Abstract
Some thirty years ago, work on mammalian tissues suggested the presence of two cytosolic hexosaminidases in mammalian cells; one of these has been more recently characterised in recombinant form and has an important role in cellular function due to its ability to cleave β-N-acetylglucosamine residues from a variety of nuclear and cytoplasmic proteins. However, the molecular nature of the second cytosolic hexosaminidase, named hexosaminidase D, has remained obscure. In the present study, we molecularly characterise for the first time the human and murine recombinant forms of enzymes, encoded by HEXDC genes, which appear to correspond to hexosaminidase D in terms of substrate specificity, pH dependency and temperature stability; furthermore, a myc-tagged form of this novel hexosaminidase displays a nucleocytoplasmic localisation. Transcripts of the corresponding gene are expressed in a number of murine tissues. Based on its sequence, this enzyme represents, along with the lysosomal hexosaminidase subunits encoded by the HEXA and HEXB genes, the third class 20 glycosidase to be found from mammalian sources.
Keywords: N-acetylgalactosaminidase, nucleocytoplasmic, GFP, hexosaminidase, mouse, human
Introduction
β-Hexosaminidases (EC 3.2.1.52) are a class of glycoside hydrolases, widespread in nature and present in species from bacteria to man, which remove terminal GalNAc or GlcNAc residues from a range of glycoconjugates such as glycoproteins and glycolipids. It is especially the relationship of human hexosaminidases to lysosomal storage diseases, particularly Tay-Sachs and Sandhoff syndromes [1], which has ensured that this class of enzyme is relatively well-studied. In mammals, ‘acidic’ lysosomal β-hexosaminidases are homo- or heterodimeric enzymes consisting of different combinations of α and β subunits (Hex A, αβ; Hex B, ββ; Hex S, αα); these subunits are encoded by the HEXA and HEXB genes [2] and are classified by sequence homology as being members of retaining glycoside hydrolase family 20, as defined by the CAZy database [3]. Other eukaryotic family 20 enzymes also include plant vacuole hexosaminidases as well as the insect FDL (fused lobes) proteins involved in N-glycan processing [4-6]; this family of enzymes adopts substrate-assisted catalysis with a transition state involving an oxazolinium ion [7]. On the other hand, some β-hexosaminidases follow a different type of catalytic mechanism featuring a covalently-linked glycosyl-enzyme intermediate [8]; these are members of glycoside hydrolase family 3 and are only found in bacteria.
Some years ago, a further β-hexosaminidase gene was molecularly identified from mammalian sources; this enzyme, O-GlcNAcase (abbreviated as OGA or NCOAT and encoded by the MGEA5 gene), is defined as a member of glycoside hydrolase family 84 and is predicted to display similarities to family 20 enzymes in terms of its mechanism and its overall protein fold [9, 10]. The biological role of O-GlcNAcase is to reverse the action of the peptide:N-acetylglucosaminyltransferase (O-GlcNAc transferase; OGT). Both these enzymes are important in intracellular signalling due to their effect on the intracellular balance of N-acetylglucosamine (GlcNAc) and phosphate modifications of nucleocytoplasmic proteins [11, 12] and they may indeed associate with each other in vivo [13]. Interestingly, though, 90% of the endogenous enzyme is cytoplasmic [14]; in the case of recombinant forms of O-GlcNAcase, the 130 kDa form is cytoplasmic [15] and the 75 kDa form is apparently nuclear [16]. However, there still remained the question as to whether O-GlcNAcase is the only hexosaminidase in the nucleus and cytoplasm.
Suggestive of yet another β-hexosaminidase gene in mammals are some studies on soluble ‘neutral’ hexosaminidases from animal sources [17-21]; two types of non-lysosomal activity were found in bovine brain, with different substrate specificities and temperature stabilities, and christened hexosaminidases C and D. Whereas hexosaminidase C is probably identical to the aforementioned O-GlcNAcase [15, 22], the origin of the hexosaminidase D activity has remained unknown; this latter enzyme has a bias towards N-acetylgalactosaminide substrates and has also been referred to as a neutral N-acetylgalactosaminidase [21]. Recently, we have identified four β-hexosaminidases from Caenorhabditis elegans, which belong to a novel branch of the glycoside hydrolase family 20 [4] and which also have a preference for aryl N-acetylgalactosaminides. Database searching indicated that their closest mammalian relatives are not the lysosomal hexosaminidases, but putatively soluble uncharacterised proteins; thus, our question was whether these novel mammalian genes encoded enzymes with characteristics similar to those of the neutral N-acetylgalactosaminidase. We have, therefore, cloned the relevant human and murine cDNAs and expressed the encoded proteins in Escherichia coli. Indeed, both these proteins are now shown to be enzymatically-active hexosaminidases with properties akin to those of the ‘historical’ hexosaminidase D activities.
