Abstract
In many invertebrates and plants, the N-glycosylation profile is dominated by truncated paucimannosidic N-glycans, i.e., glycans consisting of a simple trimannosylchitobiosyl core often modified by core fucose residues. Even though they lack antennal N-acetylglucosamine residues, the biosynthesis of these glycans requires the sequential action of GlcNAc transferase I, Golgi mannosidase II and, finally, β-N-acetylglucosaminidases. In Drosophila, the recently characterised enzyme encoded by the fused lobes (fdl) gene specifically removes the non-reducing N-acetylglucosamine residue from the α1,3-antenna of N-glycans. In the present study, we examined the products of five β-N-acetylhexosaminidase genes from Caenorhabditis elegans (hex-1 to hex-5, corresponding to reading frames T14F9.3, C14C11.3, Y39A1C.4, Y51F10.5 and Y70D2A.2) in addition to three from Arabidopsis thaliana (AtHEX1, AtHEX2 and AtHEX3, corresponding to reading frames At1g65590, At3g55260 and At1g05590). Based on homology, the Caenorhabditis HEX-1 and all three Arabidopsis enzymes are members of the same sub-family as the aforementioned Drosophila fused lobes enzyme, but either act as chitotriosidases or non-specifically remove N-acetylglucosamine from both N-glycan antennae. The other four Caenorhabditis enzymes a members of a distinct sub-family; nevertheless, two of these enzymes displayed the same α1,3-antennal specificity as the fused lobes enzyme. Furthermore, a deletion of part of the Caenorhabditis hex-2 gene drastically reduces the native N-glycan-specific hexosaminidase activity in mutant worm extracts and results in a shift in the N-glycan profile, which is a demonstration of its in vivo enzymatic relevance. Based on these data, it is hypothesised that the genetic origin of paucimannosidic glycans in nematodes, plants and insects involves highly-divergent members of the same hexosaminidase gene family.
Keywords: Arabidopsis, Caenorhabditis, hexosaminidase, paucimannosidic N-glycans
Introduction
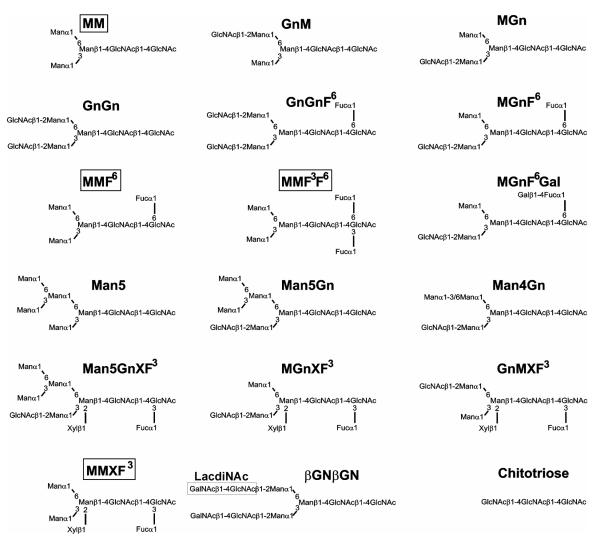
The N-linked oligosaccharides of invertebrates and plants display a number of features not observed in those of vertebrates (1); the presence of immunogenic moieties such as core α1,3-fucose and β1,2-xylose is widespread in ‘lower’ multicellular organisms, as is the tendency for core modified N-glycans to have mannose residues at the non-reducing termini. These oligosaccharides based on a trimannosyl core (see, e.g. the so-called MM, MMF6 or MMXF3 structures shown in Scheme I) have been named ‘paucimannosidic’ or ‘truncated’ in order to distinguish them from oligomannosidic, hybrid and complex N-glycans; particularly the latter are seen as a hallmark of mammals and other vertebrates.
Scheme I. Structures of selected glycans referred to in this study.
The order of the letters M (mannose), Gn (GlcNAc) and GN (GalNAc) indicates the antenna (the ‘upper’ 1,6 or the ‘lower’ 1,3) on which this sugar is the terminal residue; for modifications of the core F3 indicates a core α1,3-fucose, F6 a core α1,6-fucose, F6Gal a galactosylated core α1,6-linked fucose and, on plant-type glycans, X a β1,2-linked xylose. GnGn and GnGnF6 represent simple complex biantennary N-glycans, whereas structures with boxed names are referred to as paucimannosidic.
The reactions resulting in the biosynthesis of complex N-glycans in mammals have been well characterised (2) and begin with the transfer of Glc3Man9GlcNAc2 by oligosaccharyltransferase, followed by both removal and addition of glycan residues. As summarised in Scheme II, key events in mammalian N-glycan biosynthesis are the sequential actions of N-acetylglucosaminyltransferase I (GlcNAc-TI; EC 2.4.1.143) and Golgi mannosidase II (EC 3.2.1.114), whereby the N-acetylglucosamine residue transferred to the α1,3-antenna by GlcNAc-TI is retained in the finally processed glycan.
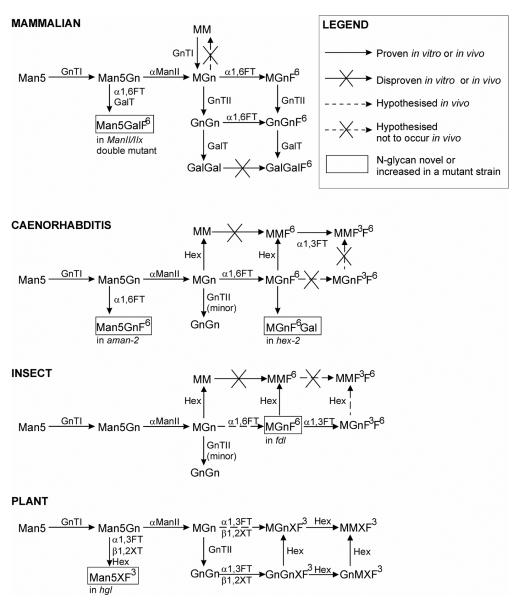
Scheme II. Putative pathways towards paucimannosidic glycans in nematodes, plants and insects.
Only the major pathways are shown for mammals (which lack paucimannosidic glycans, but have especially biantennary complex glycans), nematodes, insects and plants, as well as pathways which are revealed in strains lacking either Golgi mannosidase II from mammals (mannosidase II/IIx double mutant emybros), nematodes (aman-2) and plants (hgl) and hexosaminidase mutants from nematodes (hex-2; this study) and insects (fdl). Whereas both fucosylated and non-fucosylated paucimannosidic structures naturally dominate in nematodes and insects (especially MM and MMF6), truncated N-glycans in plants carry xylose and fucose residues; the hexosaminidases from nematodes and insects only remove the GlcNAc attached to the α1,3-antenna, whereas those from plants display no distinct arm preference. On the other hand, core α1,6-fucosyltransferases from nematodes and insects, core α1,3-fucosyltransferases from plants and insects and β1,2-xylosyltransferases from plants cannot transfer to MM (in some cases, transfer to MGn or Man5Gn has been proven in vitro), whereas the nematode core α1,3-fucosyltransferase can transfer, at least in vitro, to MM (but not MGn). The action of the hexosaminidases in nematodes, plants and insects is therefore necessary to explain the range of N-glycan structures observed. No glycans from an insect Golgi mannosidase II mutant or a plant hexosaminidase mutant have yet been analysed; thus, the in vivo affect of such defects is still unknown.
A simplistic explanation of the origin of paucimannosidic glycans in invertebrates and plants would be that truncated glycans, as in some protozoans (3), are transferred by oligosaccharyltransferase. However, it has been shown that plants and insects synthesise the full Glc3Man9GlcNAc2 structure (4,5). Furthermore, various knockouts show that normal glycan processing in these species is, as in mammals, dependent on the action of GlcNAc-TI (6-8). Enzymes dependent on GlcNAc-TI include Golgi mannosidase II from plants, insects and nematodes (9-11), core α1,3-fucosyltransferases from plants and insects (12,13) and β1,2-xylosyltransferases from plants, snails and trematodes (14-16) as well as core α1,6-fucosyltransferases from invertebrates in general (17). Also, genes encoding GlcNAc-TI and GlcNAc-TII are known to be present in plants, insects and nematodes (6,8,18-20). Thus, in order to explain the ‘paradox’ that the residue transferred by GlcNAc-TI is absent in many of the N-glycans isolated from invertebrates and plants, whereas, e.g., core fucose, is present, the concept of a ‘processing’ hexosaminidase in these lower eukaryotes must be invoked.
Some years ago, a membrane-bound hexosaminidase was enriched from a microsomal fraction of insect cells (21). This enzyme specifically removed the GlcNAc transferred by GlcNAc-TI; recently, a member of the aforementioned glycoside hydrolase family 20, encoded by the Drosophila melanogaster fused lobes (fdl) gene was verified to possess such an enzymatic activity and be localised within the secretory pathway (22). A similar activity was also found in Caenorhabditis elegans microsomes (23); on the other hand, plants are assumed to have non-specific vacuolar hexosaminidases which trim any terminal GlcNAc residue (24). However, unlike in insects, this trimming does not take place in the Golgi or secretory pathway and, therefore, secreted plant glycoproteins, such as laccase, contain extended structures with ‘Lewis a’ epitopes (25). The genetic origin of the nematode and plant hexosaminidase activities has remained unknown.
In order to investigate the hexosaminidases of both nematodes and plants (specifically Caenorhabditis elegans and Arabidopsis thaliana) in more detail, we set out to examine their family 20 glycoside hydrolases. A total of five nematode and three plant hexosaminidase cDNAs were engineered for expression of soluble forms of the corresponding proteins in the yeast Pichia pastoris. Whereas two A. thaliana hexosaminidases possessed the expected plant-type ‘random’ specificity for terminal GlcNAc residues, it was found that two members of the novel nematode hexosaminidase subfamily removed specifically the α1,3-antennal GlcNAc residue from N-glycans. Thus, it appears that non-orthologous members of glycosidase family 20 are responsible for the appearance of paucimannosidic or truncated N-glycans in insects, plants and nematodes.
Materials and Methods
Cloning and expression of hexosaminidase homologues
Total RNA was prepared from a mixed population of C. elegans (wild-type N2) with Trizol (Invitrogen, Carlsbad, USA) reagent and subject to reverse transcription using Superscript III (Invitrogen) with T18 as primer. Partial reading frames of hexosaminidase cDNAs suitable for soluble expression in P. pastoris were generated by PCR using Expand polymerase with C. elegans cDNA and the primers: CeHex1/1/PstI (aaactgcaggcagagacgacccgga) with CeHex1/2/KpnI (ggggtaccttaaagctctgtcttcgttg), CeHex2/1/PstI (aaactgcaggacatatgacctcatcatacccg) with CeHex2/2/SacII (tccccgcggtcatttcttgattgggaaatgc), CeHex3/1/PstI (aactgcagagttcaaatcgacgacaact) with CeHex3/2/KpnI (gggtacctatgtacaagttttttcgctt), CeHex4/1/PstI (aactgcagccaatgatcgatccagttat) with CeHex4/2/KpnI (gggtacctcaattagtaatctctgttc) and CeHex5/1/PstI (aactgcaggacaaaggtctatagttcattttg) with CeHex5/2/ KpnI (ggggtacctcacctccggaaccacg).
