Abstract
Photonic crystal surfaces can be designed to provide a wide range of functions that are used to perform biochemical and cell-based assays. Detection of the optical resonant reflections from photonic crystal surfaces enables high sensitivity label-free biosensing, while the enhanced electromagnetic fields that occur at resonant wavelengths can be used to enhance the detection sensitivity of any surface-based fluorescence assay. Fabrication of photonic crystals from inexpensive plastic materials over large surface areas enables them to be incorporated into standard formats that include microplates, microarrays, and microfluidic channels. This report reviews the design of photonic crystal biosensors, their associated detection instrumentation, and biological applications. Applications including small molecule high throughput screening, cell membrane integrin activation, gene expression analysis, and protein biomarker detection are highlighted. Recent results in which photonic crystal surfaces are used for enhancing the detection of Surface-Enhanced Raman Spectroscopy, and the development of high resolution photonic crystal-based laser biosensors are also described.
Keywords: Biosensors, fluorescence, high-throughput screening, GPCR assays, label-free detection
INTRODUCTION
Genomic, epigenomic, and proteomic data is now providing an explosion of new information about how individuals may be susceptible to a particular disease, which people are most likely to benefit from a pharmaceutical treatment, and who is likely to suffer from adverse side effects. These trends are introducing a new era of personalized medicine in which pharmaceutical companies will no longer produce treatments that are intended to be effective for the entire population, but instead will be effective only for patients with identifiable gene expression or presence of a protein biomarker in their blood. In pharmaceutical research, the ability to efficiently screen the biochemical interaction of potential pharmaceutical drug compounds with a wide array of proteins and cells before the clinical trial stage is an increasingly important capability for avoiding costly failures when the drug is introduced to animals and humans. Likewise, testing of patients' blood or tissue samples for expression of a gene profile or presence of a biomarker will become common practice to aid in decisions regarding the most promising course of treatment. New tools are required to address these challenges. These tools take the form of sensors and assay methods that are capable translating biochemical interactions, modifications of cell membranes, and cell differentiation into measurable signals that provide a window into how these structures operate. The study of transducers, fluorescent tags, and nanoparticle “biosensors” comprises a diverse and highly active field of research and commercial activity.
Performing assays through the detection of fluorescent proteins, chemical tags, and nanoparticles are dominant methods for applications that include gene expression microarrays, gene sequencing, protein-protein interaction screening, cell imaging, and many others. A vast array of fluorescent tag materials, surface immobilization chemistries, illumination sources, and detection instruments are used for fluorescence based assays that enable these methods to achieve detection sensitivity in the ~10 pg/ml range [1, 2], sub-diffraction limit imaging [3, 4], and throughput of up to millions of assays per chip[5]. While fluorescence-based detection is very powerful, development of effective fluorescent tags and robust, quantitative assay procedures is not always simple. For example, tags can interfere with the conformation of molecules under study, and can block active binding epitopes, while quenching and photobleaching of fluorescent dyes requires strict experimental controls and limits long term tests that provide kinetic data. Therefore, label-free assay methods that enable biomolecular binding events or cellular processes to be studied through an intrinsic physical property of the analytes are desirable in some cases. Because label-free methods transduce biological activity directly to a measurable output, these assay methods are generally simpler to implement than tag-based methods, although some type of sensor surface is required upon which assays must take place.
Of the many label-free detection methods that have been demonstrated, those based upon detection of the dielectric permittivity of their analytes at optical wavelengths, collectively known as “optical biosensors” have been most widely adopted, since the introduction of Surface Plasmon Resonance (SPR) in the 1990s [6, 7]. Detection and identification of molecules through the vibrational modes of their chemical bonds using Raman spectroscopy and Surface-Enhanced Raman Spectroscopy (SERS) has also gained prominence as a chemical analysis method and more recently as a biological assay method since the discovery of SERS by Van Duynein1977[8, 9].
Since the term “Photonic Crystal” (PC) was first used by Yablonovich [10] to describe a material comprised of two different refractive index materials that alternate in a periodic fashion in 1, 2, or 3-dimensions, a wide variety of PC structures have been studied and fabricated for an enormous range of applications. The periodic modulation of refractive index within a PC, along with incorporation of intentional “defects” in the PC can be used to concentrate and direct the electromagnetic fields associated with light to produce efficient wavelength-selective reflectors, waveguides, optical circuits, beam steering devices, optical multiplexors, and much more [11]. PCs can be designed to interact strongly with particular optical wavelengths through selection of their materials and the period of their modulation. At the wavelengths of “optical resonance” (also known as “guided mode resonance”), light will couple strongly to the PC structure for a particular incident angle, resulting in electric fields inside the PC that can be many times higher than the electric field of incident radiation [12–14]. These optical resonances can be observed by an external observer simply by illuminating the PC at normal incidence with a broad band of wavelengths (such as from a light-emitting diode or a white light lamp) and observing a narrow band of wavelengths that are reflected back with nearly 100% efficiency. The precise wavelength that is back-reflected is determined by the refractive index of the PC materials and the PC period.
In order to design a PC that can serve as a biosensor, some portion of the resonant electric field must be in contact with liquid media that contains the analyte, providing a surface upon which biomolecules or cells may be adsorbed. Therefore, “surface PCs” in which the periodic modulation is open to the liquid, so that the resonant electric fields reside partially within the liquid are effective for application as biosensors. The region of high electric field magnitude near the PC surface, known as an “evanescent field,” extends up to ~200–300 nm from the surface. As described in previous publications [15–19], incorporation of material with greater dielectric permittivity than the media (which is typically either water or air) into the evanescent field region will increase the overall refractive index of the PC, resulting in a shift of the resonant wavelength to longer wavelengths. Fundamentally, all proteins, peptides, small molecules, and cell membranes have a greater dielectric permittivity than either air or water, because these biological analytes all contain electrons that are more easily displaced by the force of an externally applied optical electric field. The result is that the speed of light is slightly reduced when traveling through these analytes, compared to the speed of light through air or water. As summarized below, this small change is transduced into a measurable change in the wavelength of light that is reflected from a PC biosensor surface.
Due to the design flexibility for producing PCs by changing their period, it is not difficult to design a PC surface for which the resonant wavelength is selected to occur at the same wavelength that is used to excite a fluorescent or nanoparticle tag. In this case, any fluorophore that is adsorbed to the PC surface will experience the resonant electric fields of the evanescent field region, and will be excited to a greater extent than a similar fluorophore adsorbed on an ordinary glass surface and illuminated by the same light source. Importantly, surface-adsorbed fluorophores will be excited to a greater extent than those floating freely in solution or autofluorescent material in the sensor substrate, thereby increasing the signal-to-noise ratio (SNR) of fluorescence detection. This phenomenon, called “PC-enhanced excitation” or “evanescent resonance” (ER) has been used to increase the detection sensitivity for detection of DNA and proteins in microarray formats by up to 115x [20, 21]. More sophisticated design of PC surfaces enables resonances to occur at more than one wavelength simultaneously. Thus, it is possible for a resonance to be designed that occurs not only at the wavelength of a laser used to excite fluorescent dyes, but also at the wavelength of dye emission. This second resonance can be used to physically direct the fluorescent emission in a desired direction, such as toward a detector or microscope objective. Instead of distributing the fluorescent output into all directions in spherical coordinates, the PC enables a greater proportion of the output to be captured by the detection system, again resulting in an increase in signal-to-noise ratio. This second technique is called “PC-enhanced extraction.” The combination of PC-enhanced excitation PC-enhanced extraction was used to enhance the fluorescence from semiconductor quantum dots [22] with a magnification factor of 8x for the excitation effect and 13x for the extraction effect, for an overall sensitivity enhancement of 108x.
Together, the effects of PC-enhanced excitation and PC-enhanced extraction multiply, resulting in overall fluorescence detection signal gain of up to 550x in recently published reports [23–25]compared to performing the same assay on a glass microscope slide.
The use of enhanced resonant electric fields of a PC surface can be used to excite molecules beside fluorophores. We will briefly review how metal nanostructures can be integrated with the PC surface and how the nanostructures can couple with the PC enhanced fields to increase the local electric fields experienced by surface-adsorbed molecules. By designing the PC surface to provide a resonant electric field at the same wavelength of light that excites molecular vibrations in a SERS instrument, the SERS signal may be increased by over an order of magnitude. SERS represents yet another mechanism for label-free detection that may have the ability to compete with fluorescence detection for many assays, as available sensitivity improves.
The goal of this paper is to briefly review the demonstrated capabilities of PC surfaces in label-free and fluorescence detection, and to familiarize the reader with some of the initial applications of the technology. For full details, the reader is directed to full articles on each topic.
