Abstract
The mechanisms that determine localized formation of reactive oxygen species (ROS) via NADPH oxidases (Nox) in nonphagocytic cells are unknown. We show that the c-Src substrate proteins Tks4 and Tks5 are functional members of a p47phox-related organizer superfamily. Tks proteins selectively support Nox1 and Nox3 (vs. Nox2 and Nox4) activity in reconstituted cellular systems, and interact with the NoxA1 activator protein through an SH3-mediated interaction. Endogenous Tks4 is required for Rac GTPase-dependent ROS production by DLD1 colon cancer cells. Tks4 recruits Nox1 to invadopodia that form in DLD1 cells in a Tks- and Nox-dependent fashion. We propose that Tks organizers represent novel members of an organizer superfamily that link Nox to localized ROS formation.
Introduction
The NADPH oxidase (Nox) family, consisting of the homologous enzymes Nox1-4 and the more distantly related Nox5, Duox1 and Duox2, catalyzes the regulated formation of reactive oxygen species (ROS) (1–4). These homologs of the phagocyte gp91 cytochrome b subunit are found in virtually all tissues, and have been implicated in such biological processes as cell growth, apoptosis and cancer, angiogenesis and blood pressure regulation, innate immunity and inflammation, cell signaling, motility, and transcription. ROS generated via Nox also contribute to a growing number of diseases, including atherosclerosis, hypertension, arthritis, Alzheimer’s disease and other neurological disorders, stroke, respiratory syndromes, cancer, and inflammation (5, 6). Many of these activities appear to require the compartmentalized and/or spatially regulated formation of ROS (7). Very little is known about how the localized formation of ROS by Nox proteins is regulated under normal, much less pathological, conditions.
A subfamily of Nox proteins, Nox1-3, are regulated by the action of cytosolic cofactors, including the Rac1 or Rac2 GTPase (8–11) and an “activator” protein (either p67phox or NoxA1) (1, 12). In addition, although not absolutely required for activity, a number of “organizer” proteins have been identified, including p47phox, p40phox, and NoxO1 (1, 12). These proteins make up a structurally similar “p47phox organizer superfamily”, possessing an N-terminal ‘phox’ domain (PX) responsible for binding to anionic phospholipids, and protein-protein interaction domains that include Src Homology-3 (SH3) and Proline Rich Regions (PRR) (13–16). The organizers serve as regulatory response elements for NADPH oxidase assembly through different signaling mechanisms. In the phagocyte Nox2 system, p47phox is phosphorylated upon inflammatory stimulation, thereby promoting plasma membrane translocation of the cytosolic oxidase components and assembly of the functional NADPH oxidase enzyme (13–16). Similarly, both p40phox and p47phox are involved in the recruitment and assembly of the Nox2 system on phagocytic vesicles during the internalization of particulate stimuli (17–19). Such evidence suggests that the organizer subunits could play roles in the differential recruitment of functional NADPH oxidases to specific subcellular compartments. This would likely occur through the binding of differentially expressed anionic phospholipids to their conserved PX domains, as well as through specific protein-protein interactions mediated via their conserved SH3 domains and PRR. Finally, accumulating evidence also indicates that these organizer proteins act through undefined mechanisms post-assembly to promote full Nox activity (20–22).
The protein Tks5 (tyrosine kinase substrate with five SH3 domains) was originally identified in a cDNA library screen for c-Src substrates (23). Subsequently, a close ortholog with four SH3 domains, termed Tks4, was also described (24, 25). The Tks proteins are widely distributed in mouse and human tissues, with the notable exception of low abundance in neutrophils. In Src-transformed cells, Tks4 and Tks5 localize to invadopodia (26, 27). These are dynamic phosphotyrosine-rich structures with an actin core and abundant actin regulatory proteins (e.g. cortactin) capable of proteolytically degrading the extracellular matrix (ECM). Invadopodia are found in many invasive cancer cells. Some invasive normal cells, such as macrophages, osteoclasts, endothelial- and vascular smooth muscle cells, contain related structures called podosomes (28, 29). There is substantial evidence for the importance of c-Src in invadopodia/podosome formation – e.g. mice lacking c-Src develop osteopetrosis because of a podosome defect in osteoclasts (30). Many Src substrates are obligate invadopodia/podosome components, including cortactin, Tks4 and Tks5 (26, 27, 31). Consistent with this, Tks4 and Tks5 have been shown to be required for the formation of invadopodia, and to promote cancer cell invasion (27, 32, 33). However, despite much investigation, the signals regulating the formation of invadopodia remain unclear.
In the accompanying manuscript by Diaz, et al., it is demonstrated that ROS formation via NADPH oxidases is a critical component in the formation and stabilization of Tks-dependent invadopodia. Tks4 and Tks5 show a similar domain architecture and composition as do members of the p47 organizer superfamily (25, 34). Here we show that Tks4 and Tks5 represent novel functional members of this family of NADPH oxidase organizers. Further, our results suggest that the interaction of Nox with distinct organizers might be a key element in localizing ROS formation to different subcellular compartments to selectively affect cellular function.
Results
Tks4 and Tks5 are novel Nox organizers
The identified c-Src substrates Tks4 and Tks5 show a similar domain architecture and composition as other known NADPH oxidase organizer proteins (Figure 1A). In addition to containing two N-terminal SH3 domains structurally similar to those of p47phox (up to 47% identity) and arranged in the same orientation, Tks proteins also contain a phosphoinositol lipid-binding phox (PX) domain that is highly conserved among the members of the p47 organizer superfamily (25, 34). p47phox, p40phox and NoxO1 have been previously shown to serve as regulatory organizer subunits for assembly and full activity of different NADPH oxidase (Nox) family members, including Nox2 (p47phox and p40phox) (16, 35, 36), and Nox1 and Nox3 (NoxO1 and p47phox) (1, 12, 37, 38). To assess the ability of Tks4 and Tks5 to serve as organizer subunits in Nox1-mediated ROS formation, HEK293 cells were transfected with expression vectors for various organizer subunits along with other components (Nox1, NoxA1, Rac1QL) required for Nox1-dependent ROS generation. After 24hrs, ROS formation was measured by luminol-based chemoluminescence assay (CL-assay) (39). As shown in Figure 1B, both Tks4 and Tks5 supported Nox1-dependent ROS production to a higher level as compared to mock-transfected cells or to cells transfected with Nox1 system components in the absence of organizer subunit. Figure S1A verifies that all transfected proteins were expressed at similar levels.
Figure 1. Tks4 and Tks5 are novel members of the p47 organizer superfamily and support Nox1-dependent ROS generation in a reconstituted HEK293 cell system.

(A). Schematic diagram of the members of the p47 organizer superfamily. The white squares indicate PX domains, while white circles indicate SH3 domains. (B). Time course of ROS generation examining the ability of Tks4 and Tks5 to support Nox1-dependent ROS formation in a reconstituted HEK293 system. HEK293 cells were transfected as indicated with expression vectors for Nox1, NoxA1 and RacQL, and with different organizers, including NoxO1, Tks4, Tks5 or empty vector. After 24hrs, ROS production was measured by a luminol-based chemiluminescence (CL)-assay continuously for 30 minutes. One representative experiment from three separate experiments is shown. All transfected proteins were expressed to similar levels, as per Fig. S1.
Tks4 and Tks5 can selectively support Nox1- and Nox3-dependent ROS generation
We next examined whether Tks4 and Tks5 were also able to support ROS generation by other members of the NADPH oxidase family, and compared this activity to the other known organizer subunits (NoxO1, p47phox and p40phox). HEK293 cells were transfected with expression vectors for different organizer subunits and with all the known components required for Nox1-, Nox2-, Nox3- and Nox4-dependent ROS generation. Figure S1 (A to D) shows that all transfected proteins were expressed at similar levels. As shown in Figure 2A, Tks4 and Tks5 could selectively support Nox1-and Nox3-dependent ROS generation, and the levels of Nox activity were similar to that obtained with NoxO1 or p47phox. Interestingly, Tks4 and Tks5 were unable to act as organizer subunits for Nox2-dependent ROS formation, and did not enhance Nox4 activity. Although Nox2 activity is organizer subunit-dependent, its activity was only supported by p47phox and NoxO1. The lack of effect of Tks proteins on Nox4 activity is perhaps not surprising, given that this Nox was already highly active and is not known to require regulatory subunits for its constitutive activity (1).
Figure 2. Tks4 and Tks5 selectively support Nox1- and Nox3-dependent ROS generation in a Rac GTPase-dependent manner.
