Abstract
Peptide O-xylosyltransferase (EC 2.4.2.26) is the first enzyme required for the generation of chondroitin and heparan sulphate glycosaminoglycan chains of proteoglycans. Cloning of cDNAs has previously shown that, whereas invertebrates generally have a single xylosyltransferase gene, vertebrate genomes encode two similar proteins, XT-I and XT-II. To date, enzymatic activity has only been demonstrated for the human XTI, Caenorhabditis SQV-6 and Drosophila OXT isoforms. In the present study, we demonstrate that a soluble form of human XT-II expressed in the xylosyltransferase-deficient pgsA-745 (S745) Chinese hamster ovary cell line is indeed capable of catalysing the transfer of xylose to a variety of peptide substrates; its enzyme activity was also proven using a Pichia- expressed form of XT-II. Its pH, temperature and cation dependencies are similar to those of XT-I expressed in either mammalian cells or yeast. Our data suggest that XT-I and XT-II are, at least in vitro, functionally identical.
Keywords: biosynthesis, glycosaminoglycan, proteoglycan, mass spectrometry, protein expression, xylosyltransferase
Morphogenesis is a highly complex process in animal development and requires the creation of growth factor gradients for its control. Central to the correct formation of such gradients and to the associated signal transduction pathways are the glycosaminoglycan chains, both chondroitin and heparan sulphates, of various proteoglycans (1-4). In the past few years, a number of mutations of glycosaminoglycan biosynthesis have shown the key role of these macromolecules. Particularly striking is the series of Caenorhabditis elegans squashed vulva (sqv) mutants (5,6); each of these mutants is defective in a different gene involved in the formation of chondroitin sulphate. In mammals, the EXT1 and EXT2 genes involved in heparan sulphate biosynthesis are implicated in brain morphogenesis (7) and in the bone disease, hereditary multiple exostoses (8,9), whereas a form of heparan sulphate, heparin, acts as an anticoagulant by activating antithrombin (10).
The initial step in the biosynthesis of both chondroitin and heparan sulphates begins with the transfer of xylose to serine residues of a proteoglycan core protein (11). Although this enzyme activity was first described in the 1960s (12-14), it was first purified some thirty-five years later (15) and the first cDNAs were cloned from mammalian species (16). Subsequently, homologues were identified in Drosophila, encoded by the oxt gene (17), and in Caenorhabditis, encoded by the sqv-6 gene (18,19). Both of these invertebrate species only possess a single xylosyltransferase gene; however, vertebrates, starting with fish, appear to have two genes (XYLT1 and XYLT2) encoding highly homologous enzymes named XT-I and XT-II1. Of vertebrate forms, enzymatic activity has been demonstrated for human XT-I (16).
Recently, it has been reported that polymorphisms exist within both human xylosyltransferase genes. Various of these have been correlated with blood pressure levels in diabetic patients, with nondiabetic nephropathy, osteoarthritis and symptoms in pseudoxanthoma elasticum (20-22). Both genes are, as judged by RT-PCR with mRNA from eight tissues, widely expressed (15) and GFP-tagged forms of both enzymes were found to be targetted to the Golgi (23), a location consistent with some earlier cell fractionation studies (24). However, the enzymatic function of XT-II has remained elusive. Preliminary data in this laboratory suggested a rather minor activity in the culture supernatant of Pichia pastoris transformed with an XT-II construct; thus, as an alternative, we decided to attempt the expression in a mammalian cell line lacking xylosyltransferase activity (25). Indeed, for the first time, we report that human XT-II is an active enzyme with properties not significantly different from those of XT-I.
EXPERIMENTAL PROCEDURES
Vector construction and transfection
Partial reading frames of human XT-I and XT-II were isolated by PCR using the forward primers HsXTI/1/FseI (cccacgaaggccggcctaagtgtgacatctcag) or HsXTII/1/FseI (cccacgaaggccggcccaagtgcgagatcgt) and the reverse primers HsXTI/2/NotI (ttttccttttgcggccgcactcctcggtgcccag) or HsXTII/2/NotI (ttttccttttgcggccgctggggccctgctac) with pGEM-T vectors previously prepared using fragments generated by PCR of human embyronic kidney (HEK-293) cell line cDNA (19) as templates and KOD polymerase (Novagen). Purified PCR fragments were then cut with FseI and NotI and ligated into the pPA-TEV vector (26), which was the kind gift of Dr. James Rini, University of Toronto. This vector, derived from pIRESpuro3 (Clontech), is intended for the expression of secreted protein A fusion proteins under control of the cytomegalovirus promoter.
The inserts of selected clones were sequenced in their entirety using the BigDye kit (Applera). Verified clones encoding XT-I and XT-II, as well as an empty vector control, were transiently transfected into the pgsA-745 (S745) Chinese hamster ovary cell line (kindly supplied by Dr. Jeffrey Esko, University of California, San Diego; Ref. 25) using Lipofectamine 2000 reagent (Invitrogen). Independent transfections of XT-II were performed on separate days. Cells were cultivated for 24 hours and the culture supernatants were collected, stored at 4 °C and, as required, concentrated using Ultrafree microfuge devices with an Mr 30,000 cut-off (Millipore). Protein content in the supernatants was determined using the MicroBCA protein assay kit (Pierce).
