify molecular fragments likely to be responsible for liver toxicity using a dataset of 400 drugs. Cruz-Monteagudo et al. (16) employed a Linear Discriminant Analysis (LDA) to build models capable of classifying correctly 74 drugs, of which 33 drugs were known as idiosyncratic hepatotoxicants and 41 did not cause this effect. Their models afforded impressive external prediction accuracies ranging from 78 to 86%. Egan et al. (3) have compiled a dataset of 244 molecules from published data and derived a series of 74 computational alerts based on specific molecular functional groups. However, there still remains a challenge in developing accurate predictors of DILI based on compound structure for several reasons: (i) the relationships between in vitro assays and in vivo hepatotoxicity are not yet well understood, (ii) the available Quantitative Structure-Activity Relationships (QSAR) models, obtained using small datasets of congeneric molecules, are not applicable to enable prediction of DILI for chemically diverse external sets.
The Safety Intelligence Program (SIP) is an industry-sponsored initiative that aims to build the world's most comprehensive intelligence resource to support drug safety assessments (http://www.biowisdom.com/content/safety-intelligence-program). SIP exploits BioWisdom's Sofia™ platform (http://www.biowisdom.com) to generate assertional meta-data, which comprises thousands of highly accurate and comprehensive observational statements. These statements are represented in triple constructs: concept_relationship_concept, e.g. Cafestol_suppresses_Bile acid biosynthesis, Azathioprine_induces_Cholestasis etc. Each assertion is derived from and evidenced by a variety of electronic data sources. Behind each assertion is a rich vocabulary (developed by BioWisdom) that renders that assertion semantically consistent with the other assertions around the same concept. For example, the liver pathology term, cholestasis can be described across the literature as bile stasis, biliary stasis, cholestasia, cholestatic injury, biliary stases. Assertional meta-data generated in this manner facilitates the semantically consistent integration of disparate observations across historic literature. The Program has set a high standard of accuracy for the generation of the assertional meta-data (>97% chance that the statement accurately represents that made by the author). This enables the meta-data to be used for sophisticated assertional meta-analyses, such as that described in this study. Given the widespread issues associated with DILI, this study uses a subset of SIP assertions which focus on chemicals known to produce effects in the liver.
Recently, SIP initiated an ambitious study to use assertional meta-data to determine the level of concordance across various species for drug-induced liver effects (http://www.biowisdom.com/files/SIP_Board_Species_Concordance.pdf). Using assertions generated from MEDLINE abstracts and the European Medicines Agency European Public Assessment Reports (EMEA EPARs; http://www.emea.europa.eu/htms/human/epar/eparintro.htm), the study showed that 38–51% of drug-induced liver effects in humans are not detected in preclinical species. These findings are in general agreement with the previous work of others (17).
In this study, we exploit the assertional meta-data derived from this initial concordance study to investigate the relationship between chemical structure and species-selectivity of liver effects. Specifically, a set of 1061 compounds was derived from the assertional meta-data referenced by MEDLINE abstracts. Each compound in this dataset was reported in the literature to induce or modify one or more liver effects. The liver effects included in the study are both pathological and physiological events, owing to the fact that interruption of normal function and the development of pathology are tightly associated. For example, cholestasis can arise from the inhibition of bile transport; the development of cancer may result from the dysregulation of apoptosis or cell cycle; and compounds that interrupt collagen metabolism are associated with liver fibrosis. Importantly, the assertional meta-data has been collected across different species, i.e., human, rodent (mostly rat and mouse) and non-rodent animals (mostly dog), and this allows consideration of species-dependent effects in this analysis. To our knowledge, this is the largest available molecular dataset containing information on chemically-induced liver effects and, thus, it allows one to ask important questions about the relationship between chemical structure and species-selectivity of liver effects.
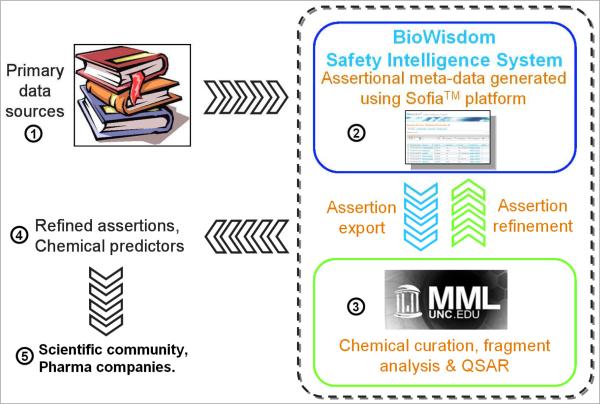
The study was conducted in an ordered and iterative workflow (see inside hash lines in Figure 1) starting with chemical curation of the molecular dataset, and re-assessment of the liver effect concordance across species. Next, we have applied standard cheminformatics procedures such as clustering by chemical similarity to the set of 951 compounds (left after the thorough chemical curation). This allowed us to identify multiple clusters of congeneric compounds and explore the concordance between chemical similarity and liver effect profiles across species. Finally, we have transformed the assertion data into binary liver effect profiles across species and applied binary QSAR modelling approaches to the resulting dataset. We show that the use of cheminformatics approaches in the analysis of toxicity assertions mined from the literature can identify apparent gaps in this dataset, leading to its refinement, as well as afford statistically robust and externally predictive models of chemical effects on the liver. To the best of our knowledge studies reported in this paper present the first instance of successful application of cheminformatics based analytical approaches to non-traditional chemical toxicity data (i.e., assertions) generated by ontology-based semi-automated text mining.
Figure 1.
The general study design workflow.
2. Methods
2.1. Data preparation
2.1.1. Generation of relevant assertions in MEDLINE abstracts using the Sofia platform
A collection of MEDLINE abstracts relevant to drug-induced liver effects was defined by querying all MEDLINE titles and abstracts with a list of terms relating to hepatobiliary anatomy and pathology, e.g. liver, hepatic, cholestasis and biliary. Assertion generation was carried out on the resulting corpus, comprising approximately 650,000 MEDLINE records, using BioWisdom's Sofia™ platform. A combination of lexical and linguistic tools was used to extract relationships that exist between any therapeutic compound (i.e. that has been or is used in the clinic) and a range of liver pathologies, e.g. hepatitis and focal necrosis, and hepatic physiological observations such as gluconeogenesis and cell growth. The use of several extraction methods, which differ in their level of recall and accuracy, ensures that the assertion generation process provides the most rapid, systematic and unbiased coverage possible. The extraction procedure was directed at the injurious effects of compounds by focusing on relationships that imply causation or regulation, such as Clozapine_induces_Hepatic Necrosis and Propofol_influences_Hepatic Lipid Metabolism, rather than the associations that imply therapeutic benefit such as treats. This yielded a set of “proto-assertions”, composed of concept_relationship_concept triplets, together with species, tissue or cell type/cell line information. All of these elements were manually validated to achieve an accuracy level of greater than 97%, confirmed by random sampling and quality control testing. For this study, we used 14,609 assertions, which contained 1061 therapeutic compounds and 2099 biomedical observations (comprising 424 pathological and 1675 physiological effects).