For isolation of the full open reading frames, the forward primers CeHex1/5 (atgcgacttttaattcccatac), CeHex2/5 (atgttcccgatgcggtgta), CeHex3/5 (atgcttcgaggttttttcgg), CeHex4/5 (atgcataaaatgtcgaaactc) and CeHex5/5 (atgtgcaacatttttcagattg) were used in conjunction with the aforementioned reverse primers and the fragments were sub-cloned into the pGEM-T vector (Promega, Madison, USA) and sequenced.
In the case of the A. thaliana hexosaminidases, partial cDNAs were cloned after RT-PCR of Trizol-extracted RNA from the Columbia ecotype with the primers AtHex1/1/EcoRI (cggaattcatagagaggttgaggatt) with AtHex1/2/XbaI (gctctagatcactgagcgagacaagaac), AtHex2/1/EcoRI (cggaattctcgccggctgattctcct) with AtHex2/2/XbaI (gctctagatcactgagcatagcaagagc) and AtHex3/1/EcoRI (cggaattcaacatttggccaaagccg) with AtHex3/2/XbaI (gctctagattattgatcttgaagagcacc).
The cDNA fragments were purified from the PCR reactions using the GFX DNA purification kit (GE Healthcare). Both fragment and vector were digested with the relevant restriction enzymes prior to ligation of the fragments into pPICZαB or C, either in the form purchased from the supplier (Invitrogen) or in a form in which the vector was modified by inverse PCR to include a region encoding a FLAG-tag just upstream of the multiple cloning site but downstream of the region encoding the Ste13 signal cleavage site. Ligation products were transformed into E. coli TOP10F' prior to selection on zeocin, plasmid preparation and sequencing. The expression vectors were linearised and transformed into P. pastoris (GS115 strain), colonies were selected on zeocin and expression performed with methanol induction at 16 or 30 °C as previously described (26). N-terminally FLAG-tagged forms of the C. elegans hexosaminidases HEX-1, HEX-3 and HEX-4 were detectable after Western blotting using the anti-FLAG M2 monoclonal antibody (1:10000) and alkaline-phosphatase conjugated anti-mouse IgG (1:10000).
In silico analysis
The in silico analysis of protein sequences was performed using the DIALIGN alignment program at the BibiServ server of the Universität Bielefeld3 (27) using default parameters.
Hexosaminidase assays
Total nematode extracts were prepared as previously described from wild-type or mutant C. elegans (28), whereas the microsomal preparation resulted from a low-percentage Triton wash of wild-type nematode membranes followed by high-speed centrifugation (23). Crude supernatants of P. pastoris expressing recombinant C. elegans and A. thaliana hexosaminidases, as well as ammonium sulphate fractions (between 50 and 90% saturation) of these supernatants, were also assayed using a range of enzymatic assays. The majority of the recombinant enzymes were stable for several months at either 4 °C or −80 °C, but not at −20 °C. A supernatant of Pichia expressing C. elegans Golgi mannosidase II was prepared during a previous study (10).
For the standard assay with p-nitrophenylglycosides, 5 μl of sample was incubated in a microtitre well with 25 μl McIlvaine citrate-phosphate buffer (pH 3.5-8.0), 2.5 μl of 100 mM p-nitrophenylglycoside in dimethylsulphoxide and 17.5 μl water for 1 hr at 37 °C; in some cases, diluted enzyme was used in order to attain linear substrate turnover over a period of up to two hours. The reaction was stopped by addition of 250 μl 0.4 M glycine-NaOH, pH 10.4, and the A405 was read using an ELISA plate reader. Three hexosaminidase inhibitors were employed in this study: N-acetylcastanospermine (6-acetamido-6-deoxycastanospermine; Industrial Research Ltd, New Zealand), 2-acetamido-1,2-dideoxynojirimycin (2-acetamido-1,2,5-trideoxy-1,5-imino-D-glucitol; the kind gift of Dr. Arnold Stütz) and O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate (PUGNAc; Toronto Research Chemicals, Canada). For oligosaccharide substrates, dabsylglycopeptides (0.25 nmol) or pyridylaminooligosaccharides (0.1 nmol) were used. The incubation mixture was then subject to either MALDI-TOF MS (dabsyl) or RP-HPLC (pyridylamino) as previously described (10). The dabsyl-βGNβGN substrate was prepared using a dabsylated fibrin glycopeptide remodelled sequentially with sialidase, galactosidase and finally bovine milk β1,4-galactosyltransferase, using UDP-GalNAc as donor (29). For an explanation of the abbreviations for the glycan substrates, see Scheme I.
Analyses of wild-type and mutant glycans
N-glycans were prepared by peptide:N-glycosidase A-mediated release from peptic peptides and a portion of each preparation was subject to pyridylamination, RP-HPLC and MALDI-TOF MS (28,30). Pyridylaminated glycans of the tm2350 mutant were also subject to single or double enzymatic digestions with recombinant Streptococcus β-hexosaminidase (Sigma, St. Louis, USA; 50 mU, using the manufacturer’s reaction buffer), bovine kidney α-fucosidase (Sigma; 15 mU, in 50 mM ammonium acetate, pH 5) or Aspergillus β1,4-galactosidase (27 mU, in 50 mM sodium citrate, pH 4.6) further purified from a cruder commercial preparation (31), prior to re-analysis by RP-HPLC. Glycans were analysed by MALDI-TOF-MS (Ultraflex Tof/Tof, BrukerDaltonics, Bremen) using a 6-aza-2-thiothymine (ATT) matrix (5 mg/ml ATT in H2O) either in the MS or MS/MS mode. Normally 100 - 500 shots were summed. For external calibration, a peptide standard mixture (Bruker) was used.
Tissue-specific promoter analysis
The 2000 bp upstream of the ATG start codon of each C. elegans hex gene was isolated by PCR from genomic DNA using Expand polymerase and the following primer pairs: CeHex1/prom3/BamHI (cgggatcctaatcataataaggcttcgaac) with CeHex1/prom4/BamHI (cgggatccatcaccttaacttcatagatg), CeHex2/prom5/BamHI (cgggatccggcaattttatgatctatggta) with CeHex2/prom6/BamHI (cgggatccatttttttacatttgaggctaa), CeHex3/prom1/BamHI (cgggatccgagtatcacttcccgtcc) with CeHex3/prom2/BamHI (cgggatccatggcgacgtttattgca), CeHex4/prom11/BamHI (cgggatccggtttgccgttaacgttt) with CeHex4/prom8/BamHI (cgggatccattattgtatttgtattgtacaa) and CeHex5/prom9/BamHI (cgggatcctggaatttccatagcctg) with CeHex5/prom10/BamHI (cgggatccataatcataatcaaaaaagattaaa).
The reporter plasmid used was a modified form of the pPD95.67 (L2459) promoterless gfp vector modified, as previously described (10), by insertion of a HindIII/XbaI fragment carrying the unc-119 gene into its multiple cloning site to generate pPD95.67/unc-119. PCR fragments and the vector were digested with BamHI and ligated. Selected clones were sequenced and verified to contain the expected upstream regions in the correct orientation and were used to transform unc-119 (ed3) mutants with a particle gun (Bio-Rad). Individuals from integrated lines were examined by confocal laser scanning microscopy (Leica TCS SP2) with Ar-laser excitation at 488 nm and emission 500-540 nm using an HC Plans 10×/25 occular and HC PL Fluotar 63×/0.30 objective.
Results
Identification of hexosaminidase homologues in C. elegans and A. thaliana
Examination of the genome of C. elegans allows prediction of five putative hexosaminidases (T14F9.3, C14C11.3, Y39A1C.4, Y51F10.5 and Y70D2A.2), which we define as HEX-1, HEX-2, HEX-3, HEX-4 and HEX-5 respectively. These proteins have been defined in the CAZy database (32) as being members of glycoside hydrolase family 20; based on actual cloning of the cDNAs, they are predicted to be of Mr 55,000-70,000 and contain potential N-glycosylation sites. Indeed, HEX-1 was proven in a large survey of glycoproteins to be glycosylated at positions 351 and 460, whereas HEX-5 is glycosylated in vivo at position 218 (33,34). HEX-2, HEX-3 and HEX-5 also have dibasic motifs just N-terminal of the single hydrophobic domain, suggestive of a Golgi localisation or, at least, export from the ER (35), whereas HEX-1 has a C-terminal KTEL sequence, which could, of course, be a variant of the typical ER (KDEL) retrieval sequence.
The three A. thaliana hexosaminidase sequences identified in this study were previously included in the CAZy database (36); we hereby define them as AtHEX1 (At1g65590), AtHEX2 (At3g55260) and AtHEX3 (At1g05590). All three genes encode proteins of Mr ~60,000 with five predicted, generally non-conserved, N-glycosylation sites and N-terminal hydrophobic regions, which may constitute signal sequences or transmembrane domains; AtHEX2 has been identified in a proteomic study as being a vacuolar resident (37) whereas both AtHEX1 and AtHEX3 are predicted by the WoLF PSORT program to have a vacuolar localisation. AtHEX1 and AtHEX2 are 51% identical at the amino acid level, whereas AtHEX3 is more distantly related and displays only 29% identity to AtHEX1. Interestingly, the rice genome contains not only two genes homologous to the AtHEX1/AtHEX2 ‘pair’, but also two genes homologous to AtHEX3.
The hexosaminidase family 20 has two major branches
In order to align the hexosaminidase-related protein sequences from a variety of species, which appear not to be globally related but are only similar to one another in certain, narrow regions, the DIALIGN alignment program (27) had to be employed. Standard alignment algorithms (e.g., ClustalW using BLOSUM30 or PAM matrices) under variety of conditions failed to align the conserved regions of the sequences. Using DIALIGN, a His/Asn-Xaa-GlyAla/Cys/Gly/Met-Asp-Glu-Ala/Ile/Leu/Val sequence was found in all sequences (see Fig. 1A). The glutamate residue of this motif corresponds to the general acid/base involved in the catalytic mechanism of hexosaminidases (38).
Figure 1. Alignment and phylogenetic analysis of class 20 hexosaminidase sequences.