LABEL-FREE PC BIOSENSORS
Sensor Structure and Optical Fiber Detection Instrument
The sensor structure used most commonly for label-free detection contains a one-dimensional surface grating structure with a period of 550 nm (Figure 1a). It is produced via a room-temperature replica molding process using a UV-curable polymer on a transparent polyester sheet. The low refractive index polymer grating structure is subsequently coated with a film of high refractive index TiO2 to achieve the final sensor structure. The completed sensor is cut from the polyester sheet and attached to the bottom of a standard 96, 384, or 1536-well microplates (Figure 1b). The readout instrument (SRU Biosystems BIND Reader) [26–28] illuminates microplate wells from below with a broadband light source coupled to 8 optical fibers, each illuminating a ~1 mm diameter region of the PC surface at normal incidence. Reflected light is collected by a second optical fiber, bundled next to the illuminating fiber, and measured by a spectrometer. An automated motion stage enables parallel collection of reflectance data at timed intervals to acquire kinetic information from all 384 wells. When illuminated with broadband light, appropriately configured PCs are able to reflect narrow band light whose wavelength is directly dependent on the local density of adsorbed biomolecules. Association of macromolecules to the sensor surface modulates the peak wavelength value (PWV) of the reflected light, allowing for detection of binding by a shift in the PWV. In Figure 1c illustrates the general experimental setup of DNA-binding assays performed using PC biosensors.
Figure 1.
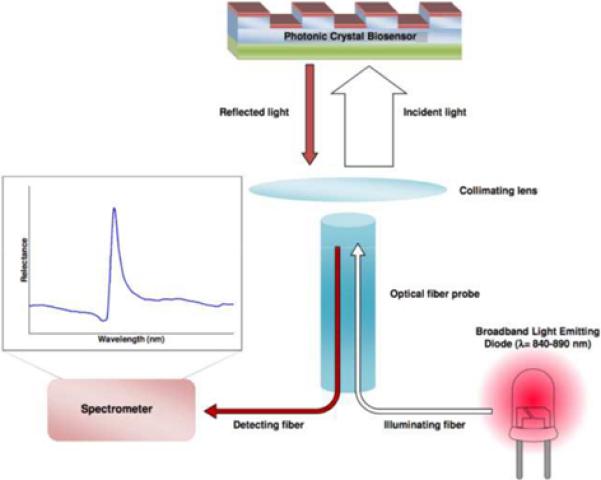

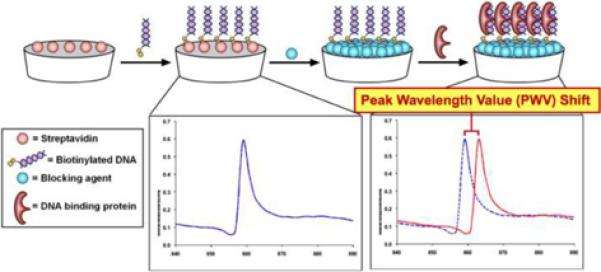
a). Schematic cross section of the PC biosensor surface structure comprised of a low refractive index polymer with periodic tooth pattern (light blue material) that is overcoated with a high refractive index film of TiO2 (red material). The PC surface is illuminated from below at normal incidence with an LED coupled to an optical fiber. The PC is illuminated with a broad band of wavelengths from the LED, but reflects only a narrow band of wavelengths. The reflected light is gathered into a second optical fiber that brings the reflected light to a spectrometer for measurement of the Peak Wavelength Value (PWV) of the resonantly reflected light, b). Photo of PC biosensors incorporated into 96, 384, and 1536-well microplates. c). Schematic for performing a typical biosensor assay. Each biosensor well is prepared with an immobilized capture probe that enables the sensor to specifically adsorb a target analyte to the surface. In this case, a streptavidin-coated biosensor is used to immobilized biotinylated double stranded DNA. Capture of a DNA-binding protein to the immobilized DNA results in a positive shift in the PWV. (Reprinted with permission from the American Chemical Society. ACS Chem. Biol., Vol. 3, No. 7, p. 437, 2008.)
Application in Screening Assays for Protein-DNA Interaction Inhibitors
Protein-DNA interactions are essential for fundamental cellular processes such as transcription, DNA damage repair, and apoptosis. As such, small molecule disruptors of these interactions could be powerful tools for investigation of these biological processes, and such compounds would have great potential as therapeutics. Unfortunately, there are few methods available for the rapid identification of compounds that disrupt protein-DNA interactions. Here we show that PC technology can be utilized to detect protein-DNA interactions, and can be used in a high-throughput screening mode to identify compounds that prevent protein-DNA binding. In a recently published report, the PC technology was used to detect binding between protein-DNA interactions that are DNA sequence-dependent (the bacterial toxin-antitoxin system MazEF), and those that are DNA sequence-independent (the human Apoptosis Inducing Factor (AIF)). The PC technology was further utilized in a screen for inhibitors of the AIF-DNA interaction, and through this screen aurin tricarboxylic acid (ATA) was identified as the first in vitro inhibitor of AIF. The generality and simplicity of the PC method should enable this technology to find broad utility for identification of compounds that inhibit protein-DNA binding.
Our goal was to develop a high-throughput screen that could be used to identify compounds that prevent the AIF-DNA interaction [29]. A 1 μM solution of biotinylated DNA was immobilized on streptavidin coated PC biosensors, and Starting Block was then added to reduce non-specific interactions between AIF and the biosensor surface. AIF (3.51 μM) and putative small molecule inhibitors (25 μM) were incubated together for 15 min at 25°C in a clear 384-well plate (Falcon); reference wells for each compound were also prepared in the same 384-well plate; these solutions were then transferred to the DNA-containing 384-well biosensor plate. Compounds that inhibit the AIF-DNA interaction would prevent the PWV shift observed in the AIF-DNA binding event. In this fashion, approximately 1000 compounds (obtained from an in-house compound collection[30]) were screened in duplicate at a concentration of 25 μM. All experimental wells were normalized against the following two reference wells: AIF with no biotinylated DNA (to account for the nonspecific interactions of AIF with the streptavidin coated biosensor), and biotinylated DNA with compounds (to account for nonspecific interactions with the DNA or biosensor surface). Most wells showed very little variation in the PWV shift, implying no prevention of the AIF-DNA interaction (Figure 2). However, one compound in this collection, aurin tricarboxylic acid (ATA), was found to inhibit the AIF-DNA interaction. In the screen ATA displayed ~80% inhibition of AIF-DNA binding, and was the only compound to exhibit significant inhibition out of the ~1000 compounds screened. The PC biosensor was then used to assess the effect of a range of concentrations of ATA to determine its IC50 value for DNA binding. This method has recently been extended to screen a 200,000-molecule library for additional inhibitors of AIF-DNA binding, resulting in 6 additional compounds that are currently undergoing optimization and validation through cell-based assays.
Figure 2.
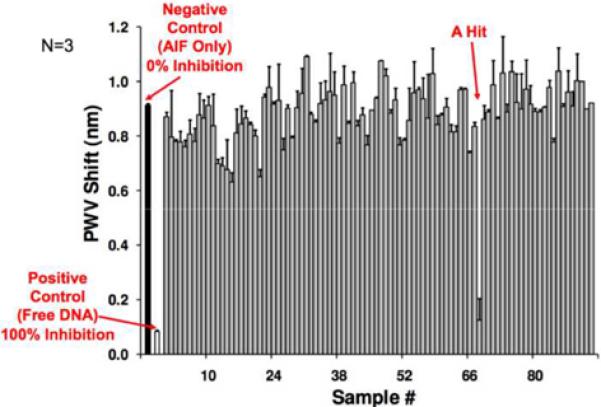
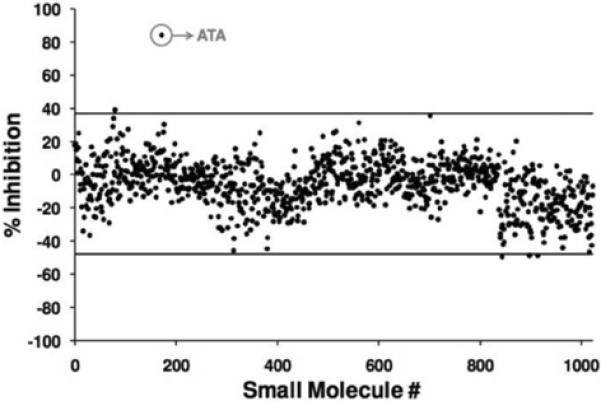
a). Results from a small section of a screen in which we seek to identify chemical compounds that have the ability to inhibit the binding of AIF to immobilized DNA. Chemical compounds are added to PC biosensor microplate wells along with the protein AIF. Each microplate well incorporates multiple negative control wells with no compound present, and positive controls where free DNA serves as a strong inhibitor of AIF binding to immobilized DNA. Here, a single hit is identified. b). A larger portion of the same screening campaign with the PWV shift data scaled to percent inhibition based upon the positive/negative controls. One molecule, ATA, was identified as an inhibitor from a group of ~1000 compounds shown here. Negative inhibition values represent compounds that enhance the binding of AIF to DNA. (Reprinted with permission from the American Chemical Society. ACS Chem. Biol., Vol. 3, No. 7, p. 437,2008.)
