Abstract
In the present study we examined expression of four real-time quantitative RT-PCR reference genes commonly applied to rodent models of brain injury. Transcripts for β-actin, cyclophilin A, GAPDH, and 18S rRNA were assessed at 2–15 days post-injury, focusing on the period of synaptic recovery. Diffuse moderate central fluid percussion injury (FPI) was contrasted with unilateral entorhinal cortex lesion (UEC), a model of targeted deafferentation. Expression in UEC hippocampus, as well as in FPI hippocampus and parietotemporal cortex was analyzed by qRT-PCR. Within-group variability of gene expression was assessed and change in expression relative to paired controls was determined. None of the four common reference genes tested was invariant across brain region, survival time, and type of injury. Cyclophilin A appeared appropriate as a reference gene in UEC hippocampus, while β-actin was most stable for the hippocampus subjected to FPI. However, each gene may fail as a suitable reference with certain test genes whose RNA expression is targeted for measurement. In FPI cortex, all reference genes were significantly altered over time, compromising their utility for time-course studies. Despite such temporal variability, certain genes may be appropriate references if limited to single survival times. These data provide an extended baseline for identification of appropriate reference genes in rodent studies of recovery from brain injury. In this context, we outline additional considerations for selecting a qRT-PCR normalization strategy in such studies. As previously concluded for acute post-injury intervals, we stress the importance of reference gene validation for each brain injury paradigm and each set of experimental conditions.
Key words: genomics, molecular biological approaches, neuroplasticity, synaptic loss and deafferentation, traumatic brain injury
Introduction
Real-time quantitative RT-PCR (qRT-PCR) is a widely preferred method for the sensitive measurement of mRNA in tissue. The number of studies using qRT-PCR to quantify gene expression after injury to the central nervous system has grown over the past decade. Given that traumatic brain injury (TBI) induces a variety of transcription-regulated processes (e.g., reactive gliosis, inflammation, activation of cell death pathways, neuroplasticity, and regeneration), it is clear that alteration in gene expression is part of the tissue response. The quantitative assessment of specific mRNAs is therefore an essential tool in TBI research and an important complement to measures of protein expression. There are several advantages of using qRT-PCR to assess mRNA transcripts (Gibson et al., 1996; Heid et al., 1996; Giulietti et al., 2001; Bustin, 2002; Nolan et al., 2006). Foremost, it offers rapid, high-throughput screening of genes with less opportunity for RNA contamination than other quantitative methods. It also enables the use of small amounts of starting material, an advantage that presents opportunities for more specific sampling with TBI studies. For example, Dash and colleagues (2004) used qRT-PCR to assess mRNA from small punches of cortical tissue harvested at varying distances from the injury core, permitting high spatial accuracy in analysis of injury-induced gene expression. Second, qRT-PCR provides a more sensitive measure of gene expression than other semi-quantitative assays (e.g., Northern blots or in-situ hybridization), particularly for the detection of small changes in genes of interest. Furthermore, relative to microarray analysis, qRT-PCR may also be preferred for the detection of less than twofold changes in gene expression. In a direct comparison between two microarray platforms and TaqMan RT-PCR, Wang and colleagues (2006) showed that the arrays displayed a significant reduction in detection accuracy at low transcript expression levels and relatively poor sensitivity in detecting small-fold changes. This difference in sensitivity is likely to be especially important with TBI, where significant changes in gene expression are often observed within a range of less than twofold. One TBI study by Li and associates (2004) showed that the two approaches produce comparable results for many genes, but for some transcripts, such as HSP70, qRT-PCR was far more sensitive, detecting a significant 1.5-fold increase, while microarray sampling showed no change.
Despite the advantages of qRT-PCR, the increased use of this technique has revealed certain technical concerns. In particular, the identification of suitable reference genes has been discussed in several publications (Dheda et al., 2004; Bustin et al., 2005; Huggett et al., 2005; Meldgaard et al., 2006; Johansson et al., 2007; Coulson et al., 2008; Bonefeld et al., 2008). In order to control for nonspecific variations caused by differences in RNA input among samples, qRT-PCR data are typically normalized to an endogenous reference gene whose transcription is expected to be stable. Commonly used reference genes include cyclophilin A (also known as PPIA), β-actin, tubulin, glyceraldehyde phosphate dehydrogenase (GAPDH), and 18S or 28S rRNA. An ideal reference gene should be stably expressed across all tissues, developmental stages, and experimental conditions, a requirement that is difficult to confirm in real practice. Prior studies have shown that CNS expression of some of these widely used reference genes may be altered in models of hypoxia/ischemia (Zhong and Simons, 1999; Bond et al., 2002; Kobayashi et al., 2004), seizure (Chen et al., 2001), hippocampal deafferentation (Phillips et al., 1987; Phillips and Steward, 1990), demyelination (Meldgaard et al., 2006), and TBI (Thal et al., 2008; Rhinn et al., 2008; Cook et al., 2008). To make the choice more difficult, other brain trauma paradigms report some of these reference genes to be invariant and useful for qRT-PCR normalization. For example, GAPDH and cyclophilin A were both stably expressed during the first 24 h after middle cerebral artery occlusion in rats (Harrison et al., 2000). Furthermore, in the human post-mortem brain, optimal reference genes vary with disease state and often differ in choice ranking for a given brain region relative to those reported for animal models of brain injury (Coulson et al., 2008).
Normalization to total input RNA mass is an alternative to the use of a reference gene control (Bustin, 2005). This approach depends on the ability to recover high-quality RNA from tissue and to accurately measure its concentration and integrity. Several studies report that, if these conditions are met, normalization to total RNA produces valid results (Tricarico et al., 2002; Dheda et al., 2004; Bustin, 2005; Meldgaard et al., 2006). The concomitant use of multiple reference genes is also an option, for which several algorithms for data analysis have been proposed (Vandesompele et al., 2002; Pfaffl et al., 2004; Andersen et al., 2004). A recent report by Cook and colleagues (2008) has taken this approach to analyze the suitability of several candidate reference genes for the impact acceleration model of TBI at 3 days post-injury, comparing neocortex and hippocampus tissue. In that case, the stability of three novel and four previously applied reference genes was compared using an algorithm that employs average pair-wise comparison of variance for each gene relative to the other control genes tested. This approach requires determination of reference efficacy for each candidate gene, and while it can help to identify optimal choices for RNA normalization, the examination of a targeted gene of interest in this context may result in reduced detection of experimental effect, particularly if the change in transcript is small.
Two additional studies have examined qRT-PCR normalization after TBI (Thal et al., 2008; Rhinn et al., 2008); however, these have also been limited to acute 1- to 3-days-post-injury intervals, and focused on single rodent models. TBI presents a particular challenge for the accurate quantification of mRNA due to the fact that heterogeneous cell populations are injured, generating a multifaceted pathology (primary tissue damage, progressive secondary degeneration, ischemic/hypoxic damage, edema, hemorrhage, and inflammation). Moreover, there can be considerable group variability due to inter-animal differences in response to brain injury. Unfortunately, each of these parameters may impact the expression of common qRT-PCR reference genes.
