Abstract
Background
The functions of proteins are strongly related to their localization in cell compartments (for example the cytoplasm or membranes) but the experimental determination of the sub-cellular localization of proteomes is laborious and expensive. A fast and low-cost alternative approach is in silico prediction, based on features of the protein primary sequences. However, biologists are confronted with a very large number of computational tools that use different methods that address various localization features with diverse specificities and sensitivities. As a result, exploiting these computer resources to predict protein localization accurately involves querying all tools and comparing every prediction output; this is a painstaking task. Therefore, we developed a comprehensive database, called CoBaltDB, that gathers all prediction outputs concerning complete prokaryotic proteomes.
Description
The current version of CoBaltDB integrates the results of 43 localization predictors for 784 complete bacterial and archaeal proteomes (2.548.292 proteins in total). CoBaltDB supplies a simple user-friendly interface for retrieving and exploring relevant information about predicted features (such as signal peptide cleavage sites and transmembrane segments). Data are organized into three work-sets ("specialized tools", "meta-tools" and "additional tools"). The database can be queried using the organism name, a locus tag or a list of locus tags and may be browsed using numerous graphical and text displays.
Conclusions
With its new functionalities, CoBaltDB is a novel powerful platform that provides easy access to the results of multiple localization tools and support for predicting prokaryotic protein localizations with higher confidence than previously possible. CoBaltDB is available at http://www.umr6026.univ-rennes1.fr/english/home/research/basic/software/cobalten.
Background
Determining the subcellular localization of proteins is essential for the functional annotation of proteomes [1,2]. Bacterial proteins can exist in soluble (i.e free) forms in cellular spaces (cytoplasm in both monoderm and diderm bacteria and periplasm in diderms), anchored to membranes (cytoplasm membrane in monoderms, inner- or outer membrane in diderms) or cell wall (in monoderms). They can also be released into the extracellular environment or directly translocated into host cells [3]. All protein synthesis takes place in the cytoplasm, so all non-cytoplasmic proteins must pass through one or two lipid bilayers by a mechanism commonly called "secretion". Protein secretion is involved in various processes including plant-microbe interactions [4,5]), biofilm formation [6,7] and virulence of plant and human pathogens [8-10]. Two main systems are involved in protein translocation across the cytoplasmic membrane, namely the essential and universal Sec (Secretion) pathway and the Tat (Twin-arginine translocation) pathway found in some prokaryotes (monoderms and diderms) and eukaryotes alike [11-16]. The Sec machinery recognizes an N-terminal hydrophobic signal sequence and translocates unfolded proteins [12], whereas the Tat machinery recognizes a basic-rich N-terminal motif (SRR-x-FLK) and transports fully folded proteins [13,14]). In addition to these systems, diderm bacteria have six further systems that secrete proteins using a contiguous channel spanning the two membranes (T1SS, [17,18], T3SS, T4SS and T6SS [19-24]) or in two steps, the first being Sec- or Tat-dependent export into the periplasmic and the second being translocation across the outer membrane (T2SS, [25-27] and T5SS, [28,29]). Other diderm protein secretion systems exist: they include the chaperone-usher system (CU or T7SS, [30,31]) and the extracellular nucleation-precipitation mechanism (ENP or T8SS, [32]). It is worth mentioning that the terminology T7SS has also been proposed to describe a completely different protein secretion system, namely the ESAT-6 protein secretion (ESX) in Mycobacteria, now considered as diderm bacteria [33]. Beside Sec and Tat pathways, monoderm bacteria have additional secretion systems for protein translocation across the cytoplasmic membrane, namely the flagella export apparatus (FEA [34]), the fimbrilin-protein exporter (FPE, [35,36]) and the WXG100 secretion system (Wss, [37,38]).