The construction of the pPICZαC vector with the XT-II insert, transformation of Pichia pastoris strain GS115 and induction of expression with methanol has been previously described (19). Supernatants were concentrated ten-fold using Ultrafree microfuge devices, diluted with 10 mM HEPES, pH 8, containing 0.01% (w/v) sodium azide and 1 mM phenylmethylsulphonyl fluoride and again concentrated prior to storage at 4 °C.
Western blotting
Aliquots of concentrated supernatants, equivalent to 12.5 μl of original culture medium, were subject to SDS-PAGE (10% gel) and Western blotting using rabbit anti-protein A (Sigma, 1:2000) and alkaline phosphatase-conjugated goat anti-rabbit (Vector Laboratories, 1:2000). SigmaFAST tablets were used to prepare the chromogenic substrate for detection. Aliquots of the supernatants were also digested overnight with PNGase F (1 U, Roche) at 37 °C in 10 mM HEPES, pH 8, prior to SDS-PAGE.
Assay of xylosyltransferase activity
Culture supernatants were assayed using either the unmodified syndecan peptide (DDDSIEGSGGR), the bikunin peptide (QEEEGSGGGGQR), or a dansylated form of the perlecan peptide (DSISGDDLGSGDLGSGDFQR), as substrates. Dansylation was performed as follows: 3 mg peptide were dissolved in 7 ml Li2CO3 (40 mM, pH 9.5); thereafter, 40 mg dansyl chloride in 16 ml acetone was added and the mixture was incubated in a water bath for 60 min at 37°C. After addition of 14 μl ethylamine (70% (w/v)), the sample was subject to rotary evaporation and purified by gel filtration (Sephadex G25 fine, 1 × 50 cm in 25% methanol) and RP-HPLC. Detection of the dansylated product was performed using fluorescence at 315/550 nm. Either the syndecan (1 mM final concentration), bikunin (1 mM) or dansylated perlecan (30 μM) peptides were then incubated with supernatants of xylosyltransferase-expressing cells in the presence or absence of 1 mM UDP-Xyl and the products were analysed by MALDI-TOF MS (Perspective Biosystems Voyager-DE STR workstation) as previously described (17,19) or by RP-HPLC at a flow rate of 1.5 ml/min (Hypersil ODS); in the case of syndecan, the RP-HPLC gradient was of 11.4 - 13.3% acetonitrile from 5 - 13 minutes with UV detection at 214 nm (19), whereas for dansylated perlecan a gradient of 34.2 - 41.8% acetonitrile was applied from 6 - 22 minutes with fluorescence detection at 315/550 nm.
Further characterisation of XT-I and XT-II
The enzymatic parameters of both human xylosyltransferases expressed in pgsA-745 cells were examined using the aforementioned RP-HPLC method with the syndecan peptide as substrate. For the pH dependency, HEPES and AMPD buffers of various pH values (6.5-9.0) were used; various cations were tested at a final concentration of 10 mM; the effects of temperature during the reaction and of substrate concentration were examined using incubations containing 40 mM HEPES, pH 8, and 10 mM MnCl2. All assays were performed in duplicate and within the linear range of product formation with respect to time. Relative activities (for pH, temperature and cation dependency) were determined based on HPLC peak areas (for the product peak and the sum of product and substrate peaks); the percentage conversion of product was then converted to an activity relative to that under the condition giving the highest rate of conversion to product. Specific activities were also calculated based upon the amount of substrate, as determined by the relative HPLC peak areas for product and substrate, converted within the linear time range, in the presence of Mn(II) at 37 °C, and are expressed in terms of mU/mg, where 1 mU/mg corresponds to 1 nmol substrate turned over per minute per mg of protein in the culture supernatant.
RESULTS AND DISCUSSION
Expression of xylosyltransferases
In previous studies, we have found that forms of Caenorhabditis SQV-6, Drosophila OXT and human XT-I could be expressed using Pichia pastoris in an active form (17,19). These forms were truncated so as to delete the cytosolic, transmembrane and stem regions, but retained the Pro-Xaa-Cys-Asp/Glu motif noted as being the most N-terminal conserved region in invertebrate and vertebrate peptide O-xylosyltransferases (11); the two human xylosyltransferase sequences display 61% identity (75% similarity) in the region beginning with this motif. However, although the expression in Pichia was easy to perform, the HPLC and MS-based assays using the culture supernatants were sometimes complicated by the peptones in the medium and, furthermore, a barely-detectable activity was found in preliminary trials with human XT-II. Considering there was no published record of XT-II displaying activity, we decided that another system should be used to determine whether this protein is indeed enzymatically active. Previously, human XT-I has been expressed in an active soluble form in wild-type Chinese hamster ovary cells (15) and insect cells (27), as well as in a full-length form in HEK-293 cells (23). On the other hand, others have used the Caenorhabditis enzyme to rescue the defect in the pgsA-745 xylosyltransferase-deficient Chinese hamster ovary cell line (18); specifically, an unknown genetic defect in this cell line results in an xylosyltransferase activity in crude cellular extracts at least 15-fold lower than in the parental line and, as a result, in an incorporation of sulphate into glycosaminoglycan which is 2% of normal levels (25). Therefore, as an alternative to Pichia whilst still using a xylosyltransferase-deficient host, we generated mammalian expression plasmids, containing the same insert sequences as the previously-used yeast vectors, and then employed these to transfect pgsA-745 cells.