Several prior studies of gene expression during injury-induced synaptic plasticity (Kerber et al., 2003; Price et al., 2003; Molteni et al., 2002; Blain et al., 2004) employ qRT-PCR reference genes recently reported to vary with brain trauma (Thal et al., 2008; Rhinn et al., 2008; Cook et al., 2008). In the present study we have extended TBI reference gene analysis, now focusing on 2–15 days post-injury, the period of functional synaptic recovery. We examined the expression of four genes widely used for qRT-PCR normalization (β-actin, cyclophilin A, GAPDH, and 18S rRNA), comparing their temporal profiles following either diffuse closed-head trauma produced by moderate central fluid percussion injury (FPI) or targeted deafferentation induced by unilateral entorhinal cortex lesion (UEC).
Methods
Experimental animals
Adult male Sprague-Dawley rats (300–390 g) were used in this study. The rats were randomly divided into three experimental groups: UEC (n = 18), FPI (n = 30), and sham-injured FPI (n = 17). Rats were housed in pairs in individual cages with food and water ad libitum, and subjected to a 12-h dark-light cycle at 22°C. All protocols for injury and use of animals were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University.
Unilateral entorhinal cortex lesion
Rats were subjected to UEC using the method of Loesche and Steward (1977). All animals were surgically prepared under isoflurane anesthesia delivered via nose cone (2% in carrier gas of 70% N2O and 30% O2). To ensure a safe surgical plane of anesthesia, and to prevent hypoxia or hypercapnia, the rats were monitored during the course of surgery with a pulse-oximetry system (MouseOx, Starr Life Sciences, Corp., Oakmont, PA). Body temperature was maintained at 37°C via a thermostatically controlled heating pad (Harvard Apparatus, Holliston, MA). The rats were placed in a stereotaxic frame, and an area of skull was removed above the entorhinal cortex of the right hemisphere. Lesion current was passed through a polytetrafluoroethylene-insulated wire electrode angled at 10° from vertical. Current was delivered (1.5 mA for 30 sec) at a total of nine stereotaxic sites: 1.5 mm anterior to the transverse sinus; 3, 4, and 5 mm lateral to the midline; and at 2, 4, and 6 mm ventral to the brain surface. After the lesions were completed, the electrode was removed, the scalp was sutured closed over the surgical site and Bacitracin applied to the wound. The animals were monitored during full recovery from anesthesia and returned to their home cages.
Central fluid percussion injury
All animals were surgically prepared under isoflurane anesthesia (2% in carrier gas of 70% N2O and 30% O2) delivered via a nose cone and monitored with the MouseOx pulse-oximetry system as described above. After placement in a stereotaxic frame, the scalp was sagittally incised and a 4.8-mm hole trephined into the skull over the sagittal suture, midway between the bregma and the lambda. Two steel screws were placed 1 mm rostral to the bregma and 1 mm caudal to the lambda. A modified Luer-Lock syringe hub with an inner diameter of 2.6 mm was placed over the exposed dura and bonded to the skull with cyanoacrylate adhesive. Dental acrylic was then poured around the syringe hub and skull screws and allowed to harden. The hub was packed with gelfoam, the scalp was sutured closed, and Bacitracin was applied to the surgical site. Animals were monitored for full recovery from anesthesia and returned to their home cages.
The model used for injury was the same as that described by Dixon and associates (1987). The fluid percussion injury device consists of an acrylic glass cylinder 60 cm long and 4.5 cm in diameter, filled with double-distilled water. The cylinder is closed at one end with a rubber-covered acrylic glass piston mounted on O rings. The opposite end of the cylinder is closed by an 8-cm metal extracranial pressure transducer (model EPN-0300-100A; Entram Devices, Inc., Fairfield, NJ). Attached to the end of the metal transducer is a 5-mm tube (2.6 mm inner diameter) that terminates with a male Luer-Lock fitting. This fitting is connected to the surgically implanted female Luer-Lock at the time of injury. The injury is created when a metal pendulum strikes the piston of the injury device, injecting a small volume of water into the closed cranial cavity, directly onto the exposed dura. This causes a brief (20-millisecond) displacement and deformation of the underlying cortex resulting in brain injury. The magnitude of the pressure pulse is recorded extracranially by an in-line transducer, is expressed in atmospheres (atm) of pressure, and is visualized on a storage oscilloscope (Tektronix 5111; Tektronix, Inc., Beaverton, OR). The severity of the injury is controlled by adjusting the height of the pendulum.
Twenty-four hours after the surgical preparation, the rats were anesthetized with isoflurane (4% in carrier gas of 70% N2O and 30% O2) for 4 min in a 2-l chamber. Once anesthetized, the animals were removed from the chamber, the surgically prepared site was exposed, and the animal was connected to the FPI device. The animals were injured at 2.0 ± 0.05 atm. This is equivalent to a moderate level of injury that has previously been demonstrated to produce consistent pathophysiological responses. Sham-injured controls received the same surgical preparation, anesthesia, and connection to the injury device, but no injury was delivered. All the animals were monitored until spontaneous breathing resumed, the injury site was sutured closed, and Bacitracin was applied. Behavioral assessments were performed on the injured animals to assure a controlled level of injury, including pinna, corneal, paw, and tail reflexes, as well as righting and head-support responses. Once all responses had been recorded and the animal was conscious, the injured rat was placed in a recovery chamber, monitored for signs of bleeding, respiratory difficulty, or problems with cardiovascular circulation for 2–3 h, and then returned to its home cage.
RNA isolation and qRT-PCR
Rats were sacrificed 2 (n = 6), 7 (n = 6), and 15 days (n = 6) after UEC lesion, or 2 (n = 6), 7 (n = 5), and 15 days (n = 3) after FPI. Sham-injured animals for FPI comparison were sacrificed at the 7-day time point only (n = 6). Whole hippocampi were rapidly dissected from UEC rats, while hippocampi and parietotemporal cortices were removed from FPI animals. Because of the midline FPI location, tissue blocks dissected from both hemispheres were pooled for analysis. Samples (∼100 mg) were homogenized in 1 mL Trizol reagent (Invitrogen, Carlsbad, CA), mixed with 0.2 mL chloroform, and centrifuged for 15 min at 12,000 × g. RNA in the upper phase was removed and precipitated with 0.5 mL isopropyl alcohol. After centrifugation for 10 min at 12,000 × g, supernatant was removed and RNA pellets were washed in 75% ethanol. The pellets were then dissolved in PCR-grade water (Ambion, Austin, TX) and incubated at 55°C for 10 min. All samples were then treated with two cycles of DNAse treatment to remove residual DNA contamination using the DNA-free DNase kit (Ambion), according to the manufacturer's protocol. Briefly, samples were incubated in a mixture of DNAse buffer and DNAse 1 for 20 min at 37°C, followed by 10 min at room temperature with DNAse Inactivation Reagent. After centrifugation at 10,000 × g for 2 min to pellet inactivation reagent, supernatants were removed and stored at −80°C.