Materials and Methods
Cloning, expression and purification of hexosaminidase homologues
Mouse liver and human embryonic kidney (HEK 293) cells were homogenised in Trizol reagent (Invitrogen, Carlsbad, USA) in order to isolate RNA, which was subsequently subject to reverse transcription using Superscript III (Invitrogen). In order to clone the full predicted open reading frames, the primer pairs MouseHex/1/EcoRI CCGAATTCATGTCATCACCCACGCC and MouseHex/2/XhoI CCGCTCGAGTCAGGGGTTCTGCCCTG or HumanHex/1/EcoRI CGGAATTCATGTCAGGTTCCACTCCATTTC and HumanHex/2/XhoI CCGCTCGAGGCATGAGCTCTCCCCTCA were used together with Expand polymerase mix (Roche) at an annealing temperature of 58 °C in the presence of the GC-rich PCR reaction additive (Roche). After digestion with the relevant enzymes, the fragments were ligated into the pET30a expression vector (Novagen), which encodes a His-tag. Selected transformed E. coli TOP10F′ colonies were screened and the purified plasmids subject to sequencing with the BigDye kit (Applera); subsequently, verified plasmid DNA was used to transform E. coli BL21(DE3)pLysS cells and, after overnight cultivation at 37 °C in LB medium containing kanamycin and chloramphenicol, expression was induced in cultures with OD600 ~ 0.6 at room temperature in the presence of 1 mM isopropyl-β-D-thiogalactoside (IPTG). In the case of trial expression, aliquots were removed at various timepoints prior to lysis, enzymatic assay and Western blotting with 1:10000 mouse anti-His antibody (HIS-1; Sigma). For larger-scale cultures, lysis and His-tag purification were performed as previously described for GDP-Fuc biosynthesising enzymes [23]. Peptide mass fingerprinting by MALDI-TOF mass spectrometry was performed after tryptic digestion of SDS-PAGE separated proteins. Gel filtration analysis of the His-tag purified murine hexosaminidase was performed on a Sephacryl S-200 Superfine column equilibrated with 40 mM Tris, pH 7.8, 400 mM NaCl, 100 mM KCl, 250 mM imidazole, 10% glycerol, 0.5% Triton X-100. Approximately 0.75 mg of each of the standard proteins (human IgG, human apo-transferrin, chicken ovalbumin and bovine ribonuclease B) were used to calibrate the column. A modified Lowry protein determination kit (Sigma; TP0300) was used to analyse the fractions; 4 μl aliquots of each fraction were then diluted with 44.75 μl of 0.4 M sodium citrate, pH 5.5, prior to assaying hexosaminidase activity for 1 hr using p-nitrophenyl-β-N-acetylgalactosamine (see below).
Hexosaminidase assays
Samples of crude E. coli lysates (5 μl) or of purified enzymes (0.25-0.5 μl) were assayed with p-nitrophenylglycosides: samples were incubated in microtitre wells with 25 μl McIlvaine citrate-phosphate buffer (pH 3.5-8.0), 1.25 μl of 100 mM p-nitrophenylglycoside in dimethylsulphoxide (final concentrations being 2.5 mM glycoside and 2.5% dimethylsulphoxide) and water (up to a final volume of 50 μl) for 1-2 hr at 37 °C; in some cases, another assay duration, pH or temperature was employed. The reaction was stopped by addition of 250 μl 0.4 M glycine-NaOH, pH 10.4, and the A405 was read using an microtitre plate reader. The effect of typical hexosaminidase inhibitors were also tested in this study: 2-acetamido-1,2-dideoxynojirimycin (2-acetamido-1,2,5-tri-deoxy-1,5-imino-D-glucitol; the kind gift of Dr. Arnold Stütz), N-acetylcastanospermine (6-acetamido-6-deoxycastanospermine; Industrial Research Ltd, New Zealand), O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate (PUGNAc; Toronto Research Chemicals, Canada), streptozotocin (STZ; Sigma), mannosamine (2-amino-2-deoxymannose; Industrial Research Ltd), N-acetylglucosamine and N-acetylgalactosamine (Sigma). For tests with complex substrates, biantennary N-glycans were utilised as before [4], whereas H-Thr-Ala-Pro-Thr-(O-GlcNAc)Ser-Thr-Ile-Ala-Pro-Gly-OH, corresponding to the O-GlcNAc modification site found within the primary sequence of the transcription factor CREB, was purchased from Invitrogen and H-Pro-Gly-Gly-Ser-Thr-Pro-Val-Ser-(O-GlcNAc)Ser-Asn-Met-Met-Ser-Gly-OH, corresponding to a site in casein kinase II, was synthesised by Dr. Nicolas Laurent (University of Manchester).
Tissue-specific expression of murine hexosaminidase
For analysis of tissue specific mRNA expression, adult wild type mice with a mixed B6/129 background were sacrificed; tissues (tibialis anterior, soleus, atrium, ventricle, uterus, lung, liver, kidney and brain cortex) were dissected and immediately frozen at −80 °C until further use. RNA was extracted from tissues using Trizol (Invitrogen) following the manufacturer's instructions. Concentrations of RNA were measured with an UV-spectrophotometer. 2 μg RNA were then mixed with 0.5 μg d(T)18 and 0.5 μg d(N)10 primers. Nuclease-free water was added to a final volume of 15 μl. The mix was incubated at 75°C for 5 minutes and subsequently cooled on ice. Then 5 μl of 5× reverse transcriptase buffer, 1.5 μl of 10 mM dNTPs, 0.5 μl RNasin (RNase Inhibitor), 2 μl nuclease-free water and 1 μl of reverse transcriptase (M-MLV, Promega, 100 units/μl) were added. The mix was incubated at 42°C for 90 minutes. Finally the enzyme was denatured by incubating at 75°C for 10 minutes. The resulting cDNAs were normalised against the levels of GAPDH transcripts. The following primer pairs were used which cross intron/exon boundaries: 5MmHexoRT_1 (ATACAGCCCGTCCGAGGTTAC) and 3MmHexoRT_1 (TCAGGGTGTTGGGGAAGAG). PCR was performed for 40 cycles at 98° for 30 sec, 67° for 20 sec and 72° for 15 sec. Furthermore, the primer pair (MouseHex/1/EcoRI and MouseHex/2/XhoI) designed to clone the full open reading frame (see above) was also employed with the various cDNAs using an annealing temperature of 56 °C and an elongation time of 80 sec.
Intracellular localisation
For localisation studies, a vector encoding a N-terminal myc-tag (pSG9M) was employed. Murine liver cDNA was subject to PCR using the primers Mouse/pSG9M/1/XhoI CCGCTCGAGATGTCATCACCCACGCC and Mouse/pSG9M/2/XbaI GCTCTAGATCAGGGGTTCTGCCCTG; the resulting fragments were, after digestion, ligated into the vector; resulting bacterial colonies were screened and plasmid DNA was prepared. Murine 3T3 fibroblasts were transfected using Lipofectin and subsequent to staining with murine anti-myc, followed by FITC or Cy2-labelled anti-mouse IgG, were examined by confocal laser scanning microscopy (Leica TCS SP2 or Zeiss LSM 510) with Ar-laser excitation at 488 nm and emission 500-540 nm or He-Ne-laser excitation at 633 nm and emission > 650 nm using an HC Plans 10×/25 occular and HC PL Fluotar 63×/0.30 or 63x/1.4 objective. Co-labelling with AlexaFluor 633 phalloidin or 4′,6-diamidino-2-phenylindole (DAPI) was also performed to detect the cytoskeleton and nucleus, respectively. The cells were also examined by enzymatic assay and Western blotting with mouse anti-myc (9E10, Sigma); in the latter case, a myc-tagged form of Saccharomyces cerevisiae Alg1p β1,4-mannosyltransferase expressed in E. coli [24] was used as a positive control.
