Abstract
DNA double-strand breaks are repaired by different mechanisms, including homologous recombination and nonhomologous end-joining. DNA-end resection, the first step in recombination, is a key step that contributes to the choice of DSB repair. Resection, an evolutionarily conserved process that generates single-stranded DNA, is linked to checkpoint activation and is critical for survival. Failure to regulate and execute this process results in defective recombination and can contribute to human disease. Here, I review recent findings on the mechanisms of resection in eukaryotes, from yeast to vertebrates, provide insights into the regulatory strategies that control it, and highlight the consequences of both its impairment and its deregulation.
The repair of double-strand breaks
DNA is constantly challenged both by exogenous agents such as mutagenic chemicals and radiation and by endogenously arising compounds such as reactive oxygen species1. To minimize the impact of these threats, cells have evolved various DNA repair mechanisms. DNA double-strand breaks (DSBs) are the most cytotoxic forms of DNA damage. Inaccurate DSB repair leads to mutations and/or gross chromosomal rearrangements (GCRs)1. Moreover, the controlled repair of programmed DSBs occurs during physiological processes such as meiosis or the diversification of immunoglobulins. Therefore, inherited defects in DSB repair genes cause embryonic lethality, sterility, developmental disorders, immune deficiencies, and predisposition to neurodegenerative diseases and cancer.
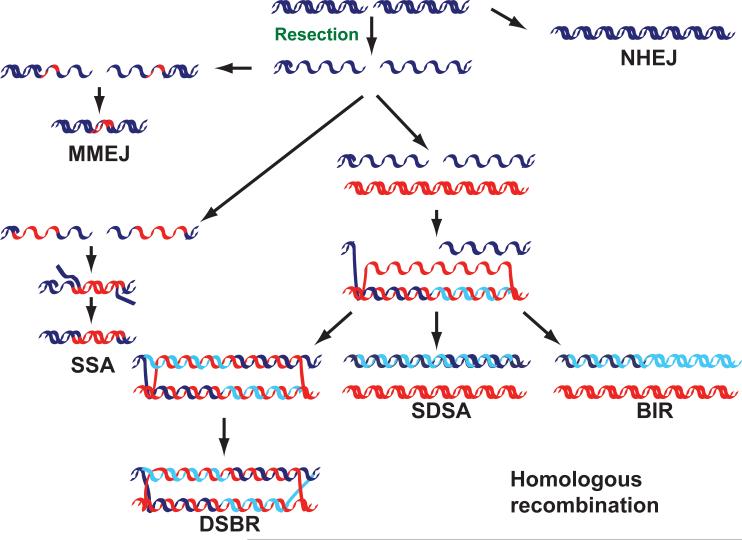
There are two major ways of repairing DSBs1. Nonhomologous end-joining (NHEJ) ligates together the two DNA ends with little or no processing2 (Fig. 1); it is highly efficient but prone to generating mutations at the sites of joining. Furthermore, because there is no apparent mechanism to ensure that the two ends being joined were originally contiguous, NHEJ can yield GCRs such as inversions and translocations. The second DSB repair mechanism is a set of pathways that use an undamaged homologous DNA sequence as a template for accurate repair, collectively known as homologous recombination (HR)3 (Fig. 1). Although HR has been primarily studied as a response to DSBs, its primary function is probably to deal with stalled or collapsed replication forks1.
Figure 1.
The repair of DNA double-strand breaks (DSBs). DSBs can be repaired using several different mechanisms. Both ends can be simply rejoined with little or no further processing (nonhomologous end-joining; NHEJ) or can be repaired using homologous sequences (red DNA; homologous recombination) after 5′–3′ degradation has occurred (resection). The 3′-OH group exposed after resection can be used to prime DNA synthesis using a homologous region as a template after DNA strand invasion. The newly synthesized DNA (light blue) can then be joined with the 5′ end of the resected strand forming a double Holliday junction (double-strand break repair; DSBR), or can be displaced and reannealed (synthesis-dependent strand annealing; SDSA); or DNA synthesis can continue to the end of the chromosome (break-induced replication; BIR). If two homologous regions flank the DSB, they can anneal after being exposed by DNA resection (single-strand annealing; SSA), which causes the deletion of the intervening region. An additional mechanism that shares components with both SSA and NHEJ, and uses short homology stretches (usually 2–3 bp) flanking the DSB, can also be used (microhomology-mediated end-joining; MMEJ). The chromosomal consequences of aberrant use of each repair mechanism are shown in the red boxes.