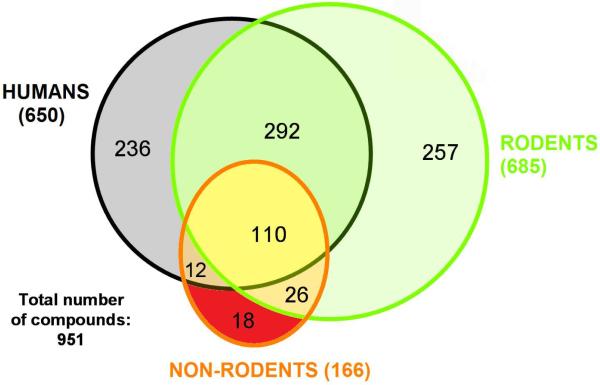
To allow the correspondence of compound-induced liver effects to be assessed in human, rodents and non-rodents, the non-human species were classified as either rodent (rat, mouse, hamster, guinea pig and rodent) or non-rodent (dog, cat, pig, monkey, goat, rabbit and sheep). Thereafter, compounds inducing liver effects across the various species were assigned to the appropriate group; for instance, the assertion “Acetaminophen induces Acute Liver Failure (rat)” resulted in the addition of Acetaminophen to the rodent species group. A profile of compounds for each group was thus created, and the concordance of the three groups was visualised in the form of a pivot chart and Venn diagram (Table 1, Figure 2).
Table 1.
The drug-induced liver effect profiles across species: humans (A), rodents (B) and non-rodents (C).
| ID | Name | HUMAN (A) | RODENT (B) | NON-RODENT (C) | (A) Only | (B) Only | (C) Only | AB | AC | BC | ABC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (R)-Roscovitine | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2 | 17-Methyltestosterone | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1-alpha-Hydroxycholecalciferol | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 2,3-Dimercaptosuccinic acid | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 5 | 2,4,6-Trinitrotoluene | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 2-Deoxy-D-glucose | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 7 | 2'-fluoro-5-methylarabinosyluracil | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 2-Methoxyestradiol | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 9 | 4-aminobenzoic acid | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 10 | 4-Hydroxytamoxifen | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 11 | 5 fluorouracil | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 12 | 5-Azacitidine | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 13 | 5-Bromouracil | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 14 | 5-fluoro-2'-deoxyuridine | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 15 | 6-Mercaptopurine | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 16 | Acadesine | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 17 | Acarbose | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 18 | Acebutolol | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 19 | Acenocoumarol | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | Acetamide | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 21 | Acetaminophen | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 22 | Acetazolamide | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 23 | Acetic acid | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 24 | Acetohexamide | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | Acetohydroxamic acid | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 26 | Acetrizoate Sodium | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 27 | Acetylcholine | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 28 | Acetylcysteine | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 29 | Acetyl-L-carnitine | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 30 | Acetylsalicylic acid | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 31 | Acitretin | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
[Note: Only a subset of the data is shown for illustration purposes. The complete table is available in the supporting information]
Figure 2.
Venn diagram representing 951 compounds classified according to their liver effects for humans (650 compounds), rodents (685) and non-rodents (166).
2.2.2. Chemical data curation
Data curation is a critical procedure in the analysis of any chemical dataset, as is particularly evident from the recent study on the detrimental effect of the incorrect representation of chemical structure on the quality of QSAR models (18). We have employed both automatic and manual procedures to the initial dataset of 1061 molecular structures defined by the assertional meta-data as follows:
- First, all inorganic compounds have been removed since our data analysis strategy includes the calculation of molecular descriptors for organic compounds only. This is an obvious limitation of our analysis since inorganic molecules are definitely known to induce diverse liver injuries; however the total fraction of inorganics in our dataset was relatively small. Thus, the following compounds have been removed from our dataset: activated charcoal, cobalt dichloride, ferrous sulphate, zinc chloride, sulphur, cis-diaminedichloroplatinum, manganese chloride etc. Moreover, additional compounds were removed because (i) their corresponding SMILE strings were impossible to retrieve despite many efforts, or (ii) they corresponded to mixtures of compounds (for example, Gramicidin, which is a product containing six different antibiotic molecules).
- 2D molecular structures (chemical connectivity maps) were generated from SMILE strings using the JChem 5.0 program of ChemAxon (http://www.chemaxon.com). We also used the Standardizer module of JChem to remove all counter-ions, clean records including multiple compounds, clean the 2D molecular geometries and normalize bonds (aromatic, nitro groups etc.) of the 989 remaining compounds.
- Duplicate molecular structures were automatically detected and deleted using the ISIDA/Duplicates (http://infochim.u-strasbg.fr) program, followed by careful manual inspection of the entire dataset. Finally, 951 compounds remained out of the initial 1061 molecules. Once again, it should be pointed out that such laborious but necessary steps of data curation are one of the critical stages of this study, making the following results more pertinent and valid (18).
2.2. QSAR modeling
2.2.1. Substructural Molecular Fragments (SMFs)
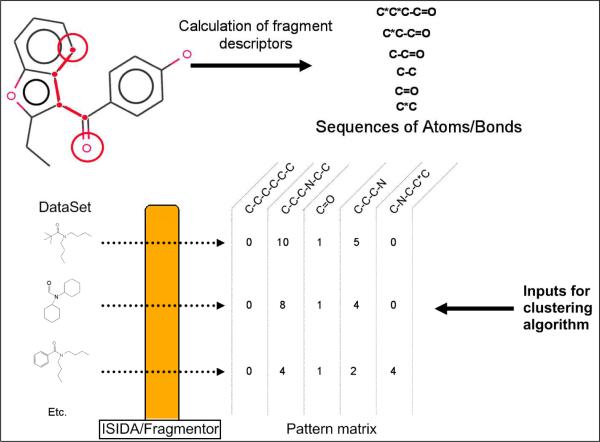
The ISIDA/Fragmentor program (19) (freely available from http://infochim.u-strasbg.fr) was employed to calculate 2D SMFs for all compounds. Briefly, each molecular structure is split into small chemical patterns (see Figure 3). Two different types of fragments were considered: “sequences” (I) and “augmented atoms” (II). Three sub-types AB, A and B are defined for each class. For the fragments I, they represent sequences of atoms and bonds (AB), of atoms only (A), or of bonds only (B). Only shortest paths from one atom to another are used. For each type of sequences, the minimal (nmin) and maximal (nmax) number of constituted atoms are defined (for this study, nmin = 2 and nmax = 7). An “augmented atom” represents a selected atom with its environment including either both neighbouring atoms and bonds (AB), or atoms only (A), or bonds only (B). Atomic hybridization (Hy) has been taken into account for augmented atoms. Unlike structural keys, there is no predefined library of fragments. In this study involving 951 compounds, fragment descriptors were rapidly calculated by the ISIDA/Fragmentor program: 1466 fragments remained after the deletion of invariants, low variance and highly correlated ones. Moreover, fragment descriptors are directly linkable to the chemical structure of compounds. As a consequence, QSAR models involving fragments are highly suitable for virtual screening of large sets of compounds as well as for defining structural alerts. Cluster analysis in cheminformatics as well as QSAR modeling is also commonly performed using molecular fingerprints like structural keys or such types of fragment descriptors (20;21). Small fragments do not represent the complexity of chemical compounds by themselves. However, in combination, multiple descriptors do reflect the structure complexity. In this work, the use of an ensemble of fragment descriptors afforded better segregation between compound classes using clustering and machine learning techniques.
Figure 3.
Examples of substructural fragment descriptors, and the corresponding pattern matrix for the clustering of compounds in chemistry space.