Application in Screening Assays for Cell Surface Interactions
In addition to registering a positive shift in reflected wavelength due to the adsorption of biomolecular layers, the PC surface also is able to detect the adsorption of large biological structures, including cells. In the same manner that the dielectric permittivity of the PC surface is increased when biomolecules displace water molecules within the evanescent field region, the lipid bilayer of cells and the protein contents of cells will also cause an increase in dielectric permittivity when the cell forms a tight association with the PC surface. While cells that simply rest upon the PC surface will not increase the reflected PWV, those that adhere to the surface produce a large PWV increase at the location of the cell. Because the PC surface may be prepared with biomolecular coatings of antibodies that specifically recognize proteins expressed on the outer surface of cells, or with extracellular matrix (ECM) coatings that promote adhesion, it is possible to specifically gather target cells from a mixed population, and to bind cells that would ordinarily be suspended in solution[31]. The advantage of using a PC biosensor as a method for monitoring cell attachment is that no staining of the cells is necessary, so that cells need not be killed for measurement. As a result, cells can be measured directly in their culture environment multiple times over the course of several days (if necessary), and the biosensor microplate may be returned to an incubator between measurements. Using the optical fiber-based detection instrument (Figure 1a), only a small number of cells are required to register a measurable PWV shift, although measurements represent an averaged PWV shift over the ~1 mm2 illuminated area. However, a high resolution imaging detection instrument [32] is capable of measuring the attachment of individual cells and monitoring them over many measurements. The biosensor provides information that is distinct from that provided by optical microscopy from the standpoint that the biosensor is a tool for measuring cell attachment to the surface, and how cell adhesion is modulated by changes in the cells' chemical environment over time.
For example, PC biosensors are used to measure the attachment of cancer cells to the biosensor surface, and to quantify how cancer cell proliferation is modulated by the presence of a variety of drugs. Used in this way, the sensor serves as a screening tool that is capable of rapidly identifying drugs from a compound library that induce apoptosis, and can subsequently determine the IC50 values of “hit” compounds through a dose-response measurement[33, 34]. Cell signaling from membrane receptors such as G-Protein Coupled Receptors (GPCRs) to integrin proteins that mediate attachment between cells and surrounding tissues is an important area of pharmaceutical research, as GPCRs represent important targets for drug discovery. For example, it has been shown recently that GPCR activation/inactivation can regulate integrin activity, resulting in gross changes in cell morphology and function [35]. One can use the PC biosensor to monitor changes in cell adhesion by first growing a layer of cells on the sensor surface (or on an ECM coating on the sensor surface), and then exposing the cells to drugs that selectively activate a targeted sub-class of GPCR. When cells become more thoroughly attached to the biosensor surface (for example, through a flattening of the cell structure), a positive shift in PWV is measured, while a negative shift in PWV is measured when the cells become less attached (for example, through rounding of the cell structure). As shown in Figure 3, selective activation of different classes of GPCR results in different kinetic characteristics of the cell adhesion as a function of time. For example, activation of the Gq receptors of Human Embryonic Kidney (HEK) cells results in a rapid increase in cell attachment, while activation of the Gi receptor results in a more moderated increase in cell attachment followed by a slow return to the baseline level. In a similar fashion, distinct kinetic responses have been recorded for several cell types in response to muscarinic, opioid, beta-arrestin, and P2Y ligands. The sensor response provides more subtle information than simply measuring proliferation or cytotoxicity by recording how cell adhesion, and its response over time, is modulated by receptor stimulation. As a general-purpose tool, it can be used to study the effects of full agonists, partial agonists, inverse agonists, and receptor desensitiziation. The system can be used with any type of cell, including overexpressed, endogenous, primary, or stem.
Figure 3.
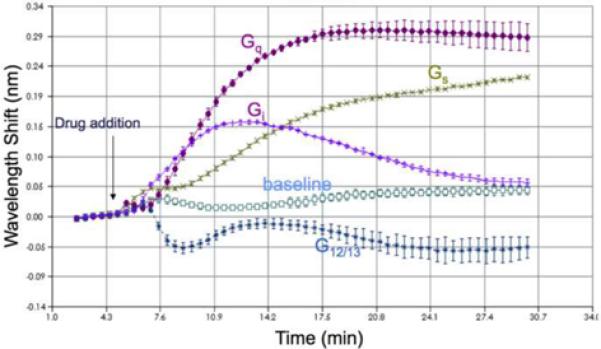
Kinetic monitoring of the PVW for immobilized HEK cells on the PC biosensor surface after exposure to four different drugs that specifically target different GPCR receptors (Gq, Gi, Gs, and G12/13). The response profile represents measurement of changes in the adhesion of the cells induced by GPCR signaling to integrins on the cell surface. Data courtesy of Jason Brown at GlaxcoSmithKline.
PC FLUORESCENCE ENHANCEMENT
The simultaneous quantitation of multiple proteins in a patient's fluid sample promises to aid researchers seeking to understand protein interaction networks and may potentially be clinically useful for diagnosis and prognosis with serum biomarkers[36]. One approach to multiplexed protein detection has been an adaptation of the DNA microarray format to immunoassays. Fluorescence-based protein microarrays have demonstrated detection limits comparable to their enzyme-based counterparts, enzyme linked immunosorbent assays (ELISA), while measuring multiple proteins within each array. These protein microarrays have been adapted and optimized for detection of cancer biomarkers[37] and cytokines[38–40]. Cytokines are a particularly promising class of analytes for multiplexed detection because they rarely act alone and rely on the upregulation or downregulation of multiple cytokines simultaneously to achieve a particular physiological effect. Cytokines are associated with immune responses to infection, but may be associated with non-infectious diseases. Because the immune system is integrated with other physiological systems such as the cardiovascular and gastrointestinal systems and cytokines often act as a signaling system throughout the body, these proteins may be a valuable tool in understanding and diagnosing disease. While protein microarrays on optically passive surfaces such as glass slides have been useful in multiplexed cytokine quantitation, the utility of these arrays can be expanded by a more accurate determination of protein levels as well as lowered limits of detection.
In recently published reports, we have demonstrated how PC surfaces can be used to achieve improved detection sensitivity and more accurate quantification of a representative protein biomarker compared to performing the same immunoassay on a glass surface [20]. Using PC enhanced excitation, we have demonstrated fluorescence enhancement from the fluorescent dye Cyanine-5[41] and detailed the spectral characteristics of the PC-fluorophore interaction[42] as well as the dependence on distance from the PC on enhanced excitation[43].
We performed a microspot fluorescence immunoassay for the cytokine TNF-a simultaneously on glass slides and PC surfaces under identical experimental conditions to evaluate the impact of enhanced fluorescence on the assay. The PC used in this work was similar to a combined label-free biosensor and enhanced fluorescence device described previously[41]. While this PC is capable of label-free detection of proteins that could enable spot density quantitation, we focus on the impact of enhanced fluorescence on the signal-to-noise ratio (SNR) of the assay since this can allow more accurate quantitation of protein levels at the lowest concentrations assayed. Using a nanoreplica molding process, PCs the size of microscope slides were fabricated for compatibility with commercial microarray spotters and scanners. A layer of SiO2 was added to the PC so an identical surface chemistry interaction can be achieved on both the PC and glass slides. A microspot immunoassay was performed on both substrates using a fluorescent Cyanine-5 label. By evaluating the immunoassay over a concentration series on glass and PCs, the impact of PC enhanced fluorescence on the assay resolution and detection limit was assessed.
Fluorescence measurements were taken using a commercially available confocal microarray scanner with user-adjustable angle of incidence laser excitation (LS Reloaded, Tecan) in order to allow alignment of the PC resonance with the incident wavelength. The PC slides and glass slides were scanned with identical conditions (photomultiplier tube (PMT) gain, incidence angle). PC slides were scanned at an angle that fulfills the resonant condition at 633 nm (3.2°) and an angle at which no resonance occurs at this wavelength (20°). Array Pro Analyzer software was used to quantify spot and background fluorescent intensities. ImageJ software was used to generate spatial profiles of the fluorescence data. ProMAT (http://www.pnl.gov/statistics/ProMAT) was used to fit fluorescence data to a four-parameter logistic model and to calculate the lower limit of detection for the immunoassay. Figure 4a illustrates the enhanced SNR for spots incubated with the lowest concentration of TNF-a (1.6 pg/ml), with an estimated SNR enhancement of over 8 times. This SNR enhancement is one component in lowering the detection limit of the immunoassay, which can be addressed in more detail by an analysis of the complete concentration series.
Figure 4.
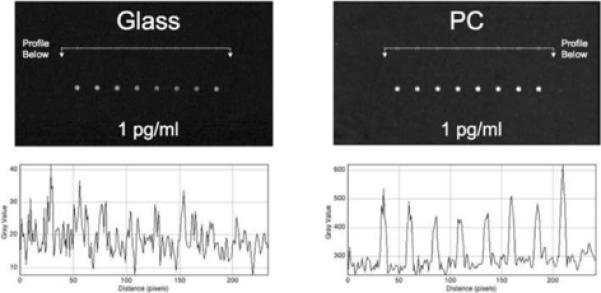
a). Fluorescent intensity images gathered with a confocal laser scanner for ~100 μm diameter spots of anti-TNFa capture probe after exposure to a 1 pg/ml solution of TNFα and subsequent labeling with Cy5-labeled secondary antibody. At this concentration, detection is near the detection limit when the assay is performed on a glass surface, but the PC surface shows robust signal-to-noise ratio. b). Dose-response characteristic for the TNFα assay, showing enhanced signal using the PC surface. Full assay details are provided inAnal. Chem. Vol. 80, No. 23, p. 9013, 2008.
