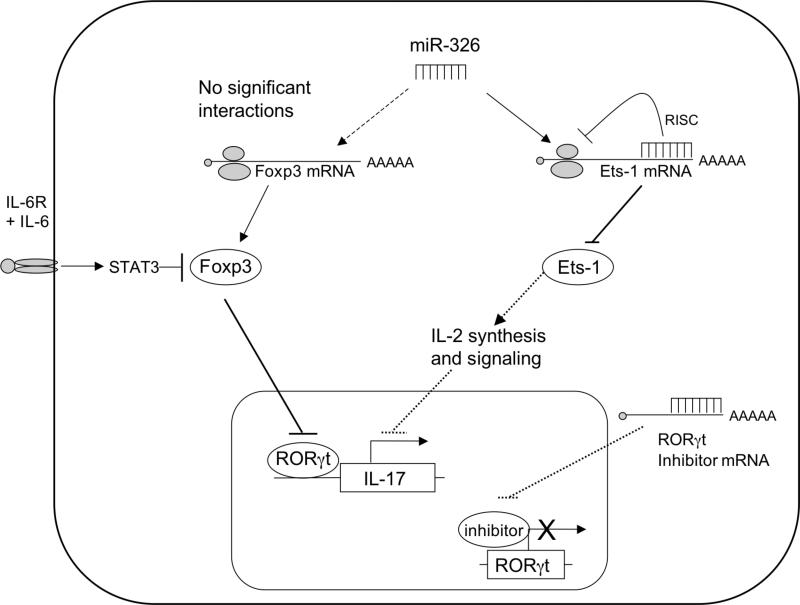
The differentiation of naïve CD4+ helper T cells (TH) into the various lineages of effector populations drives immune responses against pathogens or against self-antigens in the case of autoimmune disorders. Pro-inflammatory TH effector lineages are defined by the secretion of characteristic profiles of cytokines: interferon-γ is secreted by TH1 cells and interleukin-17 is secreted by TH-17 cells. Both of these populations of TH cells, along with CD25+Foxp3+ regulatory T cells (Tregs), control tissue destruction in multiple sclerosis (MS) – an autoimmune disorder in which the myelin sheaths of central nervous system neurons are destroyed by inflammatory and degenerative processes. Considerable effort has been devoted to elucidating the mechanisms by which autoimmunity and TH differentiation are regulated, and illuminating descriptions of several key cytokines, signaling molecules, and transcription factors have paved the way for the design of TH-17-specific diagnostic and therapeutic agents. In this issue of Nature Immunology, Du and colleagues1 establish a role for microRNA (miRNA) particles in the regulation of TH-17 cell differentiation and autoimmune disease. Their work positively associates levels of the miRNA species, miR-326, with active disease relapses in MS patients and employs overexpression and interference tools to manipulate miR-326 levels in the animal model of MS, experimental autoimmune encephalomyelitis (EAE), in which TH-17 cells play a critical pathologic role1,2. They show that forced expression of miR-326 worsens EAE symptoms and enhances TH-17 cell differentiation while miR-326 neutralization improves clinical outcome and impairs TH-17 cell differentiation. Finally, they demonstrate that miR-326 binds to and prevents translation of Ets-1 mRNA, a known inhibitor of TH-17 cell differentiation (Fig. 1).
Fig. 1. miR-326 encourages TH-17 cell differentiation.
miR-326 is transcribed from non-coding regions of the β-arrestin gene. In the cytoplasm, miR-326 binds to a complimentary UTR in the 3’ region of the Ets-1 message and prevents translation of the message. This prevents Ets-1 protein from accumulating and interfering with TH-17 cell differentiation by way of IL-2 synthesis and signaling pathways. Interestingly, miR-326 was not found to target Foxp3 mRNA to perturb Foxp3 protein expression required for the inhibition of RORγt activity. Signaling events downstream of the IL-6R are still required to prevent TH-17 cell lineage antagonism by Foxp3. RORγt expression was upregulated by miR-326, suggesting that miR-326 may somehow enhance the expression of the TH-17 cell lineage transcription factor via interference with an unknown inhibitor of RORγt.
miRNAs are short (approximately 22 nucleotides in length) molecules of RNA that are transcribed from noncoding regions of the genome and exhibit significant secondary structure3. The nucleases Drosha, Pasha, and Dicer are involved in processing the miRNA from a hairpin configuration into a short RNA duplex and finally into a single-stranded miRNA which is then loaded into the mRNA-induced silencing complex (RISC). The RISC and associated miRNA then bind complementary sequences in 3’ untranslated regions (termed seed regions) of mRNA species and inhibit their translation by two distinct mechanisms: degradation of the message by the RISC protein argonaute; and prevention of ribosomal binding and translation initiation3. Notably, Du et al.1 showed that miR-326 interferes with Ets-1 synthesis primarily via the inhibition of translation initiation because miR-326 overexpression decreased Ets-1 protein but not Ets-1 message levels.