(A). CL-assay to compare the ability of Tks4 and Tks5 to support ROS generation by different members of the NADPH oxidase family in an HEK293 cell reconstituted system. HEK293 cells were transfected with expression vectors for the indicated organizer subunits and with the known components required for Nox1- (=Nox1, NoxA1, RacQL), Nox2- (=Nox2, p67phox, RacQL), Nox3- (=Nox3, NoxA1, RacQL) and Nox4- (=Nox4)-dependent ROS generation. After 24hrs, ROS formation was monitored by CL-assay. One representative experiment from three separate experiments is shown, and results are given as mean of triplicates +/− S.D. (B) Treatment with the flavoenzyme/Nox inhibitor diphenyliodonium (DPI) blocks Tks4- and Tks5-mediated ROS generation by Nox1 in the HEK293 reconstituted system. HEK293 cells were transfected with expression vectors for different Myc-tagged organizer subunits as indicated, and with all of the components required for Nox1 activity. 24 hrs after, cells were treated with 10μM DPI or DMSO control for 30 minutes and ROS production was measured by CL-assay (upper panel). Comparable expression of Myc-tagged proteins was verified by Western blot (lower panel). One representative experiment from three separate experiments is shown, and results of CL-assay are given as mean of triplicates +/− S.D. (C) Tks4- and Tks5-mediated ROS formation by Nox1 is Rac-dependent in the reconstituted HEK293 cell system, as demonstrated using dominant negative Rac1T17N, the non-Rac-binding R103E NoxA1 mutant, and the non-Rac-binding Nox1 K421A, Y425A, K426E triple mutant (Nox1TM). HEK293 cells were transfected as indicated and after 24hrs ROS production was monitored by CL-assay (upper panels). The expression of Myc-tagged proteins was verified by Western blot (lower panels). One representative experiment from three separate experiments is shown, and results of CL-assays are given as mean of triplicates +/− S.D.
As shown in Figure 2B (upper panel), treatment with the Nox/flavoenzyme inhibitor (40) diphenyliodonium (DPI) efficiently blocked ROS generation induced by Tks4 and Tks5 (as well as NoxO1) without affecting the expression level of the transfected proteins (lower panel), establishing that the Tks protein-mediated ROS production observed was likely due to NADPH oxidase activity. Rac1 is known to be required along with NoxO1 and NoxA1 for full Nox1 activity (9). To test whether Tks4-and Tks5-mediated ROS generation by Nox1 is also Rac-dependent, HEK293 cells were transfected with expression vectors for Tks4, Tks5 or NoxO1, along with constitutively active Rac1 (Rac1-Q61L) or dominant negative Rac1 (Rac1-T17N) and with the Nox1 system components, Nox1 and NoxA1. Figure 2C (upper left panel) shows that Tks4-and Tks5-mediated ROS formation was effectively blocked in the presence of dominant negative RacN17. To confirm that Tks4- and Tks5-mediated ROS production is Rac-dependent, we also monitored ROS generation in HEK293 cells reconstituted with Tks4, Tks5 or NoxO1, with Nox1 and Rac1-Q61L, and with NoxA1 wild-type or NoxA1 R103E, a mutant form of NoxA1 unable to bind Rac because of the mutation in its TPR domain (41). As shown in Figure 2C (upper middle panel), the presence of NoxA1 R103E completely abrogates the induction of Nox1-dependent superoxide formation caused by Tks4 or Tks5. Finally, HEK293 cells were transfected with expression vectors for various organizer subunits, with NoxA1 and Rac1-Q61L, and with Nox1 wild-type or Nox1 TM, a mutant form of Nox1 that we previously demonstrated to be unresponsive to Rac1 because of a triple mutation in its Rac binding site (42). As indicated in Figure 2C (upper right panel), the presence of Nox1 TM dramatically abolished Tks4- and Tks5-mediated ROS generation. In each experimental manipulation, we observed no effect on the expression levels of the other transfected proteins (shown in lower panels).
Tks4 and Tks5 bind NoxA1 through their SH3 domains in a Rac-independent manner
In the Nox1 and Nox3 system, NoxO1 is required for full oxidase activity at least partially due to its role in the plasma membrane recruitment of the NoxA1 activator protein. To establish whether the ability of Tks4 and Tks5 to support Nox1-dependent ROS formation involved their interaction with NoxA1, co-immunoprecipitation experiments were performed in either HEK293 cells reconstituted with all components of Nox1 pathway or in human DLD1 colon cancer cells that endogenously express Nox1, NoxA1 and Tks4, but not NoxO1 (Figure S2 A to C). As indicated in Figure 3A (left panel), in reconstituted HEK293 cells Tks5 bound NoxA1 to a similar extent as did the NoxO1 positive control. The interaction of NoxA1 with Tks4 was also detected, although to a lesser extent. The interaction of NoxA1 with endogenous Tks4 was confirmed by co-immunoprecipitation analysis of human DLD1 cells (Figure 3A, right panel).
Figure 3. Support of Nox1-dependent ROS generation by Tks4 and Tks5 requires the SH3 domain-mediated binding of NoxA1 and is independent of Rac.
(A) Co-immunoprecipitation analysis indicates that Tks4 and Tks5 interact with NoxA1 in reconstituted HEK293 cells and with endogenous NoxA1 in DLD1 cells. In the left panel, HEK293 cells were transfected as indicated with Myc-tagged organizer subunits and Flag-tagged NoxA1. After 24hrs, cells were lysed and immunoprecipitation was carried out (see Methods) using anti-Myc antibody. Interaction of Myc-tagged adaptors with NoxA1 was analyzed using anti-NoxA1-specific antibody, while expression of transfected proteins in cell lysates and equal loading of proteins on gels was verified by re-blotting the membranes with anti Myc- and actin-antibody, respectively. One representative experiment from three separate experiments is shown. In the right panel, DLD1 cells were lysed in RIPA buffer and immunoprecipitation was performed using NoxA1-specific antibody or pre-immune serum. An additional control with protein G-Sepharose beads only was also carried out. Specific interaction between endogenous Tks4 and NoxA1 was confirmed using Tks4-specific antibody, while the presence of NoxA1 in cell lysates was verified by re-blotting the membrane with NoxA1-specific antibody. One representative experiment from three separate experiments is shown. (B) Co-immunoprecipitation analysis indicates that the interaction between NoxA1 and Tks5 is not dependent of the GTP-bound state of Rac1. HEK293 cells were transfected as indicated with Myc-tagged Tks5, Flag-tagged NoxA1, and alternatively with GFP-tagged Rac1-Q61L or Rac1-T17N. After 24hrs, cells were lysed and immunoprecipitation was carried out using anti-Flag antibody. Interaction of Tks5 with NoxA1 was analyzed using anti-Tks5-specific antibody, while expression of transfected proteins in cell lysates and similar amount of immunoprecipitated NoxA1 protein was verified by re-blotting the membranes with anti NoxA1- and GFP-antibody. One representative experiment from three separate experiments is shown (C) Tks5 binds to NoxA1 through its SH3 domains. HEK293 cells were transfected with expression vectors for Nox1, NoxA1, RacQL and with the Myc-tagged organizer subunits NoxO1, Tks5 wild-type, or Tks5 M1M5 mutant in which point mutations were made to inactivate all of the SH3 domains. After 24hrs, ROS generation was monitored by CL-assay (left panel) and expression of transfected proteins was checked by Western blot using Tks5-specific antibody (right panel). The loss of the ability of the Tks5 M1M5 mutant to bind NoxA1 was shown by co-immunoprecipitation analysis using Flag antibody for immunoprecipitation and Tks5 antibody to detect specific interaction (lower panel). One representative experiment from three separate experiments is shown, and results of CL-assays are given as mean of triplicates +/− S.D. (D) The deletion of the PX domain of Tks5 prevents the activation of Nox1-mediated ROS formation, but does not abrogate the ability of Tks5 to bind NoxA1. HEK293 cells were transfected with expression vectors for the Myc-tagged organizer subunits NoxO1, Tks5 wild-type, and Tks5ΔPX, along with Flag-tagged NoxA1. After 24hrs, ROS generation was measured by CL-assay (left panel), while expression of Myc-tagged organizers was verified by Western blot using Myc antibody (right panel). The ability of Tks5ΔPX to bind NoxA1 was determined by co-immunoprecipitation analysis (lower panel) using Flag antibody for immunoprecipitation and Tks5 antibody to detect interaction with NoxA1. One representative experiment from three separate experiments is shown, and results of CL-assays are given as mean of triplicates +/− S.D.