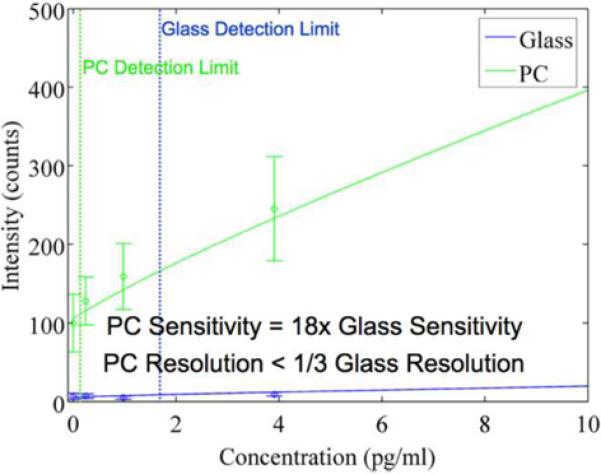
The SNR enhancement allows for an increased fluorescence signal that is higher than a concurrent increase in noise - improving the resolution and detection limit. The quantitative lower limit of detection for the resonant PC was 1/3 the limit of the glass slide. Furthermore, the lowest concentration assayed, 1.6 pg/ml, could be detected (but not quantified) above noise on the PC and not on the glass slide. The improvement of these metrics can be attained without an alteration of the basic instrumentation used for a protein microarray experiment, so this technology should be straightforward to implement by labs currently pursuing microarray research. A typical dose-reponse curve for the TNF-a assay performed upon both a PC surface and an ordinary glass microscope slide surface is shown in Figure 4b, showing the gain in fluorescence signal that is achieved for low concentration analyte.
The cytokine TNF-a plays an integral role in the immune response during infection and has been studied as a biomarker for sepsis, a state of acute inflammation throughout the body that is responsible for more than 100,000 deaths a year in the U.S[44]. An evaluation of cytokine concentrations in sepsis diagnosis determined that TNF-a could be a useful biomarker with a cutoff value of 11.5 pg/ml[45], which is below the limit of detection for the glass slide but not the PC in this study. This cutoff is not much higher than normal physiological TNF-a serum concentrations of 5–10 pg/ml[46, 47], which underscores the importance of resolution - a metric that was significantly improved in the PC relative to the glass slide. While the sensitivity and specificity of TNF-a in sepsis diagnosis is not high enough to warrant its use as a lone biomarker, the addition of other cytokines in the immunoassay may be powerful and is a subject of future study. The protein microarray format lends itself to quantitation of multiple serum proteins at once by spotting capture antibodies to multiple analytes, and accurate, multiplexed cytokine measurement would be a valuable tool in understanding the modulation of the immune system during sepsis.
LABEL-FREE AND ENHANCED FLUORESCENCE DETECTION COMBINED
Recently, we reported on the design and demonstration of an optical imaging system capable of combining high resolution label-free imaging of a PC surface with PC-enhanced fluorescence. With these two capabilities combined within a single detection system, we demonstrated label-free images self-registered to enhanced fluorescence images with 328x more sensitive fluorescence detection relative to a glass surface. This technique was applied to a DNA microarray where label-free quantification of immobilized capture DNA enables improved quality control and subsequent enhanced fluorescence detection of dye-tagged hybridized DNA yields 3x more genes to be detected versus commercially available microarray substrates.
In general, label-free biosensor imaging methods such as surface plasmon resonance (SPR) imaging [48, 49] and PC imaging do not offer the same level of sensitivity as fluorescence-based methods. However, for most fluorescent surface-based assays, there is no mechanism for quantification of the density of immobilized capture ligand. This capability is especially important for the production of DNA and protein microarrays that are produced by pin-spotting or piezoelectric jet spotting because a large variability exists in spot size and density [50]. The ability to perform LF imaging of immobilized ligand spots would potentially provide a quality control tool for elimination of missing spots or spots with poor characteristics (in terms of density, size, or uniformity) to improve the reliability of microarrays for disease diagnostics based upon detection of expressed genes or protein biomarkers. Because the immobilized spot density is typically high, detection by LF methods does not pose a difficult sensitivity challenge, although sufficient spatial resolution is required for imaging capture spots that are 50–200 mm in diameter. Therefore, one application of this technology is the detection of capture spots in LF mode and subsequent detection of fluorescent-tagged analytes in EF mode, where the highest possible level of sensitivity is desired.
A schematic drawing of the LF/EF imaging microscope is shown in Figure 5. A 35mW HeNe laser chosen to align with the excitation spectrum of the fluorescent dye cyanine-5 (Cy5) passes through a half-wave plate (for polarization control), a variable neutral density filter, a rotating diffuser (to reduce speckle and fringes at the imaging plane due to spatial coherence), a 10x beam expander, an aperture, and a motorized angle-tunable mirror. The gimbal-mounted mirror sits on top of a motorized linear stage in order to maintain a constant illumination area on the device as the mirror rotates. The remainder of the imaging path makes use of an Olympus BX-51 upright microscope with a Cy5 filter cube (Semrock) and a variety of objectives. Several important features make this implementation ideal for combined EF and LF imaging. First, it uses a common beam-path for both imaging modes, facilitating acquisition of spatially registered images of fluorescence and surface-bound molecular density. Second, the use of a charge-coupled device (CCD) rather than laser scanning imaging simplifies the optical setup and enables large-area, high-resolution and high-throughput analysis. Third, a high-resolution motorized gimbal-mounted mirror and beam-expanded laser provide efficient and selective light coupling to the PC, especially crucial for the narrow resonances that provide optimal fluorescence enhancement and sensitive LF detection. Lastly, other imaging techniques available on the microscope, including reflected brightfield and differential interference contrast, can be overlaid with EF and LF images.
Figure 5.
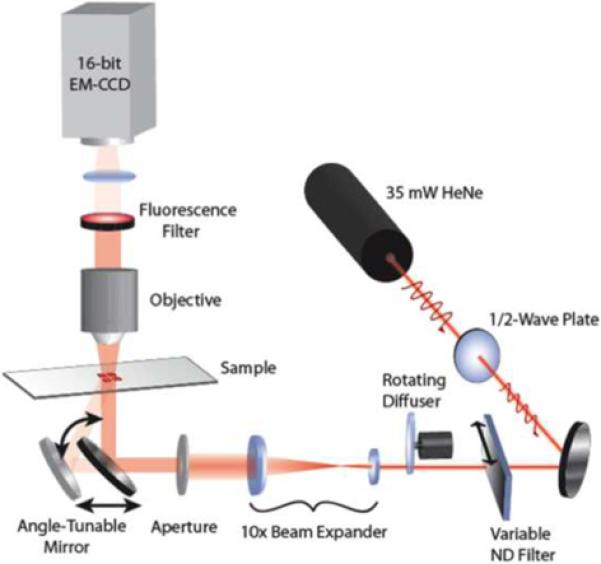
Schematic diagram of the instrument capable of providing high resolution label-free images on the PC surface and for providing PC-enhanced fluorescence images. The system is comprised of an ordinary upright fluorescence microscope, but used a collimated laser illumination source with a computer controlled mirror that allows tuning the incident illumination angle to match the PC resonance condition. (Reprinted with permission from the Optical Society of America. Optics Express, Vol. 17, No. 15, p. 13222,2009)
For LF imaging, the laser passes through the neutral density filter, rotating diffuser, and a blank in the filter wheel on the fluorescence microscope. A custom data acquisition and control program (LabView, National Instruments) translates the motorized mirror mount over a small range of angles and captures a single image for each discrete illumination angle. The resonant angle is computed for each pixel in the image stack by fitting the transmission versus angle data with a Lorentzian lineshape and finding the resonant angle. This resonant angle map is the LF image and can be used to quantify and visualize surface-bound density, as well as to find the appropriate resonant angle for EF imaging. The EF mode is generally used without the neutral density filter, without the rotating diffuser, and with an appropriate fluorescence filter set centered at 690 nm to match the Cy5 spectral emission maximum. For EF imaging, the desired angle of illumination is set in the control software and single images, sequences, or video can be captured.
Before biologically relevant experiments can be performed with a PC sensor, the surface must be functionalized with appropriate reactive groups. We chose to use a vapor-deposited monofunctional epoxysilane monolayer for its low autofluorescence, high density, and excellent uniformity [51]. The silane group provides covalent linkage to the surface oxide through free OH-groups, while the epoxide is highly reactive with proteins through the amino terminus or exposed lysine residues. Upon successful surface functionalization, 300 pL spots of strepavidin-cyanine-5 (SA-Cy5) at a range of concentrations from 50 mg/ml down to 24 ng/ml in 0.1% trehalose in PBS were deposited onto the PC using a non-contact piezoelectric spotting system (Perkin Elmer Piezorray). Following overnight incubation, the SA-Cy5 spots were washed in 0.1% Tween in PBS and then dH20 with gentle agitation and subsequently dried under nitrogen.
A LF image was constructed by first imaging laser transmission through the SA-Cy5-immobilized PC for a range of angles, fitting the resonance profile pixel-by-pixel for 100 angles, and then generating a spatial map of the resonant angle across the imaged area. A 4x 0.10 NA objective (Olympus) was used to yield a 4 mm2 imaged area. For each LF image captured, on-resonance fluorescence images were also gathered. Figs 4(a) and (b) show precise registration between the EF and LF images, respectively.