(A) Alignment of the region surrounding the His/Asn-Xaa-Gly-Ala/Cys/Gly/Met-Asp-Glu-Ala/Ile/Leu/Val motif conserved in all class 20 hexosaminidases; (B) phylogenetic tree analysis of class 20 hexosaminidases showing the division into two major subfamilies, one including the novel C. elegans hexosaminidases examined in this study and the other including previously-studied human, fungal and insect hexosaminidases. The following protein sequences were analysed by DIALIGN and Treeview programs: MmHexD: M. musculus hexosaminidase homologue D; CeHex1-5: C. elegans hexosaminidases 1-5; OiHex: Oceanobacillus iheyensis hexosaminidase homologue; CpHex: Clostridium perfringens hexosaminidase; SpmHex: Streptococcus pneumoniae StrH hexosaminidase; AtHex1-3: A. thaliana hexosaminidases 1-3; OsHex 1-4: O. sativa (rice) hexosaminidase homologues 1-4; NagA: Penicillium chrysogenum hexosaminidase; CpoHex: Coccidioides posadasii hexosaminidase; AoHexA: Aspergillus oryzae hexosaminidase; TnHEX: Trichoplusia ni hexosaminidase; HsHEXA/B: H. sapiens hexosaminidases A/B; DmFDL: D. melanogaster Fdl hexosaminidase; DmHEXO1/2: D. melanogaster hexosaminidases 1/2; DmHEX4: D. melanogaster hexosaminidase homologue 4; SfHEX1/2: S. frugiperda hexosaminidases 1/2; SpHEX: Streptomyces plicatus hexosaminidase; EsHEX: Enterobacter sp. Hexosaminidase; CfHEX: Cellulomonas fimi hexosaminidase
The phylogenetic analysis (Fig. 1B) suggests that the nematode HEX-1 belongs to the same sub-family as the human lysosomal α and β subunits and the plant AtHEX1, AtHEX2 and AtHEX3; this sub-family also includes three sequences (Hexo1, Hexo2 and Fdl) from D. melanogaster (22) as well as ascomycete hexosaminidases such as Penicillium NagA (39). Indeed, C. elegans HEX-1 and AtHEX1 display 36-38% identity over approximately 500 residues to the human hexosaminidase α and β subunits respectively. On the other hand, the other four C. elegans hexosaminidase homologues (see also Supplementary Data for an alignment) can be grouped together with one sequence each from D. melanogaster, mouse and man; members of this sub-family poorly align with the human and plant sequences. The six selected bacterial sequences, such as the well-studied Streptomyces plicatus SpHex (40), also clearly associate with one or the other subfamily (Fig. 1A), suggestive of an ancient origin for the two branches.
Expression of recombinant Caenorhabditis and Arabidopsis hexosaminidase homologues
cDNA fragments encoding the putative luminal domains of each C. elegans and A. thaliana hexosaminidase homologue (i.e., lacking putative cytosolic and transmembrane domains) were engineered into Pichia expression vectors as non-tagged and/or FLAG-tagged forms. In the case of the nematode HEX-1, HEX-3 and HEX-4, both Coomassie staining and anti-FLAG Western blotting of crude supernatants and ammonium sulphate fractions indicated expression of proteins of Mr 80000, 70000 and 55000, respectively; HEX-2 was only ever detected after SDS-PAGE by Coomassie staining (Mr ~60000) and it was considered possible that the FLAG-tag was removed by proteolysis, potentially at dibasic sites in the stem region. The expression of HEX-5 was, however, apparently below the detection limits of Western blotting and Coomassie staining, although a number of independent clones reproducibly showed an activity absent from control ‘empty vector’ supernatants. AtHEX1, AtHEX2 and AtHEX3 were seen as FLAG-tagged bands of around Mr 65000 in either 50% or 70% ammonium sulphate fractions of the supernatants of recombinant yeast (data not shown).
The initial enzymatic tests were performed with artificial chromogenic substrates (p-nitrophenyl-β-N-acetylgalactosaminide and p-nitrophenyl-β-N-acetylglucosaminide) and showed that the homologues were indeed enzymatically active and that no enzyme activity was present in the supernatants of Pichia transfected with ‘empty’ vector. Whereas HEX-1 was approximately equally active towards both substrates, HEX-2, HEX-3, HEX-4 and HEX-5 were most active (or only active) towards p-nitrophenyl-β-N-acetylgalactosaminide (see also Fig. 2); the latter hexosaminidases are indeed, other than the Streptococcus pneumoniae StrH enzyme (41), the first members of a new sub-division of family 20 to be enzymatically studied. The Km value of HEX-1 for p-nitrophenyl-β-N-acetylglucosaminide was estimated to be 1.1 mM, which can be compared to a previously-reported value of 0.45 mM with 4-methylumbelliferyl-β-N-acetylglucosaminide for a hexosaminidase activity in crude C. elegans extracts (42). The plant hexosaminidases AtHEX1, AtHEX2 and AtHEX3 were active with p-nitrophenyl-β-N-acetylglucosaminide as substrate (see also Fig. 3), but displayed only a minor activity towards p-nitrophenyl-β-N-acetylgalactosaminide (data not shown).
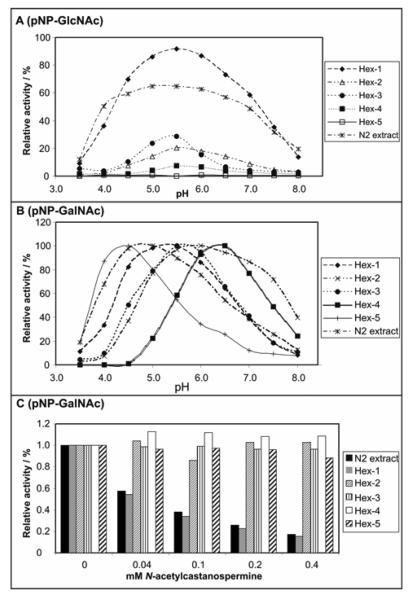
Figure 2. Enzymatic properties of Caenorhabditis class 20 hexosaminidases.
(A) pH dependency of activity towards p-nitrophenyl-β-N-acetylglucosaminide (pNP-GlcNAc) of native and recombinant C. elegans hexosaminidases assayed at 37 °C over a one-hour period using a range of McIlvaine buffers; (B) pH dependency of activity towards p-nitrophenyl-β-N-acetylgalactosaminide (pNP-GalNAc) of native and recombinant C. elegans hexosaminidases assayed at 37 °C over a one-hour period using a range of McIlvaine buffers; (C) sensitivity of native and recombinant C. elegans hexosaminidases assayed at 37 °C over a one-hour period at optimal pH using p-nitrophenyl-β-N-acetylgalactosaminide as substrate and various concentrations of N-acetylcastanospermine as potential inhibitor. Crude culture supernatants of yeast transformed with hex partial open reading frames (1:10 diluted in the case of hex-1 and hex-2) or an extract of wild-type N2 worms were used as the enzyme sources; in all cases, data was recalculated with the condition yielding the highest absorbance at 405 nm with p-nitrophenyl-β-N-acetylgalactosaminide as substrate being normalised to 100%.
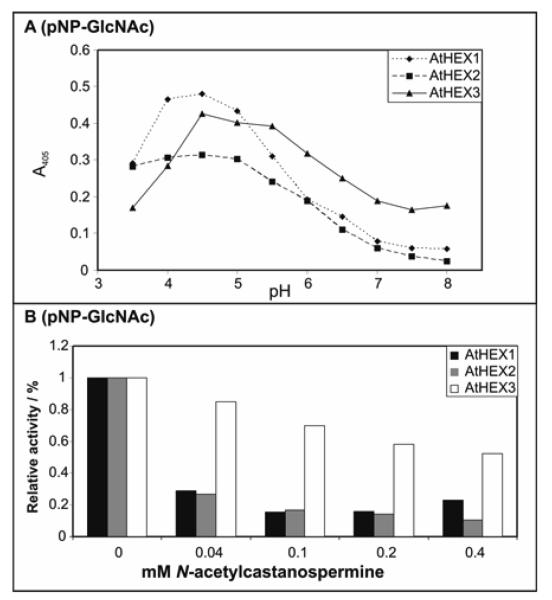
Figure 3. Enzymatic properties of Arabidopsis class 20 hexosaminidases.
(A) pH dependency of activity towards p-nitrophenyl-β-N-acetylglucosaminide (pNP-GlcNAc) of recombinant A. thaliana hexosaminidases assayed at 37 °C over a one-hour period using a range of McIlvaine buffers; (B) sensitivity of recombinant A. thaliana hexosaminidases assayed at 37 °C for one hour at optimal pH using p-nitrophenyl-β-N-acetylglucosaminide as substrate and various concentrations of N-acetylcastanospermine as potential inhibitor. Ammonium sulphate fractions of yeast culture supernatants were used as enzyme sources (70% saturation for AtHEX1 and AtHEX2, 50% saturation for AtHEX3).
Effects of pH, temperature and inhibitors on recombinant hexosaminidase activities
Using the artificial p-nitrophenyl substrates, the activities of the recombinant C. elegans HEX-1, -2, -3, -4 and -5 were compared with the activity in native C. elegans extract in terms of pH optimum within the linear range of product formation with respect to time. As measured in the presence of McIlvaine buffers, C. elegans HEX-1 to HEX-4 have pH optima of pH 5.5-6.5 (Fig. 2A and 2B), whereas HEX-5 was most active at pH 4.5 (only with p-nitrophenyl-β-N-acetylgalactosaminide; Fig. 2B). The pH dependence curve for the activity in native extract followed most closely that of recombinant HEX-1. In comparison, the Arabidopsis hexosaminidases had optimal activity at pH 4-5 (Fig. 3A); according to the literature, the well-known jack bean hexosaminidase has optimal activity at pH 5 with a citrate buffer (43).
Three inhibitors specific for hexosaminidases were employed with the nematode enzymes: N-acetylcastanospermine, 2-acetamido-1,2-dideoxynojirimycin and PUGNAc. For the first compound IC50 values of 0.4-1.6 μM have been reported with jack bean and various animal hexosaminidases (44), whereas for the latter two, respective Ki values of 140-600 μM and 0.1 μM have been reported (45,46). N-acetylcastanospermine and 2-acetamido-1,2-dideoxynojirimycin were previously both found to inhibit the Drosophila Fdl processing hexosaminidase (22). However, whereas HEX-2, -3, -4 and -5 did not show sensitivity to any of these inhibitors at the concentrations used (up to 0.4 mM), only the activity of HEX-1 and of the native worm p-nitrophenyl-β-N-acetylhexosaminidase activity was particularly affected when using 5 mM p-nitrophenyl substrates (for results with N-acetylcastanospermine using p-nitrophenyl-β-N-acetylgalactosaminide as substrate, see Fig. 2C). The Ki values of HEX-1 for N-acetylcastanospermine (5 μM), 2-acetamido-1,2-dideoxynojirimycin (1 mM) and PUGNAc (2 μM) were estimated using Dixon plots.
Finally, the three recombinant plant enzymes were tested for their sensitivity towards N-acetylcastanospermine and particularly AtHEX1 and AtHEX2 were found to be inhibited (see Fig. 3B). AtHEX1 was also tested for its sensitivity to 2-acetamido-1,2-dideoxynojirimycin and PUGNAc; about 50% inhibition with the former was achieved at 0.2 mM and 90% with the latter at 0.1 mM (data not shown). Overall, therefore, the inhibition data (i.e., the lack of inhibition of HEX-2, -3, -4 and -5 as compared to other hexosaminidases) indicate a distinct functional difference between the two major sub-families of the class 20 glycoside hydrolases, which reflects the aforementioned divergence at the primary structural level.
Activity of Caenorhabditis and Arabidopsis hexosaminidase towards glycan substrates
The various hexosaminidases were tested with typical N-glycan substrates. Under the conditions used, C. elegans HEX-2 was able to remove three HexNAc residues from the biantennary dabsyl-glycopeptide βGNβGN, which carries non-reducing terminal GalNAc residues (see Scheme I for structure); on the other hand, HEX-4 and HEX-5 removed only up to two HexNAc residues from βGNβGN, whereas HEX-3 did not detectably cleave residues from this substrate (Fig. 4). Of the five enzymes, only HEX-2 and HEX-3 cleaved one residue from the asialoagalacto-dabsyl glycopeptide GnGn (see Supplementary Data). In contrast, HEX-1 cleaved no residues from either βGNβGN or GnGn even after extended incubation times of up to three days.
Figure 4. Assay of recombinant Caenorhabditis hexosaminidases with a dabsylated GalNAc-modified biantennary N-glycopeptide substrate.
