Abstract
Background
Recognition of microorganisms by the innate immune system is mediated by pattern recognition receptors, including Toll-like receptors and cytoplasmic RIG-I-like receptors. Chlamydia, which include several human pathogenic species, are obligate intracellular gram-negative bacteria that replicate in cytoplasmic vacuoles. The infection triggers a host response contributing to both bacterial clearance and tissue damage. For instance, type I interferons (IFN)s have been demonstrated to exacerbate the course of Chlamydial lung infections in mice.
Methods/Principal Findings
Here we show that Chlamydia pneumoniae induces expression of IFN-stimulated genes (ISG)s dependent on recognition by nucleotide-sensing Toll-like receptors and RIG-I-like receptors, localized in endosomes and the cytoplasm, respectively. The ISG response was induced with a delayed kinetics, compared to virus infections, and was dependent on bacterial replication and the bacterial type III secretion system (T3SS).
Conclusions/Significance
Activation of the IFN response during C. pneumoniae infection is mediated by intracellular nucleotide-sensing PRRs, which operate through a mechanism dependent on the bacterial T3SS. Strategies to inhibit the chlamydial T3SS may be used to limit the detrimental effects of the type I IFN system in the host response to Chlamydia infection.
Introduction
Chlamydia are gram negative obligate intracellular bacteria and among the smallest bacteria known [1], [2]. Chlamydia pnuemoniae causes respiratory tract infections like pneumonia, pharyngitis, and sinusitis. All chlamydial species share a unique biphasic developmental cycle in which the chlamydiae alternate between two morphologically distinct forms. The elementary body (EB) is the infectious but metabolically inert form whereas the reticulate body (RB) is the non-infectious but metabolically active form [3].
Sensing of microbial infections via specific innate immune receptors plays a pivotal role in the proper functioning of the host innate immune system. The germline-encoded recognition receptors (PRR)s act as a molecular switch triggering the innate immune activation and in turn tightly regulate the downstream immune responses to microbial infections [4]. The microbial structures recognized by PRRs are termed pathogen-associated molecular patterns (PAMP)s, and include lipid-rich structures (e.g. lipopolysaccharide) and nucleotides (e.g. RNA). Two important classes of PRRs are the membrane-bound Toll-like receptors (TLR)s, which are localized in the plasma membrane and endosomal compartments, and the Retinoid acid induced gene (RIG)-I-like receptors (RLR)s, which are localized in the cytoplasm [5]. While TLRs recognize a wide range of different structures, the RLRs uniquely recognize RNAs [5].
Intracellular pathogens propagate inside host cells due to dependence on cellular factors for completion of the life cycle. In addition to viruses, certain bacteria and protozoa reside inside cells. Chlamydia replicates in cytoplasmic vacuoles in the host cells [2], [3], which triggers an inflammatory host response, contributing to both clearing of the infection and tissue damage [6]. Both in vitro and in vivo studies on C. pneumoniae revealed the predominant role of TLR2 over TLR4 following infection [7]. Recent studies reported that in addition to the TLR pathway yet another recognition system involving the cytoplasmic PRRs NOD1 and NOD2 is vital for recognition and clearance of C. pneumoniae [8], [9].
Pathogen recognition activates a intracellular signaling programs [5]. One of these leads to activation of the transcription factor interferon (IFN) regulatory factor (IRF)3, which plays a critical role in transcriptional activity of Type I IFN genes and IFN-stimulated genes (ISG)s. IRF3 is expressed constitutively and resides in the cytosol in a latent form [5]. When receiving the appropriate signal, IRF-3 undergoes phosphorylation, dimerization and nuclear translocation. TANK binding kinase (TBK) 1 and IκB kinase (IKK)ε are the essential serine/threonine kinases for phosphorylation of IRF3 and IRF7 [10], [11]. Bacterial pathogens replicating in the cytoplasm of the host cell (e.g. Listera monocytogenes, Legionella pneumophilla) or releasing bacterial material into the host cytoplasm (group B streptococcus) have recently been reported to induce expression of IFNs and ISGs via cytoplasmic PRRs and the TBK1/IRF-3 axis [12]–[14]. Thus, interaction between bacterial and host cell cytoplasm seems to be sensed and to trigger expression of IFNs and ISGs.