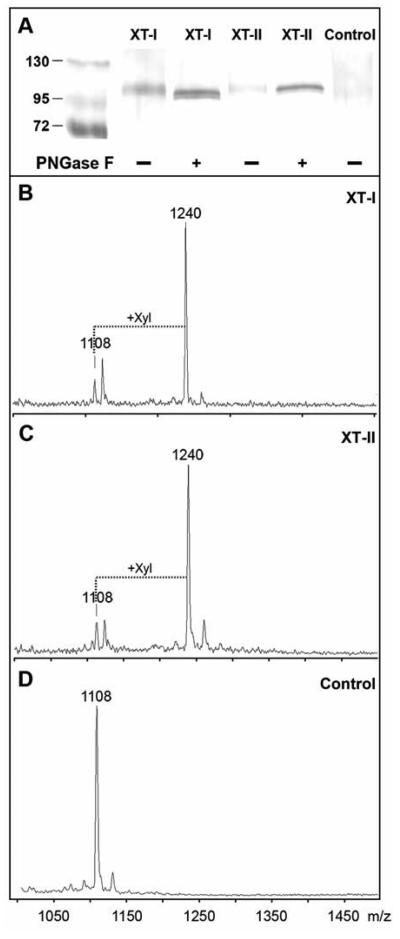
Upon transient expression, Western blotting of culture media indicated that for both XT-I and XT-II bands of Mr ~105000 were detected after PNGase F digestion. This is consistent with the expected size of the tagged soluble enzymes (Figure 1A); the tag contains residues 53-306 of protein A corresponding to Mr ~30000 and the soluble form of either enzyme theoretically confers Mr ~80000 to the overall size. The reduction in apparent Mr and the narrowing of the bands after PNGase F would be consistent with the presence of predicted N-glycosylation sites within the sequences of both the xylosyltransferase and protein A domains; both the XT-I and XT-II constructs contain the same number of predicted N-glycosylation sites. However, due to the low percentage change in molecular mass at this high range, no conclusion can be made as to the exact numbers of occupied N-glycosylation sites.
Figure 1. Expression of recombinant human xylosyltransferases in pgsA-745 cells.
(A) Western blotting of aliquots of culture supernatants of pgsA-745 cells transfected with either empty vector (Control), human XT-I or human XT-II were concentrated and, where indicated, subject to PNGase F digestion (+). Bands were detected using an anti-protein A antiserum as described. On the control blot, bands of Mr 26000 - 34000 were observed (region not shown) consistent with the expression of protein A alone in the control samples. Protein standards are indicated in kDa. (B-D) The culture supernatants were assayed overnight using a MALDI-TOF MS-based method. The syndecan peptide substrate (m/z 1108) was incubated in the presence of UDP-Xyl with culture supernatants of pgsA-745 cells transiently transfected with plasmids with either B) human XT-I, C) human XT-II or D) no insert. The increase in m/z of 132 is indicative of the transfer of xylose.
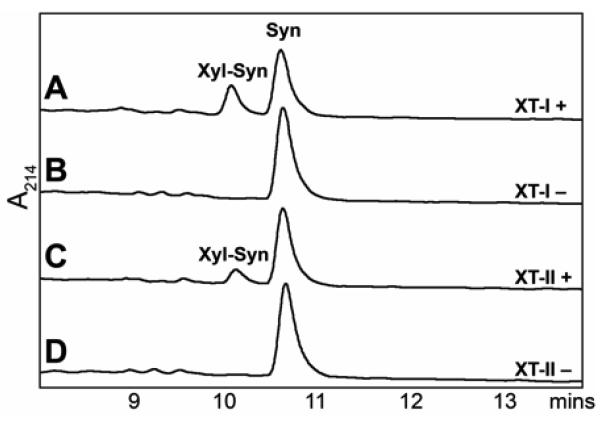
As expected, transfection with either the vectors encoding human XT-I (Figure 1B) or Drosophila OXT (data not shown) resulted in the appearance of xylosyltransferase activity in the media as judged by MALDI-TOF MS using the syndecan peptide; this activity was absent from the empty vector control (Figure 1D). Furthermore, transfection of human XT-II into these cells also resulted in a distinct xylosyltransferase activity (Figure 1C). Transfer of xylose to the peptide was also observed by RP-HPLC (Figure 2); based on the latter assays, the specific activities in the supernatants were estimated to be 0.55 and 0.23 mU/mg for XT-I and XT-II respectively. The lower specific activity of XT-II is consistent with the somewhat lower intensity of the XT-II band in the Western blot.