Culture supernatants of yeast expressing C. elegans HEX-1, -2, -3, -4 or -5 were incubated overnight at 37 °C with dabsyl-βGNβGN (m/z 2468) and analysed by MALDI-TOF MS. Laser-induced degradation of the dabsyl moiety results in a loss of m/z 132 (species indicated by an asterisk), whereas loss of m/z 203 corresponds to removal of one HexNAc residue; corresponding data for GnGn is shown as Supplementary Data. The volume of culture supernatant added was equal in each case: approximately three-times more HEX-1 than HEX-4 and ten-times more HEX-1 than HEX-3 was present, as measured by enzyme-linked immunosorbent assays with anti-FLAG, whereas as judged by Coomassie staining the amount of HEX-1 and HEX-2 are approximately equal; HEX-5 was not detected by ELISA or Coomassie staining. Extended incubation times (3 days) verified that a total of three HexNAc residues were removed by HEX-2, whereas HEX-4 and HEX-5 never removed more than two and that the spectrum with HEX-3 was unchanged in comparison to that shown. In comparison, commercial jack bean hexosaminidase removes four HexNAc residues from this substrate. The two glycan structures are depicted according to the nomenclature of the Consortium for Functional Glycomics.
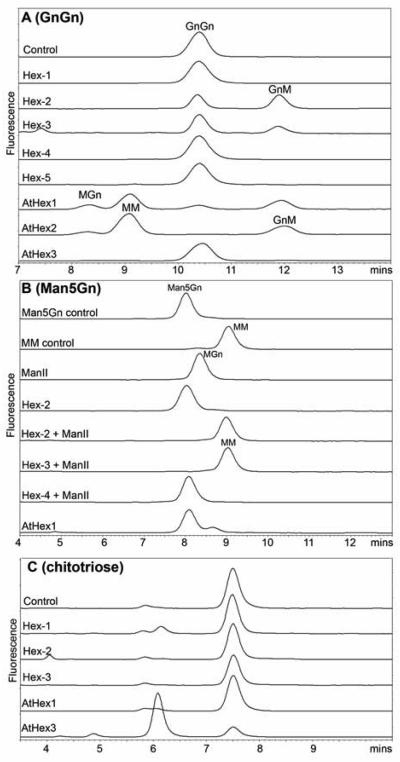
In order to determine the specificity of HEX-2 and HEX-3 further, pyridylamino-GnGn was used as a substrate; digestion of GnGn to either MGn or GnM is associated with diagnostic differences in retention time when using RP-HPLC (47). A shift in the retention time to that indicative of GnM, but not of MGn, was observed with both HEX-2 and HEX-3 (Fig. 5A). The specific cleavage by HEX-2 and HEX-3 of GnGn to GnM is a property shared with the D. melanogaster FDL hexosaminidase and with an activity in native worm extracts (see below); HEX-1, HEX-4 and HEX-5 did not digest this substrate. In contrast, AtHEX1 and AtHEX2 displayed no distinct preference for a specific terminal GlcNAc residue and indeed cleaved GnGn to a mixture of products (i.e., overnight to MGn, GnM and MM; Fig. 5A), as is also the case with Sf9 hexosaminidases (48,49).
Figure 5. Assay of recombinant Caenorhabditis and Arabidopsis hexosaminidases with pyridylaminated oligosaccharide substrates.
Culture supernatants of yeast expressing C. elegans hexosaminidases or mannosidase II and ammonium sulphate fractionated supernatants of yeast expressing A. thaliana hexosaminidases were incubated for 20 hours at 37 °C using pyridylaminated forms of GnGn, Man5Gn or chitotriose prior to RP-HPLC using a gradient of 4.2-5.7% methanol over 15 minutes. (A) Assays with GnGn-PA as substrate showing that only HEX-2 and HEX-3 displayed the activity specifically converting GnGn to GnM (as shown by the shift to higher retention time), whereas AtHEX1 and AtHEX2 converted GnGn into a mixture of MGn, MM and GnM. (B) Assays with Man5Gn-PA as substrate showing that conversion of Man5Gn to MM requires the action of both C. elegans mannosidase II (AMAN-2) and either HEX-2 or HEX-3; AMAN-2 alone catalysed the shift to MGn, but HEX-2 and HEX-3 did not catalyse a shift to Man5, whereas AtHEX1 was capable of the latter reaction. (C) Assays with chitotriose-PA showing that HEX-1 and, especially, AtHEX3 catalyse the conversion of chitotriose to a species of lower retention time.
In order to reproduce a potential in vivo pathway, Man5Gn was incubated with recombinant C. elegans mannosidase II together with HEX-2, -3 or -4. While the mannosidase II digested the Man5Gn to MGn, only incubation with HEX-2 or HEX-3 resulted, as shown by co-elution with a known standard, in further digestion to MM (Fig. 5B); indeed, the digestion in these two cases was complete. On the other hand, in the absence of mannosidase II, HEX-2 and HEX-3 did not digest Man5Gn, whereas incubation of this substrate with AtHEX1 resulted in the appearance of Man5.
In the tests with dabsylated or pyridylaminated N-glycan substrates, C. elegans HEX-1 and AtHEX3 displayed no obvious activity; thus, we considered the possibility that other GlcNAc-containing compounds may be substrates. In these tests, chitotriose-PA was digested almost completely overnight by a 50% ammonium sulphate fraction of a supernatant yeast expressing AtHEX3 and by C. elegans microsomes as well as, to a lesser extent, by a supernatant of yeast expressing C. elegans HEX-1 (Fig. 5C). Supernatants from other recombinant or control yeast showed no or low chitotriosidase activity.
Native hexosaminidases in wild-type and mutant Caenorhabditis extracts
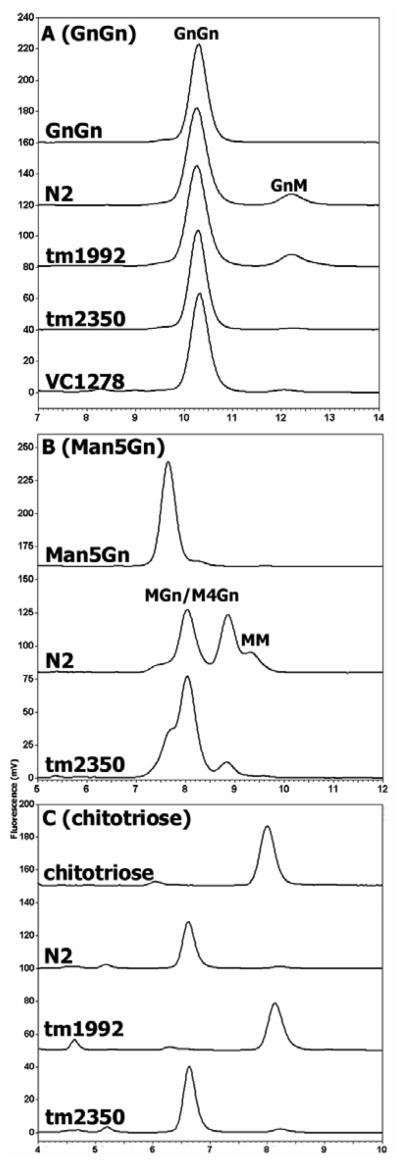
In the course of our previous studies assaying core fucosyltransferase activity in crude worm extracts, we noticed a significant hexosaminidase activity at pH 6 when using GnGn or GnGnF6 with a preference also for the α1,3-arm (see also Fig. 6A). As discussed above in connection with recombinant HEX-2 and HEX-3, this specificity is reminiscent of the one determined to be present in microsomal membrane fractions of insect cells and of recombinant fruitfly FDL (21,22). However, previous studies on C. elegans suggested that only M4Gn and MGn were substrates for a microsomal hexosaminidase (23). In preliminary trials, preparing the same type of Triton-washed microsomal fraction, however, did not result in observing any difference in the specificity; this procedure merely removed the small activity resulting in the presence of MM in the incubations. As with HEX-2 or HEX-3 when using GnGn-PA as a substrate, the native GnGn-digesting activity is reduced by 80% in the presence of 200 μM N-acetylcastanospermine and has an optimum at pH 5.5 (data not shown).
Figure 6. Assay of mutant and wild-type Caenorhabditis extracts with pyridylaminated oligosaccharide substrates.
(A) Extracts (8 μg, as measured by Pierce micro BCA protein assay) of wild-type (N2), hex-1 (tm1992) and hex-2 (tm2350 or VC1278) worms were incubated overnight with GnGn-PA prior to RP-HPLC and detection by fluorescence; conversion to GnM-PA was barely detected in the hex-2 extracts and the similar ‘biochemical phenotype’ of these two independently-isolated mutants indicates a role for HEX-2 as the major processing hexosaminidase in C. elegans. (B) Extracts of N2 or tm2350 worms were incubated overnight with Man5Gn-PA prior to RP-HPLC; although the tm2350 extract converts Man5Gn to Man4Gn and MGn (as verified by MALDI-TOF MS and indicative that mannosidase II activity is not affected by the mutation), conversion of these intermediates to MM by a hexosaminidase was relatively minor. (C) Extracts of N2, tm1992 or tm2350 worms were incubated overnight with chitotriose-PA prior to RP-HPLC; the result with the tm1992 extract is indicative that deletion of part of the hex-1 gene results in an abolition of chitotriosidase activity.
To prove more directly, than by the use of recombinant enzymes, whether our hypothesis as regards the in vivo functions of HEX-2 and HEX-3 was correct, mutants carrying deletions in selected C. elegans hexosaminidase genes were obtained and these were analysed for changes in enzyme activities and N-glycan structure. None of these mutants displayed obvious and consistent biological defects under standard laboratory conditions, a situation reminiscent of other C. elegans glycomutants (8,10), but alterations in enzymatic activities were observed.
The hex-1 (tm1992) mutant had a normal ability to digest GnGn to GnM (Fig. 6A). On the other hand this mutant displayed no chitotriosidase activity (Fig. 6C) and a much reduced ability to degrade p-nitrophenyl-β-N-acetylglucosaminide (data not shown). Thus, considering the properties of recombinant HEX-1, it is concluded that this enzyme is both the major p-nitrophenylhexosaminidase and the major chitotriosidase in the worm; on the other hand, despite its relationship to fruitfly FDL, it has no obvious role in N-glycan processing.
The hex-2 (tm2350) mutant was of particular interest, because, of the four ‘novel’ worm hexosaminidases, the corresponding gene is the one with the most ubiquitous expression (see below); also, as mentioned above, recombinant HEX-2 displayed the ability to specifically degrade GnGn to GnM as well as aid, in the presence of mannosidase II, the processing of Man5Gn to MM. Incubation of the extract of the hex-2 mutant with glycan substrates showed a greatly reduced activity towards GnGn (Fig. 6A) and a block in the processing of Man5Gn to MM, with only the products of mannosidase II (Man4Gn and MGn) being significantly present in the incubations (Fig. 6B). The latter HPLC elution data were verified by MALDI-TOF MS of the pooled fractions as shown by the presence of species with m/z 1214.497, 1376.584 and 1538.636 which correspond to pyridylaminated [M+Na]+ forms of MGn, Man4Gn and Man5Gn. In contrast, the hex-2 mutant extract displayed normal activity towards chitotriose (Fig. 6C). The normal chitotriosidase and mannosidase II activities were considered to be an indication that only the N-glycan processing hexosaminidase was affected by the deletion. We, therefore, conclude that HEX-2 is an enzyme responsible for part of the specific hexosaminidase-mediated degradation of N-glycans observed in native nematode extracts in vitro.