2.2.2. Hierarchical Cluster Analysis
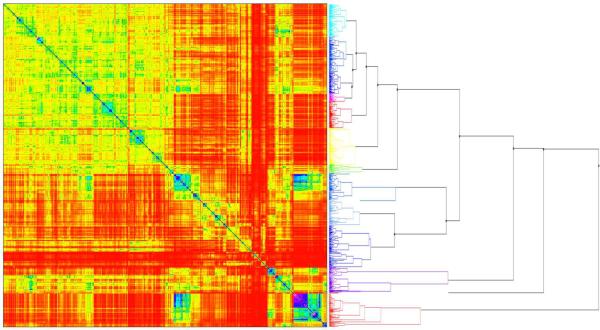
The clustering of a chemical dataset consists of merging compounds into independent clusters that include chemically similar molecules (see recent publications (22;23) for the review of the most popular clustering approaches used in computational chemistry). In this study, we have employed the Sequential Agglomerative Hierarchical Non-overlapping (SAHN) method implemented in the ISIDA/Cluster program (http://infochim.u-strasbg.fr) (24). Briefly, each compound represents one cluster at the start. Then, the m compounds are merged iteratively into clusters using their pairwise Euclidean distances stored in a squared m * m symmetric distance matrix. At each iteration, the two closest objects (molecules or clusters) are merged to form a new cluster and then, the distance matrix is updated with the distances between the newly formed cluster and the others, according to the user-specified type of linkage (single, average, complete or Ward type). The process is repeated until one cluster remains. The parent-child relationships between clusters result in a hierarchical data representation, or dendrogram. SAHN algorithms are only appropriate to treat relatively small datasets (several thousands) in high dimensional space due to their O(N3) time and O(N2) space requirements. ISIDA/Cluster allows visualization of molecular structures in each cluster, directly on the dendrogram, and has multiple options to facilitate the analysis of results and export contents of selected clusters. In particular, we used ISIDA/Cluster to obtain the heat map of the proximity matrix, as well as the dynamic dendrogram of compound clusters (see Figure 4).
Figure 4.
Heat map representing the distance matrix (left) between compounds and the corresponding dendrogram (right): the map is colored according to the chemical similarity between compounds (blue-violet: high similarity; yellow-red: low similarity); small clusters with high levels of chemical similarity can be identified on the diagonal of the matrix.
2.2.3. Support Vector Machines (SVM) approach
The description of the original SVM algorithm could be found in many publications (25). Briefly, molecular descriptors are first mapped onto a high dimensional feature space using various kernel functions, and then a linear model is constructed in this feature space to segregate compounds with different activities. Models built with this machine learning technique allow the prediction of a target property using a set of descriptors solely calculated from the structure of a given compound. Best practices to derive and select the best models with high predictive abilities from a modeling set have been detailed elsewhere (26). For SVM classification, we used WinSVM program developed in our group at UNC (available upon request) implementing the open-source libsvm package (http://www.csie.ntu.edu.tw/~cjlin/libsvm/). The WinSVM program provides users with a convenient graphical interface to prepare input data, to split compounds into training, test and validation sets, to set up parameters for SVM grid calculations (iterative and simultaneous grid optimization of SVM parameters), to launch and follow calculation progress in a powerful graphical interface, to select models with the best prediction performances on both training and internal test sets, and then apply them for the external test set as an ensemble consensus model with its defined applicability domain. The program also allows one to visualize molecular structures and various plots, making the use of SVM easier and more appropriate for QSAR modeling, in order to obtain robust and predictive models and apply them to virtual libraries as well.
3. Results
3.1. Assessment of the concordance of liver effects across species
As described in section 2.2, 951 compounds remained after the chemical curation of the dataset obtained from assertions generated from MEDLINE abstracts by Sofia. We have reorganised these data in the form of a set of liver effect profiles for each compound (see Table 1): if a compound was found to have a reported liver effect in a species (human - A, rodent - B or non-rodent - C), its corresponding cell in Table 1 was given the value of “1”. To enable the concordance analysis, we have made an assumption that every compound was tested in all three species. Thus, if no effect was reported for a compound in the assertional meta-data for a given species, we have assumed that the compound does not exert liver effects in that species. Consequently, the cell is given the value “0”. Following this, molecules have been classified as having a liver effect for one species only (A only or B only or C only), two species (AB, AC or BC) or all three species (ABC) (Table 1, right hand side). Note that by design, the dataset did not include any compound that reported no liver effect in any species. To visualize this dataset and illustrate the overlaps between species categories, a Venn diagram was generated (Figure 2).
All 951 compounds are represented on the Venn diagram: 650 molecules among them have been identified as causing liver effects in humans; 685 showed liver effects in rodents, and 166 in non-rodents. 110 molecules were reported to have liver effects in all three species. Furthermore, a total of 402 (292 + 110) compounds reported liver effects in both humans and rodents, whereas a total of 122 compounds reported liver effects in both humans and non-rodents. We provide additional comments concerning the validity and the limits of such splitting in the discussion part.
These data were used to address the important question of concordance between drug-induced liver effects reported in different species. We have defined the concordance between two species, e.g., humans (A) and rodents (B), as CONC(A,B), according to the following formula:
| (Eq. 1) |
Where the sum of the numbers of toxicants and non-toxicants for both species A and B, is divided by the total of tested chemicals. We have applied this formula to calculate the concordance between species with the following results:
-
(i)
Humans-A and rodents-B: CONC(A,B) = (292 + 110 + 18)/951 = 44.2 %,
-
(ii)
Humans-A and non-rodents-C: CONC(A,C) = (110 + 12 + 257)/951 = 39.9 %,
-
(iii)
Rodents-B and non-rodents-C: CONC(B,C) = (110 + 26 + 236)/951 = 39.1 %.
The difference between concordances (less than 5 %) was found to be very small. Certainly, we have to stress that these results are valid if and only if our underlying assumption is correct, i.e., that each compound has been effectively tested in all species groups (A, B and C), and in each case, where liver effects are found, it is reported in the assertional meta-data. Nevertheless, with this assumption in mind, the results suggest that animal testing in many cases may be inconclusive with respect to the expected liver effects in humans. Central to these species-specific effects may be differences in the specificity and affinity of chemicals interacting with their targets in different species. There are many examples of species differences in ligand binding to key receptors, enzymes and transporters between species, all of which could influence physiological and ultimately pathological outcomes. One should also consider the differences in doses of compounds administered in rodents vs. humans.
It is also useful to examine conclusions about the concordance between species-specific liver effects using prior probabilities, which are conceptually more simple and perhaps more intuitively obvious than the Conc(A,B) function described above. The prior probabilities could be formally established by calculating the fraction of compounds reporting liver effects in one species that are also known to produce liver effects in another species. However, as we demonstrate below, the use of prior probabilities with these unbalanced datasets could lead to opposite (and therefore, confusing) conclusions. Indeed,
-
(i)
Among compounds that report liver effects in humans (650), 62 % (312/650) also report liver effects in rodents, while only 19 % (112/650) report liver effects in non-rodents.
-
(ii)
On the other hand, if we consider compounds with reported liver effects in non-rodents (166), 73 % (112/166) of them also have reported liver effects in humans, while only 59 % (312/685) also have reported liver effects for humans.
These two series of statements, based on prior probability calculations, lead to opposite conclusions about the concordance between species. Specifically, prior probabilities tend to show a significant concordance for liver effects between non-rodents and humans (73%); on the contrary, that concordance value (for non-rodents and humans) calculated using Eq.1 is equal to 40% only. However, recent experimental studies (4;27;28) support the concordance results (40–44%) calculated using Eq. 1. Indeed, it was recently noted (4) that “hepatotoxicity has the poorest correlation with regulatory animal toxicity test. In only approximately half of the new pharmaceuticals that produced hepatotoxicity in clinical drug development was there any concordance with animal toxicity studies.” These considerations as well as earlier studies cited above suggest that the definition of concordance as in Eq. 1 is indeed robust. We are currently investigating, using additional data and cheminformatics approaches, if there are possible chemical determinants of concordance, i.e., if there are distinct classes of chemicals for which animal data could be significantly more indicative of the expected effects in humans. The results of these studies will be reported elsewhere.