RNA concentration and integrity were first assessed for all samples with the Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA), after which concentration was verified by NanoDrop spectrophotometry (ND-1000; NanoDrop Products, Wilmington, DE). Subsequent electropherogram analysis (Agilent Technologies, Santa Clara, CA) indicated high-quality RNA (RIN ≥ 7.0), clear peaks for 18S and 28S rRNA, and minimal sample degradation. Total RNA was prepared for each sample in PCR-grade water (20 ng/μL) and submitted for TaqMan One-Step RT-PCR on the ABI prism 7900 Sequence Detection system (Applied Biosystems; Foster City, CA). Specific TaqMan primers were designed to span exon-exon boundaries for all reference and target genes using Primer Express software (Applied Biosystems). Sequences for specific primers and TaqMan probes are listed in Table 1. A constant reference calibrator sample of total RNA was serially diluted and used to generate a standard CT-versus-quantity plot for each run. In all cases, the plot was linear with R2 > 0.98. PCR reactions were performed in triplicate and relative mRNA expression was determined from the CT-versus-quantity plots. By this standard curve method, change in target RNA was expressed in arbitrary units relative to the standard calibrator. These units were used for RNA data analysis and were plotted as relative RNA quantity. Reactions had an overall efficiency between 90% and 100% as determined by the slope of the standard curves (range −3.1 to −4.2), calculated as E = 10(−1/slope) − 1. Negative controls on each plate included no-amplification and no-template conditions.
Table 1.
TaqMan Sequences Applied for qRT-PCR of Reference and Test Genes Phosphacan and sRPTP-β
| Gene | Oligo sequence |
|---|---|
| GAPDH forward | 5′ AATGTATCCGTTGTGGATCTGACA 3′ |
| GAPDH reverse | 5′ CTCGGCCGCCTGCTT 3′ |
| GAPDH probe | 5′ CCTGGAGAAACCTGCCAAGTATGATGACATC 3′ |
| Cyclophilin A forward | 5′ CTGTTTGCAGACAAAGTTCCAAA 3′ |
| Cyclophilin A reverse | 5′ AGGAACCCTTATAGCCAAATCCTT 3′ |
| Cyclophilin A probe | 5′ CAGCAGAAAACTTTCGTGCTCTGAGCACT 3′ |
| β-Actin forward | 5′ CCCTGGCTCGCACCAT 3′ |
| β-Actin reverse | 5′ GAGCCACCAATCCACACAGA 3′ |
| β-Actin probe | 5′ ATCAAGATCATTGCTCCTCCTGAGCGC 3′ |
| 18S rRNA | Measured using TaqMan pre-developed primers and probe (Applied Biosystems) |
| Phosphacan forward | 5′ GGGCATTCAGGAGTATCCAACA 3′ |
| Phosphacan reverse | 5′ TCCGTGACTCTTCTATTTTTACTTTCAT 3′ |
| Phosphacan probe | 5′ TCAGCACATCTCGTTCTATCCCTTTGCTCA 3′ |
| sRPTP-β forward | 5′ ACAATGAGGCCAGTAATAGTAGCCAT 3′ |
| sRPTP-β reverse | 5′ TAGATGAGAATACCAACAAGAACCACTAG 3′ |
| sRPTP-β probe | 5′ AGACACGATCACAAGGGGTATAACCGCCT 3′ |
Protein extraction and Western blotting
Rats were sacrificed 2 (n = 6), 7 (n = 6), or 15 days (n = 4) after FPI, and paired sham-injured controls at 2 (n = 4), 7 (n = 4), or 15 days (n = 3). Whole hippocampi were dissected from fresh brain, rapidly homogenized in a 250-μL volume of T-PER (Thermo Scientific, Rockford, IL), and centrifuged for 5 min at 8000 × g and 4°C. Supernatant was removed and stored at –80°C. Protein concentration of each sample was determined using the Bio-Rad Protein Assay Reagent and spectrophotometry (Shimadzu UV-160, Shimadzu Scientific Instruments, Columbia, MD). Then 5 μg of protein from each sample was mixed with reduced sample buffer and electrophoresed on 4–12% Bis-Tris Criterion XT gels (200 V for 45 min) and subsequently transferred to a PVDF membrane. Membranes were blocked in 5% milk-TBST (Tris-buffered saline containing 0.05% Tween 20) for 1 h before being probed with mouse monoclonal antibody raised against phosphacan (3F8; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) in milk-TBST (1.5 μg/mL) overnight at 4°C. The blots were subsequently washed with milk-TBST and then incubated for 1 h at room temperature in peroxidase-conjugated goat anti-mouse secondary antibody (1:20,000; Rockland, Gilbertsville, PA) prior to a final washing in TBST and immunopositive signal visualization using Super Signal West Dura Extended Duration Substrate (Thermo Scientific). Parallel blots were incubated without primary antibody to confirm signal specificity. All blots were then imaged digitally with the G:Box ChemiHR system for densitometric analysis using GeneSnap software (SynGene, Frederick, MD). After phosphacan antibody binding data were captured, all blots were stripped and re-probed for β-actin (1:3000 mouse monoclonal; Sigma, St. Louis, MO) as a load control. Signal was visualized as described above for the 3F8 antibody, and blots were imaged with the same G:Box ChemiHR system. No differences in load between the lanes were detected.
Statistical analysis
Changes in gene and protein expression following UEC were evaluated by comparison of ipsilateral deafferented samples to those from the uninjured contralateral hemisphere, providing within-subject matched controls. In rats subjected to FPI, injured cases were compared to paired sham-injured control animals. For both injury models, results were expressed as percentage of control value. The significance of UEC-induced changes in RNA and protein levels was evaluated using mixed-design ANOVA (MANOVA) (SPSS v11; SPSS, Inc., Chicago, IL) with survival interval as between-subjects factor and hemisphere as within-subjects factor, and evaluations at single survival intervals were implemented as a-priori comparisons using simple main effects (Keppel, 1991; Levine, 1991). FPI-induced changes in RNA and protein levels were evaluated using between-subjects ANOVA (SPSS v11), and Bonferroni post-hoc comparisons. The normality of sample data distributions was confirmed with normal probability plots prior to ANOVA analyses. The significance of densitometric values from Western blots was analyzed using the Student's t-test. A probability of < 0.05 was considered statistically significant for all tests.
In order to assess within-group variability of reference gene expression, the coefficient of variation (CV) was determined for each group, calculated as the ratio of standard deviation to group mean. CV is a dimensionless parameter which permits the comparison of variability of gene expression between groups with different mean values.
Results
Differential reference gene expression after UEC and FPI
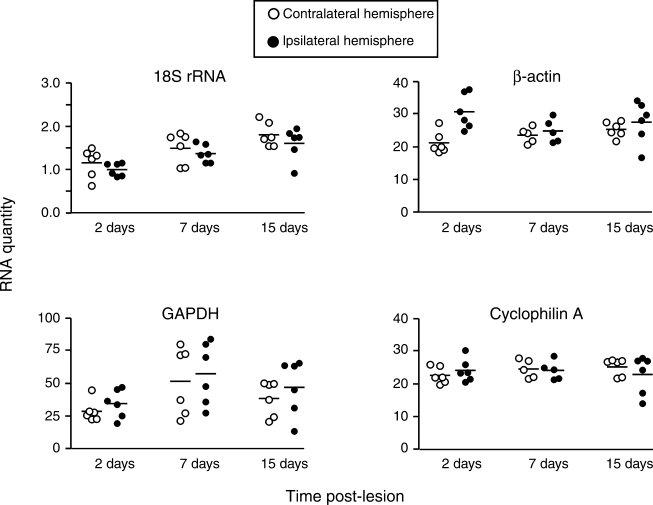
Transcripts for β-actin, GAPDH, 18S rRNA, and cyclophilin A were measured in tissue collected 2, 7, or 15 days after UEC or FPI. Results for β-actin were mixed in the UEC model. At 2 days post-injury there was a significant increase in hippocampal β-actin (147.1 ± 12.9%, p < 0.05) and, although the 7- and 15-day cases showed no change relative to controls, 15-day UEC transcripts exhibited notable variability (Table 2 and Fig. 1). GAPDH mRNA expression was highly variable at all time points after UEC, in both ipsilateral and contralateral hippocampi (note that it had the highest CV values in Table 2 and Fig. 1). While 18S rRNA showed lower variability and no difference between ipsilateral and contralateral expression at any time point (Table 2 and Fig. 1), there was a significant ipsilateral increase in 18S rRNA between 2 and 15 days after UEC (61.5 ± 13.9%, p < 0.05). Only the cyclophilin A transcript showed stable ipsilateral and contralateral expression after UEC and relatively low variance (Fig. 1 and Table 2), making it the best choice as a reference gene among the four we tested in this injury paradigm.