N-glycans of hexosaminidase mutants
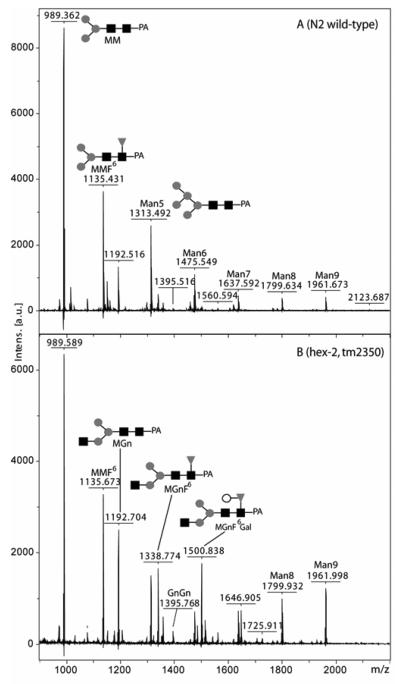
The effects of deletions within hexosaminidase genes on the worm N-glycome were also appraised; considering the results of the enzymatic assay data, it was assumed that the hex-2 mutant would show an altered N-glycan profile. Indeed, even though paucimannosidic glycans still dominated the MALDI-TOF MS spectrum of glycans derived from the hex-2 mutant, species with the composition Hex3-4HexNAc3Fuc1-2 were present either in greater amounts than in the N2 wild-type or were novel to hex-2 worms (compare Fig. 7A and 7B; see also Table 1).
Figure 7. MALDI-TOF MS analysis of N-glycans from N2 wild-type and hex-2 mutant worms.
Pyridylaminated PNGase A-released N-glycans from N2 or tm2350 strains were pooled after RP-HPLC and subject to monoisotopic MALDI-TOF MS. The spectra are annotated with the m/z values of selected species and the major putative structures (as deduced by the HPLC, exoglycosidase digestion and MS-MS data shown in Figs. 8 and 9) are depicted using the nomenclature of the Consortium for Functional Glycomics. A shift to structures containing an extra HexNAc is apparent in the hex-2 mutant; the relative peak intensities for these data are summarised in Table 1.
Table 1. Summary of MS data for pyridylaminated glycans from the N2 and tm2350 strains.
Monoisotopic m/z values [M+H+] from the spectra presented in Figure 7 and the corresponding hypothesised compositions are shown for pyridylaminated PNGase A-released N-glycans from wild-type N2 and mutant hex-2 tm2350 strains. Abundances for detected glycans refer to percentage intensities relative to the peak of highest intensity (m/z 989). ND, not detected; NR, not resolved (the isotopic distribution of a neighbouring strong signal does not allow any potential Fuc1Hex3HexNAc3PC1 to be distinguished). In the hex-2 tm2350 strain, small amounts of novel structures with extra HexNAc or HexNAcPC moieties appear; the relative amounts of Hex3HexNAc3, Fuc1Hex3HexNAc3 and Fuc1Hex4HexNAc3 are much increased in keeping with the proposed role of C. elegans HEX-2 as a processing hexosaminidase.
| Putative composition | N2 | tm2350 | ||
|---|---|---|---|---|
| m/z | abundance | m/z | abundance | |
| Hex2HexNAc2 | 827.28 | 5 | 827.48 | 8 |
| Fuc1Hex2HexNAc2 | 973.35 | 4 | 973.57 | 6 |
| Hex3HexNAc2 | 989.33 | 100 | 989.56 | 100 |
| Fuc2Hex2HexNAc2 | 1119.37 | 1 | 1119.65 | 1 |
| Fuc1Hex3HexNAc2 | 1135.42 | 43 | 1135.66 | 51 |
| Hex4HexNAc2 | 1151.40 | 10 | 1151.59 | 4 |
| Hex3HexNAc3 | 1192.44 | 16 | 1192.68 | 36 |
| Fuc2Hex3HexNAc2 | 1281.46 | 1 | 1281.71 | 1 |
| Fuc2Hex3HexNAc2Me1 | 1295.48 | 1 | 1295.70 | 1 |
| Fuc1Hex4HexNAc2 | 1297.45 | 3 | 1297.72 | 1 |
| Hex5HexNAc2 | 1313.58 | 31 | 1313.72 | 23 |
| Fuc1Hex3HexNAc3 | 1338.49 | 4 | 1338.75 | 26 |
| Hex3HexNAc3PC1 | 1357.49 | 3 | 1357.76 | 9 |
| Hex3HexNAc4 | 1395.52 | 1 | 1395.79 | 4 |
| Fuc2Hex4HexNAc2 | 1443.54 | 1 | ND | |
| Fuc2Hex4HexNAc2Me1 | 1457.54 | 2 | 1457.78 | 1 |
| Fuc1Hex5HexNAc2 | 1459.52 | 3 | 1459.81 | 1 |
| Fuc1Hex5HexNAc2Me1 | 1473.58 | 5 | 1473.76 | 4 |
| Hex6HexNAc2 | 1475.58 | 13 | 1475.78 | 12 |
| Fuc2Hex3HexNAc3 | ND | 1484.81 | 6 | |
| Fuc1Hex4HexNAc3 | 1500.54 | 1 | 1500.82 | 26 |
| Fuc1Hex3HexNAc3PC1 | 1503.57 | 1 | NR | |
| Fuc1Hex4HexNAc3Me1 | ND | 1514.83 | 8 | |
| Fuc1Hex3HexNAc4 | 1541.57 | 1 | 1541.84 | 3 |
| Hex3HexNAc4PC1 | 1560.57 | 1 | 1560.83 | 4 |
| Fuc3Hex4HexNAc2Me1 | 1603.57 | 1 | ND | |
| Fuc2Hex5HexNAc2 | 1605.60 | 1 | ND | |
| Fuc2Hex5HexNAc2Me1 | 1619.6 | 3 | 1619.83 | 2 |
| Hex7HexNAc2 | 1637.83 | 6 | 1637.83 | 11 |
| Fuc2Hex4HexNAc3 | ND | 1646.87 | 11 | |
| Fuc1Hex3HexNAc4PC1 | ND | 1706.91 | 2 | |
| Hex3HexNAc4PC2 | ND | 1725.91 | 2 | |
| Fuc3Hex5HexNAc2Me1 | 1765.64 | 1 | 1765.92 | 1 |
| Fuc2Hex6HexNAc2 | 1767.65 | 1 | ND | |
| Hex8HexNAc2 | 1799.61 | 5 | 1799.91 | 15 |
| Fuc1Hex3HexNAc4PC2 | ND | 1872.00 | 2 | |
| Fuc4Hex5HexNAc2Me1 | 1911.70 | 1 | 1910.98 | 1 |
| Fuc3Hex6HexNAc2 | 1913.71 | 1 | 1908.95 | 1 |
| Fuc3Hex6HexNAc2Me1 | 1927.67 | 1 | 1926.97 | 1 |
| Fuc3Hex7HexNAc2 | 1929.65 | 1 | 1929.02 | 1 |
| Hex9HexNAc2 | 1961.64 | 5 | 1961.95 | 17 |
| Fuc4Hex6HexNAc2Me1 | 2073.65 | 1 | 2073.99 | 1 |
| Hex10HexNAc2 | 2123.66 | 1 | 2124.18 | 1 |
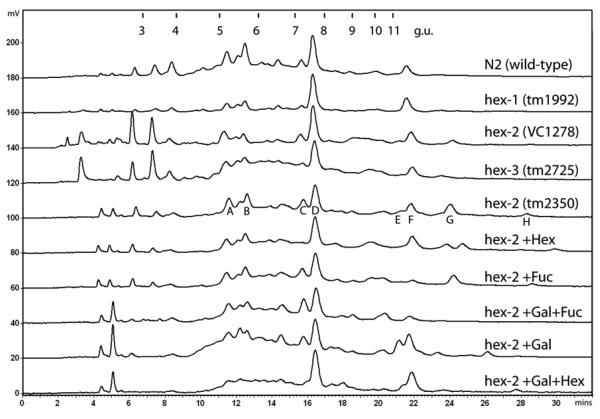
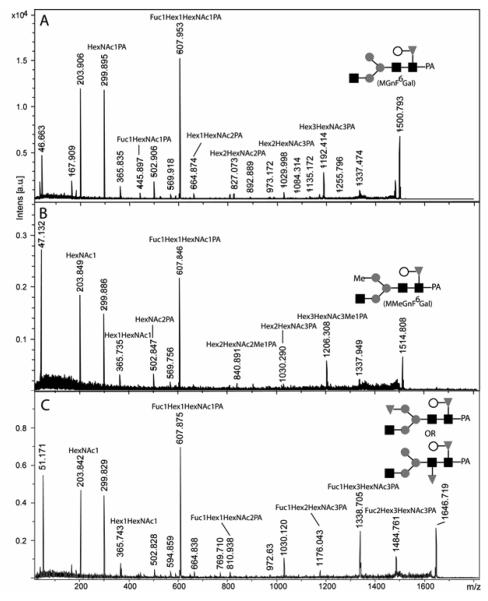
A further comparison was made with RPHPLC analysis of the pyridylaminated glycans from, N2 wild-type, the hex-1 mutant (tm1992), two hex-2 mutants (tm2350 and VC1278) and a hex-3 mutant (tm2725). These analyses showed that both hex-2 mutants had similar glycan profiles and verified the appearance of peaks (designated G and H) absent from the wild-type, hex-1 and hex-3 chromatograms (Fig. 8). Digestions of the ‘whole N-glycome’ of the tm2350 mutant with glycosidases were then performed. Particularly, a peak eluting at 24 minutes (peak G) was considered to be specific to the hex-2 profile; the major species in this peak was of m/z 1500, which would correspond to Fuc1Hex4HexNAc3-PA. This peak was not sensitive to fucosidase digestion alone; however, incubation with β1,4-galactosidase and either fucosidase or hexosaminidase resulted in a shift to peaks of lower retention time (peaks with the same retention times as MGn and MMF6 were, respectively, increased). Thus, our model was that peak G contains mainly an MGnF6 glycan carrying a β1,4-linked galactose (‘MGnF6Gal’; see Scheme I) on the core fucose residue, akin to a ‘complex core modification’ found by Reinhold and co-workers in wild-type C. elegans (50) and by others on squid rhodopsin (51) and keyhole limpet haemocyanin (52). This presumption was confirmed by MALDI-TOF MS-MS (Figure 9A), which resulted in the presence of a fragment of m/z 607, consistent with a Gal-Fuc-GlcNAc-PA structure. Another component of peak G with the composition Fuc2Hex4HexNAc3-PA was also examined by MS-MS (Figure 9C), but no unambiguous assignment of the position of the second fucose was possible; the second fucose may either be on the terminal mannose or ‘second’ core GlcNAc residue, as seen for other structures in previous reports on nematode glycans (50,53). Finally, a putatively methylated form of MGnF6Gal eluting at 28 minutes (m/z 1514; peak H) was also sensitive to hexosaminidase and galactosidase digestion; due to the presence of fragments corresponding to Hex3HexNAc3Me1-PA and Hex2HexNAc3-PA, it is hypothesised that the methyl group is attached to the unsubstituted non-reducing mannose residue (Figure 9B). Such a modification of mannose by a methyl group has been reported for gastropod paucimannosidic N-glycans (54).