Results
Identification and expression of novel mammalian hexosaminidase homologues
During our recent characterisation of four novel β-hexosaminidases (HEX-2, HEX-3, HEX-4 and HEX-5) from the nematode Caenorhabditis elegans [4], we became aware of previous uncharacterised homologous human and murine cDNA sequences, which have been annotated in genome databases with the name HEXDC. These two mammalian and four nematode sequences fall into the same subfamily, previously designated by us as ‘subfamily 1′ [4], of the glycohydrolase family 20 hexosaminidases. This subfamily 1 has been recently described by others, in a bioinformatic and phylogenetic study, as ‘clade B’ of the glycohydrolase family 20 [25] and their results confirm our previous hypothesis of an evolutionary split in this group of enzymes.
Unlike the four nematode genes mentioned above, which encode proteins with predicted transmembrane domains, the two new human and murine cDNAs putatively encode proteins lacking any signal or other hydrophobic sequences and so were considered to possibly encode cytoplasmic proteins. The relevant cDNAs were isolated after RT-PCR as full-length open reading frames and introduced into E. coli expression vectors. In the case of the murine homologue, two different cDNA clones were isolated (one short form and one longer form), whereas in the case of the human one, two identical cDNAs were cloned, both of which lacked an 86 nt region present in the predicted transcript NM_173620, but which are equivalent to the newly-deposited sequence with the Genbank accession number BC018205. The short murine form and the human form, both of 486 amino acids, are encoded by twelve exons and the longer murine form by thirteen exons stretching over some 15-17 kbp of genomic sequence. Only the shorter form of the murine sequence fully corresponds to the human one; the insertion in the longer murine form, encoding an additional 73 amino acids after residue 328, displays no homology to any mammalian sequence and so is presumed to be a splicing artefact.
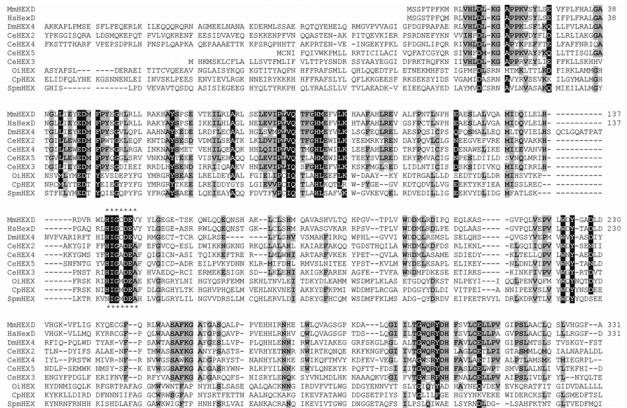
Both the mouse and human proteins contain the His/Asn-Xaa-Gly-Ala/Cys/Gly/Met-Asp-Glu-Ala/Ile/Leu/Val sequence typical of class 20 hexosaminidases (Figure 1); in other members of this family, the glutamate residue of this motif has been shown to be involved in the catalytic mechanism [26]. Presumed orthologues of HEXDC sequences occur in other mammals (horse, cow, dog; 78-80% identity with the murine sequence), chicken (63% identity), frog (58% identity) and fish (44% identity) genomes (data not shown). More distantly-related homologues exist in bacteria, such as the StrH β-hexosaminidase from Streptococcus pneumoniae [27]. Submission of the murine HEXDC sequence to the Phyre server [28] indicated that the closest relationships, to proteins with known three-dimensional structure, are to family 20 glycosidases from subfamily 2 (human lysosomal and Streptomyces plicatus hexosaminidases, Actinobacillus actinomycetemcomitans dispersin B and Serratia marcescens chitobiase; E values of 1.8e−32 - 1.6e−25) and more distantly to family 84 glycosidases (Bacteroides thetaiotaomicron GlcNAcase and Clostridium perfringens NagJ; E values 3e−06 – 9.9e−06).
Figure 1. Alignment of hexosaminidase D and other hexosaminidases.
The first 331 (of 486) residues of the predicted protein sequences of the murine (short; MmHEXD) and human (HsHexD) forms of hexosamindase D were aligned with the sequences of the four ‘subfamily 1’ class 20 hexosaminidases from Caenorhabditis elegans (CeHEX-2, -3, -4 and -5), one homologue from Drosophila melanogaster (CG7985; designated here as DmHEX4) and three bacterial homologues (from Clostridium perfringens, Oceanobacillus iheyensis and Streptococcus pneumoniae). The region conserved with respect to ‘subfamily 2’ class 20 hexosaminidases (e.g., human lysosomal hexosaminidases; also designated as clade A in Ref. 25) is highlighted by asterisks. Residues identical in 9/10 sequences are highlighted in black; those which are conserved in at least 5/10 sequences are shown with a grey background.