The binding of other organizer subunits to activator proteins has been shown to be independent of the GTP-binding form of Rac (41). To ascertain whether the interaction between NoxA1 and Tks5 was dependent on the presence of active Rac, we performed co-immunoprecipitation experiments in HEK293 cells reconstituted with all the components of the Nox1 pathway and with constitutively active Rac1-Q61L or with inactive Rac1-T17N. Figure 3B shows that neither the presence of Rac1-Q16L, nor Rac1-T17N affected the interaction between NoxA1 and Tks5.
In order to identify the structural basis for the interaction of Tks proteins with NoxA1, we generated a Myc-tagged Tks5 expression construct, Tks5M1M5, in which the function of all five SH3 domains was ablated by point mutations, and Tks5ΔPX, in which the PX domain was deleted. These mutant Tks5 expression constructs were assayed in HEK293 cells reconstituted with all components of Nox1 pathway for their ability to support Nox1-mediated ROS generation. As shown in Figure 3C, Tks5M1M5 was unable to support ROS production by Nox1 (upper panel) when expressed at comparable levels as the wild-type protein (right panel), and was also unable to bind NoxA1 in co-IP experiments (lower panel). This result indicates that the interaction between NoxA1 and Tks5 is mediated through one or more of the SH3 domains of Tks5. We were unable to identify a single SH3 domain of Tks5 responsible for interaction with NoxA1. We observed that the single SH3 domain point mutants (Tks5M1 to M5) and the double mutant (Tks5M1M2) were each able to support ROS generation and interact with NoxA1 as well as Tks5 wild-type under the assay conditions (Figure S3 A to C).
The Tks5 PX domain has been shown to be necessary for Tks5 localization to invadopodia in Src-transformed fibroblasts due to its ability to bind PI3,4-P2 (26, 43). To test the requirement for the PX domain in Nox1-dependent ROS generation by Tks5, Tks5ΔPX mutant lacking the PX domain was transfected in reconstituted HEK293 cells and ROS production monitored. As shown in Figure 3D (left panel), Tks5ΔPX was unable to support ROS generation by Nox1 when expressed at comparable levels as Tks5 wild-type (right panel). However, this mutant was still able to bind NoxA1, as revealed by co-IP analysis (lower panel). Thus, these data suggest that the Tks PX domain may contribute to the membrane recruitment of the functional Nox1 complex.
Human DLD1 colon cancer cells produce ROS and degrade the ECM in a Tks4-dependent manner
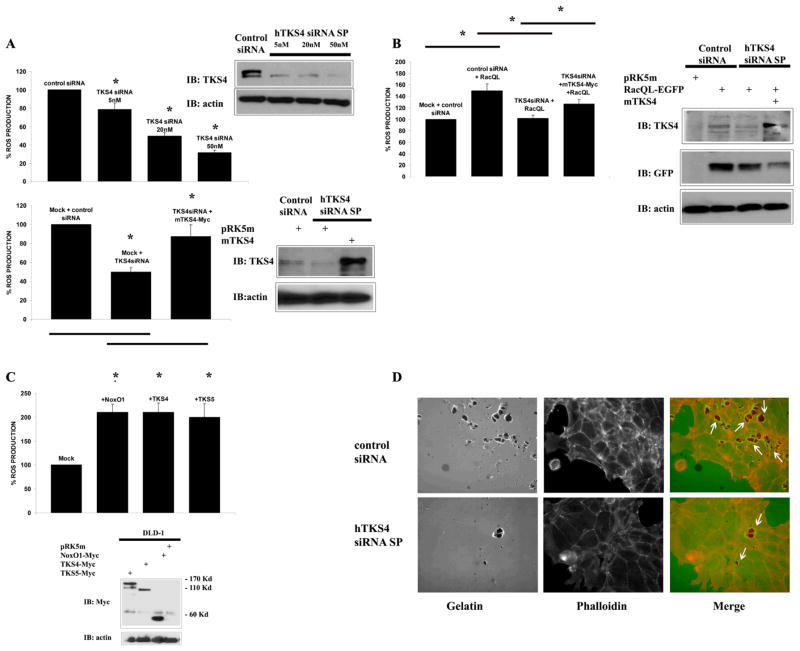
We previously established that human DLD1 colon cancer cells express a higher level of active Src compared with other colon cancer cells, and that this correlated with the increased ability of DLD1 cells to produce ROS (39). DLD1 cells only express Nox1, NoxA1, Tks4 and low levels of Tks5, with no other known organizer proteins present (Figure S2 A to C), representing a suitable system for the study of Tks-dependent, Nox1-mediated ROS generation. DLD1 cells were transfected with control siRNA or with increasing concentrations of Tks4-specific siRNA mixture (SmartPool, Dharmacon) for 72hrs, which we have shown to be sufficient time to significantly decrease Tks4 protein levels (Figure S4A), and ROS production was determined by CL-assay. As shown in Figure 4A (upper panels), the Tks4-specific siRNA efficiently reduced Tks4 protein levels (right panel) while decreasing ROS generation in a concentration-dependent manner (left panel). The loss of ROS formation observed upon siRNA-mediated depletion of Tks4 in DLD1 cells was rescued by overexpression of the siRNA-resistant mouse Tks4 (mTks4) (Figure 4A, lower panels). Similarly, to confirm that the depletion of Tks4 protein blocks Nox1-mediated, Rac1-dependent ROS generation in DLD1 cells, the cells were transfected with Rac1-Q61L and with control or Tks4-specific siRNA. The presence of active Rac1-Q61L caused a modest, but consistent 50% increase in ROS production over baseline (Figure 4B, left panel). Tks4-specific siRNA reduced the Rac1-Q61L-induced ROS generation to the level of controls, and this reduction could be partially rescued by expression of the siRNA-resistant mTks4. The Western blot in the right panels shows the extent of Tks4 protein knockdown and mTks4 overexpression, and confirms similar expression levels of the GFP-tagged Rac1-Q61L in each condition.
Figure 4. Human DLD1 colon cancer cells produce ROS and degrade the ECM in a Tks4-dependent manner.
(A) Tks4 knockdown by siRNA reduces ROS generation in a concentration-dependent fashion and this effect can be rescued by a siRNA-insensitive mouse Tks4. In the upper panels, DLD1 cells were transfected with different concentrations of Tks4-specific siRNA mixture or control-siRNA and after 72hrs, ROS generation was determined by CL-assay (left panel), while the extent of Tks4 protein knockdown was checked by Western blot using Tks4-specific antibody (right panel). In the lower panels, DLD1 cells were transfected with 20nM of Tks4-specific siRNA mixture or control siRNA and with mouse Tks4 (mTks4) expression vector or empty vector. After 72hrs, ROS generation was measured by CL-assay (left panel) while Tks4 knockdown and mTks4 expression were checked by Western blot using Tks4-specific antibody (right panel). One representative experiment from three separate experiments is shown and results of CL-assays are given as mean of triplicates +/− S.D. (B) Tks4 knockdown by siRNA reduces Rac1-induced ROS generation and this effect can be rescued by overexpression of mTks4. DLD1 cells were transfected with Tks4-specific siRNA or control siRNA and with expression vector for GFP-tagged RacQL or empty vector. After 72hrs, ROS generation was monitored by CL-assay (left panel). Tks4 protein knockdown and mTks4 expression, as well as comparable expression levels of GFP-tagged RacQL were demonstrated by Western blot using Tks4- or GFP-specific antibody, respectively (right panel). One representative experiment from three separate experiments is shown and results of CL-assays are given as mean of triplicates +/− S.D. (C) Tks4 and Tks5 overexpression in DLD1 cells induces ROS generation to a similar extent as NoxO1 overexpression (2-fold over control). DLD1 cells were transfected with expression vectors for Myc-tagged organizer subunits (NoxO1, Tks4 or Tks5) or with empty vector. After 24hrs, ROS formation was measured by CL-assay (left panel), while similar expression levels of the Myc-tagged adaptors were verified by Western Blot using anti-Myc antibody (right panel). One representative experiment from three separate experiments is shown and results of CL-assays are given as mean of triplicates +/− S.D. (D) Tks4 knockdown reduces the ability of DLD1 cells to degrade the extracellular matrix (ECM). DLD1 cells were transfected with Tks4-specific or control siRNA and after 24hrs plated on FITC-labeled gelatin-coated coverslips. After 20hrs, cells were fixed in 4% PFA, stained with Alexa-Fluor-568 phalloidin as described in Methods, and visualized by epifluorescence microscopy (40X). White arrows indicate areas in which F actin-positive structures (in red) degrade the ECM. Scale bars, 45μm. One representative picture from two separate experiments is shown.