Figure 8. RP-HPLC analysis of N-glycans from N2 wild-type and hexosaminidase-defective mutant worms.
Pyridylaminated PNGase A-released N-glycans were subject to RP-HPLC using a gradient of 0.3% methanol per minute and detected by fluorescence (response in mV); the column was calibrated in terms of glucose units (g.u.). Analysis of glycans of wild-type N2 and hex-1 tm1992 worms resulted in similar chromatograms, whereas the analysis of N-glycans from the two hex-2 mutants (VC1278 and tm2350) showed the appearance of new peaks of high retention time (over 11 g.u.); other than in the region below 4 g.u. (for which there is no clear data as to the nature of the peaks), the hex-3 (tm2725) chromatogram is similar to the wild-type one, suggesting a minor role for HEX-3 in N-glycan processing. The N-glycans from the hex-2 tm2350 mutant were subject to fractionation; the peaks A-H were collected and analysed by MALDI-TOF MS and found to contain species with the following m/z values [M+H]+: A (11 mins), 1637.669 and 1799.742; B (12.5 mins), 1961.751 and 1637.655; C (16 mins), 1192.587; D (17 mins), 989.552 and 1313.725; E (21.5 mins), 1338.623 and 1135.531; F (22 mins) 1135.465; G (24 mins), 1500.616 and 1646.669; H (29 mins), 1514.673. The whole pool of N-glycans from the hex-2 tm2350 mutant was also subject to various single and double exoglycosidase digests with Streptococcus hexosaminidase (Hex; resulting in removal of peaks C and E and shifts of peaks G and H), bovine kidney fucosidase (Fuc; resulting in reduction of peaks E and F), Aspergillus galactosidase and bovine fucosidase (Gal+Fuc; resulting in the near abolition of peaks E, F and G and an apparent increase in peak C), Aspergillus galactosidase alone (Gal; resulting in an increase in peak E at the expense of peak G and a shift forward of peak H), Aspergillus galactosidase and Streptococcus hexosaminidase (Gal+Hex; resulting in removal of peaks C and G and a shift in peak H). The elution position of peak C and its sensitivity to hexosaminidase is indicative that this glycan is the MGn isomer of Hex3HexNAc3-PA. The fucose residue of the glycans in peaks E and F is, as judged by the relative retention time and sensitivity to bovine fucosidase, core α1,6-linked. On the other hand, the putatively α1,6-linked fucose residues of peaks G and H are only accessible to digestion when fucosidase is used in combination with Aspergillus β1,4-specific galactosidase, while bovine testes galactosidase, which is β1,3-specific, has no effect on the glycan profile (not shown). The composition of peak H (m/z 1514) is compatible with the presence of a methyl group which makes the glycan more hydrophobic than the related major glycan of peak G (m/z 1500).
Figure 9. MALDI-TOF MS-MS analysis of putatively core galactofucosylated N-glycans from mutant hex-2 worms.
The major species from peaks G (panels A and C; m/z 1500 and 1646) and H (panel B; m/z 1514) isolated by RP-HPLC fractionation of pyridylaminated N-glycans derived from the hex-2 tm2350 mutant (see Fig. 8) were subject to MALDI-TOF MS-MS analysis. In conjunction with the necessity to co-incubate the glycans with galactosidase in order that they become fucosidase-sensitive, the Fuc1Hex1HexNAc1-PA fragment (m/z 607) in both MS-MS spectra is indicative of a galactose-substituted core fucose residue. The presence of a Hex3HexNAc3Me1-PA (m/z 1206) fragment and low abundance Hex2HexNAc3-PA (m/z 1030) and Hex2HexNAc2Me1-PA (m/z 841) fragments is suggestive that the methyl group of the m/z 1514 glycan (panel B) is present on the free mannose residue which is not substituted by a HexNAc. In the case of the m/z 1646 glycan (panel C), the position of the second fucose residue is ambiguous, but the MS/MS offers verification that the composition is Fuc2Hex4HexNAc3-PA. Based on the enzyme digestion data (Fig. 8), it is concluded that the second fucose is not attached to the galactose or GlcNAc residues, whereas the lack of an effect on the retention time as compared to the ‘parent’ glycan suggests that the second fucose is not attached to the reducing-terminal GlcNAc; in comparison to other data on nematode glycans, it is, therefore, hypothesised that the second fucose is either linked to the ‘distal core’ GlcNAc or on the free non-reducing mannose residue. Both potential models for the structure are shown. The proposed glycan structures are depicted according to the nomenclature of the Consortium for Functional Glycomics.
Overall, it is concluded that glycans with non-reducing terminal N-acetylhexosamine residues, absent from the wild-type, are present in hex-2 worms, consistent with its proposed enzymatic function of the corresponding gene product. The remaining hexosaminidase activity (as measured with GnGn in vitro; see above), though, may still be sufficient to account for the presence of MM and MMF6 glycans in the hex-2 worm and, on the basis of the data with recombinant enzymes, is putatively due to HEX-3. In the future the effect of double mutants should be appraised in order to make final conclusions as to the relative contribution of the different GnGn-digesting hexosaminidases towards the wild-type nematode N-glycan spectrum.
Differential expression of nematode hexosaminidase promoters
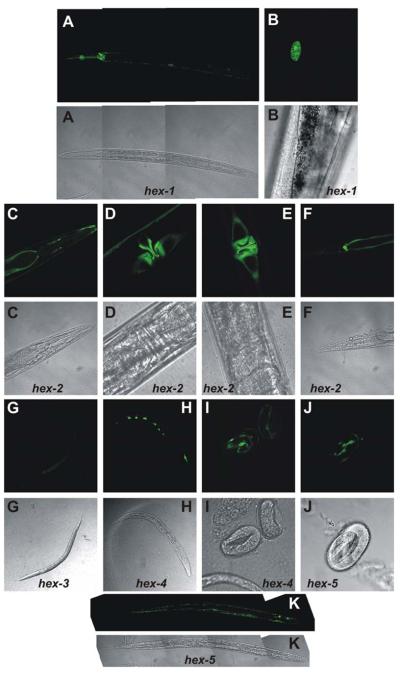
Considering the multiplicity of hexosaminidases in C. elegans, it was sought to examine whether they are expressed temporally and spatially in a differential manner. Therefore, promoter gfp reporter constructs were generated, which contained, in each case, the ~2000 bp upstream of the start codon. These vectors, containing a copy of the unc-119 gene, were stably integrated into unc-119(ed3) mutants. Confocal microscopy of at least two independent lines for each gene showed a range of locations for the expression of these constructs. The hex-1 promoter was particularly active in coelomocytes as well as in neurons of the pharyngeal region and nerve cord (Fig. 10A,B), as compared to the head and tail pattern observed in strain BC14144 (also carrying a hex-1::gfp construct) as part of a large-scale screen (55). The hex-2::gfp construct appeared to be active in the hypodermal cells, vulval toroids and various adult head and tail neurons (Fig. 10C-F), whereas the hex-3 construct was expressed in gut granules (Fig. 10G). The hex-4::gfp line displayed staining of seam cells in late embryos and L1 larvae (Fig. 10H,I). Expression of hex-5 was restricted to certain cells at the three-fold stage, but was also present in the vulval, head (muscle) and tail regions in larval and adult worms (Fig. 10J,K). Of the five promoters, hex-1, hex-2, hex-3 and hex-5 were expressed throughout the life-cycle, whereas hex-4 was restricted to the embryonal and L1 stages. Thus, to varying degrees, the results with these gfp constructs were indicative of both temporal and spatial differential expression of these genes and are in accordance with the results showing that a single hexosaminidase is not responsible for the presence of high amounts of paucimannosidic species in the N-glycan profile of wild-type worms.
Figure 10. Analysis of green fluorescent protein expression driven by Caenorhabditis hexosaminidase promoters.
Fluorescence microscopy of worms transformed with various hex::gfp constructs indicates that there is differential expression driven by the putative promoter regions of the hex-1, -2, -3, -4 and -5 genes. Pairs of confocal and fluorescent micrographs for hex-1::gfp (whole worm, A, and tail coelomocyte, B), hex-2::gfp (head region, C, vulval region lateral view, D, vulval region top view, E, and tail region, F), hex-3::gfp (L1 larvae, G), hex-4::gfp (L1 larvae, H, and embryos, I) and hex-5::gfp (embryo, J, and whole worm, K) transgenic worms are shown.
Discussion
Comparing insect and plant N-glycan processing
As discussed in the Introduction, the hexosaminidase-mediated removal of the GlcNAc transferred by GlcNAc-TI, after modification by core fucosyltransferases, has been invoked in order to explain the paucimannosidic N-glycan structures observed in insects and plants. The identity of such a ‘processing’ hexosaminidase, proposed to be located in either the insect Golgi or plant vacuole (21,24), has been a stumbling block in our understanding and exploitation of N-glycan biosynthesis in these species. Indeed, the presence of paucimannosidic N-glycans on the glycoproteins of plants and of insect cells means that these may be less valuable as biotechnological products due to the lack of mammalian-type features and could, if used therapeutically, be immunogenic (56) and be cleared from the circulation faster, although the latter property can be an advantage for enzyme replacement therapy of lysosomal storage diseases (57). Thus, the cloning of hexosaminidase genes from plants and insects is considered, in conjunction with other approaches (58), to be necessary in the development of knock-out/down strategies for attaining a more human-like glycosylation in biotechnologically-relevant systems.
A recent advance was the identification of the fdl hexosaminidase gene from D. melanogaster, which was shown to be important for the normal processing of N-glycans in this organism and to encode an enzyme, present in the secretory pathway, with the same specificity towards the α1,3-antennal GlcNAc residue on N-glycans as a previously-characterised Golgi hexosaminidase found in extracts of Sf9 cells (22). On the other hand, the two recombinant Sf9 insect cell hexosaminidases characterised to date can remove both GlcNAc residues from GnGn (48,49).
This latter property is shared with the AtHEX1 and AtHEX2 described in the present study, since these two putatively vacuolar enzymes also have no especially-strict arm preference; the ability of AtHEX1 to cleave a plant-type GnGnXF3 glycan to a mixture of products was also observed (data not shown). In this respect, these recombinant plant enzymes mimic the activity of the commercially-available jack bean hexosaminidase and the products reflect that plant glycomes contain N-glycans with a GlcNAc on either the α1,3- or α1,6-arms, e.g., the MGnXF3 and GnMXF3 structures found in pollens (30) (see Scheme I for structures and Scheme II for a putative processing pathway). Another contrast to the insect (FDL) and nematode (HEX-2, HEX-3 and native) hexosaminidases is that the plant hexosaminidases will remove the non-reducing terminal GlcNAc from Man5Gn-type structures both in vitro, as shown in this study with AtHEX1, and in vivo, exemplified by the presence of Man5XF3 as in a mannosidase II knock-out plant (11) and in papaya (59).