3.2. Clustering of compounds in chemistry space and gap-spotting in assertional meta-data
An initial quick examination of the 951 study compounds suggested a high level of dissimilarity between their molecular structures. Nevertheless, we have applied clustering procedures to this dataset to identify small groups of structurally similar compounds and assess whether they possessed similar liver effect profiles. To this end, compounds were clustered using fragment descriptors (see Figure 3) and the hierarchical algorithm of ISIDA/Cluster, as described in the Methods section. The resulting dendrogram and the associated distance matrix represented by the heat map are given in Figure 4. Analysis of this map revealed small clusters (located on the diagonal of the map) with fairly high levels of chemical similarity between compounds.
We have focused on the assertional meta-data analysis of different clusters and asked the following question: given that compounds within clusters have similar molecular structures, how similar are they in terms of liver effect profiles across the three species? We should note here that, as we began to review the similarity of liver effects, it became clear that a considerable amount of non-rodent data was reporting 0 (i.e., according to our assumptions, the compounds showed no liver effect in this species group). Given that the numbers of compounds reported in the assertional meta-data for non-rodents was sparse, we felt that our initial assumption of data completeness was perhaps weak in this case in terms of making any statistically significant conclusions. For this reason, the non-rodent data were not considered for this analysis. The following five case studies (CS) focus on liver effect profiles in humans and rodents; they are also summarized in Table 2.
- CS 1: four compounds (barbital derivatives) make up this cluster. They have the same chemical scaffold and are highly similar, and in fact, their liver effect profiles are exactly the same: these compounds have been reported as producing liver effects in rodents only. Follow-up assertion generation using MEDLINE did not identify any evidence for these compounds having human liver effects. In addition, basic searches in Google (e.g. barbital, human, hepatotoxicity) did not reveal evidence for these compounds having human liver effects either. The apparent lack of human liver effects may be due to these compounds being used for sedation/anaesthesia where lower doses and shorter exposures may be used than in animal studies.
- CS 2: Both cladribine and clofarabine are anticancer drugs and have been identified as producing liver effects in humans only. In contrast, cordycepin was found to report liver effects in rodents only. Iterative assertion generation from MEDLINE failed to identify any new evidence for cladribine and clofarabine having rodent liver effects. However, a recent assertion in SIP, referenced by an EMEA EPAR did identify clofarabine as having rodent liver effects (but still no rodent liver effects were identified for cladribine). It should also be noted that follow-up assertion generation from MEDLINE did identify an effect of cordycepin in a human hepatocellular cell line, as expected from our chemical similarity analysis.
- CS 3: Four derivatives of 3-benzoyl-1-benzofuran are clustered together: amiodarone (antiarrhythmic agent), benzarone (used for treatment of peripheral vascular disorders), benzbromarone (uricosuric agent, used for gout), benziodarone (vasodilator). Three of these compounds are reported to induce liver effects in humans and rodents. However, benziodarone reported liver effects in rodents only, despite the high level of chemical similarity between the four structures. As a result, one could suppose that this compound should be tested (or re-tested) in human in vitro assays (e.g. hepatocytes) to confirm that it does not produce liver effects. Follow-up assertion generation from MEDLINE did not identify any new evidence for benziodarone having human liver effects. However, a basic search in Google (e.g. benziodarone, human, hepatotoxicity) did reveal that the drug was reported to cause hepatotoxicity in humans (Table 10 in http://pubs.acs.org/doi/abs/10.1021/tx600260a). In fact, additional literature mining identified a report indicating that benziodarone was indeed withdrawn from the market in the UK since 1964 for hepatotoxicity (29). This finding, prompted by our chemical similarity analysis, illustrates the potential power of cheminformatics analysis to identify possible gaps in the assertional meta-data derived from the literature.
- CS 4: This cluster is composed of five estrogen-like molecules. Once again, our analysis allowed us to identify an interesting case: four among the five molecules were reported to have liver effects in both humans and rodents: 2-methoxyestradiol, estradiol, estrone and ethinyl estradiol. In contrast, estriol was not identified as producing liver effects in humans, despite its high similarity to the other four molecules of this cluster. Follow–up assertion generation from MEDLINE and a basic search in Google (e.g. estriol, human, hepatotoxicity) did not identify any new evidence for estriol having human liver effects. This observation of the apparent disparity between chemical similarity and liver effect profiles remains puzzling.
- CS 5: The last example concerns six antibiotics with good structural similarity: ciprofloxacin, gatifloxacin, levofloxacin, moxifloxacin, norfloxacin and sparfloxacin. All compounds report liver effects in humans. However, their liver effect profiles are different for rodents. Follow-up assertion re-generation from MEDLINE did identify new evidence for moxifloxacin having rodent liver effects. Basic searches in Google did not identify evidence for gatifloxacin and levofloxacin to have rodent liver effects.
To briefly summarize these results, some clusters have been identified in which compounds share similar molecular motifs, corresponding to similar liver effect profiles in humans and rodents. However, in some clusters, one can observe that similar molecules possess different liver effect profiles. These cases may correspond to missing or unreported data, and highlight areas for gap-spotting or additional experimental investigation. Indeed, we have demonstrated that additional, focused assertion re-generation using public sources led to modification of the original profiles towards greater similarity, as expected for highly chemically similar compounds. These results demonstrate the generally high accuracy of the assertional meta-data retrieved by Sofia, while confirming the importance and validity of the cheminformatics analysis of the links between chemical structure and assertions. Thus, our findings illustrate the power of cheminformatics in spotting possible data gaps.
Table 2.
Examples of five clusters identified within the entire library of 951 compounds. For each compound, its assertional meta-data profile is also shown (“1” = liver effect; “0” = no liver effect).
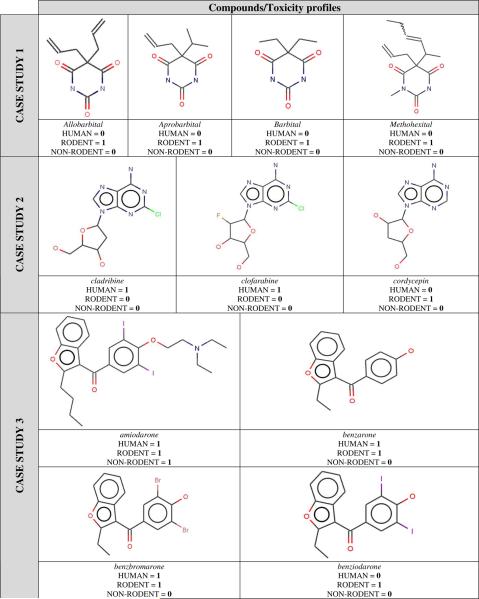
|
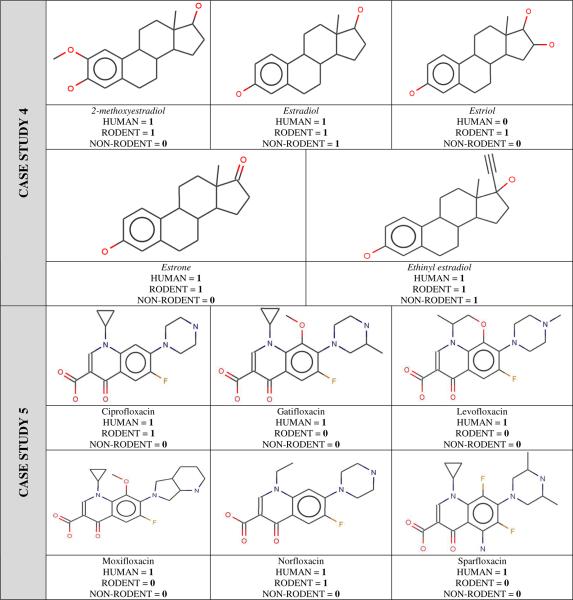
|
3.3. Analysis of chemical descriptor biases for species-specific assertions
Focussing on human and rodent data only, 248 molecules (236 + 12) – Class 1, have been identified as reporting liver effects for humans only (see Figure 2). Yet, 283 different molecules (257 + 26) – Class 2, showed liver effects for rodents only (i.e., these compounds have not been reported as having liver effects in humans). In this section, we have examined if certain chemical features are present in different proportions within these two datasets (classes 1 and 2), which could help identifying potential species-specific chemotypes. We used 2D fragment descriptors of each compound and then, we determined important variations in the fragment distributions within class 1 and class 2 separately.