Further evidence that ROS generation in DLD1 cells is regulated by Tks protein levels is shown in Figure 4C, in which Tks4 and Tks5 proteins were overexpressed. As shown in the left panel, the overexpression of either Tks4 or Tks5 (as well as of NoxO1) increased ROS formation to a similar maximum (2-fold over control) when the transfected organizer proteins were expressed at similar levels (right panel).
Tks4 has been reported to be required for cancer cells to successfully degrade the ECM (27). We first established the ability of DLD1 cells plated on FITC-labeled gelatin-coated coverslips to degrade the ECM. The siRNA-mediated knockdown of Tks4 reduced the ability of the DLD1 cells to induce pericellular proteolysis as compared to control siRNA-transfected cells (Figure 4D, and its quantification in Figure S4B), indicating that Tks4 plays a key role in enabling DLD1 cells to successfully degrade ECM.
Nox1 localizes to invadopodia in DLD1 cells
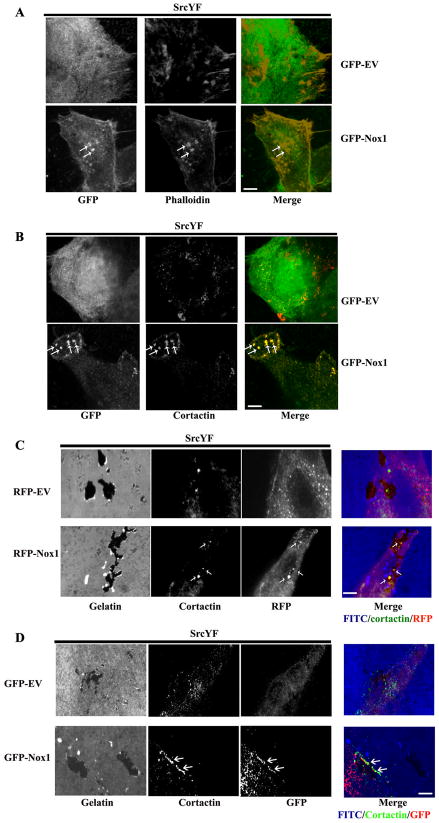
Several types of invasive human cancer cells have been reported to form invadopodia, including breast cancers and melanomas. We have shown in Figure 4D that human DLD1 colon cancer cells are able to degrade the ECM in a Tks4-dependent fashion. In the accompanying paper by Diaz et al, the formation of stable invadopodia and ECM degradation is shown to require the formation of ROS. We have found that Nox1-specific siRNAs (SmartPool, Dharmacon) reduce ROS generation, invadopodia formation and ECM degradation in DLD1 cells and that this effect can be partially rescued by the overexpression of mNox1 (Figure S5 A to C). We hypothesized that Tks4 would localize Nox1 to invadopodia in DLD1 cells. Given the lack of good working antibody for the detection of endogenous Nox1, we tested this hypothesis by investigating the localization of transfected GFP-tagged Nox1 (or GFP-empty vector control) in DLD1 cells by confocal microscopy. By comparing the localization of Nox1-GFP (Figure 5A and B, lower left panel) with the localization of known markers for invadopodia such as F-actin (Figure 5A, middle panels) and cortactin (Figure 5B, middle panels), we showed that the majority of Nox1-GFP was localized to invadopodia, where it co-localized with F-actin (Figure 5A, merge) and cortactin (Figure 5B, merge). In contrast, no co-localization was observed in GFP control-transfected cells. Additional evidence that these Nox1-positive structures are invadopodia is given in Figure S6A. Here we show Z-stacks with 0.2μm steps of the merged images shown in Figure 5A and B which clearly demonstrate that both Nox1/F-actin-positive (upper panel) and Nox1/cortactin-positive (lower panel) structures are ventral organelles, as previously reported for invadopodia (31).
Figure 5. Nox1 localizes to ECM-degrading invadopodia in DLD1 cells.
(A) and (B) Nox1 localizes to F actin- and cortactin-rich structures in DLD1 cells. DLD1 cells were plated on glass coverslips and after 24hrs cells were transfected with active SrcYF and GFP-tagged Nox1 or GFP empty vector. After 48hrs the cells were fixed in PFA4% and stained with Alexa-Fluor-568 phalloidin (A) or cortactin antibody, followed by Alexa-Fluor 568-conjugated secondary antibody (B) and visualized by confocal miscroscopy (100X). White arrows indicate areas in which Nox1- (lower left panels) and F actin in (A) or cortactin in (B) (middle panels) colocalize in structures identified as invadopodia (merge in yellow). Scale bars, 5μm. One representative picture from three separate experiments is shown. (C) and (D) Nox1 localizes to cortactin-rich structures capable of degrading the ECM in DLD1 cells. DLD1 cells were transfected with SrcYF and with RFP-tagged Nox1 or RFP empty vector in (C) and with GFP-tagged Nox1 or GFP empty vector in (D). After 24hrs, cells were trypsinized and plated on FITC-labeled gelatin-coated coverslips. 48hrs later, cells were fixed in 4%PFA in (C) or methanol in (D) and stained with mouse cortactin antibody, followed by anti mouse Alexa-Fluor 647-conjugated secondary antibody in (C) and (D) and with rabbit polyclonal GFP antibody, followed by anti-rabbit Alexa-Fluor 568-conjugated secondary antibody in (D). Successively, cells were visualized by confocal miscroscopy (60X). White arrows indicate areas in which Nox1 (in red) and cortactin (in green) colocalize in invadopodia (yellow in the merge) capable of degrading the ECM (in blue). Scale bars, 4μm. One representative image from three separate experiments is shown.
To additionally confirm that Nox1 localizes to ECM-degrading invadopodia in DLD1 cells, we examined the localization of transfected RFP-tagged Nox1 (or RFP-empty vector control, RFP-EV) in SrcYF-overexpressing DLD1 cells when plated on FITC-labeled gelatin-coated coverslips. As shown in Figure 5C, Nox1-RFP (red) co-localizes with cortactin (green) to form Nox1/cortactin structures (yellow) which efficiently degrade the ECM (indicated in blue in the merge), a hallmark of invadopodia function. Similar results were obtained using the approach illustrated in Figure 5D, where SrcYF-overexpressing DLD1 cells were transfected with Nox1-GFP and plated on FITC-labeled gelatin-coated coverslips. After 48hrs, the cells were fixed in methanol to quench GFP fluorescence and stained for GFP and cortactin (indicated in red and green respectively in the merge of Figure 5D). Under these conditions, we observed a partial co-localization of Nox1-GFP with cortactin in Nox1-GFP/cortactin structures able to degrade ECM (in blue) by confocal microscopy. Importantly, both the Nox1-GFP and Nox1-RFP plasmids used in these experiments were able to support ROS production (Figure S6B).
The overexpression of NoxO1 in DLD1 cells reduces invadopodia formation and ECM degradation
We have established that Nox1 localizes to ECM-degrading invadopodia that form in a Tks4-dependent manner in DLD1 cells, and that Tks4 is able to recruit NoxA1 to this complex. We further propose that a consequence of this interaction is the local generation of ROS by Nox1 to support the formation of invadopodia. In order to test this idea, we reasoned that the expression of a different organizer subunit, such as NoxO1, should compete with endogenous Tks4, consequently displacing Nox1 from the invadopodia and thereby reducing their ROS-dependent formation. We therefore examined the effect of NoxO1 overexpression on Nox1 localization, invadopodia formation and localized ROS generation. SrcYF-overexpressing DLD1 cells were transfected with Nox1-GFP (1x) and increasing amounts of NoxO1 (1x to 3x) or equal amounts of empty vector (mock). After 48hrs, the cells were fixed and stained for invadopodia markers, including F-actin (Figure 6A, middle panels) and cortactin (Figure 6B, middle panels), and compared with Nox1GFP localization (Figure 6A and B, left panels) by confocal microscopy. As shown both in the merge of Figure 6A and B (upper panels) and in the respective quantification of three independent biological experiments (Figure 6E, first two panels), NoxO1 overexpression strongly reduced Nox1 co-localization with invadopodia and, significantly, reduced the overall formation of invadopodia. Consistent with this, the analysis of NoxO1-GFP overexpression in SrcYF-overexpressing DLD1 cells indicates that NoxO1 does not localize to invadopodia, whose formation is strongly reduced in NoxO1-GFP positive cells (Figure S7A).
Figure 6. The overexpression of NoxO1 in DLD1 cells reduces invadopodia formation and ECM degradation.