HR has been extensively reviewed3. Briefly, all HR subpathways are initiated by a 5′–3′ degradation of one strand at both sides of the break, generating stretches of single-stranded DNA (ssDNA) that is then coated by the ssDNA binding protein complex RPA—the so-called DNA-end resection. Three of the HR subpathways use the ssDNA molecule to invade a homologous DNA region situated elsewhere in the genome (donor sequence), which is used as a template for DNA synthesis. After this the three mechanisms diverge (Fig. 1)3. In double-strand-break repair (DSBR), the second end is captured and extended and then the newly synthesized DNA is ligated to the end of the resected strands to form two cruciform structures known as Holliday junctions, which can be resolved by different mechanisms3. In break-induced replication (BIR), after one-end invasion, replication simply proceeds until the end of the chromosome. Synthesis-dependent strand annealing (SDSA) can follow either one-end or two-end invasion events (one-ended invasion shown in Fig. 1); the partially replicated strands reanneal and are ligated. The fourth subpathway (single-strand annealing; SSA) is used only when two homologous regions flank the DSB site. In this case, the homologous regions are exposed, and after annealing and cleavage of the DNA overhang, the ends are ligated, resulting in the deletion of the intervening region. A mechanism that shares some genetic requirements with both NHEJ and SSA—microhomology-mediated end-joining; MMEJ—has recently been described as well (Fig. 1; for review see ref. 4).
A key feature of HR-based repair, except for SSA, is the preservation of the genetic material, as the donor sequence is usually the sister chromatid. However, when the donor sequence used is not the sister chromatid but another homologous region, HR can yield GCRs such as deletions, inversions or loss of heterozygosity1.
The choice between different DSBs repair pathways is tightly regulated, and resection represents a primary regulatory step. Resection is needed for MMEJ and all HR pathways3,4, and resected DNA decreases NHEJ efficiency, likely as a result of poor binding of the NHEJ factor Ku70–Ku80 to ssDNA5. Indeed, the balance between HR, MMEJ and NHEJ has been shown to be controlled by key DNA resection factors such as Sae2 (refs. 6,7) and CtIP8,9. Furthermore, formation of RPA-coated ssDNA after DNA-end resection is a critical intermediate of checkpoint activation10 and is key in the switch from the ATM-driven to the ATR-controlled checkpoint11. Consequently, DNA resection is a highly complex and regulated process.
Mechanism of resection
The core resection machinery is conserved in all kingdoms of life (Table 1)3,9,12-21. An important component is the Mre11 complex, composed of Mre11, Rad50, and a third protein known as Xrs2 in the budding yeast Saccharomyces cerevisiae and as Nbs1 in most other eukaryotes6,22-27. Mre11 is a nuclease related to bacterial SbcD, whereas Rad50 is homologous to bacterial SbcC. By contrast, Nbs1/Xrs2 is less conserved and is restricted to eukaryotes. The C terminus of Nbs1/Xrs2 possess an interaction motif for ATM (in budding yeast, Tel1), a protein kinase that controls DNA damage–induced events28,29. The entire Mre11 complex acts as a single functional unit, because loss of any of the three subunits results in similar phenotypes3: hypersensitivity to DNA-damaging agents, impaired HR and defective meiosis. In vitro, the Mre11 complex shows both endonuclease and exonuclease activities23. However, budding yeast mre11 nuclease mutants have a much milder phenotype than cells lacking Mre11, which have only partial defects in resection of endonuclease-induced DSBs24. This reflects additional roles for the Mre11 complex in checkpoint activation or maintenance of chromosome structure3,22,25,26 but also argues against the idea that Mre11 is the main nuclease for resection. Moreover, Mre11 exonuclease activity in vitro operates in the 3′–5′ direction, opposite to the direction of resection in vivo. Mre11 is a poor nuclease, both endo- and exo-, and thus is unlikely to be responsible for generating the extensive ssDNA observed in vivo23,27.