The type 3 secretion system (T3SS) is primarily characterised by the translocation of the bacterial “effectors” into the eukaryotic cell for the manifestation of pathogenesis. The T3SS is important for many gram-negative bacterial species like Shigella, Salmonella and Yerisinia in delivering the effector proteins into the eukaryotic cells directly [15]. Chlamydia uses a T3SS through the developmental cycle and may possibly employ this to translocate the different effectors into the host cell depending on the phase of the bacterial developmental cycle [16]. More recently, it has been demonstrated that T3SS may also play a part in triggering the innate immune response to infections with different bacteria including C. pneumoniae [17], [18].
While Type I IFNs and expression of ISGs has classically been known to stimulate antiviral activity, the recent appreciation of the IFN response in bacterial infections has led to increasing attention on their role in the anti-bacterial defense [19]–[22]. Although the IFN response plays a protective role during some bacterial infections, studies in murine models have suggested that IFNs play a deleterious role in defense against Chlamydia infections. Therefore, knowledge on the mechanisms governing expression of ISGs during infection with C. pneumoniae may provide important evidence on the pathology of Chlamydia infections. In this work we demonstrate that expression of ISGs during infection with C. pneumoniae is mediated by nucleotide-sensing PRRs residing in endosomes and the cytoplasm, and this response is induced through signalling by TBK1/IKKε and is dependent on bacterial replication and the bacterial type III secretion system.
Materials and Methods
Cells and infection
Murine embryonic fibroblasts (MEF) cell lines were used, C57BL/6 and mavs−/− were obtained from Z.J.Chen, Department of Molecular Biology, University of Texas, Dallas,TX,USA. myd88−/−trif−/− and tbk1−/− ikke−/− were obtained from S. Akira, Department of Host Defense, Research Institute for Microbial Diseases, Osaka University Japan and atg +/− and atg5−/− cells were procured from RIKEN Bio Resource Center, Japan. All cell types were grown in RPMI 1640 (Gibco BRL, Grand Island, NY, USA) containing 25 mM HEPES, 15% (w/v) FCS, 0.3%,10 µg/ml gentamicin in growth medium and gentamicin 2 µg/ml in infection medium at 37°C in presence of 5% CO2. Cells were infected with Chlamydia pneumoniae strain TWAR CWL029 obtained from American Type Culture Collection (Rockville, MD, USA) at MOI 5 by centrifugation for 30 minutes, 2400 rpm, at 35°C.
Reagents
Cycloheximide (Sigma Aldrich), a eukaryotic protein biosynthesis inhibitor was used at a concentration of 1 µg/ml. The LPS/TLR4 inhibitor Polymyxin B was purchased from Invivogen and used at a working concentration of 50 µg/ml. The endosomal TLR inhibitor chloroquine (used at a concentration of 10 µM) was obtained from InvivoGen. Ampicillin was used at a concentration of 20 µg/ml. The RNA polymerase III inhibitor ML-60218 (Calbiochem) was used at a concentration of 30 µM and the T3SS inhibitor INP0341, a derivative of salicylidene acylhydrazine and its analogue control INP0406, were kindly donated by Innate Pharmaceuticals, Umeå, Sweden, and used at a concentration of 60 µM.
Immuno-fluorescence and Microscopy
To analyse chlamydial inclusions and development inside the fibroblasts, cells were cultivated on glass cover slips and infected with 5 culture forming units (IFU) per cell of C. pneumoniae were fixed at 36 hr time post challenge. Infected cells were treated with methanol fixed and stained with MAb 15.1 against Chlamydia LPS ([23], [24]).