(A) and (B) NoxO1 overexpression reduces invadopodia formation in a concentration-dependent manner in DLD1 cells. DLD1 cells were transfected with SrcYF, GFP-tagged Nox1, and with increasing amounts of NoxO1 (1x or 3x) or with equal amount of empty vector (mock). After 48hrs, cells were fixed in 4%PFA, stained with Alexa-Fluor-568 phalloidin in (A) or antibody followed by Alexa-Fluor 568-conjugated secondary antibody in (B) and visualized by confocal miscroscopy (100X). In the upper panels, the white arrows indicate areas in which Nox1- (left panels) and F actin in (A) or cortactin in (B) (middle panels) colocalize in invadopodia (merge in yellow). Scale bars, 5μm. One representative image from three separate experiments is shown. (C) The overexpression of NoxO1 in DLD1 cells reduces ECM degradation. DLD1 cells were transfected with SrcYF, RFP-tagged Nox1 and with NoxO1 (3x) or with equal amount of empty vector (mock). 24hrs later, cells were trypsinized and plated on FITC-labeled gelatin-coated coverslips, and after 48hrs the cells were fixed in 4%PFA and visualized by epifluorescence microscopy (40X). In the left panel, the white arrows indicate areas in which RFP-tagged Nox1-expressing cells (in red) degrade the ECM (in green). Scale bars, 45μm. One representative image from three separate experiments is shown. (D) The overexpression of NoxO1 in DLD1 cells reduces the number of ROS-positive invadopodia-like structures. DLD1 cells were transfected with SrcYF and with NoxO1 or empty vector (mock). 72hrs later, cells were incubated with 2.5μM of the ROS-sensitive probe PY1-AM in HBSS for 30 minutes at 37C (see Methods) and visualized by confocal microscopy (100X). One representative image from three separate experiments is shown. Scale bars, 5μm. (E) Quantifications of experiments illustrated in (A) to (D). In the first two panels quantification from three independent biological experiments shown in (A) and (B) is given: the number of Nox1/phalloidin positive-structures in (A) or Nox1/cortactin positive-structures in (B) was counted and averaged from 25 cells for each experiment. Error bars represent SEM. *p<0.008 **p<0.002. In the third panel, quantification from three independent biological experiments as shown in (C): for each experiment, the total degradation area was obtained as sum of degradation areas calculated using Metamorph software from 25 images and reported as percentage (mock set as 100%). In the graph, error bars represent SEM. *p<0.02. In the right panel, quantification from three independent biological experiments, as shown in (D): the number of ROS-positive structures was counted and averaged from 10 pictures for each experiment. Error bars represent SEM. *p<0.005
The overexpression of NoxO1 was accompanied by loss of the ability of the Nox1-RFP expressing cells (Figure 6C, in red) to degrade the ECM (Figure 6C, in green) - see merge of Figure 6C and the quantification of three independent biological experiments (Figure 6E, third panel). In keeping with an antagonistic function of NoxO1 on ROS-dependent formation of invadopodia, the presence of NoxO1 in SrcYF-overexpressing DLD1 cells incubated with the ROS-specific probe PY1-AM (see Figure S7B and Methods) strongly reduced the formation of ROS-positive invadopodia-like structures, as shown by the confocal microscopy images in Figure 6D and in the respective quantification of three independent biological experiments (Figure 6E, rightmost panel). These experiments strengthen the hypothesis that localized Tks-mediated ROS generation by Nox1 in DLD1 cells contributes to invadopodia formation and function.
Discussion
Tks4 and Tks5 are new members of a p47 organizer superfamily that direct localized ROS generation by Nox1
The NADPH oxidase (Nox) family, consisting of Nox1-5 and Duox1-2, catalyzes the regulated formation of reactive oxygen species (ROS). Although excessive amounts of ROS are toxic, it has been demonstrated that ROS function physiologically as signaling molecules to mediate various biological responses. The signaling properties of ROS are to a large extent due to their ability to catalyze the reversible oxidation of redox-sensitive target proteins. For example, the activities of several protein tyrosine phosphatases (i.e. PTPB1, LMW-PTP, PTP-PEST and Shp-2) and receptor tyrosine kinases (i.e. insulin receptor, EGFR, PDGFR) are modulated by a reactive cysteine with a low pKa at their active site which is susceptible to reversible oxidation by H2O2 (44). We have recently shown that ROS regulate the dephosphorylation and activation of the actin regulatory protein cofilin through the redox-dependent activation of the Slingshot cofilin phosphatase (45).
Given that ROS are diffusible and short-lived, the localization of ROS production at precise subcellular compartments may be essential for redox-mediated signaling events (46). In particular, emerging evidence that ROS are important in regulating the dynamic behavior of the actin cytoskeleton during such highly spatially and temporally regulated processes as cell motility (47–52), angiogenesis (53), and neuronal extension (54) makes the localized formation of ROS of critical importance. The NADPH oxidases are recognized to exist in specific subcellular localizations, thus making them ideal candidates for the localized production of ROS, with consequent activation of specific redox signaling pathways (46, 55). Much evidence has demonstrated that NADPH oxidase family members are activated within discrete cellular compartments, including caveolae/lipid rafts (56), focal adhesions (57), cell-cell contacts (58), phagosomes (36), lamellipodia/leading edge (59), membrane ruffles (60), endosomes (61) and the nucleus (62). We propose that one important mechanism by which NADPH oxidase enzymes are spatially regulated within specific subcellular compartments is through interaction with organizer subunits. These Nox regulators have distinct PX domains that are known to bind to specific acidic phospholipids present in distinct membranous compartments (63). In addition, they contain multiple protein interaction domains that can facilitate recruitment to various compartments.
We show that Tks4 and Tks5 represent novel functional members of the ‘p47 organizer superfamily’ that can selectively support Nox1- and Nox3-dependent ROS generation (Figure 1B). Interestingly, Tks4 and Tks5 were unable to act as organizer subunits for Nox2, nor to enhance Nox4-dependent ROS formation (Figure 2A). The former may suggest an inability of Tks to functionally recruit p67phox. The latter contrasts with the Tks-dependent regulation of Nox4 activity observed by Diaz et al (accompanying paper), raising the possibility that Nox4 activity may be organizer-dependent under certain cellular conditions. Nox1 activation mediated via Tks4- and Tks5 was blocked by the NADPH-oxidase flavoenzyme inhibitor DPI (Figure 2B) and was Rac-dependent (Figure 2C). The interaction between Tks organizers and NoxA1 is not affected by GTP-binding state of Rac (Figure 3B). Moreover, this interaction, as well as Tks-mediated ROS generation, can be prevented by the disruption of all five Tks SH3 domains (Figure 3C). We did not observe any significant change in the ability of Tks5 single- or double-SH3 domain mutants to support ROS generation and to bind NoxA1 (Figure S3 A to C), possibly due to the high expression level of these constructs in HEK293 cells. We note that a similar result was previously reported for the SH3-mediated interaction between Tks5 and N-Wasp (43). Interestingly, the basis for the interaction between Tks5 and NoxA1 differs from the interaction of the p47phox organizer with p67phox, which occurs via the binding of a C-terminal proline-rich sequence on p47phox to the C-terminal SH3 domain of p67phox (41). Tks proteins also appear to be capable of binding the p22phox subunit usually associated with the Nox1-Nox4 proteins via its first two SH3 domains (see Diaz et al). Finally, we showed that the PX domain of Tks5 was required for Nox1 activation, although not for binding to NoxA1. Overall, these observations presumably reflect the need for multiple protein- and lipid-interactions for proper membrane assembly of the Nox1 complex, analogous to those documented for Nox2 assembly by the p47phox organizer in neutrophils (14).