In order to demonstrate the potential utility of combined EF and LF imaging, a DNA microarray experiment was carried out on PC and control glass slides. PC slides were functionalized with (3-glycidoxypropyl)trimethoxysilane at 185 mTorr overnight, and were run in the experiment in parallel with commercially available glass microarray slides (Corning UltraGAPS). Both PCs and glass slides were spotted with a set of 192 different 70mer oligonucleotide sequences representative of known Glycine max genes. Using a QArray2 (Genetix) contact pin-spotter, slides were spotted with 40 replicates per oligo for a total of 7680 spots on a single slide. After rinsing unbound DNA from the array, LF images of the immobilized 70mers were captured.
Comparing fluorescence images on the PC to those captured on the control glass slide allows for a clear demonstration of the significant fluorescence enhancement that can be achieved for DNA microarrays using the instrumentation and PCs described and characterized in this work. Figure 7a shows an EF image of an area containing 64 spots where the laser illumination has been aligned to the resonance angle. A second image was captured of the same area for laser illumination several set degrees off of the resonance angle (not shown). Figure 7b contains a third image that was captured of the same array pattern on a glass slide substrate. The spot size variation between the glass and PC surface is due to differences in the hydrophobicity between the commercially and laboratory prepared surface chemistries. Gain and exposure settings were held constant for the acquisition of these three images. Registered cross section profiles through five replicate spots are given for the three devices in Figure 7c, demonstrating 11.5x excitation enhancement (from off to on-resonance on the PC) and 9.3x extraction enhancement (from the glass slide to the PC illuminated off-resonance). The effects of these multiply to provide up to 109x total fluorescence enhancement (from glass slide to PC on-resonance). Using the greater sensitivity afforded by the PC surface, three times as many genes on the PC versus the glass slide exceed the detection limit, resulting the capture of gene expression information from genes that ordinarily would have been classified as unexpressed [24].
Figure 7.
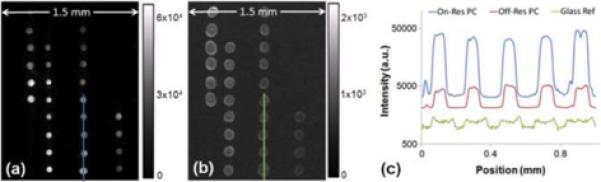
Fluorescence image of hybridized DNA microarray on a PC illuminated on-resonance a) and on a control glass slide b). Intensity cross-sections through five replicate spots for the PC illuminated on-resonance, illuminated off-resonance, and for the glass slide c). (Reprinted with permission from the Optical Society of America. Optics Express, Vol. 17, No. 15, p. 13222,2009.)
PC-ENHANCED SURFACE ENHANCED RAMAN SPECTROSCOPY
We have recently demonstrated that the resonant near-fields of a large-area replica molded Photonic Crystal (PC) slab can efficiently couple light from a laser to SiO2-Ag “post-cap” nanostructures deposited on the PC surface by a glancing angle evaporation technique for achieving high SERS enhancement factor. Although enormous enhancement factors have been achieved using metal structures, further enhancement of Raman signals is still desirable to reduce laser power and accumulation time for detection of trace quantities of analytes.
We used the Glancing Angle Deposition (GLAD) technique[52] to create a high density coating of electrically isolated Ag nanoparticles that are supported vertically from the PC surface upon 50 nm-tall SiO2 dielectric posts (Figure 8). The GLAD technique has been demonstrated to be a simple method for fabrication of metal structures with high SERS enhancement factor because the randomly distributed and sized Ag nanostructure have numerous interconnections and strong EM field within the gaps between the nano-particles[53]. Figure 8a shows a cross sectional schematic of the PC-SERS substrate, comprised of a 1-dimensional PC slab and a SiCh-Ag “post-cap” nanostructure coating.
Figure 8.
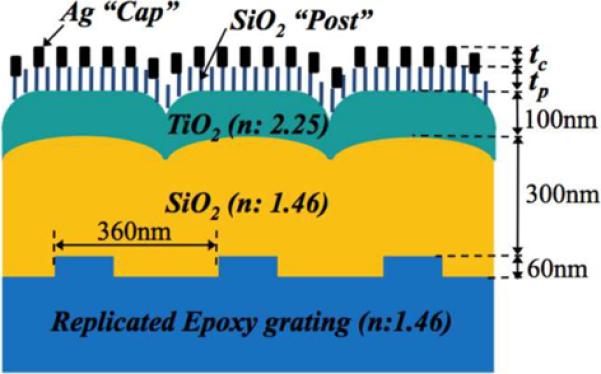
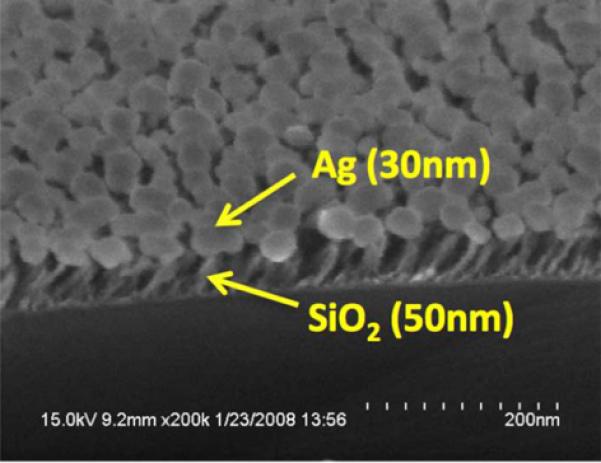
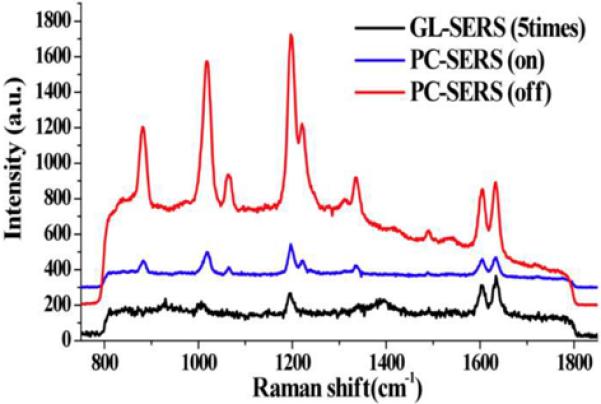
a). Cross section schematic of the PC surface structure used for PC-SERS. The PC surface is covered with a layer of dielectric nanorod “posts” that each support a 20–30 nm diameter Ag “cap” for obtaining the SERS effect. The design goal is for the Ag caps to reside within the evanescent field region of the PC when it is illuminated with a laser at a combination of wavelength and incident angle that excites an optical resonance. b). Scanning electron microscope photo of the post-cap structures, c). Measured SERS signals from a layer of BPE, comparing the SERS intensity with the post-cap structures without a PC (GL-SERS curve), with a PC illuminated at an angle that does not excite resonance (PC-SERS (off)) and at an angle that excites resonance (PC-SERS (on)). (Reprinted with permission from the American Physical Society, Appl. Phys. Lett., Vol. 93, p. 143122,2008.)
For experimental verification of the enhancement effects of PC-SERS, Raman spectra of trans-l,2-bis(4pyridyl)ethane (BPE) on ordinary microscope glass-coated SERS (GL-SERS) and PC-SERS substrates were measured and compared. The Raman detection instrument was comprised of a TE polarized Argon-dye laser excitation source (Coherent, INNOVA-90 and CR-500, λ = 600 nm, output power = 82 mW), a sample holder with a rotational stage, a power meter to measure laser transmittance through the sample, imaging optics, a spectrometer (SPEX-Triplemate), and a cooled CCD (Princeton Instruments). Raman scattered light was collected from a 235.4 × 51.8 μm2 area, and the diameter of the laser beam spot was ~470 μm. A 2 μL droplet of BPE dissolved in methanol (1×10–4 M) was applied to each substrate by pipette. The BPE/methanol droplet spread out to form a circular region with a diameter of ~1.0 cm on the substrate, resulting in a density of ~ 1.53 × 1014 molecules/cm2 and a number of exited molecules of ~ 31 femto moles. For a laser wavelength of λ = 600 nm, the PC resonance could be excited by illuminating at an angle of ~ ±13 °, and precise tuning to the on-resonance condition for any substrate could be achieved by adjustment of the rotation stage to obtain a minima in laser transmitted intensity through the PC.
Figure 8c shows the Raman spectra obtained from BPE using both GL-SERS and PC-SERS substrates at on/off resonance conditions, where the spectrum for the GL-SERS substrate was multiplied by a factor of 5 to enable visualization. The Raman signal was defined as the integrated intensity of the 1200 cm 1 peak after subtraction of the background signal. Our results show that the Raman signal from the PC-SERS substrate increased significantly. The enhancement effect upon the Raman signal due to the PC is observed throughout the entire concentration range, with an enhancement factor of 10–30x between GL-SERS and on-resonance PC-SERS.