Real-time PCR (qRT-PCR)
ISG expression at different points in time post infection of C. pneumoniae was quantified by quantitative real-time PCR (qRT-PCR) analysis of the generated cDNAs. Total RNAs were then isolated using the High Pure RNA Isolation Kit Protocol (Roche) and subjected to RT-PCR to generate cDNA using p(dT)15 primer (Roche) and Expand Reverse Transcriptase (Roche), allowing reverse transcription of total sample mRNA. The real time PCR was performed using specific primers for murine ISG56 forward primer 5′-ACC ATG GGA GAG AAT GCT GAT-3′, reverse primer: 5′-GCC AGG AGG TTG TGC-3′; murine CCL5 forward primer 5′- ACT CCC TGC TGC TTT GCC TAC-3′, reverse primer 5′- GAG GTT CCT TCG AGT GAC A-3′; murine CXCL10 forward primer 5′- CGA TGA CGG GCC AGT GAG AAT G-3′, reverse primer 5′- TCA ACA CGT GGG CAG GAT AGG CT-3′; murine β-actin forward primer: 5′- TAG CAC CAT GAA GAT CAA GAT -3′, reverse primer: 5′-CCG ATC CAC ACA GAG TAC TT -3′ and quantified using QuantiTect SYBR Green PCR Kit (Qiagen) and the cycle number at detection threshold (crossing point (cp)) weas determined using the Lightcycler (Roche) analyzer and the LightCycler Software 3.0.
Statistics
The data are presented as means ± SD. The statistical significance was estimated with the Wilcoxon rank sum test (p-values of <0.05 were considered to be statistically significant).
Reproducibility of data
The results shown in this work are derived from data that are representative for the results obtained. For each series of experiments, two to five independent repetitions were performed.
Results
Chlamydia pneumoniae induces expression of ISGs with a delayed kinetics compared to viruses
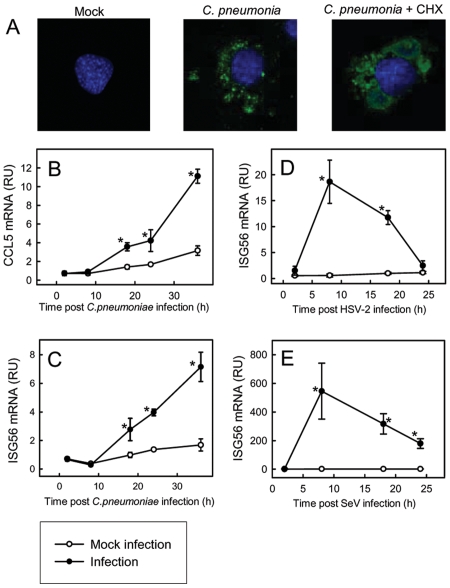
In order to test whether C. pneumoniae establishes productive infection in murine fibroblasts, we treated the cells with the bacteria for 36 h and looked for intracellular vacuoles by confocal microscopy. As shown in Fig. 1A, C . pneumoniae infection did lead to development of small bacterial inclusions, and this was strongly enhanced if the cells were treated with the eukaryote protein synthesis inhibitor, cyclohexamide. This shows that the cell can mount a defence against the infection. All the following experiments were done without cyclohexamide treatment. To evaluate the expression of ISGs in response to C. pneumoniae infection, we harvested total RNA from cells receiving bacterial or mock infection for the indicated time intervals and looked for expression of CCL5, ISG56, and CXCL10. All three ISGs were induced by C. pneumoniae infection, with a slow kinetics, where the mRNA levels did not exceede background levels for the first 8 h post bacterial challenge and subsequently were increasingly steady through the 36 h of the experiment (Fig. 1B–C and data not shown). For comparison, we examined the expression of ISG56 in response to two viruses, namely herpes simplex virus (HSV, a DNA virus) and Sendai virus (SeV, an RNA virus). Interestingly, while both viruses induced a potent ISG56 response, this occurred through a rapid and transient kinetics (Fig. 1D–E). Thus, C. pneumoniae induces expression of ISGs with a delayed kinetics compared to viruses.