The differentiation of TH-17 cells from naïve precursors is governed by a complex network of cytokines, signaling pathways, and transcription factors. It is well accepted that signaling through the IL-6 and TGF-β receptors by STAT-3 and Smads, respectively, and transcriptional regulation by RORγt are required for the establishment of this lineage4. IL-21 and IL-23 are also known as pro-TH-17 factors that function to stabilize the lineage against antagonism by opposing networks of cytokines and transcription factors. The list of anti- TH-17 factors includes, but is not limited to IL-2R-STAT5 signals, IL-27, IL-25, IFN-γ, IL-4, Foxp3, and Ets-1. Du and colleagues demonstrate here that in addition to cytokines and related receptor signals, miRNAs can promote TH-17 cell differentiation by inhibiting TH-17 cell lineage antagonists. While it has been previously shown that miRNAs can affect developmental outcomes in thymic T cell precursors, influence regulatory T cell development, and affect the production of antibodies against thymus-dependent antigens, this is the first demonstration that a specific miRNA directly influences the TH-17 cell effector subset and contributes to autoimmune pathology. Work continuing to catalog miRNA species and associate them with various TH lineages, immune functions, and autoimmune pathology would be of great interest.
An intriguing observation made in this manuscript is that miR-326 interferes with the protein synthesis of one TH-17 cell antagonist (Ets-1), but not another (Foxp3) (Fig. 1). It is therefore a challenge to determine if miR-326 contributes to the commitment of naïve precursors to the TH-17 cell lineage, as does IL-6-TGF-β, or if it functions to stabilize TH cells that are already producing IL-17 in the manner of IL-23-IL-21. TGF-β, IL-2R, and T cell receptor signals converge to upregulate Foxp3 expression and initiate Treg differentiation, and that STAT5 and Foxp3 discourage TH-17 cell differentiation by interfering with RORγt expression or function and thus preventing the transactivation of IL-174. Due to the involvement of Ets-1 in IL-2 synthesis and IL-2R signaling, it is tempting to speculate that miR-326 functions to regulate IL-2R signals through Ets-1 while allowing TGF-β signals to continue unperturbed. This may allow TGF-β signaling to contribute to both Foxp3 and IL-17 expression, which is consistent with recent reports indicating that both Foxp3 and IL-17 are expressed at early times in TH-17 cell differentiation – when the naïve precursors exhibit considerable plasticity5,6. If this is true, then miR-326 serves to uncouple TGF-β signaling from IL-2R signals and promote TH-17 over Treg cell differentiation at an early developmental stage. Regardless, due to the presence of the TGF-β-induced Foxp3, IL-6R signals transduced by STAT3 will still be required to antagonize Treg differentiation and commit naïve precursors to a TH-17 cell lineage. In fact, data presented by Du and colleagues supports this idea, as interference with miR-326 with a “sponge” vector decreases, but does not completely ablate IL-17 synthesis. It is therefore likely that miR-326 contributes to the TH-17 cell differentiation, but is not required for lineage commitment. It was shown that Ets-1 regulated IL-2 mRNA expression, but not RORγt transcript levels. Upregulation of RORγt transcripts by forced expression of miR-326 suggests that besides Ets-1, miR-326 may also perturb a potential inhibitor of the Rorc(γt) gene.. Further work in describing the precise role of this miRNA relative to the other known molecules in TH-17 cell differentiation is therefore of considerable interest.