PHOTONIC CRYSTAL LASER BIOSENOSRS
A wide variety of optical resonator structures have been used for label-free detection of chemical compounds, biomolecules, and cells.[54, 55] Several approaches have been commercially developed for application in life science research, environmental monitoring, quality control testing, and diagnostic testing.[18, 56] Label-free resonant optical sensors generally detect shifts in resonant wavelength or coupling angle caused by the interaction between the target molecule and the evanescent portion of the resonant modes. The narrow spectral linewidth achieved by using high 𝑄 factor (>105) passive optical resonators enables sensor systems to resolve smaller wavelength shifts associated with the detection of analytes at low concentration, or detection of biomolecules with low molecular weight, such as drug compounds.[57–61] While detection resolution can be substantially improved through the use of high 𝑄 factor passive resonators, the sensitivity and dynamic range of the system is generally decreased, although certain examples of passive resonators have achieved high 𝑄 factor and high sensitivity simultaneously. [62] In addition, implementation of high 𝑄 factor optical resonators typically requires high precision alignment for evanescent light in/out coupling, providing potential limits to their practical application. Active resonator sensors, such as laser-based optical biosensors[63–65] have been drawing special interest because they generate their own narrow linewidth stimulated emission, while retaining simple instrumentation and eliminating the requirement for high precision evanescent coupling to waveguides or tapered optical fibers. While our first demonstration of laser-based biosensors used glass-based materials [63], practical biosensor applications demand an inexpensive fabrication method that can be performed over large surface areas, a plastic-based sensor that can be inexpensively manufactured is more desirable. Recently, we demonstrated that a PC biosensor surface structure that incorporates a laser dye can be excited to emit high intensity, narrow linewidth laser emission, in which the laser output wavelength is modulated by the attachment of biomolecules to the sensor surface. The laser structure is commonly known as a Distributed Feedback (DFB) structure because the optical feedback required to produce stimulated emission is provided by partial reflections that occur for light with a propagation vector perpendicular to the grating lines. Like the PC label-free biosensor, the DFB laser label-free biosensor is fabricated with a plastic-based process on a flexible plastic substrate using a high surface-area nanoreplica molding process.[66],[67]
A schematic cross-sectional diagram of the designed DFB laser structure is shown in Figure 9 [68, 69]. The low refractive index polymer layer applied to the substrate functions as a cladding layer, upon which a thin film of high refractive index polymer provides vertical light confinement and feedback along the horizontal direction. Doped with laser dye, this high refractive index layer also contributes to the light amplification of the cavity oscillation mode. Altering the refractive index of the media exposed to the DFB laser surface or surface adsorption of biomolecules changes the effective refractive index associated with the resonant mode, and results in modulation of the stimulated emission wavelength. By controlling the guidance layer thickness, the DFB laser is designed to exhibit single mode radiation to facilitate determination of the laser wavelength shift.
Figure 9.
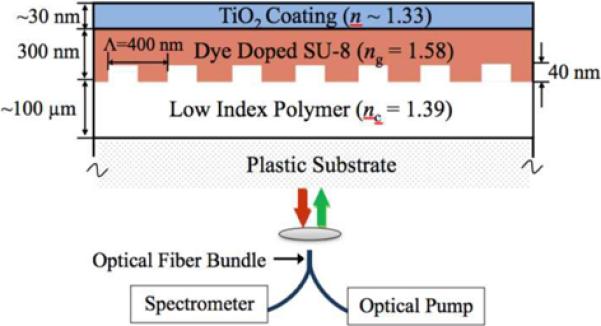
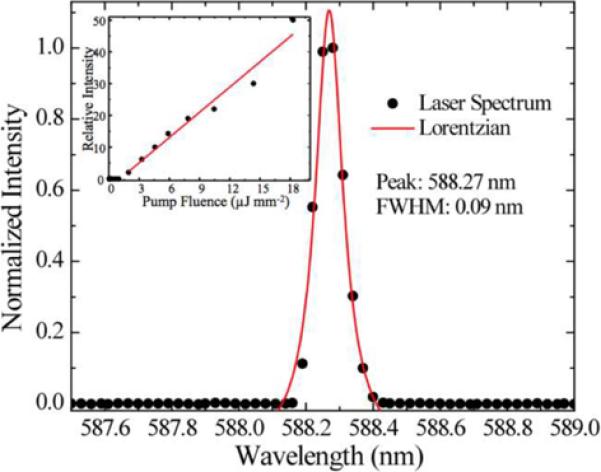
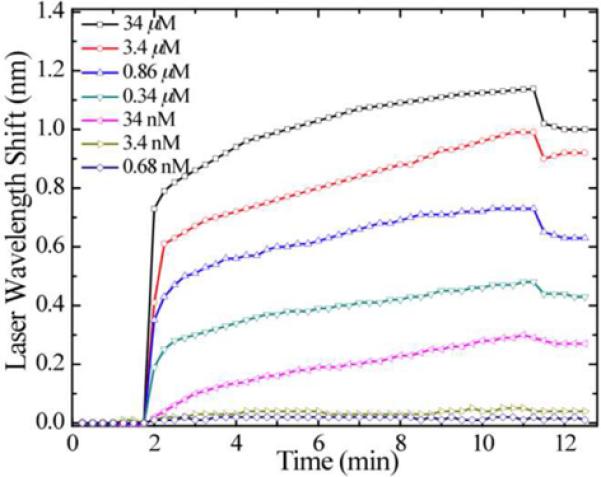
a). Cross section schematic of the surface structure used to create a photonic crystal DFB laser biosensor. Like other PC structures, a replica molded grating surface is produced on a flexible plastic substrate. A thin film of polymer that contains a laser dye is applied on top of the grating to provide a gain medium for lasing. b). Narrow bandwidth output of the laser biosensor, showing a peak that is less than 0.1 nm wide. c). Measurement of laser emission wavelength as a function of time for detection of human IgG adsorbed to immobilized Protein A for various concentration. (Reprinted with permission from the American Physical Society, Appl. Phys. Lett., Vol. 93, p. 111113,2008.)
CONCLUSION
As shown in the preceding examples, PC surfaces offer a variety of capabilities for applications in pharmaceutical high throughput screening, gene expression analysis, disease diagnostics, and life science research through the ability to detect adsorbed analytes in a label-free fashion and the ability to amplify the output of fluorescence and SERS measurements. The PC surfaces can be inexpensively fabricated from plastic-based processes and incorporated into a variety of liquid handling format used for biological assays. Detection instruments for PC-based label-free detection are simple and robust, with options that enable rapid screening of microplate wells with quantitative kinetic information, and options for high resolution imaging of adsorbed analytes for applications such as label-free microarrays, cell proliferation, and cell chemotaxis. PC surfaces for enhanced fluorescence may be designed for compatibility with commercially available confocal fluorescence laser scanners, but for optimal performance, a specially adapted fluorescence microscope with a collimated laser excitation source is most desirable. The enhanced-fluorescence microscope has the added benefit of simultaneously providing label-free detection of the PC surface, providing a means for evaluating the density of immobilized capture ligands. New applications for PC surfaces continue to emerge, including amplification of SERS, and the development of label-free biosensors with improved detection resolution though the incorporation of active dyes in the PC structure that enable the structure to provide laser output with extremely narrow bandwidth. As PC surfaces and detection instrumentation become more widely available, additional applications in the areas of cell membrane receptor imaging, single fluorophore detection, and DNA sequencing are expected to emerge.
Figure 6.