Figure 1. Chlamydia pneumoniae induces expression of ISGs with a delayed kinetics compared to viruses.
(A) MEF cells were infected with C. pneumoniae (5 UFU/cell) in presence or absence of cycloheximide (CHX, 1 µg/ml). 36 hrs post infection, the cells were fixed and stained with an antibody against chlamydial LPS and fluorescence/Nomaski images were obtained. (B–E) MEFs were infected with (B–C) C. pneumonia, MOI 5(D) HSV-2, MOI 1, or (E) SeV, MOI 1 for the indicated time intervals. Total RNA was harvested and mRNA levels of ISG56 and CCL5 were determined by real-time PCR and normalized to β-actin. The data are presented as means of triplicate cultures +/− st.dev. Similar results were obtained in 2–5 independent experiments. RU, relative units. Infection-induced gene expression significantly above the expression induced by mock infection at the same tine point (p<0.05) is marked with an asterisk, *.
Chlamydia pneumoniae-induced expression of ISGs is dependent on bacterial recognition by endosomal and cytoplasmic nucleotide-sensing PRRs
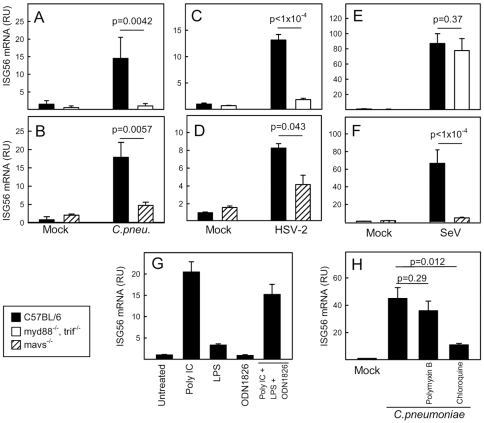
Recent evidence suggests that induction of type I IFN and ISG expression by bacteria is mediated mainly through intracellular PRRs, with RNA and DNA constituting important PAMPs [12], [14], [25]–[27]. To address the role of TLRs and RLRs in stimulation of the IFN response during infection with C. pneumoniae, we infected wildtype (WT), myd88−/−trif−/− and mavs−/− MEFs with the bacteria and measured accumulation of ISG56 mRNA 18 h post infection. Interestingly, the ability of C. pneumonia to trigger the IFN response was dependent on intact pathways from both TLRs (MyD88, TRIF) and RLRs (MAVS) (Fig. 2A and 2B). Both knock-out cells stimulated normal induction of ISGs in response to transfected virus-derived DNA (data not shown). For the viruses, which also induced expression of ISG56, HSV-2 relied on both TLRs and RLRs (Fig. 2C and 2D) whereas SeV triggered the response entirely via RLRs (Fig. 2E and 2F).
Figure 2. Chlamydia pneumoniae-induced expression of ISGs is dependent on bacterial recognition by endosomal and cytoplasmic nucleotide-sensing PRRs.
(A–F) WT, myd88−/− trif−/− and mavs−/− MEFs were infected with (A–B) C. pneumoniae, MOI 5, for 36 h (C–D) HSV-2, MOI 1, 6 hrs, or (E–F) SeV, MOI 1, 6 hrs. Total RNA was harvested and levels of ISG56 mRNA were determined by real-time PCR. (G) MEFs were stimulated with polyIC (25 µg/ml), LPS (200 ng/ml), and ODN1826 (1 µM) alone or in combination as indicated. Total RNA was harvested 6 hrs post-stimulation, and levels of ISG56 mRNA were determined by real-time PCR. (H) The cells were infected with C. pneumonia, MOI 5, for 18 h in the presence or absence of polymyxin B (50 µg/ml) or chloroquine (10 µM) before harvest of total mRNA. The measured ISG56 mRNA levels were normalized to β-actin, and data are presented as means of triplicate cultures +/− st.dev. Similar results were obtained in 2–3 independent experiments. RU, relative units.