Table 1. Proteins involved in resection in different eukaryotes.
| E. coli | P. furiosus | S. cerevisiae | S. pombe | H. sapiens | Function |
|---|---|---|---|---|---|
| RecBCD | Unk. | Unk. | Unk. | Unk. | DNA helicase, ATPase, 5′ exonuclease, 3′ exonuclease |
| SbcD | Mre11 | Mre11 | Rad32 | Mre11 | 3′–5′ exonuclease, endonuclease |
| SbcC | Rad50 | Rad50 | Rad50 | Rad50 | ATPase |
| Unk. | Unk. | Xrs2 | Nbs1 | Nbs1 | |
| Unk. | Unk. | Sae2 | Ctp1 | CtIP | ssDNA specific endonuclease |
| Unk. | Unk. | Unk. | Unk. | BRCA1 | Ubiquitin ligase |
| Unk. | Unk. | Exo1 | Exo1 | Exo1 | 5′–3′ exonuclease |
| RecQ | Hjma,b | Sgs1 | Rqh1a | RECQ1a,BLM, WRNa, RTSa, RECQ5 |
DNA helicases |
| RecJ | Several homologsa |
Unk. | Unk. | Unk. | 5′–3′ exonuclease |
| Unk. | Unk. | Dna2 | Dna2a | Dna2a | 5′ flap endonuclease, DNA helicase |
| Unk. | Unk. | Rad9 | Crb2a | 53BP1a | Checkpoint adaptor protein |
| Unk. | NurA | Unk. | Unk. | Unk. | 5′–3′ exonuclease |
| Unk. | HerA | Unk. | Unk. | Unk. | DNA helicase |
E. coli, Escherichia coli; P. furiosus, Pyrococcus furiosus; S. cerevisae, Saccharomyces cerevisiae; S. pombe, Schizosaccharomyces pombe; H. sapiens, Homo sapiens; Unk., unknown.
Not formally shown to be involved in DNA resection.
Despite the lack of RecQ sequence orthologs in Archea, Hjm can complement E. coli RecQ mutants.
The poor in vitro activity of Mre11 may reflect the lack of accessory factors. One likely candidate is the budding yeast protein Sae2. sae2 deletion phenocopies the nuclease-defective mre11 mutants and a specific family of mutations in Rad50 called rad50S (ref. 30): that is, sae2Δ strains are completely defective in processing meiotic DSBs31-33 but are mildly sensitive to DNA damaging agents and impair DNA-end resection only partially34. Sae2 is an endonuclease that cooperates with the Mre11 complex in the processing of various DNA structures35. The current model proposes that the endonuclease activities of Mre11 and/or Sae2 initiate resection (Fig. 2). This endonucleolytic processing will, theoretically, release small ssDNA oligonucleotides. Such oligonucleotides have been observed in the processing of meiotic DSBs in yeast32 and have been detected in Xenopus laevis extracts36.
Figure 2.
Mechanism of resection in budding yeast. DSBs are detected by the Mre11 complex (MRX) and Sae2. Upon activation of the endonucleolytic activity of MRX and Sae2, initial processing results in the generation of short stretches of single-stranded DNA. This partially resected DNA will then be the substrate for further nucleolytic degradation either by Exo1 or by Dna2 and Sgs1. The initial processing by Mre11 and Sae2 can be bypassed in mitotic interphase (dashed black arrow), probably by the action of Exo1 or of Sgs1 and Dna2. In the absence of Exo1 and Sgs1, several rounds of the endonucleolytic activity of Mre11-Sae2 will be sufficient for short processing close to the ends (dashed red arrow).
DNA-end resection and HR are barely affected in the absence of Sae2 and not at all in mre11 nuclease–defective mutants24,34, suggesting the existence of additional nucleases. One candidate is the 5′–3′ exonuclease Exo1, which is conserved from yeast to humans19 and is essential for DNA-end processing at uncapped telomeres37. Like deletion of sae2, exo1 deletion results in only mild DNA damage sensitivity and partial impairment of DNA-end resection38. sae2 exo1 and exo1 mre11 double mutants show a synergistic decrease in DNA-end resection and greater DNA-damage sensitivity than the single mutants38. Overexpression of EXO1 partially rescues the DNA sensitivity phenotype of mre11 mutants39, suggesting that Mre11 and Exo1 may function in parallel pathways. Surprisingly, mre11 exo1 mutants show residual DNA-end resection, suggesting that a third pathway also exists39. In bacteria, the multifunctional enzyme RecBCD, which harbors helicase and nuclease activities, does most of the resection, but in its absence, the helicase RecQ acts together with the nuclease RecJ to resect DNA ends12. Mutations in sgs1, the budding yeast homolog of RecQ, in combination with exo1 deletion completely abolish long-range DNA-end resection, and only some minimal processing close to the break can be detected in such double mutants13,16,21. Residual processing is dependent on Sae2 and Mre11 (refs. 13,16,21). As is the case for bacterial RecQ, budding yeast Sgs1 works in combination with a nuclease called Dna2 (ref. 21). Although Dna2 has both helicase and flap-endonuclease activity40, only the nuclease activity is required for DNA-end resection21.