Table 2.
Reference Gene Expression after Experimental Brain Injury: Change Relative to Control and Variability
| |
UEC hippocampus |
||
|---|---|---|---|
| 2 days | 7 days | 15 days | |
| β-Actin | ↑ 0.175 | 0.144 | 0.233 |
| GAPDH | 0.317 | 0.415 | 0.454 |
| 18S rRNA | 0.149 | 0.151 | 0.235 |
| Cyclophilin A | 0.145 | 0.120 | 0.256 |
| |
FPI hippocampus |
||
|---|---|---|---|
| 2 days | 7 days | 15 days | |
| β-Actin | 0.149 | 0.092 | 0.114 |
| GAPDH | 0.159 | ↑ 0.051 | 0.071 |
| 18S rRNA | 0.092 | ↑ 0.120 | 0.108 |
| Cyclophilin A | ↓ 0.201 | ↑ 0.050 | 0.123 |
| |
FPI parietotemporal cortex |
||
|---|---|---|---|
| 2 days | 7 days | 15 days | |
| β-Actin | ↑ 0.207 | ↑ 0.250 | 0.018 |
| GAPDH | 0.154 | ↑ 0.061 | 0.046 |
| 18S rRNA | 0.209 | ↑ 0.138 | 0.054 |
| Cyclophilin A | ↓ 0.213 | ↑ 0.051 | 0.160 |
Arrows indicate direction of significant change (p < 0.05). Numerical values represent coefficient of variation (standard deviation/ mean) for each gene.
FIG. 1.
Variance of hippocampal reference gene expression after UEC. qRT-PCR replicate averages for individual animals are plotted in arbitrary RNA units. Bars represent group means for each gene at 2, 7, and 15 days post-lesion. 18S rRNA expression did not change relative to controls, but increased over time. β-Actin transcript was increased at 2 days after UEC, with high variance at 15 days. GAPDH mRNA was the most variable, particularly at 7 and 15 days. By contrast, cyclophilin A expression was stable at all time points and exhibited low variability. These data suggest that cyclophilin A is a good reference gene for the UEC paradigm, while 18S rRNA, β-actin, and GAPDH would not serve as good choices for RNA normalization.
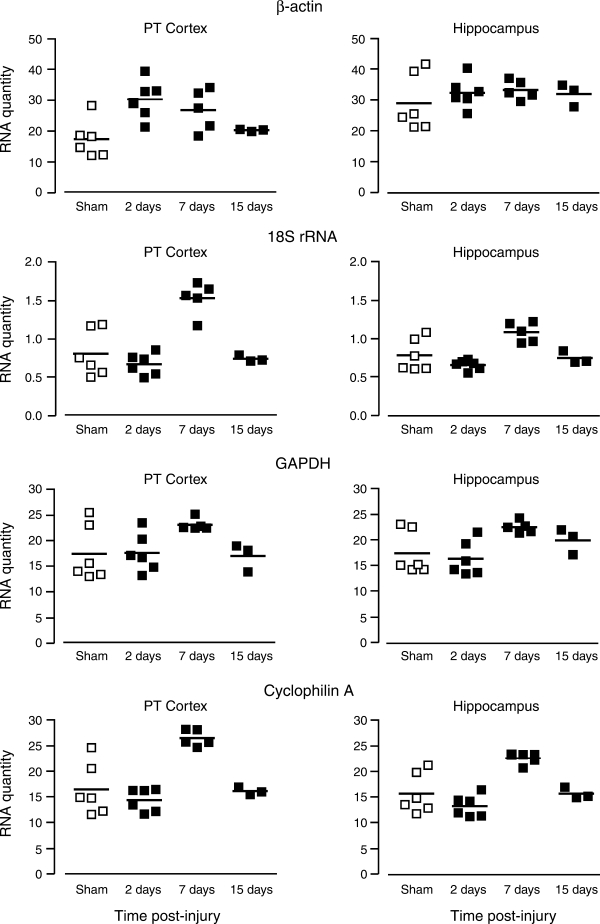
Following FPI, we examined reference gene expression in the hippocampus and parietotemporal cortex, two brain regions known to be affected in that model. At 2 and 7 days after FPI, β-actin mRNA was significantly increased in parietotemporal cortex (173.9 ± 14.6% and 154.2 ± 15.7%; p < 0.05). By contrast, expression of β-actin message in the hippocampus remained remarkably stable after injury at all time points sampled, with low CV values (Fig. 2 and Table 2). Thus β-actin may be a good reference gene choice in hippocampus, but not in parietotemporal cortex, following FPI. The commonly applied 18S rRNA had relatively low CV values for FPI; however, a trend toward reduction was seen at 2 days post-injury (82.8 ± 3.4% for hippocampus and 80.4 ± 7.4% for parietotemporal cortex), although this did not reach statistical significance. By 7 days after FPI, 18S rRNA was elevated over sham controls in both the hippocampus and parietotemporal cortex (138.8 ± 6.6% and 190.0 ± 10.8%, p < 0.01) (Fig. 2 and Table 2). GAPDH transcript expression was increased in both the parietotemporal cortex and hippocampus at 7 days after injury (132.8 ± 2.8% and 128.8 ± 2.6%, p < 0.05) (Fig. 2 and Table 2), while remaining relatively stable at 2 and 15 days. Finally, cyclophilin A mRNA was reduced in both hippocampal and parietotemporal cortical tissue at 2 days post-injury (80.4 ± 5.6% and 85 ± 5.5%, p < 0.01), and increased in both regions at 7 days post-injury (144.3 ± 3.3% for hippocampus and 161.2 ± 4.4% for parietotemporal cortex, p < 0.05) (Fig. 2 and Table 2). At 15 days after FPI, cyclophilin A transcript continued to show low CV values but no difference from sham controls.
FIG. 2.
Regional differences in reference gene expression after FPI. qRT-PCR replicate averages for individual animals are plotted in arbitrary RNA units. Bars represent group means for each gene at 2, 7, and 15 days post-injury. In the parietotemporal cortex, transcript for β-actin increased at 2 and 7 days, while 18S rRNA and GAPDH were elevated only at 7 days. Overall variance was moderate for β-actin, 18S rRNA, and GAPDH in the parietotemporal cortex. Hippocampal β-actin expression was stable over time; however, sham controls showed high variance in this region. By contrast, hippocampal transcripts for 18S rRNA and GAPDH were significantly increased over sham controls at 7 days. Cyclophilin A was decreased at 2 days and increased at 7 days in both brain regions. Thus, of the genes tested, β-actin may be the most appropriate reference for FPI hippocampus, but is a poor choice for qRT-PCR normalization in the parietotemporal cortex (PT Cortex, parietotemporal cortex).