Human DLD1 colon cancer cells represent a system for the study of the biological effect of Tks-mediated ROS generation by Nox1
In this study, we have shown that human DLD1 colon cancer cells i) express only Nox1 (among members of the NADPH oxidase family), NoxA1, Tks4 and low levels of Tks5 (among members of the p47 organizer superfamily) (Figure S2), ii) are able to form invadopodia and degrade the ECM in a Tks4-dependent manner (Figure 4D) and iii) generate ROS via Nox1 through a mechanism dependent on Tks4 protein levels (Figure 4A to C) as a result of its biochemical interaction with endogenous NoxA1 (Figure 3A). In this cell system, Nox1 co-localizes with F-actin- and cortactin-positive structures which are able to degrade the ECM (Figure 5A to C and S6A), a hallmark of invadopodia function. We demonstrated that the expression of a different organizer subunit (NoxO1) competes with endogenous Tks4 in regulating Nox1 localization to these structures, consequently reducing the ROS-dependent formation of invadopodia. Indeed, despite an overall ROS generation comparable to that observed with Tks4 expression alone (Figure 4C), the overexpression of NoxO1 organizer in these cells caused a significant reduction in invadopodia formation and function overall (Figure 6A to C). This was most likely due to the loss of local ROS generation by the displaced Nox1, as suggested by the reduction of ROS-positive invadopodia-like structures observed in NoxO1-overexpressing DLD1 cells (Figure 6D). Consistent with this, the analysis of NoxO1 localization in these cells shows that this organizer subunit does not localize to invadopodia and, importantly, its presence strongly reduces invadopodia formation (Figure S7A). Future experiments will be planned to examine the localization of different members of the p47phox organizer superfamily in various subcellular compartments and evaluate their contribution to localized ROS generation in specific biological contexts. Results from this study demonstrate how the presence of different organizer subunits may localize Nox1 complex activity to distinct subcellular locations, facilitating spatially-confined ROS production near redox-sensitive targets to initiate specific redox signaling events.
Physiological and pathophysiological implications
Nox1 is abundant in colon epithelial cells (1, 64, 65), and Nox1 overexpression and increased ROS generation has been observed in human colon cancers and colon cell lines (66, 67). ROS generated by Nox1 have been implicated in the migration of colon cancer cells (50). Increased c-Src activity is also a characteristic of both premalignant and progressively advanced colon neoplasia, and it has been correlated with the conversion of benign polyps into malignant metastatic tumors (68). We have previously shown that there is a correlation between the levels of c-Src activity in colon cancer lines and their ability to generate ROS (39), yet the exact relationships between Src activity, ROS formation, carcinogenesis and metastasis remain poorly defined.
Metastatic cancer cells have the ability to both migrate to and degrade the ECM, and invasiveness has been correlated with the presence of invadopodia (28, 29), as well as ROS production (69). In the accompanying manuscript (Diaz, et al.), ROS formation via NADPH oxidases is shown to be a critical component of the formation of invadopodia in cancer cells. Consistent with this, we have found that i) specific-Nox1 siRNAs decrease ROS generation and consequently invadopodia formation and ECM degradation (Figure S6 A to C), ii) that the formation of invadopodia in DLD1 cells is Tks4-dependent, and iii) the presence of NoxO1 organizer subunit reduces formation of invadopodia and ECM degradation in DLD1 cells. Results from this study strengthen the notion that, rather than overall ROS production, it is the localized Tks-mediated generation of ROS at invadopodia that is responsible for invadopodia maintenance and function. They also suggest that members of the p47phox organizer superfamily may be more generally important in regulating other cellular processes requiring the localized formation of ROS. A better understanding of the mechanisms underlying the compartmentalization of redox signaling may suggest new selective approaches into the use of antioxidant and/or anti-Nox-directed therapies for treatment of various redox stress-dependent diseases and/or cancer metastasis.
Materials and Methods
DNA constructs, reagents and antibodies
Expression vectors for all members of the NADPH oxidase family and p47 organizer family, along with plasmids for SrcYF, RacQL, RacN17, NoxA1, NoxA1-R103E and Nox1TM were previously described (27, 39). RFP-tagged and GFP-tagged Nox1 were created by subcloning pcDNA3.1-Nox1 into pDsRed-Monomer-C1 and pEGFP-N1 (Clontech) respectively. GFP-tagged NoxO1 was created by subcloning pRK5m-NoxO1 into pEGFP-N1. The Tks5 SH3 domain mutant constructs were generated inserting the following mutations: W188A, W260A, W441A, W827A, W1056A by using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described (42). Cell culture medium, fetal bovine serum, supplements and Hank’s Balanced Salt Solution (HBSS, Cat# 24020-117) were from Invitrogen (Carlsbad, CA). Plasmids for transfection were purified using the Qiagen Qiafilter system (Chatsworth, CA). DLD1 colonic adenocarcinoma cells (Cat #CCL-221) and other carcinoma cell lines were purchased from ATCC. DPI (D2926), horseradish peroxidase HRP (77330) and luminol (09253) were purchased from Sigma (St. Louis, USA). The following antibodies were purchased as indicated: rabbit polyclonal GFP antibody from Molecular Probes (Eugene, OR); mouse monoclonal actin antibody (691002) from MD Bioscience; mouse monoclonal Flag antibody M2 from Sigma-Aldrich, St. Louis, MO, and mouse monoclonal cortactin antibody 4F10 from Millipore, Temecula, CA. Anti-mouse Alexa-Fluor-568 and 647, anti-rabbit Alexa-Fluor-568 and Alexa-Fluor-568 phalloidin were purchased from Molecular Probes, Carlsbad, CA. Rabbit polyclonal Tks4 and Tks5 antibody have been previously described (23, 27). Rabbit polyclonal Nox1 antibody was kindly provided by Dr. David Lambeth (Emory). 9E10 anti-myc antibody and rabbit polyclonal NoxA1 were prepared in-house.
Cell cultures, transfection and siRNAs
Human HEK293, human DLD1, HT29, SW620 colonic adenocarcinoma and mouse 3T3 and Src-3T3 fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (Invitrogen), 2mM glutamine and antibiotics (100 units/ml penicillin and 100 g/ml streptomycin) at 37 °C in 5% CO2. Cells were transfected by Lipofectamine 2000 (InVitrogen) as described (39), and 24–96hrs after transfection the cells were processed accordingly. ON-TARGETplus SMARTpool Tks4, Nox1 and control siRNA mixture (J-032834-05/08, J-010193-05/08) were purchased from Dharmacon.
RT-PCR, Western blot and immunoprecipitation
RNA extraction was performed using RNeasy kit (Quiagen) and reverse transcribed with Superscript II enzyme (Invitrogen) according to the manufacturer’s instructions. Western blot and protein extracts were performed as described (39). For immunoprecipitations, 1–2ul of antibodies was incubated with 1 mg of protein lysates for 2 h at 4C, followed by 30 min incubation with 20ul of Protein G–Sepharose (Amersham). Immunoprecipitates were washed three times in lysis buffer and proteins released by boiling in Laemmli SDS sample buffer, then analyzed by Western blot.
Measurements of ROS
Reactive oxygen species were measured using a luminol-based chemiluminescence assay (CL-assay), as described (39). Chemiluminescence was recorded using 96-well plate luminometer (Berthold) 5 min after the addition of HRP/luminol mixture (with the exception of Figure 1B in which ROS generation was monitored continuously for 30 minutes). For real-time visualization of ROS generation illustrated in Figure 6D, DLD1 cells plated on glass coverslips were incubated with 2.5μM of the ROS-sensitive probe PY1-AM (Peroxy-Yellow 1 Acetoxymethyl-ester) in HBSS for 30 minutes at 37C, washed with PBS, mounted on slides and analyzed by confocal microscopy. PY1-AM (unpublished; kindly provided by Dr. Bryan C. Dickinson and Dr. Christopher J. Chang, UC Berkeley) is a selective fluorescent indicator for hydrogen peroxide related to Peroxy Green 1 (70) and Mito Peroxy Yellow 1 (71).
Confocal and epifluorescence microscopy
24–48hrs after transfection, DLD1 cells plated on glass or FITC-labeled gelatin-coated coverslips were fixed in 4% paraformaldehyde (PFA) at room temperature or in methanol at −20C for 10 minutes. Successively, cells were permeabilized in 0.5% Triton for 10 minutes and blocked in 2%BSA in PBS for 45 minutes at room temperature. Cells were then immunolabeled as indicated in the figure legends with appropriate primary and Alexa-Fluor 568- or 647-conjugated secondary antibodies. F-actin was detected by using Alexa-Fluor 568-conjugated phalloidin. Cells were mounted on slides with mowiol mounting medium (Calbiochem) according to the manufacturer’s instructions. Epifluorescence images of fixed cells were acquired on an inverted microscope (Eclipse TE 2000-U, Nikon) equipped with an electronically controlled shutter, filter wheels, and a 14-bit cooled CCD camera (Cool SNAP HQ, Photometrics) controlled by MetaMorph software (Universal Imaging Corp.) by using a 60x/1.4 NA Plan Apo DIC or a 40x/1.4 NA Plan Apo Ph3 objective lens (Nikon) (72). Confocal images were acquired on a spinning disk confocal microscope system (72), equipped with a CoolSnapHQ camera and 100x/1.4 NA Plan Apo or a 60x/1.4 NA Plan Apo objective lens (Nikon).