The following model for DNA-end resection has been proposed in S. cerevisiae13,16,21 (Fig. 2). First, the Mre11 complex and Sae2 are responsible for the initial processing through their endonucleolytic activities. The resulting partially resected DNA is further processed by the action either of Exo1 or of Sgs1 and Dna2. In the absence of Sgs1 and Exo1, the activities of Mre11 and Sae2 are responsible for short-range processing (Fig. 2). Although Sae2 and Mre11 are completely essential for resection during meiotic recombination, their functions can be bypassed during mitotic recombination24,34. This difference is probably due to the specific nature of meiotic DSBs, which requires Mre11 and Sae2 to remove the covalently bound nuclease, Spo11, that creates the breaks32,41. The nature of this bypass is unknown, but it probably involves Exo1 and Sgs1, as sae2 exo1 sgs1 mutants are unable to resect DNA and overexpression of Exo1 partially rescues mre11 mutants16,21,39 (Fig. 2).
Resection in vertebrates
For a long time, the only component of the DNA-end resection machinery known in higher eukaryotes was the Mre11 complex11,22,26. Recently, functional counterparts of Sae2 have been found in several organisms9,15,17,18,20 (Table 1). Human CtIP, as well as fission yeast Schizosaccharomyces pombe Ctp1, physically interact with the Mre11 complex and have a major role in ssDNA formation at the site of DSBs9,15,18, but it is still unknown whether they function as endonucleases like Sae2. In vitro, human CtIP together with Mre11 showed an increased nuclease activity compared with Mre11 alone18, but whether this activity relies on Mre11, CtIP or both remains to be established. CtIP downregulation completely abolishes ssDNA formation, as measured from RPA focus formation8,9,13,18, in contrast with S. cerevisiae sae2 (refs. 6,34,38) or S. pombe ctp1 (ref. 15). Whether this is due to differences in the techniques used or reflects a true impossibility of resection in the absence of CtIP is still an open question. Although in vitro Mre11, Rad50 and CtIP are sufficient to catalyze the nuclease activity18, they require additional factors in vivo. Apart from Nbs1, which is necessary for recruitment of the Mre11 complex to sites of breaks41, proper DNA resection in vertebrates requires the action of specific factors such as the tumor suppressor BRCA1 (refs. 9,42). BRCA1 is an ubiquitin ligase that physically interacts with and polyubiquitinates CtIP43. Interaction of CtIP and BRCA1 is controlled by phosphorylation and is essential for CtIP recruitment to sites of DNA damage43 and proper DNA resection9,42. However, the role of BRCA1-mediated ubiquitination in DNA-end resection remains to be determined.
Despite the strong effect of CtIP downregulation, both the Exo1 and Sgs1 resection pathways are functional in higher eukaryotes13. Although in humans there is only one ortholog of Exo1, there are five homologs of RecQ and Sgs1 (Table 1), and at least one of these (BLM) is involved in DNA resection13. As in yeast, the BLM pathway appears to be parallel and independent of Exo1, as the simultaneous downregulation of Exo1 and BLM severely impaired ssDNA formation13. However, in vitro BLM interacts with Exo1 and stimulates its activity44, arguing that BLM and Exo1 might function in the same pathway. Future work will be required to clarify these discrepancies between the in vivo and in vitro data. The role of vertebrate Dna2 is also unclear. Human Dna2 is an endonuclease45 (Table 1), and although it is primarily located in the mitochondria46, it is also present in the nucleus46. Xenopus Dna2 possesses the major activity responsible for 5′–3′ DNA processing in extracts47.