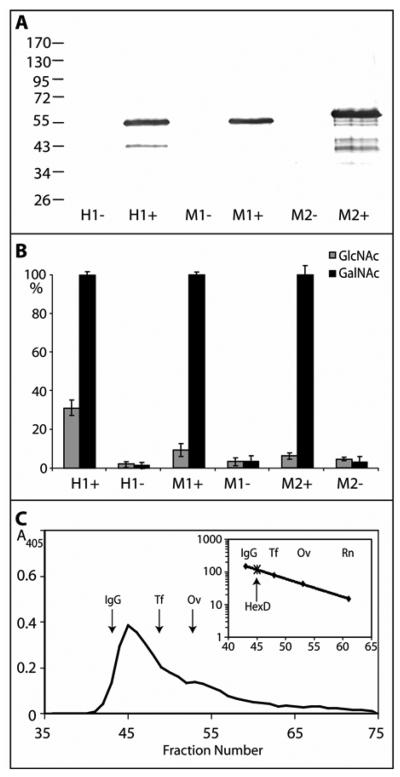
The long and short murine forms as well as the human HEXDC cDNA were then expressed in BL21(DE3)pLysS cells and, upon induction with IPTG, hexosaminidase activity was detected in lysates of the bacterial cells (Figure 2). Initial indications, when using p-nitrophenyl substrates, were that both enzymes were relatively specific for p-nitrophenyl-β-N-acetylgalactosaminide and that His-tagged proteins of ca. Mr 55,000 were expressed in only the induced cells (Mr 62,000 in the case of the murine longer form; see Figure 2), as compared to the predicted Mr values of 54575 and 53788 for the murine (short) and human proteins. The fact that the longer murine form is active is perhaps due to the insertion in its sequence being outside the region homologous to most other class 20 hexosaminidases of the subfamily 1.
Figure 2. Recombinant expression of mammalian hexosaminidase D in E. coli.
Two clones encoding His-tagged forms of either the short (M1; 486 residues) or long (M2; 559 residues) forms of murine hexosaminidase D and of one clone encoding human hexosaminidase D (H1; 486 residues) were used to transform E. coli and lysates were prepared after cultivation for three hours in the presence (+) or absence (−) of IPTG. (A) Western blotting of transformed E. coli indicates expression of proteins of the expected sizes; (B) assay with either p-nitrophenyl-β-N-acetylgalactosaminide or p-nitrophenyl-β-N-acetylglucosaminide for one hour at 37 °C shows induction of especially p-nitrophenyl-β-N-acetylgalactosaminidase activity with mammalian hexosaminidase D clones. Duplicate data were normalised with 100% corresponding to the activity of the induced cell lysate towards p-nitrophenyl-β-N-acetylgalactosaminide in that experiment and the averaged normalised data are from two independent experiments; therefore, each point corresponds to four single enzymatic assays. (C) Gel filtration analysis of the short form of the murine hexosaminidase D (M1) on a Sephacryl S-200 Superfine column calibrated (see insert with scale in terms of Mr × 10−3) with IgG (IgG; 150 kDa), apo-transferrin (Tf; 80 kDa), ovalbumin (Ov; 43 kDa), and ribonuclease B (Rn; 15 kDa). Hexosaminidase activity was recorded at 405 nm; the presence of the protein with the expected molecular mass was verified by Western blotting (data not shown).
Further characterisation of murine hexosaminidase D
In order to facilitate further characterisation, the enzymes were then partially purified by His-Tag affinity chromatography; the major portions of the activity and anti-His reactivity were in the fractions eluted with 250 mM imidazole. The identity of the purified enzymes was also verified by tryptic peptide mass fingerprinting; due to the 80% identity of the mouse short form and the human form, we performed most experiments with the murine short form, since this was more stable upon storage. Gel filtration analysis of the purified recombinant murine short form suggested that the enzyme is predominantly present as a dimer with a native molecular mass of approximately 120000 (Figure 2C), whereas under denaturing conditions SDS-PAGE and Western blotting demonstrated that a homogenous protein of ca. Mr 55000 was present in fractions containing hexosaminidase activity.
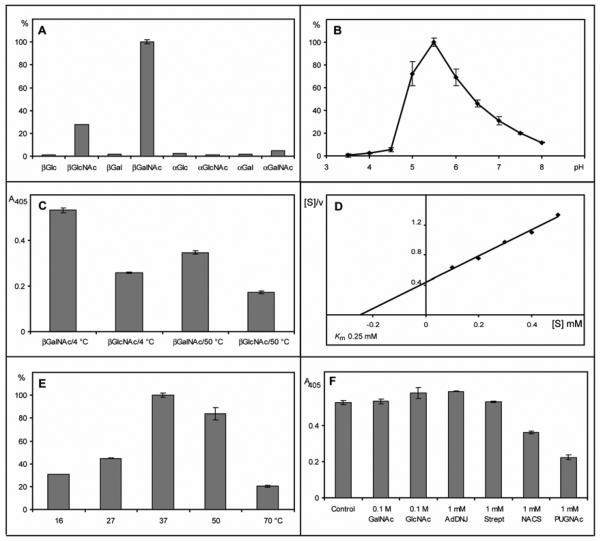
Bearing in mind the hypothesis that the newly-identified genes encode proteins which may correspond to so-called “hexosaminidase D” activities, the affinity-purified murine enzyme was then examined considering the criteria previously used to categorise the different hexosaminidase activities detected earlier in mammalian tissues [17-20]. The apparent relative specificity for p-nitrophenyl-β-N-acetylgalactosaminide (Figure 3A), the pH optimum of 5.5 (Figure 3B) and the relative stability to incubation at 50 °C (Figure 3C) appeared to agree well with previous data on hexosaminidase D [18, 21]. The Km value for the murine enzyme was also determined (~0.25 mM) and was in reasonable agreement to the value of 0.35 mM [18] also found for the hexosaminidase D (Figure 3D); the temperature optimum was found to be 37 °C (Figure 3E). The pH optimum and relative temperature stability of the human and murine enzymes in E. coli lysates were similar to those of the purified murine enzyme.
Figure 3. Enzymatic properties of the recombinant purified murine hexosaminidase D.