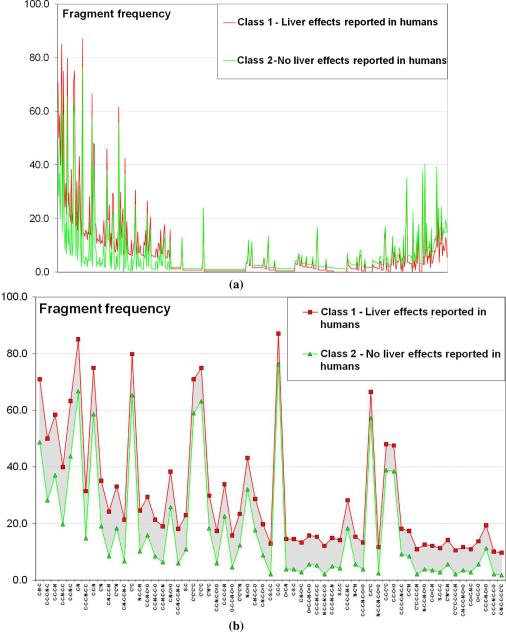
The frequency distribution of the fragment descriptors is illustrated in Figure 5a. Results indicate that two distinct categories of fragments can be identified based on their frequency differences (ΔF) between the two classes of compounds (Figure 5b) (the analysis was also done with frequency ratios leading to comparable conclusions):
-
(i)
fragments with positive ΔF (found more frequently in class 1 than in class 2; see Figure 5b): the highest ΔF values (more than 15%) are associated with amine-derived fragments (C-N-C, C-C-C-N-C, C-C-C-N, C-C-N-C-C, C-C-N-C, C-N, C-C-C-N-C-C, C-C-N) or aromatic rings including nitrogen atoms (C*N, C*C*N, * represent aromatic bonds);
-
(ii)
fragments with negative ΔF (found more frequently in class 2 than in class 1) : for example, O-C-C-C-O, C-C-C-C-C-O, C-O-C-C-C-O, C-C-C-C-O-C, O-C-C-C-C-O, C-C-C-O-C (alcohol, ether and carboxylic acid substituted alkyl groups) have ΔF smaller than – 5 %, as well as C*C*C-C-O (aromatic ring substituted by a methoxy group or carboxyl group) or C-C-C=C-C=O (alkenes with a carbonyl group in α-position).
This study is not exhaustive, and a more detailed list of chemotypes could be derived using additional descriptors, such as the number of H-bond donors or acceptors, the number of rings, etc. However, it follows from this analysis that some particular chemotypes influence certain species-specific liver effects: for instance, in our dataset, 71 % (176 out of 248) of compounds reporting liver effects in humans only (class 1) possess the fragment C-N-C (amine II), whereas that fragment is present in only 48.8 % (138 out of 283) of compounds that report no liver effects in humans (see Figure 5b) and in 53.7 % (216 out of 402) of compounds that report liver effects in both humans and rodents. This information is not sufficient to conclude that the presence of the amine group in a compound will result in a liver effect in humans. Moreover, we stressed that such statistical analysis could be relatively biased by different issues: it is obvious that certain classes of compounds known as being extremely toxic for human liver would not be tested any longer; it is also clear that drug candidates being toxic for rodent liver will be rejected early in the development process and thus will not be tested at all for humans. It finally leads to important inaccuracies in the fragment statistics because of the weak assumption of data completeness. However, we still believe that such frequency analysis is a critical preliminary step in order to create a series of novel filters (or structural alerts) that can be used to rapidly identify drug candidates likely to cause liver effects in humans.
Figure 5.
(a) Frequency analysis of fragment descriptors (X axis) in class 1 (compounds inducing liver injuries for humans) and class 2 (compounds inducing no liver injuries for humans). (b) List of fragment descriptors that are more frequent in class 1 than in class 2.
3.4. QSAR modeling of drug-induced liver effects
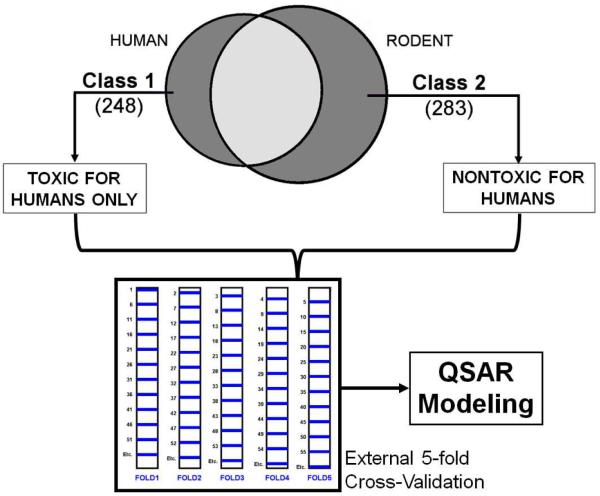
Following the simple relative descriptor frequency analysis described above, we have endeavoured to establish quantitative relationships between molecular structures and their species-specific liver effects. We limited ourselves to the analysis of data reported for humans and rodents because of scarcity of data reported for non-rodents (only 18 compounds reporting liver effects in non-rodents only; see Figure 2). Indeed, from datasets A and B (see Table 1), we could define two classes: 248 compounds inducing liver effects in humans only (class 1) and 283 compounds inducing no liver effects in humans (class 2 – reported as causing liver effects for rodents only). Based on this classification, we have derived QSAR classification models with the SVM machine learning approach and two different types of molecular descriptors: 2D fragments (described in section 3.3) and Dragon molecular descriptors (30). Datasets A and B were merged (531 molecules) and randomly split into 5 modeling/external sets (see Figure 6) to enable a 5-fold external Cross-Validation (CV; each compound is taken one time only in an external test set). QSAR models were derived for the modeling set only (i.e., internal 5-fold CV is done for each of the five modeling sets; models are selected if they have reasonable statistical accuracy of both training and internal test set) and selected based on prediction performances for the modelling set only. External test sets were never used to derive or select the models. Our recently developed WinSVM program was used to generate, select and apply models. The goal of our modelling studies was to obtain a series of predictive classification QSAR models capable of separating compounds reporting liver effects in humans from those that report no effects in humans, based on their chemical structure.
Figure 6.
QSAR modeling workflow for drug-induced liver effects in humans.