Plants, insects and nematodes possess family 20 glycosidases with chitotriosidase activity
The third plant hexosaminidase, AtHEX3, is a putative chitotriosidase and, thus, shares some properties with the Hexo1 and Hexo2 from Drosophila (22); however, it is evolutionarily closer to fungal hexosaminidases with this type of activity and may, in conjunction with family 18 and 19 chitinases (60), have roles in defence-related chitin degradation in vivo. The nematode HEX-1, the major p-nitrophenyl-β-N-acetylglucosaminide-cleaving enzyme in the worm, is also a putative chitotriosidase; such an activity has been previously reported in C. elegans extracts (61). Chitin is a component of nematode eggshells and C. elegans possesses two chitin synthase genes (62). Thus, exo- and endoglycosidases capable of degrading chitin can be expected in this organism and family 18 endochitinases have been described from other nematodes (61,63). The hex-1 promoter-driven expression of GFP in coelomocytes may correlate with the degradative and scavenging function of these cells.
The nematode genome encodes unusual N-glycan-specific hexosaminidases
The nematode HEX-2 and HEX-3 remove specifically the α1,3-antennal GlcNAc residue (i.e., that transferred by GlcNAc-TI) from the GnGn N-glycan. Thus, their activity is akin to that of FDL and this, therefore, suggests that the same basic mechanism for generation of paucimannosidic N-glycans operates in both insects and nematodes, i.e., that the residue transferred by GlcNAc-TI is removed, but that non-orthologous enzymes are responsible. On the other hand, HEX-2, HEX-4 and HEX-5 are capable of removing also the non-reducing terminal GalNAc residues from the βGNβGN N-glycan; it is unclear whether the latter reaction is physiological, but certainly C. elegans possesses a β1,4-N-acetylgalactosaminyltransferase (BRE-4) which can generate LacdiNAc moieties on N-glycans in vitro (64) and probably glycolipids in vivo (65). Although N-glycans with LacdiNAc moieties are not demonstrated on C. elegans, such structures are found in the nematode Trichinella spiralis (66).
The relative insensitivity of the nematode HEX-2, -3, -4 and -5 towards standard hexosaminidase inhibitors, when using p-nitrophenyl substrates, was another complicating factor in tracking down the processing hexosaminidase in the worm. This is due to the finding that the native GnGn-digesting activity is inhibited by 80% in the presence of N-acetylcastanospermine. However, when using this ‘natural’ substrate, N-acetylcastanospermine also inhibits both recombinant HEX-2 and HEX-3 by some 80%. The apparent discrepancy in the inhibition data may be due to the vastly lower ‘native’ substrate concentration used (20 μM), as compared to that of the p-nitrophenyl substrates (5 mM) and of the inhibitor (200 μM). As judged by the use of GFP-fusions, the hex-2 and hex-3 promoters appear to be expressed in a tissue-specific manner.
Comparing insect and nematode N-glycan processing
One consideration in respect to N-glycan processing pathways in insects and nematode is that GnGn is probably not a dominant intermediate. Insect cells have apparently low levels of GlcNAc-TII (67,68), thus reducing the potential GnGn pool within the secretory pathway; certainly, MM-related, and not GnM-related, structures dominate in both insects or nematodes. Thus, it is probable that MGn or MGnF6 are major in vivo substrates of the processing hexosaminidases in these organisms. Certainly, the D. melanogaster core α1,3-fucosyltransferase (FucTA) and the C. elegans core α1,6-fucosyltransferase (FUT-8) are capable of modifying both MGn and Man5Gn in vitro (69); indeed, these enzymes must act on these glycans prior to removal of the non-reducing terminal GlcNAc residue. On the other hand, the C. elegans core α1,3-fucosyltransferase (FUT-1) can accept MM and GnM, but not GnGn (28); this enzyme can also fucosylate Man5 (unpublished data). Thus, as previously hypothesised (10,17,70) and as summarised in Scheme II, a pathway of Man5 → Man5Gn → MGn → MGnF6 → MMF6 → MMF3F6 may operate in C. elegans, whereas in insects the order is slightly different: Man5 → Man5Gn → MGn → MGnF6 → MGnF3F6 → MMF3F6, with the potential that some Man5 is directly converted to MM by a so-called mannosidase III (71). In the absence of processing hexosaminidase activity, an accumulation of Man3-5GlcNAc3Fuc1 may result. Indeed in the fdl fruitfly mutant, MGnF6 is a dominant species in the glycan profile (22). This predicted effect, however, only occurs to a lesser extent in the nematode hex-2 mutant. This would correlate with the finding that the deletion of part of the hex-2 gene does not entirely abolish the GnGn-digesting activity in worm extracts. Thus, even though the hexosaminidase activity towards N-glycans is drastically reduced, this does not result in a lack of MM and MMF6. This effect is, though, partly reminiscent of an experiment with antisense-mediated knock-down of GlcNAc-TI in plants: a large decrease in enzyme activity had only a minor effect on the glycan profile (72). Presumably a small amount of a residual activity of a processing hexosaminidase (putatively HEX-3, whose promoter is also active in tissues in which the hex2-driven GFP expression is absent) can be sufficient to generate a glycomic profile with MM and MMF6 remaining as major structures. Nevertheless, there is an impact on the glycome of the hex-2 mutant worm: glycans with an ‘extra’ HexNAc, such as MGn and MGnF6, are more pronounced in the hex-2 N-glycome than that of wild-type N2 or hex-1 worms. Furthermore, as with the aman-2 mutant in which various non-wild-type hybrid glycans were present (10), unusual N-glycans result from knocking-out hex-2, presumably due to otherwise minor biosynthetic pathways being revealed. In particular, novel N-glycans of the form Fuc1-2Hex3-4HexNAc3Me0-1 are found in the hex-2 mutant, which are forms of MGn modified by galactosylated core fucose moieties, the latter modification having been previously found on other glycans in wild-type worms (50). Thereby, it is noteworthy how the use of nematode mutants aids identification or verification of new aspects of the glycomic capabilities of this species.
Other fates of aberrant N-glycans with terminal GlcNAc are conceivable in invertebrates since the GlcNAc can be ‘capped’ with other moieties, such as fucosylated LacdiNAc in honeybee venom (73), sialylated LacNAc in fruitfly embryos (74) or phosphorylcholine-containing glycans in nematodes (75,76). However, large shifts to these structures are not seen in the fruitfly fdl or worm hex-2 mutants, although some extra phosphorylcholine-modified glycans were observed. The biological repercussions of alterations in the N-glycome of hex-2 worms are, as yet, unknown, but it is interesting to note that mutations in the three C. elegans GlcNAc-TI genes result in alterations in nematode survival in the presence of bacterial pathogens (70).
Conclusion
Overall, the examination of recombinant forms of plant, nematode and, in an earlier study (22), insect N-glycan-modifying hexosaminidases suggests that, although the final result is similar in that paucimannosidic N-linked oligosaccharides are expressed in these organisms, the exact genetic mechanisms are different, i.e., that non-orthologous members of glycoside hydrolase family 20 are responsible for the biosynthesis of these structures. Obviously, questions remain as to the intracellular localisation of the nematode hexosaminidases, the mechanisms by which their transcription is controlled, their relevance as models for human lysosomal storage diseases and the effect of knocking-out multiple worm hexosaminidase genes as well as the effect of knocking-out the plant hexosaminidases. However, the use of mutant organisms as well as of recombinant enzymes demonstrates that the major processing hexosaminidases in C. elegans have been molecularly identified and show a remarkable example of functional convergence between enzymes displaying low homology yet still being members of the same glycosidase family.
Supplementary Material
Acknowledgements
We wish to thank Denise Kerner for technical assistance, Ramona Dumreanu, Claudia Eisenkölbl, Daniel Gregorich, Lilian Kuster and Johannes Lesigang, project students in the laboratory, for performing some of the experiments, Dr. Arnold Stütz (Technische Universität Graz, Austria) for the kind gift of 2-acetamido-1,2-dideoxynojirimycin, the National Bioresource Project for the Experimental Animal Nematode C. elegans (Tokyo Women's Medical University, Japan) for the tm1992, tm2350 and tm2725 mutant strains and the Caenorhabditis Genetics Centre for the VC1278 (being provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, part of the International C. elegans Gene Knockout Consortium); the original pPD118.25 (L3786) vector was the kind gift of Drs. Andrew Fire and Si-Qun Xu.
The abbreviations used are
- GFP
green fluorescent protein
- GlcNAc-TI
N-acetylglucosaminyltransferase I
- MALDI-TOF
matrix-assisted laser desorption ionisation/time-of flight
- MS
mass spectrometry
- PA
pyridylamino
- PUGNAc
O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate
- RP-HPLC
reverse phase high pressure liquid chromatograph
Glycan abbreviations are designated in Scheme I.
Footnotes
This work was supported by grants P15475 and P18447 (to I.B.H.W.) from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung. The nucleotide sequences encoding Caenorhabditis elegans hexosaminidases HEX-1, HEX-2, HEX-3, HEX-4 and HEX-5 have been submitted to the GenBankTM/EBI Data Bank (accession numbers AM748820, AM748821, AM748822, AM748823 and AM748824).