There are two additional exciting suggestions resulting from the paper from Du et al.: the interpretation of data previously generated from knockout and overexpression models; and biomarker discovery. This miRNA species is encoded by a nontranslated region of the β arrestin-1 gene, and both β arrestin-1 and miR-326 are positively associated with the development of CNS autoimmunity1,7. Although the mechanistic contributions of β arrestin-1 have been clearly demonstrated, it is possible that β arrestin-deficient mice also lack miR-326. Knocking out both a gene and an embedded miRNA-coding sequence that both mediate autoimmune disease could potentially affect interpretation of disease outcome and lead to assigning an improper role to the gene being studied. Going forward, it will be of importance to carefully examine candidate genes for miRNA sequences prior to performing knockout studies.
The second exciting possibility this work opens is the authors’ proposed use of miR-326 as a biomarker for the development or relapse of multiple sclerosis. Such a diagnostic tool would be of tremendous clinical importance in MS, as tissue samples of the target organ (the CNS or cerebrospinal fluid) are difficult or impossible to obtain. Conversely, miR-326 can be detected in easily-accessible circulating T cells. Studies aiming to validate miR-326 as a biomarker in multiple sclerosis should be prioritized.
The finding that miRNAs can directly regulate TH-17 cell differentiation and contribute to autoimmune pathology is a significant advance, however, several outstanding questions remain. While the authors claim that miR-326 can serve as a possible diagnostic marker is well supported, the therapeutic potential of this target is less clear. It would be significantly more clinically relevant to see the outcome of treating animals with miR-326-sponge after they presented with clinical symptoms of EAE. Since previous studies have shown that epitope spreading in the SJL/J mouse model of relapsing EAE involves activation of TH-17 cells specific for endogenously released myelin epitopes predominantly in the inflamed CNS8,9, determination of the effects of miR-326-sponge treatment during disease remission on incidence of disease relapses would be extremely interesting. Furthermore, if miRNAs are to be seriously considered as therapeutic targets in autoimmune disorders, the development of an efficient non-viral delivery method is of paramount importance particularly if activation of T cells to spread epitopes preceding disease relapse primarily occurs in the CNS. Several studies involving delivery of small interfering RNA molecules by biopolymer micro- or nano-particles have been published10, and Phase I clinical trial are active or recruiting (NCT00689065 and NCT00882180). Assuming these obstacles are overcome, a third concern involves the proper timing of any therapeutic designed to target the TH-17 cell lineage. The TH1 lineage can initiate EAE by facilitating the entry of TH-17 cells into the CNS11. Furthermore, the TH1: TH-17 cell ratio can affect the location of CNS lesions and the severity of disease12. It is therefore possible that therapeutics targeting only the TH-17 cell lineage may be effective only at certain times or under certain disease conditions. The most successful treatment strategies will likely target both the TH1 and TH-17 cell effector subsets in an autoantigen-specific manner.
Collectively, the data in Du's publication establish a role for miR-326 as a regulator of TH-17 cell differentiation and the development of autoimmune disease while identifying a promising target for use as a diagnostic or therapeutic biomarker. Furthermore, this paper lays a fertile groundwork for future research into the precise roles miRNAs play in TH differentiation, cytokine receptor signaling, and transcription factor function.
Footnotes
TH-17 cell differentiation is controlled by a complex network of cytokines, signaling pathways, and transcription factors. Regulation by microRNA particles can now be added to this list.
References
- 1.Du C, et al. Nat. Immunol. 2009 doi: 10.1038/ni.1798. In press. [DOI] [PubMed] [Google Scholar]
- 2.Langrish CL, et al. J. Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shivdasani RA. Blood. 2006;108:3646–53. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong C. Nat. Rev. Immunol. 2008;8:337–48. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 5.Yang XO, et al. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou L, et al. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, et al. Nat Immunol. 2007;8:817–24. doi: 10.1038/ni1489. [DOI] [PubMed] [Google Scholar]
- 8.McMahon EJ, Bailey SL, Castenada CV, Waldner H, Miller SD. Nat. Med. 2005;11:335–339. doi: 10.1038/nm1202. [DOI] [PubMed] [Google Scholar]
- 9.Bailey SL, Schreiner B, McMahon EJ, Miller SD. Nat. Immunol. 2007;8:172–180. doi: 10.1038/ni1430. [DOI] [PubMed] [Google Scholar]
- 10.Aouadi M, et al. Nature. 2009;458:1180–4. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connor RA, et al. J. Immunol. 2008;181:3750–4. doi: 10.4049/jimmunol.181.6.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM. Nat. Med. 2008;14:337–342. doi: 10.1038/nm1715. [DOI] [PMC free article] [PubMed] [Google Scholar]