To investigate which PRRs may be involved in innate recognition of C. pneumonia, we treated MEFs with PAMPs, known to activate IFN-inducing TLRs and RLRs, and measured ISG56 mRNA expression. The TLR3 agonist polyIC strongly induced ISG56 expression, and the TLR4 agonist LPS induced a modest but significant response (Fig. 2G). Intracellular delivery of polyIC, which activates RLRs [28], also induced ISG56 expression (data not shown). By contrast, no induction of ISG56 was observed after treatment with the TLR9 agonist ODN1826 or the TLR7 agonist ssRNA40 (Fig. 2G and data not shown). TLR2, 3, 4 have been reported to be involved in recognition of Chlamydia species [7], [29], [30], [30,31], and therefore we inhibited LPS/TLR4 action using polymyxin B and also the endosomal TLRs (TLR3, 7, 8, 9) using chloroquine. Inhibition of LPS activity did not affect the ISG response to C. pneumoniae, but treatment of the cells with chloroquine strongly inhinited the ability of the bacteria to induce ISG56 expression (Fig. 2H).
Recently it has been reported that RNA polymerase III transcribes AT-rich DNA in the cytoplasm, thus generating RNA-species recognized by RLRs (refs). To examine if the ISG response during C. pneumoniae infection was triggered through this pathway, we inhibited RNA polymerase III with ML-60218 (30 µM) and infected with the bacteria. However, this treatment did not affect C. pneumoniae-induced ISG56 expression (data not shown), thus suggesting that MAVS signaling during C. pneumoniae infection is not activated by RNA polymerase III driven transcription of AT-rich regions in the bacterial genome. Collectively, C. pneumoniae infection stimulates expression of ISGs via recognition by endosomal TLRs and cytoplasmic RLRs.
Induction of ISGs by Chlamydia pneumoniae is dependent on TBK1/IKKε, bacterial replication and the type III secretion system
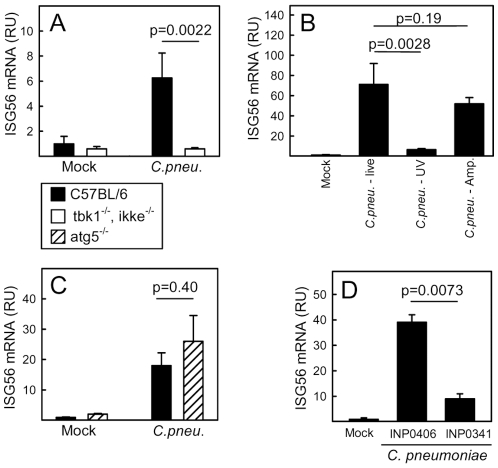
The observed essential role for intracellular nucleotide-sensing PRRs for induction of ISGs by C. pneumonia prompted us to examine the potential role for the IRF-3 kinases TBK1 and IKKε, which are involved in signaling from most of these PRRs. As expected activation of the ISG response during C. pneumonia infection was dependent on these kinases (Fig. 3A).
Figure 3. Induction of ISGs by Chlamydia pneumoniae is dependent on TBK1/IKKε, bacterial replication and the T3SS.
(A, C) WT, tbk1−/− ikke−/− and atg5−/− MEFs were infected with C. pneumoniae, MOI 5, for 36 h before harvest of total RNA. (B) WT MEFs were infected with live or UV-inactivated C. pneumoniae (CFU/cell 5) in the presence and absence of ampicillin (20 µg/ml) as indicated. (D) The cells were treated with INP0341 (60 µM), INP0406 (60 µM) or DMSO (vehicle) and infected with C. pneumoniae, MOI 5. Total RNA was harvested 36 hrs post-infection. (A–D) Levels of ISG56 mRNA were determined by real-time PCR. The measured ISG56 mRNA levels were normalized to β-actin, and data are presented as means of triplicate cultures +/− st.dev. Similar results were obtained in 2–3 independent experiments. RU, relative units.