Figure 2. Assay of recombinant human xylosyltransferases by RP-HPLC.
The syndecan peptide substrate was incubated for 90 minutes with culture supernatants of pgsA-745 cells transiently transfected with plasmids with either human XT-I (A and B) or human XT-II (C and D) in the presence (A and C) or absence (B or D) of UDP-Xyl. The peaks eluting at ~10 and ~11 minutes were verified by MALDI-TOF MS to contain the xylosylated product and the substrate respectively.
Enzymatic characteristics of XT-I and XT-II
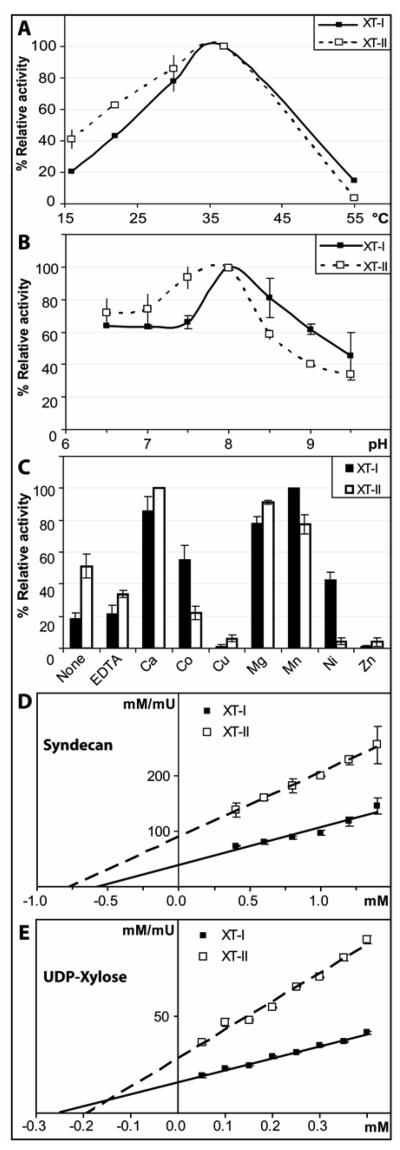
Using the HPLC-based method with the syndecan peptide, we examined the various characteristics of XT-II and compared to those of XT-I. As shown in Figure 3A, both XT-I and XT-II displayed significant activity in the range 16 - 37 °C; the optimum of 37 °C is the same as that of the Pichia-expressed XT-I (19) and is as expected for a mammalian enzyme. The pH optima of both enzymes with both buffers employed was fairly broad, but with a peak around pH 7.5-8.0 in both cases (Figure 3B); both enzymes displayed significant activity at pH 6.5, which contrasts with the apparent lack of activity of the Pichia-expressed XT-I using AMPD at pH 6.5 (19). There is no difference, though, between the actual optima of XT-I expressed in the different systems; these values, however, are still somewhat higher than that which may be expected for a Golgi enzyme, considering the pH inside this organelle has been estimated in one study as being pH 6.2 ± 0.4 (28). Others have found pH optima for natural forms of mammalian xylosyltransferases of between pH 6.5 and 7.5 (29-31), although it is not known whether these native forms were mixtures of both XT-I and XT-II or not.
Figure 3. Characterisation of recombinant human xylosyltransferases.
RP-HPLC based assays using the syndecan peptide substrate were performed for 90 minutes with culture supernatants of pgsA-745 cells transfected with XT-I and XT-II either A) at different temperatures, B) with AMPD buffers with different pH values and C) with different divalent cations. Data from duplicate experiments were normalised so that 100% corresponds to the condition resulting in the highest activity. The dependence of reaction rate for XT-I and XT-II with respect to either (D) syndecan peptide acceptor concentration or (E) UDP-Xyl donor concentration was measured using RP-HPLC based assays; the assays were performed in duplicate and the averaged data analysed using Hanes' plots are shown. For all panels, the individual data-points are indicated by the error bars.
The cation dependency was also broadly comparable for the both XT-I expressed in the Pichia or mammalian systems (Figure 3C): the highest activities were again observed with Ca(II), Co(II), Mg(II) and Mn(II). For XT-II, the pattern was similar, except that the activity in the presence of Co(II) was lower than that in the presence of EDTA and that XT-II, in contrast to the XT-I expressed in the mammalian system, is almost completely inhibited by Ni(II). Both enzymes were still active in the absence of any added cation and in the presence of EDTA. Recently, this lack of a necessity for a divalent cation has been structurally explained for an enzyme, core 2 β1,6-N-acetylglucosaminyltransferase, from the same glycosyltransferase family (CAZy family 14); indeed, all members of this enzyme family lack the degenerate ‘DXD’ sequence motif involved in metal ion binding and catalysis in other so-called ‘GT-A’ glycosyltransferases (32). Although, mammalian xylosyltransferase sequences actually contain an Asp/Glu-Trp-Asp sequence necessary for the activity of XT-I (33), it does not correspond to a canonical ‘DXD’ motif. It is located in the unique C-terminal region, which displays no homology to other glycosyltransferases, and we assume it does not play a direct role in the catalytic mechanism or in metal ion binding.