Prediction accuracies of the resulting models are given in Table 3. For each fold, the internal 5-fold CV performed on each modeling set led to SVM models with reasonable classification accuracies ranging from 61.9 % to 67.5 %. When applied to the entire modeling set, SVM models had high accuracies from 77.6 % to 99.3 %. Here, it should be pointed out that, in too many published QSAR studies, only those latter results are reported to prove the high statistical accuracy of the models. However, as we have pointed out in many publications (reviewed recently in (26)), rigorous external validation is a mandatory component of any QSAR study. Thus, our internal 5-fold CV allowed an honest assessment of the model prediction abilities: from the modeling set, we can realistically expect external predictions with an accuracy ranging from 61.9 % to 67.5 %.
Table 3.
Prediction accuracies obtained by the selected classification QSAR models for the 5-folds, using the SVM machine learning approach and two types of descriptors.
| Fold | Modeling set 5 fold CV | Modeling set Accuracy | External set Accuracy | Descriptors |
|---|---|---|---|---|
| 1 |
62.3%
62.9% |
88.2%
77.6% |
71.0%
67.3% |
fragments
Dragon |
| 2 |
64.9%
67.5% |
81.2%
81.2% |
64.2%
55.7% |
fragments
Dragon |
| 3 |
62.4%
65.2% |
91.3%
91.1% |
64.2%
61.3% |
fragments
Dragon |
| 4 |
64.9%
62.1% |
99.3%
84.9% |
72.6%
68.9% |
fragments
Dragon |
| 5 |
63.3%
61.9% |
82.6%
94.4% |
68.9%
70.8% |
fragments
Dragon |
Concerning the `blind' prediction for the 5 external test sets, our models led (as expected) to reasonable accuracies ranging from 55.7 % to 72.6 %, depending on the fold. Assessment of these results shows a slightly better prediction accuracy reached by models built with fragment descriptors (64.2 % to 72.6 %) compared to the models developed with Dragon descriptors (55.7 % to 70.8 %). Y-randomization procedure applied to the modeling set, as a matter of internal validation (discussed in (26)), was applied to the modeling set as a matter of an additional internal validation. According to this popular statistical test, it is expected that models built for the original datasets with shuffled target property (i.e., Y-randomized) should have low accuracy of prediction for both training and test sets. Indeed, in our calculations, models built with the Y-randomized target property (in our case, toxicity class assignment) led to training set models with very low accuracies (40–50%) and very poor external prediction performances. The failure of building QSAR models using this property randomization procedure demonstrates the statistical significance and robustness of our validated DILI prediction models. These `blind' predictions demonstrate the ability of our models to successfully classify and thus segregate between compounds that affect human liver and those that do not.
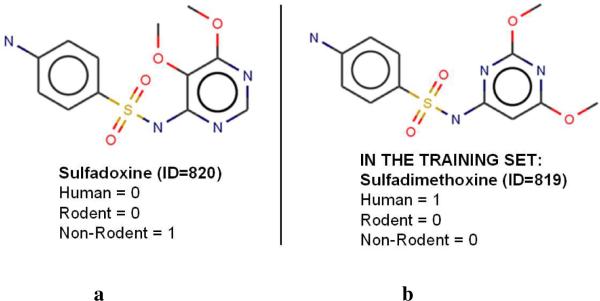
In order to test the prediction performance of our models, one may suggest applying them to the set of 18 compounds reported as liver toxicants for non-rodents only (and thus, expected to have no liver effects for humans according to our initial assumption). Indeed, we have found that 14 among the 18 were correctly classified as non-toxicants for humans, leading to a fairly good prediction accuracy of 77.8 % using SVM models built with fragment descriptors. To further this analysis, we made use of our strategy for gap spotting in assertional data that was discussed in section 3.2. To this end, we searched public data sources for additional information concerning four apparently misclassified compounds. We noticed that one of these compounds, i.e., sulfadoxine, is a very close chemical analog of sulfadimethoxine (see Figure 7), which was, in fact, reported as a liver toxicant in humans (31). Indeed, the assertion re-generation from MEDLINE did identify evidence for pyrimethamine/sulfadoxine (fansidar) causing hepatitis in patients (NB: combinations of compounds were excluded from the original analyses). This means that the prediction accuracy for the external set is likely to be 83% instead of 78%.
Figure 7.
The chemical structure of sulfadoxine (a), which was misclassified by our QSAR models (i.e., predicted to produce liver effect in humans), compared to the structure of sulfadimethoxine (b) included in the modeling set and reported later to have liver effects in humans.
4. Discussion
The main objective of our study was to explore the relationships between chemical structure and species-specific liver effects using assertions derived by Sofia from published literature. We have demonstrated that conventional cheminformatics approaches such as chemical similarity analysis and QSAR modelling can be applied in a meaningful way to an observational dataset that was generated by means of lexical, linguistic and ontological methods. In this paper, we have addressed several important questions relevant to drug safety assessment.
First, we have explored the issue of concordance of liver effects across species. Since animal testing is widely used both in environmental and drug discovery applications, as a substitute of human studies, it is very important to understand in what cases the results of animal testing are acceptable as accurate predictors of expected human effects. To this end, we have developed a simple metric to calculate the pairwise concordance between species (Eq. 1) and found that the concordance values between any two species from the three groups studied herein (humans, rodents, non-rodent animals) were relatively low (40–45%). These results are in agreement with earlier studies reported in the literature (4;27;28). We should mention that, due to the small amount of data for non-rodents, the conclusions for this species group may not be as significant as those reached for human vs. rodent concordance analysis. We are currently searching for additional species-specific DILI data and plan to revisit the issue of concordance in future studies.
To enable the statistical analysis of species-specific drug-induced liver effects, we needed to make an important (and perhaps somewhat speculative) assumption concerning the completeness of the assertional meta-data. We have assumed that each compound was effectively tested in all species and that the assertional meta-data represents a summary of all known liver effects. Thus, if the assertional meta-data reported no liver effect for a particular compound, we assumed that it has been tested and found not to produce a liver effect (see the Methods section 2.2.2). There are two observations that may support this assumption. Firstly, we only included compounds which are known active ingredients of therapeutic products that either remain on the market or have been withdrawn. One would assume that any chemical shown to be a toxicant in animal studies would be typically considered not suitable for human use; however, a large number of compounds shown to produce liver effects in both humans and rodents (as many as 292 in our studies according to the Venn diagram in Figure 2) implies that the demonstration of these effects in rodents does not prevent compounds from human exposure. It may follow then that there is a category of compounds that are toxic in rodents but not toxic in humans. Furthermore, it is even more plausible that compounds only shown to be toxic in humans were first tested in rodents and found to be safe. Thus, at least some of our primary assumptions appear logical. But, most importantly, we believe that our ability to achieve statistically significant and externally predictive QSAR models make it especially plausible that our assumption about the completeness of the assertional meta-data has merits. We did not, however, feel that we could make the same assumption for non-rodent data, given that the number of compounds reporting liver effects in this group was much smaller than the number of compounds reporting effects in humans and rodents. For this reason we decided to exclude the non-rodent data from any QSAR modeling or fragment analysis exercise, because the statistical rigor and the applicability domain of the resulting models would be very low.