ECM degradation assays
Fluorescently labeled gelatin-coated coverslips were prepared as previously described (32). Cells were transfected as indicated in figure legends and incubated on labeled coverslips for 24–48hrs, then processed accordingly for epifluorescence or confocal microscopy.
Statistical analysis
In this study representative experiments from at least three independent experiments are shown. Results for each experiment are given as the mean of triplicates +/− standard deviation (S.D.). Statistically significant differences between sample groups were determined using two-tailed t-tests (Microsoft Excel, Redmond WA). p values were <0.001 unless differently indicated in the figure legends.
Supplementary Material
Acknowledgments
We acknowledge and profoundly appreciate Dr. Bryan C. Dickinson and Dr. Christopher J. Chang (UC Berkeley) for allowing our use of the ROS-specific probe PY1-AM in this study. We thank Dr. David Lambeth (Emory) for Nox1 antibody. The authors also acknowledge the expert technical assistance of Ben Bohl, and Violaine Delorme-Walker for helping with confocal microscopy and suggestions in revising this manuscript. Tks4 constructs were provided by Paul Bromann (Biomedicum, Helsinki). We thank G. Danuser for access to the confocal microscope used in these studies. This work was supported by NIH grant HL48008 (to GMB) and The National Cancer Institute (to SC), and a postdoctoral fellowship from Fondation pour la Recherche Medicale FRM (to N.T.). This is manuscript IMM20051 from The Scripps Research Institute.
References and Notes
- 1.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 2.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 3.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch GM, Knaus UG. NADPH oxidases: not just for leukocytes anymore! Trends Biochem Sci. 2003;28:502. doi: 10.1016/S0968-0004(03)00194-4. [DOI] [PubMed] [Google Scholar]
- 5.Lambeth JD. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radic Biol Med. 2007;43:332. doi: 10.1016/j.freeradbiomed.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambeth JD, Krause KH, Clark RA. NOX enzymes as novel targets for drug development. Semin Immunopathol. 2008 doi: 10.1007/s00281-008-0123-6. [DOI] [PubMed] [Google Scholar]
- 7.Ushio-Fukai M. Compartmentalization of Redox Signaling through NADPH Oxidase-derived ROS. Antioxid Redox Signal. 2008 doi: 10.1089/ars.2008.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bokoch GM, Diebold BA. Current molecular models for NADPH oxidase regulation by Rac GTPase. Blood. 2002;100:2692. doi: 10.1182/blood-2002-04-1149. [DOI] [PubMed] [Google Scholar]
- 9.Cheng G, Diebold BA, Hughes Y, Lambeth JD. Nox1-dependent reactive oxygen generation is regulated by Rac1. The Journal of biological chemistry. 2006;281:17718. doi: 10.1074/jbc.M512751200. [DOI] [PubMed] [Google Scholar]
- 10.Miyano K, Sumimoto H. Role of the small GTPase Rac in p22(phox)-dependent NADPH oxidases. Biochimie. 2007;89:1133. doi: 10.1016/j.biochi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Ueyama T, Geiszt M, Leto TL. Involvement of Rac1 in activation of multicomponent Nox1- and Nox3-based NADPH oxidases. Mol Cell Biol. 2006;26:2160. doi: 10.1128/MCB.26.6.2160-2174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambeth JD, Kawahara T, Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic Biol Med. 2007;43:319. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLeo FR, Quinn MT. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J Leukoc Biol. 1996;60:677. doi: 10.1002/jlb.60.6.677. [DOI] [PubMed] [Google Scholar]
- 14.Groemping Y, Rittinger K. Activation and assembly of the NADPH oxidase: a structural perspective. Biochem J. 2005;386:401. doi: 10.1042/BJ20041835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nauseef WM. Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol. 2004;122:277. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 16.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci. 2002;59:1428. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suh C, Stull N, Fujii Y, Grinstein S, Yaffe M, Atkinson S, Dinauer MC. Role for p40 phox in Fc g -receptor-induced NADPH oxidase activation. Blood. 2006;104:188a. [Google Scholar]
- 18.Ueyama T, Kusakabe T, Karasawa S, Kawasaki T, Shimizu A, Son J, Leto TL, Miyawaki A, Saito N. Sequential Binding of Cytosolic Phox Complex to Phagosomes through Regulated Adaptor Proteins: Evaluation Using the Novel Monomeric Kusabira-Green System and Live Imaging of Phagocytosis. J Immunol. 2008;181:629. doi: 10.4049/jimmunol.181.1.629. [DOI] [PubMed] [Google Scholar]
- 19.Ueyama T, Tatsuno T, Kawasaki T, Tsujibe S, Shirai Y, Sumimoto H, Leto TL, Saito N. A regulated adaptor function of p40phox: distinct p67phox membrane targeting by p40phox and by p47phox. Mol Biol Cell. 2007;18:441. doi: 10.1091/mbc.E06-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissonnette SA, Glazier CM, Stewart MQ, Brown GE, Ellson CD, Yaffe MB. Phosphatidylinositol 3-phosphate-dependent and -independent functions of p40phox in activation of the neutrophil NADPH oxidase. J Biol Chem. 2008;283:2108. doi: 10.1074/jbc.M706639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian W, Li XJ, Stull ND, Ming W, Suh CI, Bissonnette SA, Yaffe MB, Grinstein S, Atkinson SJ, Dinauer MC. Fc{gamma}R-stimulated activation of the NADPH oxidase: phosphoinositide-binding protein p40phox regulates NADPH oxidase activity after enzyme assembly on the phagosome. Blood. 2008;112:3867. doi: 10.1182/blood-2007-11-126029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taura M, Miyano K, Minakami R, Kamakura S, Takeya R, Sumimoto H. A region N-terminal to the tandem SH3 domain of p47phox plays a crucial role in activation of the phagocyte NADPH oxidase. Biochem J. 2008 doi: 10.1042/BJ20082028. [DOI] [PubMed] [Google Scholar]
- 23.Lock P, Abram CL, Gibson T, Courtneidge SA. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. The EMBO journal. 1998;17:4346. doi: 10.1093/emboj/17.15.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courtneidge SA. Isolation of novel Src substrates. Biochemical Society transactions. 2003;31:25. doi: 10.1042/bst0310025. [DOI] [PubMed] [Google Scholar]
- 25.Courtneidge SA, Azucena EF, Pass I, Seals DF, Tesfay L. The SRC substrate Tks5, podosomes (invadopodia), and cancer cell invasion. Cold Spring Harbor symposia on quantitative biology. 2005;70:167. doi: 10.1101/sqb.2005.70.014. [DOI] [PubMed] [Google Scholar]
- 26.Abram CL, Seals DF, Pass I, Salinsky D, Maurer L, Roth TM, Courtneidge SA. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J Biol Chem. 2003;278:16844. doi: 10.1074/jbc.M300267200. [DOI] [PubMed] [Google Scholar]
- 27.Buschman MD, Bromann PA, Cejudo-Martin P, Wen F, Pass I, Courtneidge SA. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol Biol Cell. 2009;20:1302. doi: 10.1091/mbc.E08-09-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Current opinion in cell biology. 2008;20:235. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends in cell biology. 2007;17:107. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Lowe C, Yoneda T, Boyce BF, Chen H, Mundy GR, Soriano P. Osteopetrosis in Src-deficient mice is due to an autonomous defect of osteoclasts. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4485. doi: 10.1073/pnas.90.10.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldassarre M, Pompeo A, Beznoussenko G, Castaldi C, Cortellino S, McNiven MA, Luini A, Buccione R. Dynamin participates in focal extracellular matrix degradation by invasive cells. Molecular biology of the cell. 2003;14:1074. doi: 10.1091/mbc.E02-05-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blouw B, Seals DF, Pass I, Diaz B, Courtneidge SA. A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. European journal of cell biology. 2008;87:555. doi: 10.1016/j.ejcb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seals DF, Azucena EF, Jr, Pass I, Tesfay L, Gordon R, Woodrow M, Resau JH, Courtneidge SA. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer cell. 2005;7:155. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Kawahara T, Lambeth JD. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC evolutionary biology. 2007;7:178. doi: 10.1186/1471-2148-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matute JD, Arias AA, Dinauer MC, Patino PJ. p40phox: the last NADPH oxidase subunit. Blood Cells Mol Dis. 2005;35:291. doi: 10.1016/j.bcmd.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Quinn MT, Gauss KA. Structure and regulation of the neutrophil respiratory burst oxidase: comparison with nonphagocyte oxidases. J Leukoc Biol. 2004;76:760. doi: 10.1189/jlb.0404216. [DOI] [PubMed] [Google Scholar]