The helicase-nuclease tandem for DNA resection seems is a general theme of DNA-end processing machinery6. In addition to Sgs1 and Dna2, RecBCD and RecQJ, in the archean Pyrococcus furiosus SbcCD-mediated resection is stimulated by the action of the HerA-NurA helicase-nuclease pair48. Therefore, it is possible that in future other helicases will be found to be involved in resection. Strong candidates are the additional members of the RECQ family (Table 1). In fact, human RECQ5 has been shown to be recruited to sites of DNA damage by the Mre11 complex and has been reported to inhibit the 3′–5′ nuclease activity of Mre11 (ref. 49).
Regulation of resection
DNA-end resection has a major role in regulating the balance between HR and NHEJ4,6,8,9 and is a key modulator of checkpoint activation10. Therefore, it is highly regulated and responds to many different cellular signals. An overview of the multiple layers of regulation of DNA-end resection is shown in Figure 3 and explained in more detail below.
Figure 3.
Regulation of resection in budding yeast. Schematic representation of how DNA-end resection is regulated. Positive actions are shown as red arrows and negative regulations as light blue arrows; solid arrows represent interactions by known mechanisms and dashed arrows interactions by unknown mechanisms. Question marks indicate points at which additional layers of regulation may be acting. DNA end resection is activated in S/G2 cells by the activity of CDKs, directly by phosphorylation of Sae2 and by an unknown mechanism regulating Rad9. Although Rad9 does not bind naked DNA but rather chromatin, histones are not shown for simplification. The presence of KU and Rad9 are negative regulators of resection. Multiple or ‘ragged’ ends also stimulates DNA processing even in G1 cells.
DNA-end resection during the cell cycle
HR is a highly accurate repair process when the sister chromatid is readily available and held in close proximity after DNA replication either in S or G2. Therefore, DNA-end resection and HR are almost completely confined to S and G2 (refs. 11,50,51). Although from now on we will distinguish merely_between the G1 (little or no resection) and S/G2 (high resection) phases, resection occurs faster in S than in G2 (ref. 52). The mechanism underlying this difference remains unclear. One tempting idea is that the DNA replication machinery itself can recruit the resection machinery. Accordingly, CtIP has been shown to be recruited to active replication sites via an interaction with the replication factor PCNA53. As a result of this difference between G2- and S-phase resection, it is also difficult to compare results obtained with cycling versus G2-arrested cells, and from this point on we note when arrested cells were used.
DNA resection takes place only when cyclin-dependent kinases (CDKs), master regulators of cell cycle progression, are active (S/G2)6,11,50,51. So far Rad9 and Sae2 have been implicated in the CDK-dependent regulation of DNA resection in S. cerevisiae. Deletion of the checkpoint protein Rad9 increases DNA-end resection even when CDKs are not active (G1)54. Rad9, a large chromatin-binding protein, could pose a physical obstacle for processive DNA resection. Indeed, rad9Δ mutants resect faster and further than wild type, suggesting that CDK-mediated phosphorylation of either Rad9 itself or an unknown substrate can diminish this physical block54. Rad9, and its orthologs (Table 1) 53BP1 (higher eukaryotes) and Crb2 (fission yeast), undergo multiple CDK-dependent phosphorylations55,56, but it is unknown whether these modifications affect on resection.
Sae2 is directly phosphorylated by CDK at Ser267 (ref. 6). Impairment of this phosphorylation leads to a reduction in DNA-end resection, a delay of HR, an increase in NHEJ and an increase in DNA-damage sensitivity6. More interestingly, sae2-S267E mutants, which mimic constitutive phosphorylation, resect in the absence of CDK activity and, as a consequence, have faster HR and decreased NHEJ6. The sae2-S267E strain is not sensitive to DSBs that arise during DNA replication, but it shows enhanced hypersensitivity when DSBs appear in G1 (ref. 6). Although sae2-S267E mutants resect in the absence of CDK, such resection is limited to a few kilobases flanking the break6, suggesting a lack of activation of the Exo1 and/or Sgs1 pathways14,17,22.