(A) Assays were performed with various p-nitrophenyl α- and β-glycosides with 0.25 μl purified hexosaminidase D for 2 hours at 37 °C (pH 5.5); average absorbance values (405 nm) were re-calculated as a percentage of the activity with p-nitrophenyl-β-N-acetylgalactosaminide. (B) pH optimum of purified hexosaminidase D using 2.5 mM p-nitrophenyl-β-N-acetylgalactosaminide as substrate (2 hours, 37 °C, McIlvaine buffers); average absorbance values were re-calculated as a percentage of the activity at pH 5.5. (C) Assay of hexosaminidase pre-incubated for 30 minutes at either 4 °C or 50 °C prior to incubation for 2 hours with either 2.5 mM p-nitrophenyl-β-N-acetylgalactosaminide or p-nitrophenyl-β-N-acetylglucosaminide at 37 °C; in this experiment the stock solution of the latter substrate was 3 mM in buffer and not 100 mM in dimethylsulphoxide. (D) Hanes' plot of data with various concentrations of p-nitrophenyl-β-N-acetylgalactosaminide as substrate (0.5 μl enzyme, 30 minutes, 37 °C, pH 5.5). (E) Temperature optimum of hexosaminidase D using 2.5 mM p-nitrophenyl-β-N-acetylgalactosaminide as substrate (2 hours, pH 5.5); the data at 50 °C and 70 °C are corrected for values obtained in the absence of enzyme and average absorbance values were re-calculated as a percentage of the activity at 37 °C (pH 5.5). (F) Inhibition of hexosaminidase D using either 0.1 M GalNAc or GlcNAc or 1 mM AdDNJ, STZ, NACS or PUGNAc (0.25 μl enzyme, 2 hours, 37 °C, pH 5.5, 2.5 mM p-nitrophenyl-β-N-acetylgalactosaminide); background-corrected absolute absorbance values are presented. In assays with 0.5 mM substrate, the inhibition by 0.2 mM PUGNAc also approached 70% (data not shown). Error bars indicate the standard deviations of assays performed in triplicate or quadruplicate.
The effect of various typical hexosaminidase inhibitors was also tested: N-acetylcastanospermine (NACS), 2-acetamido-1,2-dideoxynojirimycin (AdDNJ), O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate (PUGNAc), streptozotocin (STZ), GlcNAc and GalNAc [15, 22, 29-34]. In the case of our novel murine hexosaminidase, only 60% inhibition with 1 mM PUGNAc and 30% with 1 mM N-acetylcastanospermine was observed when the enzyme was assayed with 2.5 mM substrate (Figure 3F). Thereby, the recombinant murine hexosaminidase D shows properties at variance with O-GlcNAcase, which is inhibited by some 96% by 1.25 mM PUGNAc, when assayed with 2 mM p-nitrophenyl-β-N-acetylglucosaminide [32]. It also requires far higher concentrations of PUGNAc and N-acetylcastanospermine for inhibition than Arabidopsis HEX1 and Caenorhabditis HEX-1, which are ‘typical’ (subfamily 2) class 20 hexosaminidases; these two enzymes are inhibited by some 60-90% when incubated with 0.1 mM of these inhibitors under assay conditions similar to those employed here [4].
The relatively poor inhibition of hexosaminidase D with PUGNAc and N-acetylcastanospermine, though, is similar to the results with the p-nitrophenyl-β-N-acetylhexosaminidase activity of four recently-characterised C. elegans class 20 hexosaminidases (HEX-2, -3, -4 and -5), which are part of the same sub-family (sub-family 1) as hexosaminidase D and which also prefer p-nitrophenyl-β-N-acetylgalactosamine as a substrate [4]. Considering the homology of mouse hexosaminidase D with these C. elegans enzymes displaying activity towards N-glycans, the mouse and human hexosaminidases were incubated with N-glycans carrying terminal GlcNAc or GalNAc residues; however, no digestion was observed (data not shown). Furthermore, considering the potential cytosolic localisation of the enzyme, tests were also performed with two peptides modified with O-linked β-GlcNAc; whereas the control reaction with jack bean hexosaminidase showed removal of the GlcNAc residue from the peptides, no such effect occurred in the incubation with the purified murine hexosaminidase (data not shown).
Tissue-specific expression of the novel murine hexosaminidase
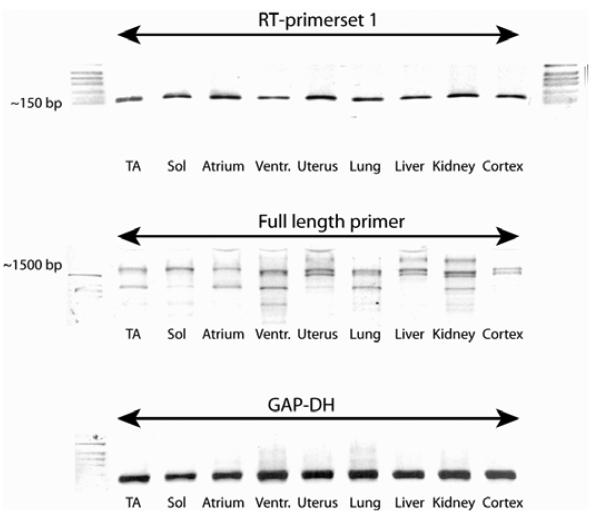
RT-PCR was performed using mRNA from a variety of murine tissues (various muscles, uterus, lung, liver, kidney and brain cortex); the cDNAs were normalised based on the levels of GAPDH transcripts and it was found that the putative hexosaminidase D is expressed in all tissues examined, with no major differences in transcript levels (Figure 4).
Figure 4. Distribution of hexosaminidase D transcripts in murine tissues.
RT-PCR reactions were performed using cDNAs from various murine tissues which were normalised based on their glyceraldehyde-3-phosphate dehydrogenase (GAP-DH) transcript levels (lower panel); primer sets were used to either detect a small internal fragment (upper panel; ca, 150 bp) or the full length transcript (middle panel; ca. 1600 bp) of murine hexosaminidase D. The data show ubiquitous expression in all tissues examined: tibialis anterior (TA), soleus (Sol), atrium, left ventricle (Ventr), uterus, lung, liver, kidney and brain cortex.