Taken together, these data show that none of the four common reference genes tested was invariant across all tissues, time points, and types of experimental injury. Selection of an appropriate reference gene can be facilitated by carefully examining the variance of the gene's expression within each experimental group, and whether the injury paradigm itself induces a shift in the gene's transcription relative to paired control samples. When this comparison is made, some genes may serve as good reference transcripts only under limited conditions, while others might be used over a wider range of post-injury recovery intervals or within different brain regions.
Reference gene selection changes for different transcripts
To investigate the effect of different normalization approaches with different genes of interest, we ran qRT-PCR for two related splice variants, phosphacan and short receptor protein tyrosine phosphatase-β (sRPTP-β), using UEC hippocampal tissue samples. Results were compared for these two test transcripts when normalized to cyclophilin A (a reference gene predicted to be stable in this injury model), or to total RNA input. Target genes showed PCR efficiencies in the same range as those observed for the reference gene. Analysis showed that the choice of reference normalization method significantly affects qRT-PCR results, depending on the target gene of interest selected (Fig. 3).
FIG. 3.
Application of selected reference genes may vary with test gene. Phosphacan and sRPTP-β mRNA level after UEC was normalized to either stably expressed cyclophilin A or to total RNA. In the left panel, significant elevation of phosphacan transcript at 7 days was confirmed by each normalization method, matching an observed 7-day elevation of phosphacan protein in the deafferented zone (data not shown). By contrast, when transcript for sRPTP-β, a related phosphacan splice variant, was normalized to cyclophilin A the data fails to replicate a significant increase detected by total RNA reference (right panel). Furthermore, this loss of effect was inconsistent with a clear rise in sRPTP-β protein at all three post-injury intervals (data not shown). These comparisons illustrate that optimized reference genes vary in utility with different transcripts. The dashed line at zero denotes the normalized level observed in contralateral (uninjured) hippocampus (*p < 0.05, **p < 0.01 for comparisons involving total RNA; §p < 0.05, §§p < 0.01 for comparisons involving cyclophilin A reference).
Phosphacan normalization to cyclophilin A and to total RNA produce a similar overall pattern of transcript expression (Fig. 3, left panel), with significant elevation occurring at 7 days (120.4 ± 3.0%, p < 0.05 for cyclophilin A; 113.6 ± 8.7%, p < 0.05 for total RNA). Since prior studies suggest that a qRT-PCR change of less than twofold is best expressed relative to total RNA content (Bustin et al., 2002; Bustin and Nolan, 2004), it would be predicted that the normalization of phosphacan transcript to both the optimal reference gene cyclophilin A and total RNA would give similar results. A 7-day increase in phosphacan mRNA is also supported by our prior immunohistochemical observation that phosphacan protein is elevated in UEC hippocampus at the same post-injury time point (Harris et al., 2005, 2006). Thus these results suggest that either cyclophilin A or total RNA may be applied as reasonable strategies for qRT-PCR normalization of phosphacan transcript.
By contrast, in the qRT-PCR analysis of sRPTP-β mRNA expression, normalization to total RNA input appeared to be a preferred strategy (Fig. 3, right panel). Normalizing sRPTP-β expression to total RNA showed a significant increase over control at 2, 7, and 15 days post-injury (111.7 ± 10%, p < 0.05; 112.8 ± 5.6%, p < 0.05; 118.8 ± 6.9%, p < 0.01). In this case, however, cyclophilin A normalization showed a significant increase only at 15 days (134.6 ± 8.2%; p < 0.01). Again, small changes in transcript were documented, favoring total RNA as a standard. This choice was supported by a further comparison of the total RNA-normalized sRPTP-β transcript with the Western blot profile of sRPTP-β protein, where we observed consistent elevation in transcript and protein after UEC (Harris et al., 2008). These differences between phosphacan and sRPTP-β show that alternative qRT-PCR normalization strategies may need to be considered, even when comparisons are made within related gene families.
Verification of reference genes with other outcome measures
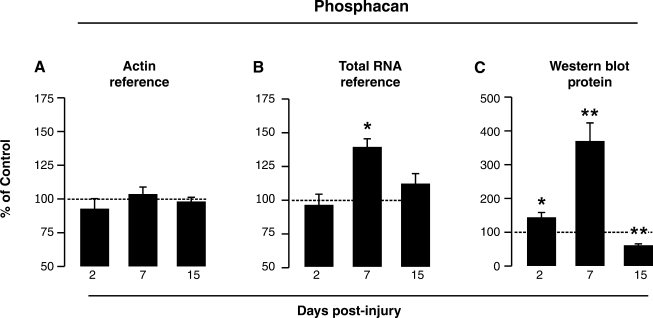
Although a potential reference gene meets initial criteria for stability over time post-injury and low variability, normalization using that reference gene still may not be the best choice. We investigated this issue in the FPI model, again assessing phosphacan transcript change over time post-injury using hippocampal tissue samples. Our above results suggested that β-actin transcript would be appropriate for qRT-PCR normalization, having low CV, similar PCR efficiency as phosphacan (≥90%) and showing no change in post-injury expression relative to sham controls. When hippocampal phosphacan mRNA was normalized to β-actin mRNA, there was no effect on the transcript after FPI (Fig. 4A). However, subsequent analysis of the same data using total RNA input as a standard (Fig. 4B) revealed a significant elevation in phosphacan transcript at 7 days post-injury (138.9 ± 6%, p < 0.05). If these results are compared with hippocampal phosphacan levels determined by Western blot, increased transcript at 7 days after FPI is supported by a prominent threefold rise (370.19 ± 53.4%, p < 0.05) in phosphacan protein (Fig. 4C). Re-probe of all blots for β-actin showed equal lane loads (data not shown). This example points to the benefit of applying more than one qRT-PCR reference method, and shows the value of using additional outcome measures to verify mRNA results. For FPI phosphacan, differences in the temporal profile of mRNA level were most likely due to the variance of β-actin transcript observed in the sham-control group (see Fig. 2). When control values are skewed due to variance of a specific reference gene, and then used to normalize injury-induced experimental effect, significant differences may be lost. Thus with selection of a reference gene, variability of transcript expression for all groups used in the analysis should be carefully examined.
FIG. 4.
Reference gene choice must consider variance in all groups sampled. The stable and low-variance reference gene β-actin was used to normalize hippocampal phosphacan mRNA, showing no change over time after FPI (A). When the same data were normalized to total RNA (B), significant elevation in transcript was observed at 7 days post-injury. Application of total RNA as a reference was consistent with the large 7-day increase in phosphacan protein seen by Western blot probe (C). Such differences in qRT-PCR effect were most likely produced by high variance in the sham β-actin group (see Fig. 2). Even when a potential reference gene meets initial criteria for stability over time post-injury, high variance in paired controls may result in the failure to detect an experimental effect (*p < 0.05; **p < 0.01).