Finally, the substrate concentration dependency of XT-I and XT-II was examined (Figure 3D and 3E); the apparent Km values for UDP-xylose were determined to be in the region of 250 μM; these values are somewhat higher than the value of 100 μM found with the Drosophila enzyme using the same method (17), but similar to the 300 μM estimated for the Pichia-expressed form of XT-I (data not shown). In comparison, values of between 10 and 180 μM were found with various native mammalian enzymes (31,34-36), whereas ~1 μM was found by others for a form of XT-I expressed in insect cells and assayed using a protein acceptor (33). For syndecan, the Km values were in the region of 1 mM, which are comparable to those found for the same substrate with Caenorhabditis and Drosophila xylosyltransferases expressed in Pichia (17,19). Considering that the various parameters, including Km values, for XT-I and XT-II are approximately the same, there is no obvious enzymological reason for the previous lack of success in detecting the latter's activity; we can only assume some aspect of the vector construction, exact insert sequence, promoter or cell line employed plays a role.
Use of alternative substrates by XT-II
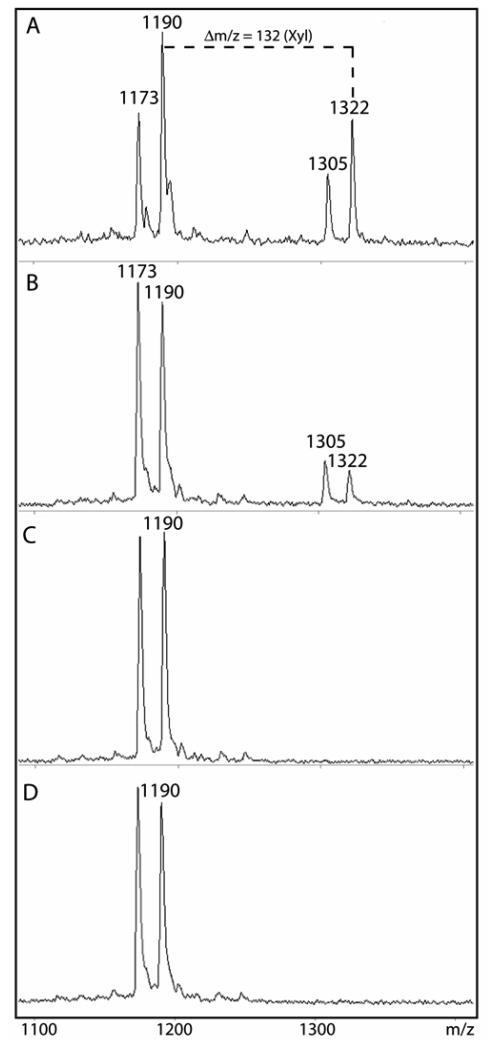
Previously, we have assayed recombinant xylosyltransferases from vertebrate (XT-I) and invertebrate (OXT and SQV-6) sources with a variety of peptide substrates: i.e., those based on portions of Drosophila syndecan, human bikunin or human perlecan (17,19). As shown above, an initial hypothesis that XT-II may require some particular unique substrate (19) was disproven in the course of the present study, since the syndecan peptide was a substrate when this enzyme was expressed in pgsA-745 cells. However, it was still of interest to test XT-II with the bikunin and perlecan substrates. Indeed, as for XT-I, transfer of xylose by XT-II was observed with the bikunin peptide as judged by the appearance of a peak with m/z 1322 (Figure 4A). Furthermore, encouraged by the results with XT-II in the mammalian expression system, we re-attempted the expression of XT-II in Pichia and, this time, a significant transfer of xylose to the bikunin peptide was also observed (Figure 4B). In controls (empty vector or no donor) no species with m/z 1322 was present (Figures 4C and D). This result is an independent indication that XT-II is indeed truly active, since yeasts are not known to possess an endogenous peptide O-xylosyltransferase.
Figure 4. Assay of recombinant human xylosyltransferase II expressed in mammalian and yeast systems using the bikunin peptide.
Xylosyltransferase activity of concentrated supernatants of either (A) pgsA-745 or (B) Pichia transformed with vectors encoding XT-II was measured overnight at 37 °C using the bikunin peptide substrate in the presence of UDP-Xyl, HEPES buffer, pH 8, and Mn(II). Controls with either (C) Pichia-expressed XT-II incubated in the absence of UDP-Xyl or of (D) Pichia transformed with empty vector were also performed. The primary xylosylation product has an m/z of 1322. In all cases, a secondary reaction resulting in a shift in m/z of −17 (species of 1173 and 1305) was also, as previously described (Ref. 19), observed and is presumed to be due to the conversion of the N-terminal glutamine residue of both the acceptor and product to pyroglutamate; this modification has no obvious effect on the xylosyltransferase reaction.