The first part of our cheminformatics study identified series of molecular chemotypes responsible for liver effects in a species-dependent manner. Such knowledge is key for pharmaceutical companies to detect and eliminate potentially toxic drug candidates in early stages of the drug development workflow. We have specifically verified the general principle that highly chemically similar compounds have similar liver effect profiles across species. Using clustering approaches frequently used in conventional cheminformatics research, we have identified multiple small clusters of compounds with high structural similarity, and confirmed that, for the most part, these compounds indeed had similar species-dependent liver effect profiles. In several cases of discrepancy between chemical similarity and liver effect similarity, we were able to revise the original assertions by additional focused re-mining of public data. Furthermore, we have conducted a detailed analysis of 2D substructural molecular fragments found both in compounds reporting liver effects in humans and those compounds that do not, and isolated key chemical fragments that are statistically more frequently associated with compounds inducing human liver effects. We are keenly interested in deepening the understanding of structural determinants of liver effects, since these determinants translate into structural alerts to filter out undesirable drug candidates in early stages of the drug discovery process. A more detailed study on differential fragment frequency in compounds reporting different liver pathologies is in progress.
The second cheminformatics part of this study was dedicated to QSAR modeling, with the goal of establishing quantitative models of chemically-induced liver effects in humans. Using a reduced and curated dataset, we have built classification models that could predict, with high accuracy, if a compound is likely to produce liver effects in rodents-only (which we interpreted as NOT producing liver effects in humans) or in humans-only. The best models were selected following an external 5-fold CV procedure. Two types of descriptors have been employed: Dragon and 2D fragment descriptors. From our internal 5-fold CV performed on the modeling set for each of the five splits, we found that our computational models can realistically reach external predictions with an accuracy ranging from 61.9 % to 67.5 %, pretty much in line with experimental in vitro data (10). Perhaps surprisingly, given the extreme diversity of chemical structures and the unconventional source of the liver data (i.e., assertions generated from the literature using automated extraction procedures), SVM QSAR models showed high external prediction accuracies ranging from 55.7% to 72.6%.
It should be pointed out that the splits in the external 5 fold cross-validation have been done randomly without any study of the distribution of chemicals into the splits (except that each compound is present in the external test set only once). So the potential overlap between modeling and external test sets in terms of chemical space is excluded by the virtue of the method used to build and validate models. Y-randomizations of the modeling set led to poor models with low accuracies (40–50%) and constant outputs whatever the compounds, demonstrating the robustness of our models. The results of these modeling studies serve to validate the consistency of the data and reassure the rigor of the data collection and curation procedures, in a sense, lending some statistical support to our main assumption about the `completeness' of the dataset (except for non-rodents). In this study, regarding the raw nature of the data provided by a literature-mining platform, it seemed to us that the application of a massive QSAR strategy involving many machine learning approaches (such as multi-linear regressions, artificial neural networks, random forest etc.) was not appropriate since our first goal was simply to demonstrate the feasibility of using cheminformatics approaches for the analysis and refinement of data generated from mining literature sources. Therefore only SVM models with two different types of chemical descriptors have been built.
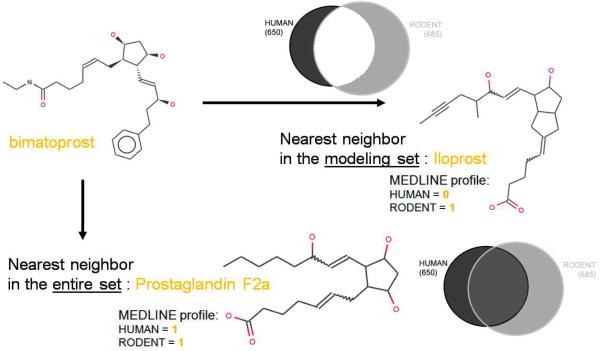
Finally, we made every attempt to prove the accuracy of our models using external datasets. As a corollary to the analysis described in this paper, we have mined the EMEA EPARs database for compounds having liver effects. This resulted in a set of 44 compounds reported to induce liver effects in humans only. After removing compounds already present in our modeling set, 34 molecules remained. The consensus SVM model employing individual validated SVM models built with both fragment and Dragon descriptors afforded a good prediction accuracy (67.6%) for this additional external validation set. Specifically, 23 out of 34 compounds were indeed predicted to produce liver effects in humans, whereas 11 compounds were classified as non-toxic. 2 out of these 11 compounds were found to fall outside of the models' domain of applicability (defined based on chemical similarity threshold in the descriptor space as described in (32;33). Moreover, as illustrated in Figure 8, analysis of apparently misclassified compounds revealed the importance in the choice of the modeling set: for instance, bimatoprost was reported to produce human liver effects in the EMEA dataset of 34 molecules, but was misclassified by our QSAR models as non-toxic. The nearest neighbor of bimatoprost (based on structural similarity in the descriptor space) in the modeling set of 531 compounds is iloprost which has not been reported to produce liver effects in humans. Considering the entire human set of 650 compounds as a potential modeling set, the nearest neighbor of bimatoprost is now prostaglandin F2a, annotated as producing liver effects in humans. Thus, beyond the modeling exercise reported in this study, and following the objective of building an efficient predictor of DILI in humans, we will have to build additional models trained on an enlarged set, including all compounds reporting human liver effects in our dataset, to avoid basic misclassification, as in the case of bimatoprost.
Figure 8.
Nearest neighbors of bimatoprost in the modeling set of 531 compounds and the entire human dataset of 650 compounds.
Additional mining of MEDLINE, conducted after this study was completed, afforded an additional external dataset of 246 compounds; 92 annotated as “causing effects in humans only” and 154 as “causing effects in rodents only”. External prediction accuracies varied from 65.4% to 67.5% using our WinSVM models and Dragon descriptors. The removal of structural outliers, using the applicability domain implementation, led to a reduced set of 222 compounds (90.2% coverage of the dataset) with a prediction accuracy of 67.6%. These results agree quantitatively with external prediction accuracies generated with the original dataset using 5-fold external CV and other external sets. This additional analysis adds weight to the robustness of the overall approach and to the power of combining the expertise in comprehensive assertion generation and cheminformatics in the analysis of chemically-induced liver effects (see Figure 1).
5. Conclusions
We have employed conventional cheminformatics approaches to analyze assertions of drug-induced liver effects in different species, retrieved by a novel approach comprising lexical and linguistic extraction and ontology-based methods. After a critical step of chemical data curation, cluster analysis of the remaining 951 compounds allowed us to identify multiple clusters in which compounds belong to structurally congeneric series. Similar liver effect profiles have been observed for most clusters although some compounds appeared as outliers. In several cases of such outliers, additional focused mining of public data sources led to revised assertions that were more in tune with liver effect profiles expected on the basis of chemical similarity. QSAR models were generated to predict liver effects of compounds in humans from chemical structure. Despite the apparent chemical diversity of the modeling set, an uncommon source of biological data, and the apparent complexity of underlying biological mechanisms the models showed good prediction power as assessed by five-fold external CV procedures. The external predictivity of models was further confirmed by applying them to different external sets of compounds.
We believe that the studies reported in this paper present the first example of combining rigorous text mining and cheminformatics data analysis towards establishing predictive models of chemical toxicity. This work suggests that cheminformatics approaches could find a prominent place in refining the compound annotations resulting from biomedical text mining. The approach could be described in two steps: (1) launch a deeper literature- and internet-mining to search for evidences concerning the compound in question; (2) if nothing is retrieved from (1), we could at least add a warning sign for this compound as a potential safety alert on a dedicated website and suggest that supplementary experimental assessments are needed.
Supplementary Material
Acknowledgements
The authors from UNC gratefully acknowledge the financial support from NIH (grant R21GM076059) and EPA (RD 83382501 and R832720). DF and AT also thank Pr. Alexandre Varnek (ULP - Strasbourg) for providing us with the ISIDA software.