Discussion
Here we describe the post-injury time course of mRNA expression for four common qRT-PCR reference genes (GAPDH, β-actin, cyclophilin A, and 18S rRNA) in two distinct models of brain injury. We have focused on how the two types of injury affect these genes during long-term synaptic recovery. A majority of TBI qRT-PCR studies use one of these reference genes for quantitative evaluation of gene expression (Rhinn et al., 2008). Expression of such genes in target tissue is assumed unchanged by experimental treatment, and differences in these genes serve to control for variance in sample quality (Bond et al., 2002; Wilson, 1997). Using high-quality RNA and a consistent RT-PCR protocol, we found that each reference gene tested was specifically altered by focal deafferentation or diffuse brain injury. It is unlikely that these changes are the result of nonspecific factors (e.g., endogenous PCR inhibition), since we observed differential alteration of these genes in the same tissue samples. Commonly applied 18S rRNA was problematic for both UEC and FPI, changing over time post-lesion, and within both TBI sample regions. Similarly, the frequently-used β-actin transcript was elevated after both types of injury, particularly in brain areas vulnerable to insult. Cyclophilin A was found to be a suitable qRT-PCR reference gene for the UEC model, but was directly altered by FPI. Finally, GAPDH transcript was highly variable after UEC and increased in expression with FPI. Together, these results reveal that CNS trauma often results in long-term modulation of commonly applied reference genes, and show that selection of appropriate qRT-PCR reference genes for experimental TBI must take into consideration model type, brain region sampled, and post-injury survival interval. In some cases, stably expressed reference genes may be inappropriate for standardization, and detection of small differences in transcript may require normalization to total RNA input.
Altered reference gene expression and CNS trauma
Several lines of evidence suggest that physiological and pathological events predisposing the brain to synaptic reorganization can induce reference genes such as 18S rRNA, β-actin, cyclophilin, and GAPDH. For example, degeneration of normal aging or simple dietary restriction will alter expression of 18S rRNA within the rodent hippocampus and neocortex (Tanic et al., 2007). In models of injured dorsal root ganglion cells (Willis et al., 2005) and hippocampal ischemia (Farwell et al., 1998), β-actin mRNA also shows alteration from control expression levels. Our present observations of reference gene change during the time course of UEC-reactive synaptogenesis are consistent with this literature. In a prior in-situ hybridization study, we found that during UEC-induced synaptogenesis, there was significant elevation in rRNA (Phillips et al., 1987), as well as both β-actin and β-tubulin message (Phillips and Steward, 1990) at the local sites of axonal sprouting. These changes match the current data on transcript variance, supporting the conclusion that both 18S rRNA and β-actin are poor choices for qRT-PCR normalization after UEC. Similarly, we have identified GAPDH as a poor reference gene for studies of injury-induced plasticity due to its high variability following UEC lesion. A previous study also demonstrated that GAPDH mRNA expression was reduced during synaptic reorganization of mouse hippocampus after perforant path lesion (Meldgaard et al., 2006).
By contrast, we found that cyclophilin A can serve as a good reference gene for long-term qRT-PCR studies after UEC deafferentation. This differs from an earlier study that reported that cyclophilin transcript is reduced during neocortical reorganization stimulated by feline retinal lesions (Arckens et al., 2003). It is possible that species or a brain-region-specific cell response could account for these differences. Notably, we observed a significant reduction in hippocampal and parietotemporal cortical cyclophilin A transcript at 2 days following FPI, which is consistent with the results of Arckens and colleagues. However, at 7 days after FPI, cyclophilin A was elevated in both brain regions sampled. Clearly, cyclophilin A may participate in synaptic reorganization after diffuse brain injury and is a poor choice for qRT-PCR reference for such models.
In the diffuse FPI model we also compared reference gene expression at two sites of tissue vulnerability, parietotemporal cortex and hippocampus. Our findings revealed low variance and absence of injury effect for only one gene and one region: hippocampus and β-actin. However, subsequent qRT-PCR experiments with a specific target gene revealed that variability of β-actin gene expression in sham hippocampus was high. It appeared that this variance contributed to a failed detection of experimental effect, making β-actin a poor choice for that particular FPI experimental design as well. In this context, prior studies have also reported rapid upregulation of β-actin and GAPDH in rodent models of cerebral ischemia (Harrison et al., 2000; Kobayashi et al., 2004). Given that secondary ischemia is often a component of TBI pathogenesis, we would predict an acute 2-day increase in parietotemporal cortical β-actin transcript after FPI as was observed. In the present study, the lack of change in hippocampal β-actin mRNA was surprising, particularly since its pyramidal cell subregions are selectively vulnerable to ischemic insult in combination with TBI. We interpret this difference as indicating that secondary ischemia was not a major pathology for the animals tested. Despite regional differences, overall CV values in FPI cases were lower than those found with UEC, suggesting that multiple reference genes (e.g., β-actin, 18S rRNA, or GAPDH) may be suitable for use with FPI qRT-PCR, provided the study is restricted to individual time points.
Finally, three recent studies have similarly explored reference gene regulation in rodent models of brain trauma. Thal and colleagues (2008) examined expression of candidate reference genes at the site of contusion over the first 24 h after controlled cortical impact (CCI) in mice. This study identified cyclophilin A and β-2-microglobulin as the best controls, while GAPDH and β-actin were poor choices for normalization due to their variability and increased expression within injured tissue. Our results with GAPDH and cortical β-actin support that same conclusion for FPI. Interestingly, a second mouse study employing the weight-drop model of TBI showed that GAPDH had low variability over the first 48 h post-injury, making it preferred as a qRT-PCR reference gene (Rhinn et al., 2008). This group also reported that 18S rRNA and total cDNA content were good normalization choices. As in the CCI model, β-actin mRNA expression was increased acutely after weight-drop injury, and thus was identified as an inappropriate reference gene. Our study had only one reference gene in common with those verified in a third report by Cook and colleagues (2008). That study showed that GAPDH was an acceptable hippocampal reference at 3 days following impact acceleration insult, a gene that we found not to be appropriate for FPI hippocampal qRT-PCR.
Clearly, optimization of a reference gene is dependent on the details of the experimental design. Each of the cited TBI studies focused on the acute (1–3 days) post-injury period, in contrast to the long-term regenerative time course we report here. In addition, the earlier studies examined gene expression at the site of the lesion, in some cases including regions with contusion and tissue cavitation. The present study models diffuse and targeted deafferentation insult, without these confounding factors. Species differences should also be considered. In contrast to the two mouse studies, we have shown that β-actin may be stably expressed in injured rat hippocampus 2–15 days after FPI. However, the potential for variable expression of β-actin in sham-control cases suggests that it is not a good reference gene for FPI studies examining relative change in certain transcripts. In contrast to the mouse CCI model (Thal et al., 2008), our results show rat cyclophilin A as a poor reference gene for diffuse TBI. Notably, it was the best of those tested for use with the UEC targeted deafferentation model. Taken together, all of these TBI reference gene analyses underscore the fact that choice of appropriate qRT-PCR control genes is specific to the brain region, survival time, and specific pathological components of the TBI model employed.
Validation of reference genes for long-term recovery studies
Given the growing use of qRT-PCR in the field of brain trauma, and the certain application to studies focusing on periods of long-term recovery, it is important to establish criteria for validation of reference genes applied over broad post-injury time courses. Based upon the present results, we suggest that several points must be met to select such control genes:
A valid reference gene should not be altered in expression under the varying experimental conditions of a temporal study. In the context of long-term TBI recovery, this means no difference in injured:control ratio for all time points included, no overall change during the post-injury time course selected, and no change in reference gene as a function of experimental treatment condition (e.g., therapeutic drug effect). Different types of reference gene alteration can profoundly affect outcome (Bond et al., 2002). For example, if the reference and target gene are similarly shifted (i.e., both increased or both decreased), normalization can produce a false-negative result. Alternatively, if the reference gene changes, but the target gene does not, normalization can lead to a false-positive finding. Injury effect on reference gene expression may also be model- and tissue-specific; it cannot be assumed that a reference gene will be stably expressed in all brain regions using the same paradigm.