Intracellular localisation of the novel murine hexosaminidases
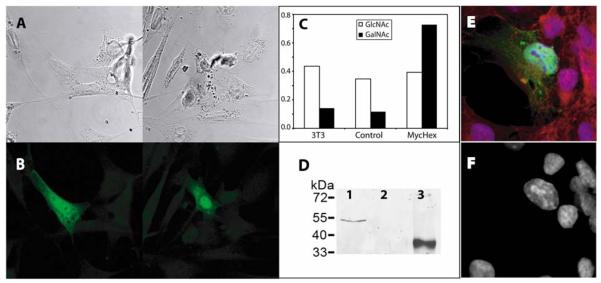
Based on the lack of any obvious hydrophobic domains, our hypothesis was that the murine and human forms of the putative hexosaminidase D are not, as other eukaryotic class 20 hexosaminidases, directed via the endoplasmic reticulum to the lysosome or secretory pathway, but that they are present in the nucleus or cytoplasm. Thus, a plasmid encoding an N-terminally myc-tagged form of the full-length murine hexosaminidase was engineered and transfected into murine 3T3 fibroblast cells, which were then subject to immunofluorescence and confocal microscopy. Those cells which were transfected showed that the tagged protein was present in the nucleus and cytoplasm (Figure 5 A and B); it is unclear whether the less elongated appearance of transfected (fluorescent) cells, as compared to the typical fibroblast morphology of the non-transfected ones, is of functional significance. Certainly, actual overexpression of the hexosaminidase was demonstrated by the appearance of a myc-tagged protein of the appropriate molecular mass and of increased N-acetylgalactosaminidase activity in the cell lysates (Figure 5 C and D). In an independent experiment, transfection of COS cells also showed a concentration of the myc-tagged protein in the nucleus (Figure 5 E).
Figure 5. Expression of myc-tagged murine hexosaminidase D.
A clone encoding a myc-tagged form of the murine hexosaminidase D (MycHexD) was used to transfect murine 3T3 cells: (A) confocal microscopy of two selected transfected cultures; (B) corresponding anti-myc fluorescence micrographs showing nucleocytoplasmic localisation of the construct; (C) comparison of N-acetylglucosaminidase and N-acetylgalactosaminidase activities of non-transfected, ‘empty vector’ transfected and MycHexD-transfected 3T3 cells showing induction of p-nitrophenyl-β-N-acetylgalactosaminidase activity in the latter; (D) demonstration of expression of MycHexD by Western blotting (lane 1, cells transfected with the vector encoding the myc-tagged form of murine hexosaminidase D; lane 2, cells transfected with ‘empty vector’; lane 3, E. coli-expressed myc-tagged β1,4-mannosyltransferase positive control). (E) fluorescent micrograph of COS cells transfected with the same MycHexD construct stained with anti-myc (green), DAPI (blue) and phalloidin (red) showing a bias towards nuclear anti-myc labelling; (F) the corresponding blue channel showing DAPI staining alone.
Discussion
A range of earlier literature suggested that, in addition to the well-known lysosomal hexosaminidases encoded by the HEXA and HEXB genes and the neutral cytosolic O-GlcNAcase, there may be an additional soluble hexosaminidase present in mammalian cells. In 1967, a β-N-acetylgalactosaminidase with optimal activity at pH 5.5 was found in a 100,000 g supernatant of a calf brain homogenate [18]. Later, another report on bovine brain hexosaminidases indicated the presence of four chromatographically-distinguishable enzymes, hexosaminidases A, B, C and D, the latter which was a heat-stable specific galactosaminidase [20]; bovine and human hexosaminidase C, on the other hand, was found as a glucosaminidase with a higher molecular weight and optimal activity in the range pH 5-7 [17, 20, 35] and, in retrospect, probably is identical to O-GlcNAcase [15]. Finally, two rat brain neutral hexosaminidases were described which, unlike the lysosomal forms and suggestive of a lack of N-linked glycosylation, did not interact with concanavalin A: one glucosaminidase with, as determined by SDS-PAGE, Mr 130000 and one galactosaminidase with Mr 55000 [19]. The former apparent molecular weight also corresponds well with that of the rather heat-sensitive O-GlcNAcase; the galactosaminidase, on the other hand, had optimal activity at pH 6 and was relatively heat-stable.
In comparison to these data on native enzymes, the recombinant enzymes reported in the present study are soluble, are of Mr 55000, are relatively stable at 50 °C, display optimal activity at pH 5.5 and prefer the galactosaminide substrate; thus, it is hypothesised that these human and murine enzymes correspond to the “hexosaminidase D” activities previously detected in mammalian brain extracts. Furthermore, the intracellular localisation data indicates that a myc-tagged form of the murine enzyme is localised to the nucleus and cytoplasm; interestingly, a large-scale yeast two-hybrid screen also suggests that the human enzyme has a high confidence interaction with the tubulin β4 subunit [36]. The recombinant murine enzyme also appears to be predominantly in a dimeric form as is the case with the lysosomal hexosaminidases.
Thus, it appears that hexosaminidase D is indeed a second nucleocytoplasmic hexosaminidase with properties, other than the general intracellular location, unlike the O-GlcNAcase proven to play a role in intracellular signalling; on the other hand, ‘D-type’ hexosaminidases are the only vertebrate members of a recently-described new sub-group within the glycoside hydrolase family 20. Hexosaminidase D does have homologues in nematodes (C. elegans HEX-2, HEX-3, HEX-4 and HEX-5; ca. 30% identity over 440 amino acids) and in insects (Drosophila CG7985; ca. 35% identity), but the predicted proteins contain potential transmembrane regions and so, unlike hexosaminidase D, are not expected to be cytosolic proteins. No function has yet been proven for the insect CG7985, but all four nematode enzymes do digest selected N-glycan substrates in vitro and, as judged by N-glycan analysis of the relevant hex-2 mutant, C. elegans HEX-2 is indeed involved in N-glycan biosynthesis and so is expected to be a Golgi enzyme [4]. Thus, whereas invertebrates possess only the family 84 O-GlcNAcase as a cytosolic enzyme, such as the C. elegans OGA-1 [37], despite having other family 20 hexosaminidases, the presence of two nucleocytoplasmic hexosaminidases appears to be confined to vertebrates. Akin to the nematode members of the subfamily 1 (clade B) of family 20 and in keeping with the phylogenetic distance [4], the recombinant hexosaminidase D is only inhibited by relatively large inhibitor:substrate ratios when using a range of compounds, such as AdDNJ, NACS or PUGNAc, typically considered to be effective inhibitors of other family 20 hexosaminidases [29-31].