Footnotes
Supporting Information Available. The entire dataset (SMILEs and species concordance of liver effects) is available via the Internet at http://pubs.acs.org.
References
- (1).Fung M, et al. Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets -1960 to 1999. Drug. Inf. J. 2009;35:293–317. [Google Scholar]
- (2).Watkins P, Seeff L. Drug-induced liver injury: summary of a single topic clinical research conference. Hepatology. 2006;43(3):618–631. doi: 10.1002/hep.21095. [DOI] [PubMed] [Google Scholar]
- (3).Egan W, Zlokarnik G, Grootenhuis P. In silico prediction of drug safety: despite progress there is abundant room for improvement. Drug Discovery Today: Technologies. 2004;1(4):381–387. doi: 10.1016/j.ddtec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- (4).O'Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, Gao B, Kaludercic N, Angeline A, Bernardi P, Brain P, Hougham C. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 2006;80(9):580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- (5).Kaplowitz N. Drug-induced liver injury. Clin. Infect. Dis. 2004;38(Suppl 2):S44–S48. doi: 10.1086/381446. [DOI] [PubMed] [Google Scholar]
- (6).Ballet F. Hepatotoxicity in drug development: detection, significance and solutions. J. Hepatol. 1997;26(Suppl 2):26–36. doi: 10.1016/s0168-8278(97)80494-1. [DOI] [PubMed] [Google Scholar]
- (7).Sutter W. Predictive value of in vitro safety studies. Current Opinion in Chemical Biology. 2006;10(4):362–366. doi: 10.1016/j.cbpa.2006.06.023. [DOI] [PubMed] [Google Scholar]
- (8).Farkas D, Tannenbaum S. In vitro methods to study chemically-induced hepatotoxicity: a literature review. Current Drug Metabolism. 2005;6(2):111–125. doi: 10.2174/1389200053586118. [DOI] [PubMed] [Google Scholar]
- (9).Elferink M, Olinga P, Draaisma A, Merema M, Bauerschmidt S, Polman J, Schoonen W, Groothuis G. Microarray analysis in rat liver slices correctly predicts in vivo hepatotoxicity. Toxicology and Applied Pharmacology. 2008;229:300–309. doi: 10.1016/j.taap.2008.01.037. [DOI] [PubMed] [Google Scholar]
- (10).Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de GD. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008;105(1):97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- (11).Blomme EA, Yang Y, Waring JF. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol. Lett. 2009;186(1):22–31. doi: 10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- (12).Elferink MG, Olinga P, Draaisma AL, Merema MT, Bauerschmidt S, Polman J, Schoonen WG, Groothuis GM. Microarray analysis in rat liver slices correctly predicts in vivo hepatotoxicity. Toxicol. Appl. Pharmacol. 2008;229(3):300–309. doi: 10.1016/j.taap.2008.01.037. [DOI] [PubMed] [Google Scholar]
- (13).Cheng A, Dixon S. In silico models for the prediction of dose-dependent human hepatotoxicity. J. Comput. Aided Mol. Des. 2003;17:811–823. doi: 10.1023/b:jcam.0000021834.50768.c6. [DOI] [PubMed] [Google Scholar]
- (14).Clark R, Wolohan P, Hodgkin E, Kelly J, Sussman N. Modelling in vitro hepatotoxicity using molecular interaction fields and SIMCA. J. Mol. Graph. Model. 2004;22:487–497. doi: 10.1016/j.jmgm.2004.03.009. [DOI] [PubMed] [Google Scholar]
- (15).Contrera J, Matthews E, Benz R, Kruhlak N, Weaver J, Hanig J. MCASE prediction of hepatotoxicity using post-market adverse effects data. Hepatotoxicity Steering Committee Meeting; Rockville, MD. 01-21-03.2003. [Google Scholar]
- (16).Cruz-Monteagudo M, Cordeiro MN, Borges F. Computational chemistry approach for the early detection of drug-induced idiosyncratic liver toxicity. J. Comput. Chem. 2008;29(4):533–549. doi: 10.1002/jcc.20812. [DOI] [PubMed] [Google Scholar]
- (17).Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regulatory Toxicology and Pharmacology. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- (18).Young D, Martin T, Venkatapathy R, Harten P. Are the chemical structures in your QSAR correct? QSAR Comb. Sci. 2008;27:1337–1345. [Google Scholar]
- (19).Varnek A, Fourches D, Hoonakker F, Solov'ev V. Substructural fragments: An universal language to encode reactions, molecular and supramolecular structures. J. Comp. -Aided Mol. Des. 2006;19 doi: 10.1007/s10822-005-9008-0. [DOI] [PubMed] [Google Scholar]
- (20).Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov'ev V, Hoonakker F, Tetko IV, Marcou G. ISIDA - Platform for virtual screening based on fragment and pharmacophoric descriptors. Current Computer-Aided Drug Design. 2008;4(3):191–198. [Google Scholar]
- (21).Baskin I, Varnek A. Building a chemical space based on fragment descriptors. Combinatorial Chemistry & High Throughput Screening. 2008;11(8):661–668. doi: 10.2174/138620708785739907. [DOI] [PubMed] [Google Scholar]
- (22).Downs G, Barnard J. Clustering methods and their uses in computational chemistry. Rev. Comp. Chem. 2002;18:1–40. [Google Scholar]
- (23).Mercier D. Clustering large datasets. Electronic review - Linacre College. 2003 [Google Scholar]
- (24).Varnek A, Fourches D, Sieffert N, Solov'ev VP, Hill C, Lecomte M. QSPR modeling of the Am-III/Eu-III separation factor: How far can we predict. Solvent Extraction and Ion Exchange. 2007;25(1):1–26. [Google Scholar]
- (25).Vapnik VN. In The Nature of Statistical Learning Theory. Springer; New York: 2000. [Google Scholar]
- (26).Tropsha A, Golbraikh A. Predictive QSAR Modeling Workflow, Model Applicability Domains, and Virtual Screening. Curr. Pharm. Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- (27).Olson H, Betton G, Stritar J, Robinson D. The predictivity of the toxicity of pharmaceuticals in humans from animal data--an interim assessment. Toxicol. Lett. 1998;102-103:535–538. doi: 10.1016/s0378-4274(98)00261-6. [DOI] [PubMed] [Google Scholar]
- (28).Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van DK, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32(1):56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- (29).Guengerich FP, MacDonald JS. Applying mechanisms of chemical toxicity to predict drug safety. Chem. Res. Toxicol. 2007;20(3):344–369. doi: 10.1021/tx600260a. [DOI] [PubMed] [Google Scholar]
- (30).Todeschini R. DRAGON for Windows (Software for Molecular Descriptor Calculations) version 5.4 Talete s. r. l; Milan, Italy: 2006. [Google Scholar]
- (31).Meier P, Schmid M, Staubli M. Acute hepatitis following administration of fansidar. Schweiz. Med. Wochenschr. 1990;120(7):221–225. [PubMed] [Google Scholar]
- (32).Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko I. Combinatorial QSAR Modeling of Chemical Toxicants Tested against Tetrahymena pyriformis. J. Chem. Inf. Model. 2008;48(4):766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- (33).Tetko IV, Sushko I, Pandey AK, Zhu H, Tropsha A, Papa E, Oberg T, Todeschini R, Fourches D, Varnek A. Critical assessment of QSAR models of environmental toxicity against Tetrahymena pyriformis: focusing on applicability domain and overfitting by variable selection. J. Chem. Inf. Model. 2008;48(9):1733–1746. doi: 10.1021/ci800151m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.