Variability of reference gene expression within each experimental group (as tested by CV or gene pair-wise variation algorithms) must be considered. For example, we show that mean GAPDH expression after UEC is no different between ipsilateral and contralateral hippocampi, and that expression levels do not change over time. However, GAPDH expression has high intra-group variability, and is therefore not a good reference gene choice. Variability of reference gene expression should be considered not only in injured groups, but in control groups as well. In this context we show that β-actin expression remains stable in FPI hippocampus, but fails as a control gene during long-term recovery due to high sham-control variability.
When TBI produces small expression shifts in genes of interest over time, total RNA input may be the best reference for normalization. The chief limitations of applying total RNA normalization are its dependence upon accurate assessment of RNA quantity and quality, as well as the potential for variation from cell to cell within a tissue sample. Prior studies have directly addressed the latter issue, measuring total RNA from individual blood cells, and found little cell-to-cell variation (Bustin, 2002). Furthermore, in tissues undergoing complex changes associated with pathological insult and recovery, total RNA levels are predicted to be less variable than most target gene transcripts (Bustin, 2002; Bustin and Nolan, 2004). If the quality of extracted RNA is high and concentration is accurately measured, normalization to total RNA mass is a good alternative. For example, after careful determination of our input RNA quality and quantity, we show that significant shifts of 12–20% in phosphacan and sRPTP-β following UEC lesion can be detected by this method of normalization. Depending on the overall data distribution, small increases or decreases in a gene may be no less biologically significant than large quantitative changes (Bond et al., 2002). Acceptable variability cannot be defined a priori, but must be dictated by the resolution required to detect injury effect. When a gene exhibits a large change (more than twofold), conclusions may only be minimally impacted by control gene variability. However, for genes exhibiting small injury change (much less than twofold), normalization to a variable reference gene may produce type II error and failure to detect a real experimental effect (Dheda et al., 2005).
Correlative outcome measures, such as in-situ assessment of mRNA, as well as protein quantification and tissue distribution, may be required to assess which qRT-PCR normalization strategy generates biologically relevant information. Assessment should include both reference genes and target genes of interest in order to make useful comparisons. Here we illustrate how additional outcome measures help to determine appropriate normalization for hippocampal phosphacan expression after FPI. While β-actin had a low CV and did not change relative to sham controls, when it was used as a reference gene, FPI had no effect on phosphacan mRNA. By contrast, when normalized to total RNA input, significant elevation in transcript was found at 7 days, which positively correlated with an increase in phosphacan protein. As indicated above, this difference appears to be due to high variance of sham β-actin transcript. While correlative studies cannot substitute for reference gene validation, they should be applied as confirmation of the normalization approach.
In conclusion, qRT-PCR is a powerful tool for research targeting recovery mechanisms induced by TBI. However, for each new set of experimental conditions it is essential that careful validation of the candidate reference genes be performed. It should also be recognized that in some cases, normalization to total RNA or multiple reference genes may be preferable. Results from qRT-PCR assays will provide the most meaningful information when combined with other outcome measures assessing mRNA and protein levels. Continued research is needed to identify additional reference genes for studies focusing on long-term recovery after TBI. Microarrays may be useful tools for screening a broad spectrum of mRNA transcripts (Dash et al., 2004), in this case, with the goal of identifying those least affected by TBI.
Acknowledgments
The authors wish to thank Lesley K. Harris, Raiford T. Black, and Nancy N. Lee for excellent technical assistance. All qRT-PCR was performed in the Nucleic Acids Research Facilities, Center for the Study of Biological Complexity, Virginia Commonwealth University. This work was supported by NIH grants NS44372 and NS57758, and Virginia Commonwealth Neurotrauma Initiative 07-302F.
Author Disclosure Statement
No conflicting financial interests exist.
References
- Andersen C.L. Jensen J.L. Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Arckens L. Van der Gucht E. Van den Bergh G. Massie A. Leysen I. Vandenbussche E. Eysel U.T. Huybrechts R. Vandesande F. Differential display implicates cyclophilin A in adult cortical plasticity. Eur. J. Neurosci. 2003;18:61–75. doi: 10.1046/j.1460-9568.2003.02726.x. [DOI] [PubMed] [Google Scholar]
- Blain J.-F. Paradis E. Gaudreault S.B. Champagne D. Richard D. Poirier J. A role for lipoprotein lipase during synaptic remodeling in the adult mouse brain. Neurobiol. Dis. 2004;15:510–519. doi: 10.1016/j.nbd.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Bond B.C. Virley D.J. Cairns N.J. Hunter A.J. Moore G.B. Moss S.J. Mudge A.W. Walsh F.S. Jazin E. Preece P. The quantification of gene expression in an animal model of brain ischemia using TaqMan real-time RT-PCR. Brain Res. Mol. Brain Res. 2002;106:101–116. doi: 10.1016/s0169-328x(02)00417-5. [DOI] [PubMed] [Google Scholar]
- Bonefeld B.E. Elfving B. Wegener G. Reference genes for normalization: A study of rat brain tissue. Synapse. 2008;62:302–309. doi: 10.1002/syn.20496. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Real-time, fluorescence-based quantitative PCR: A snapshot of current procedures and preferences. Expert Rev. Mol. Diagn. 2005;5:493–498. doi: 10.1586/14737159.5.4.493. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. Benes V. Nolan T. Pfaffl M.W. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Chen J. Sochivko D. Beck H. Marechal D. Wiestler O.D. Becker A.J. Activity-induced expression of common reference genes in individual CNS neurons. Lab. Invest. 2001;81:913–916. doi: 10.1038/labinvest.3780300. [DOI] [PubMed] [Google Scholar]
- Cook N.L. Vink R. Donkin J.J. van den Heuvel C. Validation of reference genes for normalization of real-time quantitative RT-PCR data in traumatic brain injury. J. Neurosci. Res. 2009;87:34–41. doi: 10.1002/jnr.21846. [DOI] [PubMed] [Google Scholar]
- Kerber G. Strief R. Schwaiger F.-W. Kreutzberg G.W. Hager G. Neuregulin-1 isoforms are differentially expressed in the intact and regenerating adult rat nervous system. J. Mol. Neurosci. 2003;21:149–165. doi: 10.1385/JMN:21:2:149. [DOI] [PubMed] [Google Scholar]
- Coulson D.T. Brockbank S. Quinn J.G. Murphy S. Ravid R. Irvine G.B. Johnston J.A. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol. Biol. 2008;9:46. doi: 10.1186/1471-2199-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P.K. Kobori N. Moore A.N. A molecular description of brain trauma pathophysiology using microarray technology: An overview. Neurochem. Res. 2004;29:1275–1286. doi: 10.1023/b:nere.0000023614.30084.eb. [DOI] [PubMed] [Google Scholar]