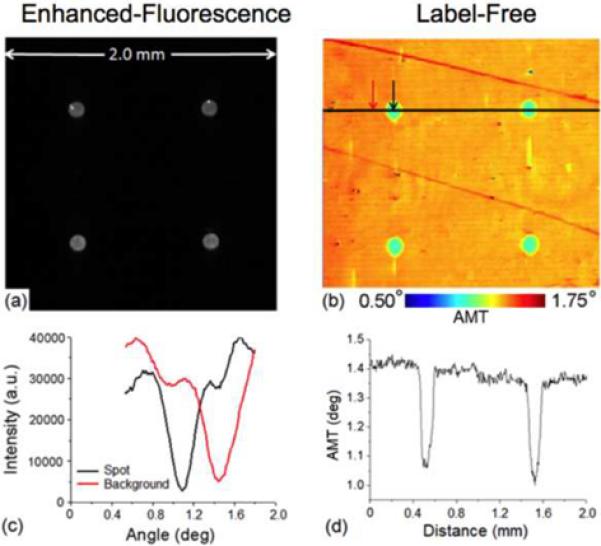
Enhanced fluorescence a) and label-free b) images of 50 μg/ml SA-Cy5 spots on a PC. Inverted transmission versus angle response for a pixel inside and outside the SA-Cy5 spot c) and cross-section of the label-free image through 2 SA-Cy5 spots, in d). Rather than measuring the PWV, the label-free imaging system measures the Angle of Minimum Transmission (AMT) by illuminating the sensor at a fixed wavelength while scanning the angle of illumination through computer-controlled rotation of the mirror. (Reprinted with permission from the Optical Society of America. Optics Express, Vol. 17, No. 15, p. 13222,2009)
ACKNOWLEDGEMENT
The author is grateful for financial support provided by NSF (NSF CBET 07-54122), NIH (R01 CA118562 and R01 GM086382), and SRU Biosystems. Any opinions, findings, and conclusions or recommendations in this work are those of the author and do not necessarily reflect the views of the funding agencies. As this paper is a review article, it represents the efforts of many talented and dedicated individuals. In particular, the author would like to acknowledge the contributions of Prof. Paul Hergenrother at the University of Illinois for his collaboration in small molecule screening, Prof. Lila Vodkin at the University of Illinois for her collaboration in gene expression analysis, and Dr. Lance Laing from SRU Biosystems for directing the cell-based assays reported here. The author gratefully acknowledges Dr. Julio Martin and Dr. Jason Brown from GSK for the GPCR data shown in Figure 3. The research of postdoc associate Seok-min Kim and graduate students Leo Chan, Maria Pineda, James Heeres, Meng Lu, Ian Block, Wei Zhang, Patrick Mathias, Vikram Chaudhery, and Nikhil Ganesh are summarized in the paper. The author discloses that he is a founder and CTO of SRU Biosystems.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Gonzalez R, Varnum SM, Zangar RC. Sandwich ELISA microarrays. Generating reliable and reproducible assays for high-throughput screening. Methods in Parmacol. Toxicol. 2008 In Press. [Google Scholar]
- [2].Gonzalez RM, Seurynck-Servoss SL, Crowley SA, Brown M, Omenn GS, Hayes DF, Zangar RC. Development and Validation of Sandwich ELISA Microarrays with Minimal Assay Interference. Journal of Proteome Research. 2008;7:2406–2414. doi: 10.1021/pr700822t. [DOI] [PubMed] [Google Scholar]
- [3].Eid J, Fehr A, Gray J, Luing K, Lyle J, Otto G, Peluso P, Rank D. Real-tiem DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- [4].Hell SW. Toward fluorescence nanoscopy. Nature Biotechnology. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- [5].Schena M, Shalon D, DAvis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- [6].Lofas S. Dextran modified self-assembled monolayer surfaces for use in biointeraction analysis with surface plasmon resonance. Pure and Applied Chemistry. 1995;67:829–34. [Google Scholar]
- [7].Lofas S, Malmqvist M, Ronnberg I, Stenberg E, Liedberg B, Lundstrom I. Bioanalysis with surface plasmon resonance. Sensors and Actuators B. 1991;5:79–84. [Google Scholar]
- [8].Haynes CL, Yonzon CR, Zhang X, Duyne RPV. Surface-enhanced Raman sensors: early history and the devleopment of sensors for qualitative biowarfare agent and glucose detection. Journal of Raman Spectroscopy. 2005;36:471–484. [Google Scholar]
- [9].Jeanmaire DL, Duyne RPV. Journal of Electroanalytical Chemistry. 1977;84:1. [Google Scholar]
- [10].Yablonovitch E. Inhibited spontaneous emission in solid-state physics and electronics. Physical Review Letters. 1987;58:2059–2062. doi: 10.1103/PhysRevLett.58.2059. [DOI] [PubMed] [Google Scholar]
- [11].Joannopoulos JD, Meade RD, Winn JN. Photonic Crystals. Princeton University Press; Princeton, NJ: 1995. [Google Scholar]
- [12].Budach W, Abel AP, Bruno AE, Neuschaefer D. Planar waveguides as high-performance sensing platforms for fluorescence-based multiplexed oligonucleotide hybridization assays. Analytical Chemistry. 1999;71:3347–3355. [Google Scholar]
- [13].Mathias PC, Ganesh N, Chan LL, Cunningham BT. Combined enhanced fluorescence and label-free biomolecular detection with a photonic crystal surface. Applied Optics. 2007;26:2351–2360. doi: 10.1364/ao.46.002351. [DOI] [PubMed] [Google Scholar]
- [14].Mathias PC, Ganesh N, Zhang W, Cunningham BT. Graded wavelength one-dimensional photonic crystal reveals spectral characteristics of enhanced fluorescence. Journal of Applied Physics. 2008;103:094320. [Google Scholar]
- [15].Block ID, Ganesh N, Lu M, Cunningham BT. A sensitivity model for predicting photonic crystal biosensor performance. IEEE Sensors Journal. 2008;8:274–280. [Google Scholar]
- [16].Cunningham BT, Laing L. Label-free detection of biomolecular interactions: applications in proteomics and drug discovery. Expert Opinions in Proteomics. 2006;3:271–281. doi: 10.1586/14789450.3.3.271. [DOI] [PubMed] [Google Scholar]
- [17].Cunningham BT, Li P, Lin B, Pepper J. Colorimetric resonant reflection as a direct biochemical assay technique. Sensors and Actuators B. 2002;81:316–328. [Google Scholar]
- [18].Cunningham BT, Li P, Schulz S, Lin B, Baird C, Gerstenmaier J, Genick C, Wang F, Fine E, Laing L. Label-Free Assays on the BIND System. Journal of Biomolecular Screening. 2004;9:481–490. doi: 10.1177/1087057104267604. [DOI] [PubMed] [Google Scholar]
- [19].Cunningham BT, Qiu J, Li P, Pepper J, Hugh B. A plastic colorimetric resonant optical biosensor for multiparallel detection of label-free biochemical interactions. Sensors and Actuators B. 2002;85:219–226. [Google Scholar]
- [20].Mathias PC, Ganesh N, Cunningham BT. Application of photonic crystal enhanced fluorescence to a cytokine immunoassay. Analytical Chemistry. 2008;80:9013–9020. doi: 10.1021/ac801377k. [DOI] [PubMed] [Google Scholar]
- [21].Zhang W, Ganesh N, Mathias PC, Cunningham BT. Enhanced fluorescence on a photonic crystal surface incorporating nanorod structures. Small. 2008;4:2199–2203. doi: 10.1002/smll.200800367. [DOI] [PubMed] [Google Scholar]
- [22].Ganesh N, Zhang W, Mathias PC, Chow E, Soares JANT, Malyarchuk V, Smith AD, Cunningham BT. Enhanced fluorescence emission from quantum dots on a photonic crystal surface. Nature Nanotechnology. 2007;2:515–520. doi: 10.1038/nnano.2007.216. [DOI] [PubMed] [Google Scholar]
- [23].Block ID. Electrical and Computer Eningeering. University of Illinois at Urbana-Champaign; Urbana, IL: 2009. Photonic crystal enhanced fluorescence and label-free bioimaging; p. 141. Ph.D. [Google Scholar]
- [24].Block ID, Mathias PC, Ganesh N, Jones ID, Dorvel BR, Chaudhery V, Vodkin L, Bashir R, Cunningham BT. A combined enhanced-fluorescence and label-free imaging instrument. Optics Express. 2009;17:13222–13235. doi: 10.1364/oe.17.013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ganesh N, Block ID, Mathias PC, Zhang W, Chow E, Malyarchuk V, Cunningham BT. Leaky-mode assisted fluorescence extraction: Application to fluorescence enhancement biosensors. Optics Express. 2008;16:21626–21640. doi: 10.1364/oe.16.021626. [DOI] [PubMed] [Google Scholar]
- [26].Cunningham BT, Li P, Schulz S, Lin B, Baird C, Gerstenmaier J, Genick C, Wang F, Fine E, Laing L. Label-free assays on the BIND system. J Biomol Screen. 2004 Sep;9:481–90. doi: 10.1177/1087057104267604. [DOI] [PubMed] [Google Scholar]
- [27].Lin B, Qiu J, Gerstenmeier J, Li P, Pien H, Pepper J, Cunningham B. A label-free optical technique for detecting small molecule interactions. Biosens Bioelectron. 2002 Sep;17:827–34. doi: 10.1016/s0956-5663(02)00077-5. [DOI] [PubMed] [Google Scholar]
- [28].Cunningham BT, Laing L. Microplate-based, label-free detection of biomolecular interactions: applications in proteomics. Expert Rev Proteomics. 2006 Jun;3:271–81. doi: 10.1586/14789450.3.3.271. [DOI] [PubMed] [Google Scholar]
- [29].Chan LL, Pineda MF, Heeres J, Hergenrother PJ, Cunningham BT. General method for discovering inhibitors of protein-DNA interactions using photonic crystal biosensors. ACS Chemical Biology. 2008;3:437–448. doi: 10.1021/cb800057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hergenrother PJ. Obtaining and screening compound collections: a user's guide and a call to chemists. Curr Opin Chem Biol. 2006 Jun;10:213–8. doi: 10.1016/j.cbpa.2006.04.005. [DOI] [PubMed] [Google Scholar]