Sae2 and CtIP share only a small stretch of sequence homology, but this short region includes Sae2 Ser267 and its equivalent CtIP Thr847 (refs. 6,8,9,17,18,20). Moreover, CtIP Thr847 phosphorylation controls DNA-end resection in human cells much as Sae2 Ser267 phosphorylation does in budding yeast. Impairment of this phosphorylation, as well as constitutive phosphorylation, leads to the appearance of GCRs due to an imbalance between NHEJ and HR8. Phosphorylation of chicken CtIP at the equivalent residue has similar functions9. As all the homologs of Sae2 and CtIP except S. pombe Ctp1 share this small region of homology6,8,9,15,17,18,20, it is tempting to speculate that a similar mechanism regulates DNA resection in most eukaryotes. In fact, S. pombe Ctp1, although lacking a residue homologous to Ser267, is controlled during the cell cycle both transcriptionally and by CDK phosphorylation15,57.
Additional layers of regulation by CDKs control CtIP function. CtIP protein levels are minimal in G1 and increase in S/G2 (ref. 42). Moreover, CDK-dependent phosphorylation of CtIP at Ser327 promotes its interaction with BRCA1 in S/G2 (ref. 58), which is essential for CtIP recruitment to sites of DSBs and CtIP-mediated DNA-end resection9,42,58. How CDK-dependent phosphorylations of Ser327 and Thr847 collaborate to regulate DNA resection and HR is not clear, but both seem to be essential. Ser327 phosphorylation, CtIP-BRCA1 interaction and CtIP recruitment to sites of damage are not affected by Thr847 phosphorylation8, and mutants that mimic constitutive Thr847 phosphorylation cannot suppress the lack of BRCA1 (ref. 9). On the contrary, a CTIP-T847E phosphomimetic mutant is able to resect DSB in G1 to a certain extent, even in the absence of an interaction with BRCA1. One possible model is that CDK-dependent phosphorylation at CtIP Thr847 is required to activate the DNA-resection machinery, but BRCA1 is required to efficiently target CtIP to sites of DSB in G2. Although phosphorylation of Sae2 Ser267 or CtIP Thr847 reflects a conserved mechanism of activation of DNA-end resection, the targeting of Sae2, Ctp1 or CtIP to sites of DNA damage has diverged throughout evolution; recruitment of CtIP requires BRCA1 (ref. 42), Ctp1 requires Nbs1 (ref. 57) and Sae2 is recruited by itself to sites of DSBs59. Despite the conservation between Sae2 Ser267 and CtIP Thr847 in the licensing of DNA resection, little is known about the molecular mechanism underlying this activation. One tempting idea is that phosphorylation at these residues stimulates the nuclease activity of Sae2 (ref. 35), but such activity has yet to be proven for CtIP. Another possibility is that such modifications affect DNA-end resection by either helping the recruitment of positive factors or blocking the action of negative modulators.
In addition to the CDK-dependent phosphorylations of Sae2, CtIP and Rad9, it is clear that other cell cycle–regulated phosphorylations or post-translational modifications of the resection machinery are required to fully activate long-range DNA resection.
DNA-end resection and the checkpoint
The nature of the breaks also regulates DNA-end resection. Low numbers of endonuclease-generated DSBs are not resected in G1 (refs. 6,50-52,60). However, similar numbers of DSBs produced by γ-irradiation result in limited DNA resection, sufficient to promote RPA focus formation in yeast59,61. γ-irradiation–induced resection, similar to that in sae2-S267E mutants, covers only a few kilobases and is probably due to the action of Mre11 and Sae2 rather than of Exo1 or of Sgs1 and Dna2. It has been proposed that cells distinguish these radiation-induced breaks as ‘ragged’ ends, as opposed to the ‘clean’ endonuclease-induced breaks, and activate processing activities to ‘clean’ them61. In addition to the type, the number of breaks also plays a role in the activation of DNA resection52. In S/G2, the more DNA breaks generated, the faster the resection takes place52. In G1, induction of up to three endonuclease-induced breaks results in no resection, but four breaks are sufficient to activate DNA-end resection52. Similar to what occurs with either “ragged” breaks or in sae2-S267E mutants, resection activated in G1 by multiple breaks is limited to the proximity of the end52. How the number or type of breaks modulate the response is not understood, but it is probably related to checkpoint activation, a process intimately connected to DNA resection. The Mre11 complex is required, independently of its resection activity, for the recruitment28 and activation25 of the apical checkpoint kinase ATM (Tel1 in budding yeast). The resection machinery is also a downstream substrate of this checkpoint kinase. Tel1 phosphorylates Sae2 and ATM phsophoryates CtIP in response to DNA damage, and these phosphorylations are essential for resection18,62. As discussed previously, the checkpoint protein Rad9 blocks DNA resection54. Thus, it is possible that activation of Rad9 by checkpoint kinases also facilitates resection54. Once resection is activated, it creates a positive feedback loop that amplifies the signal. The production of short, Mre11-generated oligonucleotides contributes to further activation of ATM in Xenopus36. In addition, DNA-end resection generates ssDNA, which activates another checkpoint kinase, ATR10,18,38 (Mec1 in budding yeast), providing a potential mechanism for ATM-mediated ATR activation11,38. Mec1 phosphorylates Sae2 at the same sites as Tel1 (ref. 62), thus further hyperactivating Sae2. Additionally, CtIP also controls the recruitment of the human PCNA-like DNA-damage sensor, the 9-1-1 complex to sites of ionizing radiation-induced DSBs63.