The observed delayed kinetics of ISG expression relative to viral infections suggested that bacterial replication was necessary for induction of the response. To test this, we inactivated the bacteria with UV treatment and examined for induction of ISG56. As shown in Fig. 3B, inhibition of bacterial replication prevented induction of ISG56 expression.
The requirement for both endosomal TLRs, cytosolic RLRs and bacterial replication led us to hypothesize that interactions between the bacterial vacuole and the host cytoplasm are important for innate immune recognition and that cytoplasmic PAMPs are delivered to endosomes through autophagy. The potential role for autophagy was examined by comparing atg5+/− and atg5−/− cells with respect to C. pneumoniae-induced expression of ISG56 and showed that a deficient autophagy machinery did not impact on the IFN response evoked by C. pneumoniae (Fig. 3C). By contrast, inhibition of the type III secretion system using the inhibitor INP0341 largely prevented induction of ISG56 expression whereas the control compound INP0406 had no effect (Fig. 3D). Importantly, ampicillin, which halts the Chlamydia replication cycle before the conversion of RBs in to EBs but leaves interaction with the host cytosol unaffected [29], did not interfere with ISG56 expression. Thus, stimulation of ISG expression by C. pneumonia is dependent on bacterial replication, signaling through TBK1/IKKε and the T3SS.
Discussion
C. pneumoniae is an obligate intracellular gram-negative bacteria responsible for about 10% of community-acquired pneumonias. C. pneumoniae has a 2-stage life-cycle involving infectious EB and metabolically active RB [3]. The host cytokine response to Chlamydia infection is involved in both protection and pathogenesis of the disease, with e.g. IFN-γ exerting bactericidal activities and interleukin 1 contributing to tissue damage [6]. Type I IFNs, which were originally believed primarily to participate in the host response to viral infections, have recently emerged as important players in host-defense and pathogenesis of bacterial infections. However, unlike the protective role of type I IFNs in antiviral defense, this class of cytokines has been reported to participate in development of disease during several bacterial infections [19]–[22]. For instance, type I IFNs enhance susceptibility of mice to genital and lung infection with Chlamydia muridarum through a mechanism involving inhibition of the CD4 T cell response and enhancement of macrophage apoptosis [19], [32]. Despite the role of type I IFN in the pathogenesis of Chlamydia infection, it remains unresolved which PRRs are responsible for induction of the IFN response during infection with Chlamydia. In this work we demonstrate that C. pneumoniae induces expression of ISGs with a delayed kinetics relative to viruses, and that this is dependent on bacterial replication, innate immune recognition by intracellular nucleotide-sensing PRRs and signaling through TBK1/IKKε. Finally, we report that induction of the ISG response is dependent on bacteria-host interaction through the T3SS.
The finding that C. pneumoniae-induced expression of ISGs is dependent on replication and occurs with a delayed kinetics relative to viruses suggests that PAMPs produced late during the bacterial life cycle (or exposed to PRRs only at late time points) are responsible for induction of the IFN response. Since the conversion of EBs to RBs occurs between 8 and 10 h after infection, it is tempting to speculate that the Chlamydial ISG-inducing nucleotide PAMP is produced or accessible only when RBs are actively dividing.