- [31].Lin B, Li PY, Cunningham BT. A label-free biosensor-based cell attachment assay for characterization of cell surface molecules. Sensors and Actuators B. 2006;114:559–561. [Google Scholar]
- [32].Li P, Lin B, Gerstenmaier J, Cunningham BT. A new method for label-free imaging of biomolecular interactions. Sensors and Actuators B. 2004;99:6–13. [Google Scholar]
- [33].Chan LL, Gosangari S, Watkin K, Cunningham BT. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis. 2007;12:1061–1068. doi: 10.1007/s10495-006-0031-y. [DOI] [PubMed] [Google Scholar]
- [34].Chan LL, Gosangari S, Watkin K, Cunningham BT. Label-free imaging of cancer cells using photonic crystal biosensors and application to cytotoxicity screening of a natural compound library. Sensors and Actuators B. 2008;132:418–425. [Google Scholar]
- [35].Chigaev A, Waller A, Amit O, Sklar LA. Ga-coupled receptor signaling actively down-regulates a4bl-integrin affinity: A possible mechanism for cell de-adhesion. BMC Immunology. 2008;9 doi: 10.1186/1471-2172-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nature Reviews Drug Discovery. 2006;5:310–321. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Woodbury RL, Varnum SM, Zangar RC. Elevated HGF Levels in Sera from Breast Cancer Patients Detected Using a Protein Microarray ELISA. Journal ofProteome Research. 2002;1:233–237. doi: 10.1021/pr025506q. [DOI] [PubMed] [Google Scholar]
- [38].Li Y, Reichert WM. Adapting cDNA Microarray Format to CytokineDetection Protein Arrays. Langmuir. 2003;19:1557–1566. [Google Scholar]
- [39].Saviranta P, Okon R, Brinker A, Warashina M, Eppinger J, Geierstanger BH. Evaluating Sandwich Immunoassays in Microarray Format in Terms of the Ambient Analyte Regime. Clinical Chemistry. 2004;50:1907–1920. doi: 10.1373/clinchem.2004.037929. [DOI] [PubMed] [Google Scholar]
- [40].Kusnezow W, Syagalio YV, Ruffer S, Baudenstie N, Gauer C, Hoheisel JD, Wild D, Goychuk I. Optimal Design of Microarray Immunoassays to Compensate for Kinetic Limitations. Molecular and Cellular Proteomics. 2006;5:1681–1696. doi: 10.1074/mcp.T500035-MCP200. [DOI] [PubMed] [Google Scholar]
- [41].Mathias PC, Ganesh N, Chan LL, Cunningham BT. Combined enhanced fluorescence and label-free biomolecular detection with a photonic crystal surface. Applied Optics. 2007;46:2351–2360. doi: 10.1364/ao.46.002351. [DOI] [PubMed] [Google Scholar]
- [42].Mathias PC, Ganesh N, Zhang W, Cunningham BT. Graded wavelength one-dimensional photonic crystal reveals spectral characteristics of enhanced fluoresence. Journal of Applied Physics. 2008;103:094320. [Google Scholar]
- [43].Ganesh N, Mathias PC, Zhang W, Cunningham BT. Distance dependence of fluorescence enhancement from photonic crystal surfaces. Journal of Applied Physics. 2008;103:083104. [Google Scholar]
- [44].Carrigan SD, Scott G, Tabrizian M. Toward Resolving the Challenges of Sepsis Diagnosis. Clinical Chemistry. 2004;50:1301–1314. doi: 10.1373/clinchem.2004.032144. [DOI] [PubMed] [Google Scholar]
- [45].Balc C, Sungurtekin H, Gurses E, Sungurtekin U, Kaptanoglu B. Usefulness of procalcitonin for diagnosis of sepsis in the intensive care unit. Critical Care. 2002;7:85–90. doi: 10.1186/cc1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aziz N, Nishanian P, Fahey JL. Levels of Cytokines and Immune Activiation Markers in Plasma in Human Immunodeficiency Virus Infection: Quality Control Procedures. Clinical and Diagnostic Laboratory Immunology. 1998;5:755–761. doi: 10.1128/cdli.5.6.755-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Scala E, Pallotta S, Frezzolini A, Abeni D, Barbieri C, Sampogna F, Pita OD, Puddu P, Paganelli R, Russo G. Cytokine and chemokine levels in systemic sclerosis: relationship with cutaneous and internal organ involvement. Clinical & Experimental Immunology. 2004;138:540–546. doi: 10.1111/j.1365-2249.2004.02642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rothenhausler B, Knoll W. Surface-plasmon microscopy. Nature. 1988;332:615–617. [Google Scholar]
- [49].Giebel F, Bechinger C, Herminghaus S, Riedel M, Leiderer P, Weiland U, Bastmeyer M. Imaging of cell/substrate contacts of living cells with surface plasmon resonance microscopy. Biophysical Journal. 1999;76:509–516. doi: 10.1016/s0006-3495(99)77219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang W, Shmulevich I, Astola J. Microarray Quality Control. Wiley; Hoboken: 2004. [Google Scholar]
- [51].Dorvel B, B. R., Block ID, Mathias PC, Clare SE, Bergstrom DE, Cunningham BT, Bashir R. Vapor phase deposition of monofunctional alkoxysilanes for sub-nanometer level biointerfacing on oxide surfaces. Advanced Functional Materials. 2009 Under Review. [Google Scholar]
- [52].Zhao Y-P, Chaney SB, Shanmukh S, Dluhy RA. Polarized Surface Enhanced Raman and Absorbance Spectra of Aligned Silver Nanorod Arrays. J. Phys. Chem. B. 2006;110:3155–3157. doi: 10.1021/jp057406o. [DOI] [PubMed] [Google Scholar]
- [53].Ueno K, Juodkazis S, Mizeikis V, Sasaki K, Misawa H. Clusters of Closely Spaced Gold Nanoparticles as a Source of Two Photon Photoluminescence at Visible Wavelengths. Advanced Materials. 2008;20:26–30. [Google Scholar]
- [54].Narayanaswamy R, Wolfbeis OS. Optical sensors : industrial, environmental and diagnostic applications. Springer; Berlin; New York: 2004. [Google Scholar]
- [55].Cunningham AJ. Introduction to bioanalytical sensors. Wiley; New York: 1998. [Google Scholar]
- [56].Jonsson U, Fagerstam L, Ivarsson B, Johnsson B, Karlsson R, Lundh K, Lofas S, Persson B, Roos H, Ronnberg I, Sjolander S, Stenberg E, Stahlberg R, Urbaniczky C, Ostlin H, Malmqvist M. Real-Time Biospecific Interaction Analysis Using Surface-Plasmon Resonance and a Sensor Chip Technology. Biotechniques. 1991 Nov;11:620–&. [PubMed] [Google Scholar]
- [57].White IM, Fan XD. On the performance quantification of resonant refractive index sensors. Optics Express. 2008 Jan 21;16:1020–1028. doi: 10.1364/oe.16.001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yalcin A, Popat KC, Aldridge JC, Desai TA, Hryniewicz J, Chbouki N, Little BE, King O, Van V, Chu S, Gill D, Anthes-Washburn M, Unlu MS. Optical sensing of biomolecules using microring resonators. Ieee Journal of Selected Topics in Quantum Electronics. 2006 Jan–Feb;12:148–155. [Google Scholar]
- [59].Chao CY, Fung W, Guo LJ. Polymer microring resonators for biochemical sensing applications. Ieee Journal of Selected Topics in Quantum Electronics. 2006 Jan–Feb;12:134–142. [Google Scholar]
- [60].Hanumegowda NM, Stica CJ, Patel BC, White I, Fan XD. Refractometric sensors based on microsphere resonators. Applied Physics Letters. 2005 Nov 14;87 [Google Scholar]
- [61].Vollmer F, Braun D, Libchaber A, Khoshsima M, Teraoka I, Arnold S. Protein detection by optical shift of a resonant microcavity. Applied Physics Letters. 2002 May 27;80:4057–4059. [Google Scholar]
- [62].White IM, Oveys H, Fan XD. Liquid-core optical ring-resonator sensors. Optics Letters. 2006 May 1;31:1319–1321. doi: 10.1364/ol.31.001319. [DOI] [PubMed] [Google Scholar]
- [63].Lu M, Choi S, Wagner CJ, Eden JG, Cunningham BT. Label free biosensor incorporating a replica-molded, vertically emitting distributed feedback laser. Applied Physics Letters. 2008 Jun 30;92 [Google Scholar]
- [64].Fang W, Buchholz DB, Bailey RC, Hupp JT, Chang RPH, Cao H. Detection of chemical species using ultraviolet microdisk lasers. Applied Physics Letters. 2004 Oct 25;85:3666–3668. [Google Scholar]
- [65].Loncar M, Scherer A, Qiu YM. Photonic crystal laser sources for chemical detection. Applied Physics Letters. 2003 Jun 30;82:4648–4650. [Google Scholar]
- [66].Rogers JA, Meier M, dodabalapur A, Laskowski EJ, Cappuzzo MA. Distributed feedback ridge waveguide lasers fabricated by nanoscale printing and molding on nonplanar substrates. Applied Physics Letters. 1999 May 31;74:3257–3259. [Google Scholar]
- [67].Cunningham B, Lin B, Qiu J, Li P, Pepper J, Hugh B. A plastic colorimetric resonant optical biosensor for multiparallel detection of label-free biochemical interactions. Sensors and Actuators B-Chemical. 2002 Jul 25;85:219–226. [Google Scholar]
- [68].Lu M, Choi SS, Irfan U, Cunningham BT. Plastic distributed feedback laser biosensor. Applied Physics Letters. 2008;93:111113. [Google Scholar]
- [69].Lu M, Choi SS, Wagner CJ, Eden JG, Cunningham BT. Label free biosensor incorporating a replica-molded, vertically emitting distributed feedback laser. Applied Physics Letters. 2008;92:261502. [Google Scholar]