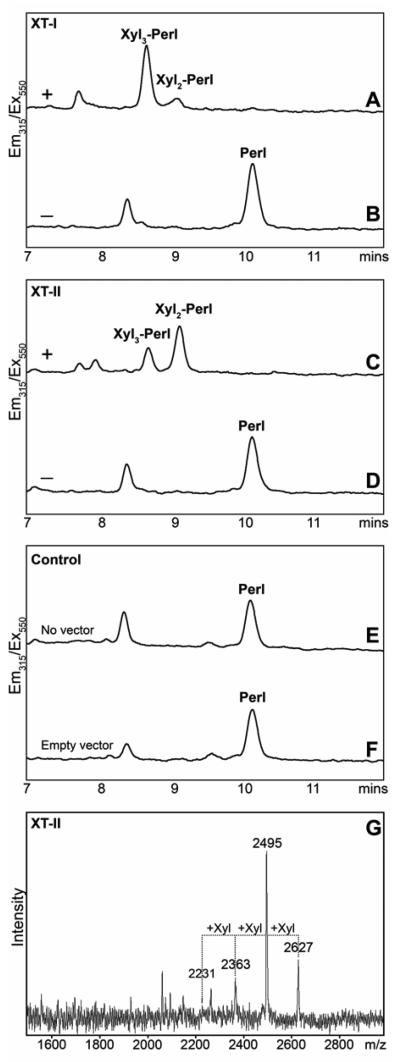
In the case of the perlecan substrate, we decided to develop an HPLC-based assay; in this case, the peptide was modified by dansylation to yield a fluorescently-tagged form, whose detection was expected not be subject to interference by other substances in the culture medium. Initial tests with recombinant Drosophila OXT showed that the dansylated perlecan peptide was a suitable substrate for sensitive detection of xylosyltransferase activity (data not shown). Thus, we also tested this substrate with supernatants of cells transfected with the XT-I and XT-II constructs. In both cases, a number of products were observed by RP-HPLC (Figure 5A and C), which were not present in incubations with the control supernatants (Figure 5E and F) or in the absence of the donor substrate (Figure 5B and D). The various fractions were collected and analysed by MALDI-TOF MS either separately (see annotations on Figure 5A and C) or as a pool (as shown for the incubation with XT-II; Figure 5G).
Figure 5. Assay of recombinant human xylosyltransferases with a peptide containing multiple acceptor sites.
Xylosyltransferase activity was measured by RP-HPLC (A-F) or MALDI-TOF MS (G) after incubation of the dansylated perlecan acceptor substrate with concentrated culture supernatants of pgsA-745 cells transfected with either human XT-I (A,B) or human XT-II (C,D,G), untransfected cells (E) or cells transfected with the pPA-TEV vector lacking an insert (F), in the presence (A,C,E,F) or absence (B,D) of UDP-xylose for 12 hours. In order to minimise the appearance of a degraded form the substrate (eluting ~1.5 minutes earlier), these assays were performed in the presence of EDTA. The xylosylation status of the annotated individual HPLC peaks was verified after collection by MALDI-TOF MS.
In contrast to transfer of only two xylose residues to the non-dansylated form of the perlecan peptide in incubations of either the mammalian-expressed XT-I and XT-II (data not shown) or the Pichia-expressed XT-I and OXT (19), up to three xylose residues were transferred by both XT-I and XT-II to the dansylated perlecan peptide; this corresponds to the number of Ser-Gly motifs in this peptide. We assume that the modification of the N-terminus by the dansyl group facilitates transfer to the first Ser-Gly motif (this serine residue being the fourth residue of the peptide); in the case of the non-dansylated form xylosylated with Pichia-expressed enzymes, this residue does not, as shown by tandem MS, serve as a xylosylation acceptor site (19). These data may be compatible with the effects of peptide length on the degree of xylosylation previously noted by others (29,37) and show that relatively small changes in a peptide substrate can have significant effects on the results obtained.