References
- 1.Wilson IBH. Curr. Opin. Struc. Biol. 2002;12:569–577. doi: 10.1016/s0959-440x(02)00367-6. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld R, Kornfeld S. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 3.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. Proc Natl Acad Sci U S A. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hori H, Elbein AD. Arch Biochem Biophys. 1983;220:415–425. doi: 10.1016/0003-9861(83)90431-9. [DOI] [PubMed] [Google Scholar]
- 5.Parker GF, Williams PJ, Butters TD, Roberts DB. FEBS Letters. 1991;290:58–60. doi: 10.1016/0014-5793(91)81225-w. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar M, Leventis PA, Silvescu CI, Reinhold VN, Schachter H, Boulianne GL. J Biol Chem. 2006;281:12776–12785. doi: 10.1074/jbc.M512769200. [DOI] [PubMed] [Google Scholar]
- 7.von Schaewen A, Sturm A, O'Neill J, Chrispeels MJ. Plant Physiol. 1993;102:1109–1118. doi: 10.1104/pp.102.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S, Hanneman A, Reinhold V, Spence A, Schachter H. Biochem. J. 2004;382:995–1001. doi: 10.1042/BJ20040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altmann F, März L. Glycoconjugate Journal. 1995;12:150–155. doi: 10.1007/BF00731359. [DOI] [PubMed] [Google Scholar]
- 10.Paschinger K, Hackl M, Gutternigg M, Kretschmer-Lubich D, Stemmer U, Jantsch V, Lochnit G, Wilson IBH. J. Biol. Chem. 2006;281:28265–28277. doi: 10.1074/jbc.M602878200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strasser R, Schoberer J, Jin C, Glössl J, Mach L, Steinkellner H. Plant J. 2006;45:789–803. doi: 10.1111/j.1365-313X.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 12.Fabini G, Freilinger A, Altmann F, Wilson IBH. J. Biol. Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- 13.Wilson IBH, Rendić D, Dumić J, Freilinger A, Altmann F, Mucha J, Müller S, Hauser M-T. Biochim. Biophys. Acta. 2001;1527:88–96. doi: 10.1016/s0304-4165(01)00151-9. [DOI] [PubMed] [Google Scholar]
- 14.Strasser R, Mucha J, Mach L, Altmann F, Wilson IBH, Glössl J, Steinkellner H. FEBS Lett. 2000;472:105–108. doi: 10.1016/s0014-5793(00)01443-5. [DOI] [PubMed] [Google Scholar]
- 15.Mulder H, Dideberg F, Schachter H, Spronk BA, De Jong-Brink M, Kamerling JP, Vliegenthart JFG. Eur J Biochem. 1995;232:272–283. doi: 10.1111/j.1432-1033.1995.tb20809.x. [DOI] [PubMed] [Google Scholar]
- 16.Faveeuw C, Mallevaey T, Paschinger K, Wilson IBH, Fontaine J, Mollicone R, Oriol R, Altmann F, Lerouge P, Capron M, Trottein F. Eur. J. Immunol. 2003;33:1271–1281. doi: 10.1002/eji.200323717. [DOI] [PubMed] [Google Scholar]
- 17.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IBH. Glycobiology. 2005;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 18.Bakker H, Lommen A, Jordi W, Stiekema W, Bosch D. Biochem Biophys Res Commun. 1997;261:829–832. doi: 10.1006/bbrc.1999.1117. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Tan J, Reinhold VN, Spence AM, Schachter H. Biochim Biophys Acta. 2002;1573:271–279. doi: 10.1016/s0304-4165(02)00393-8. [DOI] [PubMed] [Google Scholar]
- 20.Strasser R, Steinkellner H, Boren M, Altmann F, Mach L, Glössl J, Mucha J. Glycoconj. J. 1999;16:787–791. doi: 10.1023/a:1007127815012. [DOI] [PubMed] [Google Scholar]
- 21.Altmann F, Schwihla H, Staudacher E, Glössl J, März L. J. Biol. Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 22.Léonard R, Rendić D, Rabouille C, Wilson IBH, Préat T, Altmann F. J. Biol. Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Cao P, Chen S, Spence AM, Zhu S, Staudacher E, Schachter H. Biochem. J. 2003;372:53–64. doi: 10.1042/BJ20021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitale A, Chrispeels MJ. J Cell Biol. 1984;99:133–140. doi: 10.1083/jcb.99.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitchette-Lainé AC, Gomord V, Cabanes M, Michalski JC, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. Plant Journal. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- 26.Bencúrová M, Rendić D, Fabini G, Kopecky EM, Altmann F, Wilson IBH. Biochemie. 2003;85:413–422. doi: 10.1016/s0300-9084(03)00072-5. [DOI] [PubMed] [Google Scholar]
- 27.Morgenstern B. Nucl. Acids. Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paschinger K, Rendić D, Lochnit G, Jantsch V, Wilson IBH. J. Biol. Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 29.Mucha J, Domlatil J, Lochnit G, Rendić D, Paschinger K, Hinterkörner G, Hofinger A, Kosma P, Wilson IBH. Biochem. J. 2004;382:67–74. doi: 10.1042/BJ20040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson IBH, Altmann F. Glycoconjugate Journal. 1998;15:1055–1070. doi: 10.1023/a:1006960401562. [DOI] [PubMed] [Google Scholar]
- 31.Zeleny R, Altmann F, Praznik W. Anal Biochem. 1997;246:96–101. doi: 10.1006/abio.1996.9973. [DOI] [PubMed] [Google Scholar]
- 32.Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. The Royal Society of Chemistry; Cambridge: 1999. [Google Scholar]
- 33.Fan X, She YM, Bagshaw RD, Callahan JW, Schachter H, Mahuran DJ. Glycobiology. 2005;15:952–964. doi: 10.1093/glycob/cwi075. [DOI] [PubMed] [Google Scholar]
- 34.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K.-i., Takahashi N, Isobe T. Nat. Biotech. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 35.Giraudo CG, Maccioni HJ. Mol Biol Cell. 2003;14:3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henrissat B, Coutinho PM, Davies GJ. Plant Mol Biol. 2001;47:55–72. [PubMed] [Google Scholar]
- 37.Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Y, Vocadlo DJ, Leung A, Withers SG, Mahuran D. Biochemistry. 2001;40:2201–2209. doi: 10.1021/bi002018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diez B, Rodriguez-Saiz M, de la Fuente JL, Moreno MA, Barredo JL. FEMS Microbiol Lett. 2005;242:257–264. doi: 10.1016/j.femsle.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Mark BL, Vocadlo DJ, Knapp S, Triggs-Raine BL, Withers SG, James MN. J Biol Chem. 2001;276:10330–10337. doi: 10.1074/jbc.M011067200. [DOI] [PubMed] [Google Scholar]
- 41.Clarke VA, Platt N, Butters TD. J Biol Chem. 1995;270:8805–8814. doi: 10.1074/jbc.270.15.8805. [DOI] [PubMed] [Google Scholar]
- 42.Bolanowski M, Jacobson LA, Russell RL. Mech. Aging Dev. 1983;21:295–319. doi: 10.1016/0047-6374(83)90048-9. [DOI] [PubMed] [Google Scholar]
- 43.Li SC, Li YT. J Biol Chem. 1970;245:5153–5160. [PubMed] [Google Scholar]
- 44.Liu PS, Kang MS, Sunkara PS. Tetrahedron Lett. 1991;32:719–720. [Google Scholar]
- 45.Gradnig G, Legler G, Stütz AE. Carbohydrate Research. 1996;287:49–57. doi: 10.1016/0008-6215(96)00065-1. [DOI] [PubMed] [Google Scholar]
- 46.Horsch M, Hoesch L, Vasella A, Rast DM. Eur J Biochem. 1991;197:815–818. doi: 10.1111/j.1432-1033.1991.tb15976.x. [DOI] [PubMed] [Google Scholar]
- 47.Tomiya N, Awaya J, Kurono M, Endo S, Arata Y, Takahashi N. Anal Biochem. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- 48.Aumiller JJ, Hollister JR, Jarvis DL. Protein Expr Purif. 2006;47:571–590. doi: 10.1016/j.pep.2005.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomiya N, Narang S, Park J, Abdul-Rahman B, Choi O, Singh S, Hiratake J, Sakata K, Betenbaugh MJ, Palter KB, Lee YC. J Biol Chem. 2006;281:19545–19560. doi: 10.1074/jbc.M603312200. [DOI] [PubMed] [Google Scholar]
- 50.Hanneman AJ, Rosa JC, Ashline D, Reinhold V. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi N, Masuda K, Hiraki K, Yoshihara K, Huang H-H, Khoo K-H, Kato K. Eur. J. Biochem. 2004;270:2627–2632. doi: 10.1046/j.1432-1033.2003.03636.x. [DOI] [PubMed] [Google Scholar]
- 52.Wuhrer M, Robijn ML, Koeleman CA, Balog CI, Geyer R, Deelder AM, Hokke CH. Biochem J. 2004;378:625–632. doi: 10.1042/BJ20031380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haslam SM, Coles GC, Munn EA, Smith TS, Smith HF, Morris HR, Dell A. J. Biol. Chem. 1996;271:30561–30570. doi: 10.1074/jbc.271.48.30561. [DOI] [PubMed] [Google Scholar]
- 54.Gutternigg M, Ahrer K, Grabher-Meier H, Burgmayr S, Staudacher E. Eur J Biochem. 2004;271:1348–1356. doi: 10.1111/j.1432-1033.2004.04045.x. [DOI] [PubMed] [Google Scholar]
- 55.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJ, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 56.van Remoortere A, Bank CMC, Nyame AK, Cummings RD, Deelder AM, van Die I. Glycobiology. 2003;13:217–225. doi: 10.1093/glycob/cwg025. [DOI] [PubMed] [Google Scholar]
- 57.Van Patten SM, Hughes H, Huff MR, Piepenhagen PA, Waire J, Qiu H, Ganesa C, Reczek D, Ward PV, Kutzko JP, Edmunds T. Glycobiology. 2007;17:467–478. doi: 10.1093/glycob/cwm008. [DOI] [PubMed] [Google Scholar]
- 58.Betenbaugh MJ, Tomiya N, Narang S, Hsu JT, Lee YC. Curr Opin Struct Biol. 2004;14:601–606. doi: 10.1016/j.sbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Wilson IBH, Zeleny R, Kolarich D, Staudacher E, Stroop CJM, Kamerling JP, Altmann F. Glycobiology. 2001;11:261–274. doi: 10.1093/glycob/11.4.261. [DOI] [PubMed] [Google Scholar]
- 60.Iseli B, Armand S, Boller T, Neuhaus JM, Henrissat B. FEBS Lett. 1996;382:186–188. doi: 10.1016/0014-5793(96)00174-3. [DOI] [PubMed] [Google Scholar]
- 61.Wu Y, Egerton G, Underwood AP, Sakuda S, Bianco AE. J Biol Chem. 2001;276:42557–42564. doi: 10.1074/jbc.M103479200. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Foster JM, Nelson LS, Ma D, Carlow CK. Dev Biol. 2005;285:330–339. doi: 10.1016/j.ydbio.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 63.Gao B, Allen R, Maier T, McDermott JP, Davis EL, Baum TJ, Hussey RS. Int. J. Parasitol. 2002;32:1293–1300. doi: 10.1016/s0020-7519(02)00110-8. [DOI] [PubMed] [Google Scholar]
- 64.Kawar Z, van Die I, Cummings RD. J. Biol. Chem. 2002;277:34924–34932. doi: 10.1074/jbc.M206112200. [DOI] [PubMed] [Google Scholar]
- 65.Griffitts JS, Haslam SM, Yang T, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 66.Morelle W, Haslam SM, Olivier V, Appleton JA, Morris HR, Dell A. Glycobiology. 2000;10:941–950. doi: 10.1093/glycob/10.9.941. [DOI] [PubMed] [Google Scholar]
- 67.Altmann F, Kornfeld G, Dalik T, Staudacher E, Glössl J. Glycobiology. 1993;3:619–625. doi: 10.1093/glycob/3.6.619. [DOI] [PubMed] [Google Scholar]
- 68.Hollister J, Grabenhorst E, Nimtz M, Conradt H, Jarvis DL. Biochemistry. 2002;41:15093–15104. doi: 10.1021/bi026455d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rendić D, Linder A, Paschinger K, Borth N, Wilson IBH, Fabini G. J. Biol. Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- 70.Shi H, Tan J, Schachter H. Methods Enzymol. 2006;417:359–389. doi: 10.1016/S0076-6879(06)17022-6. [DOI] [PubMed] [Google Scholar]
- 71.Kawar Z, Karaveg K, Moremen KW, Jarvis DL. J Biol Chem. 2001;276:16335–16340. doi: 10.1074/jbc.M100119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strasser R, Altmann F, Glössl J, Steinkellner H. Glycoconj J. 2004;21:275–282. doi: 10.1023/B:GLYC.0000045099.29038.04. [DOI] [PubMed] [Google Scholar]
- 73.Kubelka V, Altmann F, Staudacher E, Tretter V, März L, Hård K, Kamerling JP, Vliegenthart JFG. Eur J Biochem. 1993;213:1193–1204. doi: 10.1111/j.1432-1033.1993.tb17870.x. [DOI] [PubMed] [Google Scholar]
- 74.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 75.Haslam SM, Houston KM, Harnett W, Reason AJ, Morris HR, Dell A. J Biol Chem. 1999;274:20953–20960. doi: 10.1074/jbc.274.30.20953. [DOI] [PubMed] [Google Scholar]
- 76.Pöltl G, Kerner D, Paschinger K, Wilson IBH. FEBS J. 2007;274:714–726. doi: 10.1111/j.1742-4658.2006.05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.