Other than O-linked GlcNAc, there are a variety of N-acetylhexosamine-containing compounds in mammalian cells, including mucin-type O-glycans (α-linked GalNAc linked to Ser/Thr residues), various glycolipids (such as the β-GalNAc-containing ganglioside GM2, whose catabolism is blocked in Tay-Sachs and Sandhoff syndromes), N-glycans (containing β-linked GlcNAc and, occasionally, GalNAc) and glycosaminoglycans (with both α- and β-linked hexosamine-containing repeating units); these, though, are all products of the secretory pathway. In the case of N-glycans, some cytosolic exoglycosidase-mediated degradation does occur, for instance via the action of the Man2c1 α-mannosidase on oligomannosidic glycans derived from misfolded proteins [38]. It is also noteworthy that mammalian Neu2 sialidases display a similar localisation and pH optimum as does hexosaminidase D [39]; however, unlike hexosaminidase D, Neu2 can cleave sialic acid from N-glycans [40, 41] as well as from glycolipids [41].
Although, as mentioned above, hexosaminidase D may interact with β-tubulin, a protein shown to carry O-GlcNAc residues [42], we detected no activity towards O-GlcNAc-modified peptides. Thus, it appears that hexosaminidase D may not constitute part of another route for control of the balance between O-GlcNAc and phosphorylation of vertebrate cytosolic and nuclear proteins or it may require another co-factor for such an activity or display a unique substrate specificity. On the other hand, its cytoplasmic localisation would argue against a role in metabolism of N-, O- or lipid-linked oligosaccharides originating from the secretory pathway, although non-classical secretion of this enzyme cannot be ruled out; therefore, the functional and biological significance of this enzyme remains to be resolved.
Acknowledgements
We wish to thank Andreas Rupp, project student in the laboratory, for performing Western blotting, Dr. Boris Ferko for the sample of mouse liver as well as Dr. Elisabeth Ehler for facilitating the work of T.I. on this project, Dr. Johannes Grillari for the pSG9M vector and helpful discussions, Drs. Arnold Stütz and Nicolas Laurent for the kind gifts of chemically-synthesised compounds and Dr. Katharina Paschinger for reading the manuscript. This work was supported by grants P15475 and P18447 (to I.B.H.W.) from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung.
The abbreviations used are
- AdDNJ
2-acetamido-1,2-dideoxynojirimycin
- GFP
green fluorescent protein
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MALDI-TOF
matrix-assisted laser desorption ionisation/time-of flight
- MS
mass spectrometry
- NACS
N-acetylcastanospermine
- OGT
O-GlcNAc transferase
- PUGNAc
O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate
- STZ
streptozotocin
Footnotes
REFERENCES
- 1.Mahuran DJ. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta. 1999;1455:105–138. doi: 10.1016/s0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 2.Korneluk RG, Mahuran DJ, Neote K, Klavins MH, O'Dowd BF, Tropak M, Willard HF, Anderson MJ, Lowden JA, Gravel RA. Isolation of cDNA clones coding for the α-subunit of human β-hexosaminidase. Extensive homology between the α- and β-subunits and studies on Tay-Sachs disease. J. Biol. Chem. 1986;261:8407–8413. [PubMed] [Google Scholar]
- 3.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutternigg M, Kretschmer-Lubich D, Paschinger K, Rendić D, Hader J, Geier P, Ranftl R, Jantsch V, Lochnit G, Wilson IBH. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants and insects. J. Biol. Chem. 2007;282:27825–27840. doi: 10.1074/jbc.M704235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Léonard R, Rendić D, Rabouille C, Wilson IBH, Préat T, Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J. Biol. Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 6.Geisler C, Aumiller JJ, Jarvis DL. A fused lobes gene encodes the processing β-N-acetylglucosaminidase in Sf9 cells. J Biol Chem. 2008;283:11330–11339. doi: 10.1074/jbc.M710279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams SJ, Mark BL, Vocadlo DJ, James MN, Withers SG. Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 2002;277:40055–40065. doi: 10.1074/jbc.M206481200. [DOI] [PubMed] [Google Scholar]
- 8.Vocadlo DJ, Withers SG. Detailed comparative analysis of the catalytic mechanisms of β-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry. 2005;44:12809–12818. doi: 10.1021/bi051121k. [DOI] [PubMed] [Google Scholar]
- 9.Hurtado-Guerrero R, Dorfmueller HC, van Aalten DM. Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 2008;18:551–557. doi: 10.1016/j.sbi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 11.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 12.O'Donnell N. Intracellular glycosylation and development. Biochim Biophys Acta. 2002;1573:336–345. doi: 10.1016/s0304-4165(02)00401-4. [DOI] [PubMed] [Google Scholar]
- 13.Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, Kudlow JE. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology. 2006;16:551–563. doi: 10.1093/glycob/cwj096. [DOI] [PubMed] [Google Scholar]
- 14.Wells L, Gao Y, Mahoney JA, Vosseller K, Chen C, Rosen A, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic β-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterisation of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 16.Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem Biophys Res Commun. 2001;283:634–640. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- 17.Braidman I, Carroll M, Dance N, Robinson D. Separation and properties of human brain hexosaminidase C. Biochem. J. 1974;143:295–301. doi: 10.1042/bj1430295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frohwein YZ, Gatt S. Isolation of β-N-acetylhexosaminidase, β-N-acetylglucosaminidase and β-N-acetylgalactosaminidase from calf brain. Biochemistry. 1967;6:2775–2782. doi: 10.1021/bi00861a018. [DOI] [PubMed] [Google Scholar]