- 37.Banfi B, Malgrange B, Knisz J, Steger K, Dubois-Dauphin M, Krause KH. NOX3, a superoxide-generating NADPH oxidase of the inner ear. J Biol Chem. 2004;279:46065. doi: 10.1074/jbc.M403046200. [DOI] [PubMed] [Google Scholar]
- 38.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008 doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 39.Gianni D, Bohl B, Courtneidge SA, Bokoch GM. The involvement of the tyrosine kinase c-Src in the regulation of reactive oxygen species generation mediated by NADPH oxidase-1. Molecular biology of the cell. 2008;19:2984. doi: 10.1091/mbc.E08-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross AR. Inhibitors of the leukocyte superoxide generating oxidase: mechanisms of action and methods for their elucidation. Free Radic Biol Med. 1990;8:71. doi: 10.1016/0891-5849(90)90147-b. [DOI] [PubMed] [Google Scholar]
- 41.Takeya R, Ueno N, Kami K, Taura M, Kohjima M, Izaki T, Nunoi H, Sumimoto H. Novel human homologues of p47phox and p67phox participate in activation of superoxide-producing NADPH oxidases. The Journal of biological chemistry. 2003;278:25234. doi: 10.1074/jbc.M212856200. [DOI] [PubMed] [Google Scholar]
- 42.Kao YY, Gianni D, Bohl B, Taylor RM, Bokoch GM. Identification of a conserved Rac-binding site on NADPH oxidases supports a direct GTPase regulatory mechanism. The Journal of biological chemistry. 2008;283:12736. doi: 10.1074/jbc.M801010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oikawa T, Itoh T, Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J Cell Biol. 2008;182:157. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Kim JS, Huang TY, Bokoch GM. Reactive Oxygen Species (ROS) Regulate a Slingshot-Cofiin Activation Pathway. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terada LS. Specificity in reactive oxidant signaling: think globally, act locally. J Cell Biol. 2006;174:615. doi: 10.1083/jcb.200605036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moldovan L, Moldovan NI, Sohn RH, Parikh SA, Goldschmidt-Clermont PJ. Redox changes of cultured endothelial cells and actin dynamics. Circ Res. 2000;86:549. doi: 10.1161/01.res.86.5.549. [DOI] [PubMed] [Google Scholar]
- 48.Wesley UV, Bove PF, Hristova M, McCarthy S, van der Vliet A. Airway epithelial cell migration and wound repair by ATP-mediated activation of dual oxidase 1. J Biol Chem. 2007;282:3213. doi: 10.1074/jbc.M606533200. [DOI] [PubMed] [Google Scholar]
- 49.Wu RF, Xu YC, Ma Z, Nwariaku FE, Sarosi GA, Jr, Terada LS. Subcellular targeting of oxidants during endothelial cell migration. J Cell Biol. 2005;171:893. doi: 10.1083/jcb.200507004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadok A, Bourgarel-Rey V, Gattacceca F, Penel C, Lehmann M, Kovacic H. Nox1-dependent superoxide production controls colon adenocarcinoma cell migration. Biochim Biophys Acta. 2008;1783:23. doi: 10.1016/j.bbamcr.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Schroder K, Helmcke I, Palfi K, Krause KH, Busse R, Brandes RP. Nox1 mediates basic fibroblast growth factor-induced migration of vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2007;27:1736. doi: 10.1161/ATVBAHA.107.142117. [DOI] [PubMed] [Google Scholar]
- 52.Weber DS, Taniyama Y, Rocic P, Seshiah PN, Dechert MA, Gerthoffer WT, Griendling KK. Phosphoinositide-dependent kinase 1 and p21-activated protein kinase mediate reactive oxygen species-dependent regulation of platelet-derived growth factor-induced smooth muscle cell migration. Circ Res. 2004;94:1219. doi: 10.1161/01.RES.0000126848.54740.4A. [DOI] [PubMed] [Google Scholar]
- 53.Ushio-Fukai M. Redox signaling in angiogenesis: role of NADPH oxidase. Cardiovasc Res. 2006;71:226. doi: 10.1016/j.cardiores.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 54.Munnamalai V, Suter DM. Reactive Oxygen Species Regulate F-actin Dynamics in Neuronal Growth Cones and Neurite Outgrowth. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- 56.Hilenski LL, Clempus RE, Quinn MT, Lambeth JD, Griendling KK. Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:677. doi: 10.1161/01.ATV.0000112024.13727.2c. [DOI] [PubMed] [Google Scholar]
- 57.Ushio-Fukai M, Hilenski L, Santanam N, Becker PL, Ma Y, Griendling KK, Alexander RW. Cholesterol depletion inhibits epidermal growth factor receptor transactivation by angiotensin II in vascular smooth muscle cells: role of cholesterol-rich microdomains and focal adhesions in angiotensin II signaling. The Journal of biological chemistry. 2001;276:48269. doi: 10.1074/jbc.M105901200. [DOI] [PubMed] [Google Scholar]
- 58.Nwariaku FE, Liu Z, Zhu X, Nahari D, Ingle C, Wu RF, Gu Y, Sarosi G, Terada LS. NADPH oxidase mediates vascular endothelial cadherin phosphorylation and endothelial dysfunction. Blood. 2004;104:3214. doi: 10.1182/blood-2004-05-1868. [DOI] [PubMed] [Google Scholar]
- 59.Ikeda S, Yamaoka-Tojo M, Hilenski L, Patrushev NA, Anwar GM, Quinn MT, Ushio-Fukai M. IQGAP1 regulates reactive oxygen species-dependent endothelial cell migration through interacting with Nox2. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:2295. doi: 10.1161/01.ATV.0000187472.55437.af. [DOI] [PubMed] [Google Scholar]
- 60.Wu RF, Gu Y, Xu YC, Nwariaku FE, Terada LS. Vascular endothelial growth factor causes translocation of p47phox to membrane ruffles through WAVE1. The Journal of biological chemistry. 2003;278:36830. doi: 10.1074/jbc.M302251200. [DOI] [PubMed] [Google Scholar]
- 61.Li Q, Harraz MM, Zhou W, Zhang LN, Ding W, Zhang Y, Eggleston T, Yeaman C, Banfi B, Engelhardt JF. Nox2 and Rac1 regulate H2O2-dependent recruitment of TRAF6 to endosomal interleukin-1 receptor complexes. Molecular and cellular biology. 2006;26:140. doi: 10.1128/MCB.26.1.140-154.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuroda J, Nakagawa K, Yamasaki T, Nakamura K, Takeya R, Kuribayashi F, Imajoh-Ohmi S, Igarashi K, Shibata Y, Sueishi K, Sumimoto H. The superoxide-producing NAD(P)H oxidase Nox4 in the nucleus of human vascular endothelial cells. Genes Cells. 2005;10:1139. doi: 10.1111/j.1365-2443.2005.00907.x. [DOI] [PubMed] [Google Scholar]
- 63.Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta. 2006;1761:878. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Perner A, Andresen L, Pedersen G, Rask-Madsen J. Superoxide production and expression of NAD(P)H oxidases by transformed and primary human colonic epithelial cells. Gut. 2003;52:231. doi: 10.1136/gut.52.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rokutan K, Kawahara T, Kuwano Y, Tominaga K, Sekiyama A, Teshima-Kondo S. NADPH oxidases in the gastrointestinal tract: a potential role of Nox1 in innate immune response and carcinogenesis. Antioxid Redox Signal. 2006;8:1573. doi: 10.1089/ars.2006.8.1573. [DOI] [PubMed] [Google Scholar]
- 66.Arnold RS, Shi J, Murad E, Whalen AM, Sun CQ, Polavarapu R, Parthasarathy S, Petros JA, Lambeth JD. Hydrogen peroxide mediates the cell growth and transformation caused by the mitogenic oxidase Nox1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5550. doi: 10.1073/pnas.101505898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, Lambeth JD. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 68.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 69.Okada F, Kobayashi M, Tanaka H, Kobayashi T, Tazawa H, Iuchi Y, Onuma K, Hosokawa M, Dinauer MC, Hunt NH. The role of nicotinamide adenine dinucleotide phosphate oxidase-derived reactive oxygen species in the acquisition of metastatic ability of tumor cells. Am J Pathol. 2006;169:294. doi: 10.2353/ajpath.2006.060073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol. 2007;3:263. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 71.Dickinson BC, Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc. 2008;130:9638. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Developmental cell. 2007;13:646. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.