We found an essential role for the T3SS in induction of the ISG response. This is in agreement with a recent report that C. muridarum induces expression of a subset of inflammatory cytokines in a manner dependent on the T3SS [17]. The Salmonella enterica Typhimurium T3SS has also been reported to stimulate innate immune responses, but in that case it was reported to occur directly via bacterial effector proteins independent of any known innate immune receptors [33]. TLRs are believed to interact with the cytoplasm through autophagy [34]. However, abrogation of autophagy by deletion of the atg5 gene did not affect the ability of C. pneumoniae to stimulate ISG expression, suggesting other mechanisms to be involved. Others have reported that the TLR adaptor MyD88 localizes to the inclusions during Chlamydia trachomatis infections, and there may therefore be a more direct interaction between the Chlamydia inclusion and endosomal TLRs [29]. Therefore, based on our findings, we propose that active secretion of specific molecules by the T3SS during the RB stage of the bacterial life cycle either leads to exposure of PAMPs or amplifies host cell signaling, thus augmenting the ISG response. As to the first possibility, there is no direct evidence of nucleotides being translocated across T3SS. However, one recent work reported that synthetic RNA amplified the IFN-β response induced by Yersinia pseudotuberculosis and this was dependent on the T3SS [35]. Concerning the later possibility, this would be parallel to the recent finding that C. trachomatis infection stimulates production of reactive oxygen species through the T3SS, which leads to activation of the inflammasome and production of bioactive IL-1β [18].
Bacteria are recognized through all known classes of PRRs, and specifically for Chlamydia it is known that TLR2, TLR4, NOD1, NOD2, NLRP3 and possibly also TLR3 play a role in innate recognition of the bacteria [8], [9], [18], [29]–[31], [36]. In this work we found that endosomal TLRs (which include the nucleotide-sensing TLR3, 7, and 9) and cytoplasmic RLRs (which include the RNA-sensing RIG-I and MDA5) were essential for induction of the ISG response by C. pneumonia, and that the MEFs induced ISG56 expression after stimulation with agonists for TLR3, RIG-I, and MDA5 but not after stimulation with agonists for TLR7 and 9. Therefore, the ISG response to C. pneumoniae is likely to be mediated by intracellular RNA-sensing PRRs. This is in contrast to most reports on bacterial induction of the IFN response, which is mediated primarily by cytoplasmic DNA-sensing PRRs [12], [14], [25], [26]. However, one recent study demonstrated recognition of group B streptococcus in TLR7 in conventional dendritic cells as a central mechanism of type I IFN induction [27]. Previously we found that purified DNA from different bacteria all stimulated TLR9, but only a subset of the live bacteria stimulated this PRR [37]. This suggests that the nature of the interactions between host cells and the bacterial life cycle, rather than the intrinsic immunostimulatory potential of bacterial nucleotides, are responsible for which PRRs that mediate the innate immune response to infection.
In conclusion, we report that activation of the type I IFN response during C. pneumoniae infection is mediated by intracellular nucleotide-sensing PRRs, which operate through a mechanism dependent on the bacterial T3SS. Thus, specific strategies to inhibit the chlamydial T3SS may be used to limit the detrimental effects of the type I IFN system in the host response to Chlamydia infection, and hence to limit the pathology of infections with this intracellular bacterial pathogen.