- 19.Izumi T, Suzuki K. Neutral β-N-acetylhexosaminidases of rat brain: purification and enzymatic and immunological characterisation. J. Biol. Chem. 1983;258:6991–6999. [PubMed] [Google Scholar]
- 20.Overdijk B, van der Kroef WMJ, Veltkamp WA, Hooghwinkel GJM. The separation of bovine brain β-N-acetyl-D-hexosaminidases. Biochem. J. 1975;151:257–261. doi: 10.1042/bj1510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi T, Suzuki K. Neutral hydrolases of rat brain. Preliminary characterization and developmental changes of neutral β-N-acetylhexosamindases. Biochim. Biophys. Acta. 1980;615:402–413. doi: 10.1016/0005-2744(80)90507-0. [DOI] [PubMed] [Google Scholar]
- 22.Dong DLY, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-β-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 23.Rhomberg S, Fuchsluger C, Rendić D, Paschinger K, Jantsch V, Kosma P, Wilson IBH. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006;273:2244–2256. doi: 10.1111/j.1742-4658.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 24.Revers L, Bill RM, Wilson IBH, Watt GM, Flitsch SL. Development of recombinant, immobilised β-1,4-mannosyltransferase for use as an efficient tool in the chemoenzymatic synthesis of N-linked oligosaccharides. Biochim. Biophys. Acta. 1999;1428:88–98. doi: 10.1016/s0304-4165(99)00048-3. [DOI] [PubMed] [Google Scholar]
- 25.Intra J, Pavesi G, Horner DS. Phylogenetic analyses suggest multiple changes of substrate specificity within the glycosyl hydrolase 20 family. BMC Evol. Biol. 2008;8:214. doi: 10.1186/1471-2148-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y, Vocadlo DJ, Leung A, Withers SG, Mahuran D. Characterization of the Glu and Asp residues in the active site of human β-hexosaminidase B. Biochemistry. 2001;40:2201–2209. doi: 10.1021/bi002018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke VA, Platt N, Butters TD. Cloning and expression of the β-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J. Biol. Chem. 1995;270:8805–8814. doi: 10.1074/jbc.270.15.8805. [DOI] [PubMed] [Google Scholar]
- 28.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 29.Liu PS, Kang MS, Sunkara PS. A potent inhibitor of β-N-acetylglucosaminidases: 6-acetamido-6-deoxycastanospermine. Tetrahedron Lett. 1991;32:719–720. [Google Scholar]
- 30.Gradnig G, Legler G, Stütz AE. A novel approach to the 1-deoxynojirimycin system: Synthesis from sucrose of 2-acetamido-1,2-dideoxynojirimycin, as well as some 2-N-modified derivatives. Carbohydr. Res. 1996;287:49–57. doi: 10.1016/0008-6215(96)00065-1. [DOI] [PubMed] [Google Scholar]
- 31.Horsch M, Hoesch L, Vasella A, Rast DM. N-acetylglucosaminono-1,5-lactone oxime and the corresponding (phenylcarbamoyl)oxime. Novel and potent inhibitors of β-N-acetylglucosaminidase. Eur. J. Biochem. 1991;197:815–818. doi: 10.1111/j.1432-1033.1991.tb15976.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee TN, Alborn WE, Knierman MD, Konrad RJ. The diabetogenic antibiotic streptozotocin modifies the tryptic digest pattern for peptides of the enzyme O-GlcNAc-selective N-acetyl-β-D-glucosaminidase that contain amino acid residues essential for enzymatic activity. Biochem. Pharmacol. 2006;72:710–718. doi: 10.1016/j.bcp.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Gao Y, Parker GJ, Hart GW. Streptozotocin-induced β-cell death is independent of its inhibition of O-GlcNAcase in pancreatic Min6 cells. Arch. Biochem. Biophys. 2000;383:296–302. doi: 10.1006/abbi.2000.2094. [DOI] [PubMed] [Google Scholar]
- 34.Kapur DK, Gupta GS. Purification, biochemical properties and active sites of N-acetyl-β-D-hexosaminidases from human seminal plasma. Biochem. J. 1986;236:103–109. doi: 10.1042/bj2360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besley GTN, Broadhead DM. Studies on human N-acetyl-β-D-hexosaminidase C separated from neonatal brain. Biochem. J. 1976;155:205–208. doi: 10.1042/bj1550205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 37.Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism and dauer. Proc. Natl. Acad. Sci. USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T, Hara I, Nakano M, Shigeta M, Nakagawa T, Kondo A, Funakoshi Y, Taniguchi N. Man2C1, an α-mannosidase is involved in the trimming of free oligosaccharides in the cytosol. Biochem. J. 2006;400:33–41. doi: 10.1042/BJ20060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monti E, Preti A, Nesti C, Ballabio A, Borsani G. Expression of a novel human sialidase encoded by the NEU2 gene. Glycobiology. 1999;9:1313–1321. doi: 10.1093/glycob/9.12.1313. [DOI] [PubMed] [Google Scholar]
- 40.Ishizuka A, Hashimto Y, Naka R, Kinoshita M, Kakehi K, Seino J, Funakoshi Y, Suzuki T, Kameyama A, Narimatsu H. Accumulation of free complex-type Nglycans in MKN7 and MKN45 stomach cancer cells. Biochem. J. 2008;413:227–237. doi: 10.1042/BJ20071562. [DOI] [PubMed] [Google Scholar]
- 41.Tringali C, Papini N, Fusi P, Croci G, Borsani G, Preti A, Tortora P, Tettamanti G, Venerando B, Monti E. Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ganglioside molecules. J. Biol. Chem. 2004;279:3169–3179. doi: 10.1074/jbc.M308381200. [DOI] [PubMed] [Google Scholar]
- 42.Park J, Kwon H, Kang Y, Kim Y. Proteomic analysis of O-GlcNAc modifications derived from streptozotocin and glucosamine induced β-cell apoptosis. J. Biochem. Mol. Biol. 2007;40:1058–1068. doi: 10.5483/bmbrep.2007.40.6.1058. [DOI] [PubMed] [Google Scholar]