Conclusion
In this study, we have shown that both isoforms of human xylosyltransferase, XT-I and XT-II are indeed active enzymes when expressed in the Chinese hamster ovary mutant pgsA-745 cell line, thereby showing activity of the latter for the first time; the demonstration of XT-II activity also in recombinant Pichia confirms this result and suggests that the activity of the supernatants of the transformed pgsA-745 cells is indeed due to XT-II activity and not due to some indirect rescue of the endogenous defective xylosyltransferase gene(s). Thus, now knowing that both xylosyltransferase isoforms are active, while assuming their relatively ubiquitous expression (15), it may be that many or most mammalian cells have two enzymes capable of initiating chrondroitin and heparan sulphate biosynthesis, although the situation in, e.g., wild-type Chinese hamster ovary cells is unknown. On the other hand, there is only one isoform of the next enzyme in the glycosaminoglycan biosynthetic pathway (38), β4Gal-TVII, and it is interesting to note that some cases of Ehlers-Danlos syndrome, a connective tissue disorder, are associated with mutations in the β4Gal-TVII gene, which result in reduced enzymatic activity and defective proteoglycan glycosylation (39). This raises the issue as to whether mutations in one xylosyltransferase gene will result in strong phenotypes or whether only ablation of both will result in lethality in mammals. This is in contrast to the situation in invertebrates, which generally only have a single xylosyltransferase gene, but may be akin to the situation with another Golgi enzyme pair, mannosidase II and mannosidase IIx; in the case of the latter, although the single knock-outs do show either immunological or reproduction defects, both mannosidase genes must be ablated for either in utero or postnatal lethality to occur in the mouse (40). Perhaps, the possession of two xylosyltransferases with similar activities is an evolutionary accident or is indeed a reflection of an essential role for proteoglycans in many developmental and physiological processes in mammals.
ACKNOWLEDGEMENTS
The authors thank Dr. James Rini for the pA-TEV vector and for discussion of DXD motifs, Dr. Jeffrey Esko for the pgsA-745 Chinese hamster ovary cell line, Katharina Paschinger for reading the manuscript, Dr. Dubravko Rendić for help with some figures and Martin Gutternigg for performing some of the MALDI-TOF MS measurements.
Footnotes
This work was supported in part by grants from the Hochschuljubliäumsstiftung der Stadt Wien (H-1114) and the Fonds zur Förderung der wissenschaftlichen Forschung (P18447) to IBHW.
Abbreviations: AMPD, 2-amino-methyl-1,3-propanediol; HEPES, N-(2-hydroxyethyl)-piperazine-N′-2-ethanesulfonic acid; MALDI-TOF, matrix-assisted laser-desorption ionisation/time-of-flight; MS, mass spectrometry; RP-HPLC, reverse phase high pressure liquid chromatography; XT-I, human xylosyltransferase I; XT-II, human xylosyltransferase II.
REFERENCES
- 1.Selleck SB. Trends Genet. 2000;16:206–212. doi: 10.1016/s0168-9525(00)01997-1. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, Perrimon N. Glycoconj J. 2002;19:363–368. doi: 10.1023/A:1025329323438. [DOI] [PubMed] [Google Scholar]
- 3.Mulloy B. An Acad Bras Ciênc. 2005;77:651–664. doi: 10.1590/s0001-37652005000400007. [DOI] [PubMed] [Google Scholar]
- 4.Selleck SB. Sci STKE. 2006;2006:pe17. doi: 10.1126/stke.3292006pe17. [DOI] [PubMed] [Google Scholar]
- 5.Herman T, Horvitz HR. Proc. Natl. Acad. Sci. USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berninsone PM, Hirschberg CB. Glycoconj J. 2002;19:325–330. doi: 10.1023/A:1025364820713. [DOI] [PubMed] [Google Scholar]
- 7.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 8.Duncan G, McCormick C, Tufaro F. J. Clin. Invest. 2001;108:511–516. doi: 10.1172/JCI13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Development. 2005;132:5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petitou M, Casu B, Lindahl U. Biochimie. 2003;85:83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
- 11.Wilson IBH. Cell Mol Life Sci. 2004;61:794–809. doi: 10.1007/s00018-003-3278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebner EE, Hall CW, Neufeld EF. Arch. Biochem. Biophys. 1966;116:391–398. doi: 10.1016/0003-9861(66)90045-2. [DOI] [PubMed] [Google Scholar]