Acknowledgments
The Technical Assistance of Kirsten Stadel Petersen is greatly appreciated.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by The Danish Medical Research Council Grant nos. 271-05-0488 and 09-072636, the Lundbeck Foundation (Grants no. R17-A1526 and R19-A2023), the Kathrine og Vigo Skovgards Fond, the Aase and Ejnar Danielsens Fond, and The Aarhus University Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Birkelund S, Stephens RS. Construction of physical and genetic maps of Chlamydia trachomatis serovar L2 by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2742–2747. doi: 10.1128/jb.174.9.2742-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 3.Wyrick PB. Intracellular survival by Chlamydia. Cell Microbiol. 2000;2:275–282. doi: 10.1046/j.1462-5822.2000.00059.x. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 5.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hvid M, Baczynska A, Deleuran B, Fedder J, Knudsen HJ, et al. Interleukin-1 is the initiator of Fallopian tube destruction during Chlamydia trachomatis infection. Cell Microbiol. 2007;9:2795–2803. doi: 10.1111/j.1462-5822.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- 7.Prebeck S, Kirschning C, Durr S, Da Costa C, Donath B, et al. Predominant role of toll-like receptor 2 versus 4 in Chlamydia pneumoniae-induced activation of dendritic cells. J Immunol. 2001;167:3316–3323. doi: 10.4049/jimmunol.167.6.3316. [DOI] [PubMed] [Google Scholar]
- 8.Shimada K, Chen S, Dempsey PW, Sorrentino R, Alsabeh R, et al. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 2009;5:e1000379. doi: 10.1371/journal.ppat.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz KR, Stephens RS. The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect Immun. 2008;76:3150–3155. doi: 10.1128/IAI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 12.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 13.Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, et al. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity. 2006;24:41–51. doi: 10.1016/j.immuni.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, et al. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008;4:543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slepenkin A, Motin V, de la Maza LM, Peterson EM. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect Immun. 2003;71:2555–2562. doi: 10.1128/IAI.71.5.2555-2562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prantner D, Nagarajan UM. Role for the chlamydial type III secretion apparatus in host cytokine expression. Infect Immun. 2009;77:76–84. doi: 10.1128/IAI.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdul-Sater AA, Koo E, Hacker G, Ojcius DM. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem. 2009;284:26789–26796. doi: 10.1074/jbc.M109.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu H, Fan Y, Joyee AG, Wang S, Han X, et al. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J Immunol. 2008;181:2092–2102. doi: 10.4049/jimmunol.181.3.2092. [DOI] [PubMed] [Google Scholar]
- 20.Nagarajan UM, Prantner D, Sikes JD, Andrews CW, Jr, Goodwin AM, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76:4642–4648. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkelund S, Lundemose AG, Christiansen G. Immunoelectron microscopy of lipopolysaccharide in Chlamydia trachomatis. Infect Immun. 1989;57:3250–3253. doi: 10.1128/iai.57.10.3250-3253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkelund S, Lundemose AG, Christiansen G. Characterization of native and recombinant 75-kilodalton immunogens from Chlamydia trachomatis serovar L2. Infect Immun. 1989;57:2683–2690. doi: 10.1128/iai.57.9.2683-2690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, et al. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu YH, Macmillan JB, Chen ZJ. RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol. 2009;10:587–594. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- 28.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 29.O'Connell CM, Ionova IA, Quayle AJ, Visintin A, Ingalls RR. Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J Biol Chem. 2006;281:1652–1659. doi: 10.1074/jbc.M510182200. [DOI] [PubMed] [Google Scholar]
- 30.Sasu S, LaVerda D, Qureshi N, Golenbock DT, Beasley D. Chlamydia pneumoniae and chlamydial heat shock protein 60 stimulate proliferation of human vascular smooth muscle cells via toll-like receptor 4 and p44/p42 mitogen-activated protein kinase activation. Circ Res. 2001;89:244–250. doi: 10.1161/hh1501.094184. [DOI] [PubMed] [Google Scholar]
- 31.Derbigny WA, Hong SC, Kerr MS, Temkit M, Johnson RM. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect Immun. 2007;75:1280–1290. doi: 10.1128/IAI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 33.Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, et al. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 35.Auerbuch V, Golenbock DT, Isberg RR. Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog. 2009;5:e1000686. doi: 10.1371/journal.ppat.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opitz B, Forster S, Hocke AC, Maass M, Schmeck B, et al. Nod1-mediated endothelial cell activation by Chlamydophila pneumoniae. Circ Res. 2005;96:319–326. doi: 10.1161/01.RES.0000155721.83594.2c. [DOI] [PubMed] [Google Scholar]
- 37.Mogensen TH, Paludan SR, Kilian M, Ostergaard L. Live Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis activate the inflammatory response through Toll-like receptors 2, 4, and 9 in species-specific patterns. J Leukoc Biol. 2006;80:267–277. doi: 10.1189/jlb.1105626. [DOI] [PubMed] [Google Scholar]