- Dheda K. Huggett J.F. Bustin S.A. Johnson M.A. Rook G. Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- Dheda K. Huggett J.F. Chang J.S. Kim L.U. Bustin S.A. Johnson M.A. Rook G.A. Zumla A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005;344:141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Dixon C.E. Lyeth B.G. Povlishock J.T. Findling R.L. Hamm R.J. Marmarou A. Young H.F. Hayes R.L. A fluid percussion model of experimental brain injury in the rat. J. Neurosurg. 1987;67:110–119. doi: 10.3171/jns.1987.67.1.0110. [DOI] [PubMed] [Google Scholar]
- Farwell W. Simonyi A. Scott H. Shang J.-P. Carruthers V. Madsen R. Johnson J. Sun G.Y. Effects of ischemic tolerance on mRNA levels of IP3R1, β-actin, and neuron-specific enolase in hippocampal CA1 area of the gerbil brain. Neurochem. Res. 1998;23:539–542. doi: 10.1023/a:1022486619201. [DOI] [PubMed] [Google Scholar]
- Gibson U.E. Heid C.A. Williams P.M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Giulietti A. Overbergh L. Valckx D. Decallonne B. Bouillon R. Mathieu C. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- Harris J.L. Harris L.K. Black R.T. Reeves T.M. Phillips L.L. Changes in hippocampal phosphacan expression during trauma-induced synaptic plasticity. J. Neurotrauma. 2005;22:1166. [Google Scholar]
- Harris J.L. Harris L.K. Lee N.N. Black R.T. Reeves T.M. Phillips L.L. Protein distribution and transcriptional profile of phosphacan/RPTPb during reactive synaptogenesis. J. Neurotrauma. 2006;23:988. [Google Scholar]
- Harris J.L. Harris L.K. Lee N.L. Phillips L.L. Elevation of receptor protein tyrosine phosphatase β after brain injury: Evidence for regulation of synaptic plasticity through postsynaptic substrates. J. Neurotrauma. 2008;25:857. [Google Scholar]
- Harrison D.C. Medhurst A.D. Bond B.C. Campbell C.A. Davis R.P. Philpott K.L. The use of quantitative RT-PCR to measure mRNA expression in a rat model of focal ischemia—caspase-3 as a case study. Brain Res. Mol. Brain Res. 2000;75:143–149. doi: 10.1016/s0169-328x(99)00305-8. [DOI] [PubMed] [Google Scholar]
- Heid C.A. Stevens J. Livak K.J. Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Huggett J. Dheda K. Bustin S. Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Johansson S. Fuchs A. Okvist A. Karimi M. Harper C. Garrick T. Sheedy D. Hurd Y. Bakalkin G. Ekstrom T.J. Validation of endogenous controls for quantitative gene expression analysis: Application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–28. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Keppel G. Design and Analysis: A Researcher's Hand book. Prentice Hall; Englewood Cliffs, NJ: 1991. [Google Scholar]
- Kobayashi M.S. Takahashi Y. Nagata T. Nishida Y. Murata A. Ishikawa K. Asai S. Screening for control genes in rat global cerebral ischemia using high-density oligonucleotide array. J. Neurosci. Res. 2004;76:512–518. doi: 10.1002/jnr.20094. [DOI] [PubMed] [Google Scholar]
- Levine G. A Guide to SPSS for Analysis of Variance. Lawrence-Erlbaum, Assoc.; Hillsdale, NJ: 1991. [Google Scholar]
- Li H.H. Lee S.M. Cai Y. Sutton R.L. Hovda D.A. Differential gene expression in hippocampus following experimental brain trauma reveals distinct features of moderate and severe injuries. J. Neurotrauma. 2004;21:1141–1153. doi: 10.1089/neu.2004.21.1141. [DOI] [PubMed] [Google Scholar]
- Loesche J. Steward O. Behavioral correlates of dennervation and reinnervation of the hippocampal formation of the rat: Recovery of alternation performance following unilateral entorhinal cortex lesions. Brain Res. Bull. 1977;2:31–39. doi: 10.1016/0361-9230(77)90022-3. [DOI] [PubMed] [Google Scholar]
- Meldgaard M. Fenger C. Lambertsen K.L. Pedersen M.D. Ladeby R. Finsen B. Validation of two reference genes for mRNA level studies of murine disease models in neurobiology. J. Neurosci. Methods. 2006;156:101–110. doi: 10.1016/j.jneumeth.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Molteni R. Ying Z. Gomez-Pinilla F. Differential effects of acute and chronic exercise on plasticity-related genes in the rat hippocampus revealed by microarray. Eur. J. Neurosci. 2002;16:1107–1116. doi: 10.1046/j.1460-9568.2002.02158.x. [DOI] [PubMed] [Google Scholar]
- Nolan T. Hands R.E. Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. Tichopad A. Prgomet C. Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Best Keeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Phillips L.L. Steward O. Increases in mRNA for cytoskeletal proteins in the denervated neuropil of the dentate gyrus: An in situ hybridization study using riboprobes for beta-actin and beta-tubulin. Brain Res. Mol. Brain Res. 1990;8:249–257. doi: 10.1016/0169-328x(90)90024-8. [DOI] [PubMed] [Google Scholar]
- Phillips L.L. Nostrandt S.J. Chikaraishi D.M. Steward O. Increases in ribosomal RNA within the denervated neuropil of the dentate gyrus during reinnervation: Evaluation by in situ hybridization using DNA probes complementary to ribosomal RNA. Brain Res. Mol. Brain Res. 1987;2:251–261. doi: 10.1016/0169-328x(87)90032-5. [DOI] [PubMed] [Google Scholar]
- Price M. Lang M.G. Frank A.T. Goetting-Minesky M.P. Patel S.P. Silviera M.L. Krady J.K. Milner R.J. Ewing A.G. Day J.R. Seven cDNAs enriched following hippocampal lesion: possible role in neuronal responses to injury. Mol. Brain Res. 2003;117:58–67. doi: 10.1016/s0169-328x(03)00285-7. [DOI] [PubMed] [Google Scholar]
- Rhinn H. Marchand-Leroux C. Croci N. Plotkine M. Scherman D. Escriou V. Housekeeping while brain's storming validation of normalizing factors for gene expression studies in a murine model of traumatic brain injury. BMC Mol. Biol. 2008;9:62. doi: 10.1186/1471-2199-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanic N. Perovic M. Mladenovic A. Ruzdijic S. Kanazir S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J. Mol. Neurosci. 2007;32:38–46. doi: 10.1007/s12031-007-0006-7. [DOI] [PubMed] [Google Scholar]
- Thal S.C. Wyschkon S. Pieter D. Engelhard K. Werner C. Selection of endogenous control genes for normalization of gene expression analysis after experimental brain trauma in mice. J. Neurotrauma. 2008;25:785–794. doi: 10.1089/neu.2007.0497. [DOI] [PubMed] [Google Scholar]
- Tricarico C. Pinzani P. Bianchi S. Paglierani M. Distante V. Pazzagli M. Bustin S.A. Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: Normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Vandesompele J. De Preter K. Pattyn F. Poppe B. Van Roy N. De Paepe A. Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034 Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Barbacioru C. Hyland F. Xiao W. Hunkapiller K.L. Blake J. Chan F. Gonzalez C. Zhang L. Samaha R.R. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. Li K.W. Zheng J.-Q. Chang J.H. Smit A. Kelly T. Merianda T.T. Sylvester J. van Minnen J. Twiss J.L. Differential transport and local translation of cytoskeletal injury-response, and neurodegeneration protein mRNA in axons. J. Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H. Simons J.W. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 1999;259:523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]