In budding yeast the hyperphosphorylation of the major downstream checkpoint kinase Rad53 cannot be detected on ragged IR-induced breaks in G1. However, low levels of checkpoint activation, as measured by degradation of Sml1, can be observed61. This limited checkpoint activation can trigger limited resection, for example via Mec1 or Tel1-dependent hyperphosphorylation of Sae2. In contrast, Rad53 hyperphosphorylation is readily observed when four endonuclease-induced breaks are produced in G1, in which case DNA-end resection is also observed52. The reason for this difference remains a mystery. One possibility is that a threshold of ssDNA must be surpassed in order to fully activate Mec1 and cause Rad53 hyperphosphorylation. The limited resection of four HO-induced breaks, when combined together, may fully stimulate Rad53 in a way that one or two ragged ends cannot. A similar threshold mechanism has been proposed for checkpoint activation by stalled replication forks, in which multiple uncoupled forks together provide enough ssDNA to activate the checkpoint64. Despite all that, fully processive resection is only obtained in S/G2 when CDKs are active6,8,52,61.
To add an additional layer of complexity, it has been shown that the checkpoint machinery can negatively regulate resection of uncapped telomeres. This is achieved by the phosphorylation, and consequent inhibition, of Exo1 (ref. 65). However, it is unknown whether this negative feedback loop acts on DSBs or whether it is specific to telomeres.
DNA resection and NHEJ
NHEJ and DNA-end resection machineries compete in vivo for the same substrates. NHEJ is generally initiated by the binding to the break of the heterodimer Ku70–Ku80 (ref. 2), which serves as a scaffold for other proteins that contribute to the end-joining reaction. Ku dimers have a high affinity for DSBs, but they bind poorly to ssDNA5 such as that generated by DNA-end resection. Therefore, resection reduces the ability of Ku to bind, and consequently, lack of Mre11, Rad50, Xrs2 or Sae2 lead to increased amounts of Ku bound to DSBs66. By contrast, in the absence of NHEJ proteins such as Ku or ligase IV, an increase in DNA resection and in the amount of Mre11 bound to the break is observed52,60,66. Cells lacking Ku are able to resect in G1 close to the break, in agreement with an Mre11- and Sae2-mediated resection52,60. This limited resection of a single break is enough to grant full checkpoint activation as measured by Rad53 phosphorylation60. Also, in S/G2 cells, resection is faster in the absence of Ku, and overexpression of Ku70–Ku80 reduces DNA-end resection in G2 cells52,60.