- 13.Grebner EE, Hall CW, Neufeld EF. Biochem. Biophys. Res. Commun. 1966;22:672–677. doi: 10.1016/0006-291x(66)90199-9. [DOI] [PubMed] [Google Scholar]
- 14.Robinson HC, Telser A, Dorfman A. Proc. Natl. Acad. Sci. USA. 1966;56:1859–1865. doi: 10.1073/pnas.56.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn J, Götting C, Schnolzer M, Kempf T, Brinkmann T, Kleesiek K. J. Biol. Chem. 2001;276:4940–4947. doi: 10.1074/jbc.M005111200. [DOI] [PubMed] [Google Scholar]
- 16.Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. J. Mol. Biol. 2000;304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- 17.Wilson IBH. J. Biol. Chem. 2002;277:21207–21212. doi: 10.1074/jbc.M201634200. [DOI] [PubMed] [Google Scholar]
- 18.Hwang H-Y, Olson S, Brown JR, Esko JD, Horvitz HR. J. Biol. Chem. 2003;278:11735–11738. doi: 10.1074/jbc.C200518200. [DOI] [PubMed] [Google Scholar]
- 19.Brunner A, Kolarich D, Voglmeir J, Paschinger K, Wilson IBH. Glycoconjugate J. 2006;23:543–554. doi: 10.1007/s10719-006-7633-z. [DOI] [PubMed] [Google Scholar]
- 20.Schön S, Prante C, Müller S, Schottler M, Tarnow L, Kuhn J, Kleesiek K, Götting C. Kidney Int. 2005;68:1483–1490. doi: 10.1111/j.1523-1755.2005.00561.x. [DOI] [PubMed] [Google Scholar]
- 21.Schön S, Schulz V, Prante C, Hendig D, Szliska C, Kuhn J, Kleesiek K, Götting C. J Med Genet. 2006;43:745–749. doi: 10.1136/jmg.2006.040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schön S, Huep G, Prante C, Müller S, Christ R, Hagena FW, Kuhn J, Kleesiek K, Götting C. Osteoarthritis Cartilage. 2006;14:442–448. doi: 10.1016/j.joca.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Schön S, Prante C, Bahr C, Kuhn J, Kleesiek K, Götting C. J Biol Chem. 2006;281:14224–14231. doi: 10.1074/jbc.M510690200. [DOI] [PubMed] [Google Scholar]
- 24.Nuwayhid N, Glaser JH, Johnson JC, Conrad HE, Hauser SC, Hirschberg CB. J. Biol. Chem. 1986;261:12936–12941. [PubMed] [Google Scholar]
- 25.Esko JD, Stewart TE, Taylor WH. Proc. Natl. Acad. Sci. USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pak JE, Rini JM. Methods Enzymol. 2006;416:30–48. doi: 10.1016/S0076-6879(06)16003-6. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn J, Müller S, Schnolzer M, Kempf T, Schön S, Brinkmann T, Schottler M, Götting C, Kleesiek K. Biochem Biophys Res Commun. 2003;312:537–544. doi: 10.1016/j.bbrc.2003.10.157. [DOI] [PubMed] [Google Scholar]
- 28.Wu MM, Grabe M, Adams S, Tsien RY, Moore H-PH, Machen TE. J. Biol. Chem. 2001;276:33027–33035. doi: 10.1074/jbc.M103917200. [DOI] [PubMed] [Google Scholar]
- 29.Pfeil U, Wenzel K-W. Glycobiology. 2000;10:803–807. doi: 10.1093/glycob/10.8.803. [DOI] [PubMed] [Google Scholar]
- 30.Guillaumond M, Louisot P. Int. J. Biochem. 1975;6:491–496. [Google Scholar]
- 31.Stoolmiller AC, Horwitz AL, Dorfman A. J. Biol. Chem. 1972;247:3525–2532. [PubMed] [Google Scholar]
- 32.Pak JE, Arnoux P, Zhou S, Sivarajah P, Satkunarajah M, Xing X, Rini JM. J Biol Chem. 2006;281:26693–26701. doi: 10.1074/jbc.M603534200. [DOI] [PubMed] [Google Scholar]
- 33.Götting C, Müller S, Schöttler M, Schön S, Prante C, Brinkmann T, Kuhn J, Kleesiek K. J Biol Chem. 2004;279:42566–42573. doi: 10.1074/jbc.M401340200. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz NB, Dorfman A. Arch. Biochem. Biophys. 1975;171:136–144. doi: 10.1016/0003-9861(75)90016-8. [DOI] [PubMed] [Google Scholar]
- 35.Campbell P, Jacobsson I, Benzing-Purdie L, Rodén L, Fessler JH. Anal. Biochem. 1984;137:505–516. doi: 10.1016/0003-2697(84)90119-2. [DOI] [PubMed] [Google Scholar]
- 36.Kearns AE, Campbell SC, Westley J, Schwartz NB. Biochemistry. 1991;30:7477–7483. doi: 10.1021/bi00244a016. [DOI] [PubMed] [Google Scholar]
- 37.Brinkmann T, Weilke C, Kleesiek K. J. Biol. Chem. 1997;272:11171–11175. doi: 10.1074/jbc.272.17.11171. [DOI] [PubMed] [Google Scholar]
- 38.Esko JD, Weinke JL, Taylor WH, Ekborg G, Rodén L, Anantharamaiah G, Gawish A. J Biol Chem. 1987;262:12189–12195. [PubMed] [Google Scholar]
- 39.Seidler DG, Faiyaz-Ul-Haque M, Hansen U, Yip GW, Zaidi SH, Teebi AS, Kiesel L, Gotte M. J Mol Med. 2006;84:583–594. doi: 10.1007/s00109-006-0046-4. [DOI] [PubMed] [Google Scholar]
- 40.Akama TO, Nakagawa H, Wong NK, Sutton-Smith M, Dell A, Morris HR, Nakayama J, Nishimura S-I, Pai A, Moremen KW, Marth JD, Fukuda MN. Proc Natl Acad Sci U S A. 2006;103:8983–8988. doi: 10.1073/pnas.0603248103. [DOI] [PMC free article] [PubMed] [Google Scholar]