Biological relevance of resection
Here, I have discussed how DNA-end resection plays a key role in the repair of DSBs and controls the balance between HR and NHEJ. This is especially relevant because failure to repair DSB is associated with human diseases, including cancer. Not only the lack of repair, but also the use of an inappropriate DSB repair pathway, can be a source of GCRs and the appearance of potentially deleterious mutations1. Accordingly, complete loss of any of the major players in DNA-end resection, such as Mre11, Rad50, Nbs1 or CtIP, leads to embryonic lethality in mice67-69 and increased DNA-damage hypersensitivity in yeast and mammalian cells6,8,9,18,24,67,68. In addition, point mutations in MRE11, NBS1 and RAD50 result in inherited syndromes that are associated with increased genomic instability and cancer predisposition68,70. CTIP mutations have also been detected in several cancers71,72. Moreover, haploid insufficiency of Ctip in mice also predisposes to cancer69. Additionally, hyperactive sae2-S267E and CTIP-T847E result in increased sensitivity to ionizing radiation due to a decrease in NHEJ efficiency and an increase in GCRs resulting from aberrant HR6,8,9. Along these lines, overexpression of CtIP can be detected in several breast cancers72. Bloom syndrome, caused by mutations in BLM, is associated with genomic instability and cancer predisposition73. Although it is difficult to correlate these genetic syndromes with resection defects, it is tempting to speculate that aberrant resection is at least partially responsible for the increased genomic instability and cancer predisposition observed in individuals with such conditions. Therefore, the understanding of DNA resection regulation bears great importance for the understanding of cancer development.
In addition, many cancer therapies are based on the idea that DSBs are extremely potent promoters of cell death, especially in cancer cells that divide rapidly and are usually defective in some DSB repair pathway. In fact, targeting DNA repair mechanisms to increase the lethality of endogenous damage has proven a successful way to selectively kill cancer cells74. The development of new therapeutic strategies that target the resection machinery, through either inhibition or spurious activation, could increase the effectiveness of conventional cancer treatments.
ACKNOWLEDGMENTS
I would like to apologize to all authors whose work could not be cited due to space limitations. I am grateful to all the members of S. Jackson’s laboratory in Cambridge, UK, for helpful discussions and especially to S. Jackson, A. Kaidi, J. Harrigan, K. Miller and R. Belotserkovskaya for their helpful suggestions and comments on the manuscript. I would also like to thank BBSRC and Cancer Research UK for funding my work.
Footnotes
Published online at http://www.nature.com/nsmb/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K, Zhang Y, Lee S. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 8.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 11.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 12.Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 13.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khakhar RR, Cobb J, Bjergbaek L, Hickson I, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 15.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penkner A, et al. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst.) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Uanschou C, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Chung WH, Shim E, Lee S, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 23.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 24.Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 26.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 28.Falck J, Coates JA, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 29.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Usui T, Petrini JH, Morales M. rad50S alleles of the Mre11 complex: questions answered and questions raised. Exp. Cell Res. 2006;312:2694–2699. doi: 10.1016/j.yexcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 31.McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 35.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantiero D, Clerici M, Lucchini G, Longhese MP. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8:380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae SH, et al. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 41.Horejsí Z, et al. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–3127. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JH, et al. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duxin JP, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′→3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008;36:6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng L, et al. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu B, Chen PL. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle. 2009;8:1409–1420. doi: 10.4161/cc.8.9.8322. [DOI] [PubMed] [Google Scholar]
- 54.Lazzaro F, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grenon M, et al. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24:105–119. doi: 10.1002/yea.1441. [DOI] [PubMed] [Google Scholar]
- 56.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akamatsu Y, et al. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 60.Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warmerdam DO, Freire R, Kanaar R, Smits VA. Cell cycle-dependent processing of DNA lesions controls localization of Rad9 to sites of genotoxic stress. Cell Cycle. 2009;8:1765–1774. doi: 10.4161/cc.8.11.8721. [DOI] [PubMed] [Google Scholar]
- 64.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morin I, et al. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 67.Chen L, et al. Effect of amino acid substitutions in the Rad50 ATP binding domain on DNA double strand break repair in yeast. J. Biol. Chem. 2005;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- 68.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 69.Chen PL, et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waltes R, et al. Human RAD50 deficiency in an Nijmegen Breakage Syndrome-like disorder. Am. J. Hum. Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chinnadurai G. CtIP, a candidate tumor susceptibility gene is a team player with luminaries. Biochim. Biophys. Acta. 2006;1765:67–73. doi: 10.1016/j.bbcan.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Wu G, Lee WH. CtIP, a multivalent adaptor connecting transcriptional regulation, checkpoint control and tumor suppression. Cell Cycle. 2006;5:1592–1596. doi: 10.4161/cc.5.15.3127. [DOI] [PubMed] [Google Scholar]
- 73.Cheok CF, et al. Roles of the Bloom’s syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 74.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 2008;8:363–969. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]