Establishing whole proteome subcellular localization by biochemical experiments is possible but arduous, time consuming and expensive. Data concerning predicted proteins (from whole genome sequences) is continuously increasing. High-throughput in silico analysis is required for fast and accurate prediction of additional attributes based solely on their amino acid sequences. There are large numbers of global (that yield final localization) and specialized (that predict features) tools for computer-assisted prediction of protein localizations. Most specialized tools tend to detect the presence of N-terminal signal peptides (SP). Prediction of Sec-sorting signals has a long history as the first methods, based on weight matrices, were published about fifteen years ago [39-41]. Numerous machine learning-based methods are now available [42-50]. The distinction between Tat- and Sec- sorting signals is essentially based on the recognition, in the n/h regions edge, of the twin-arginine motif [51], using regular expressions combined with hydrophobicity measures [52] or machine learning [53]. Pre-lipoproteins SP have the same n- and h- regions as Sec SP but contain, in the c-region, a well-conserved lipobox [54], recognized for cleavage by the type II signal peptidase [55]. Lipoprotein prediction tools use regular expression patterns to detect this lipobox [56,57], combined with Hidden Markov Models (HMM) [58] or Neural Networks (NN) [59]. Other attributes predicted by specialized tools are α-helices and β-barrel transmembrane segments. In 1982, Kyte and Doolittle proposed a hydropathy-based method to predict transmembrane (TM) helices in a protein sequence. This approach was enhanced by combining discriminant analysis [60], hydrophobicity scales [61-63] amino acid properties [64,65]. Complex algorithms are also available and employ statistics [66], multiple sequence alignments [67] and machine learning approaches [68-73]. β-barrel segments, embedded in outer membrane proteins, are harder to predict than α-helical segments, mostly because they are shorter; nevertheless, many methods are available based on similar strategies [74-87].
This plethora of protein localization predictors and databases [88-91] constitutes an important resource but requires time and expertise for efficient exploitation. Some of the tools require computing skills, as they have to be locally installed; others are difficult to use (numerous parameters) or to interpret (large quantities of graphics and output data). Web tools are disseminated and need numerous manual requests. Additionally, researchers have to decide which of these numerous tools are the most pertinent for their purposes, and selection is problematic without appropriate training sets. Recent work shows that the best strategy for exploiting the various tools is to compare them [92-94].
Here, we describe CoBaltDB, the first public database that displays the results obtained by 43 localization predictor tools for 776 complete prokaryotic proteomes. CoBaltDB will help microbiologists explore and analyze subcellular localization predictions for all proteins predicted from a complete genome; it should thereby facilitate and enhance the understanding of protein function.
Construction and content
Data sources
The major challenge for CoBaltDB is to collect and integrate into a centralized open-access reference database, non-redundant subcellular prediction features for complete prokaryotic orfeomes. Our initial dataset contained 784 complete genomes (731 bacteria and 53 Archaea), downloaded with all plasmids and chromosomes (1468 replicons in total), from the NCBI ftp server ftp://ftp.ncbi.nih.gov/genomes/Bacteria in mid-December 2008. This dataset contains 2,548,292 predicted non-redundant proteins (Additional file 1).
The CoBaltDB database was designed to associate results from disconnected resources. It contains three main types of data: i) CoBaltDB pre-computed prediction using 23 feature-based localization tools (Table 1), ii) CoBaltDB pre-computed prediction obtained using 5 localization meta-tools (Table 2) and iii) data collected from 20 public databases with both predicted and experimentally determined subcellular protein localizations (Table 3).
Table 1.
A summary of CoBaltDB precomputed features-tools
| Program | Reference | Analytical method | CoBaltDB features prediction group(s) | ||||
|---|---|---|---|---|---|---|---|
| LipoP 1.0 Server | [59] | HMM + NN | LIPO | SEC | |||
| DOLOP | [57] | RE | LIPO | ||||
| LIPO | [56] | RE | LIPO | ||||
| TatP 1.0 | [53] | RE + NN | TAT | ||||
| TATFIND 1.4 | [52] | RE | TAT | ||||
| PrediSi | [112] | Position weight matrix | SEC | ||||
| SignalP 3.0 Server | [45-47] | HMM + NN | SEC | ||||
| SOSUIsignal | [113] | Multi-programs | SEC | ||||
| SIG-Pred | J.R. Bradford | Matrix | SEC | ||||
| RPSP | [44] | NN | SEC | ||||
| Phobius | [48,49] | HMM | SEC | αTMB | |||
| HMMTOP | [71] | HMM | αTMB | ||||
| TMHMM Server v.2.0 | [70] | HMM | αTMB | ||||
| TM-Finder | [65] | AA FEATURES | αTMB | ||||
| SOSUI | [114] | AA FEATURES | αTMB | ||||
| SVMtm | [73] | SVM | αTMB | ||||
| SPLIT 4.0 Server | [115] | AA FEATURES | αTMB | ||||
| MCMBB | [116] | HMM | βBarrel | ||||
| TMBETADISC: | [117] | ||||||
| _COMP | AA FEATURES | βBarrel | |||||
| _DIPEPTIDE | Dipeptide composition | βBarrel | |||||
| _MOTIF | Motif(s) | βBarrel | |||||
| TMB-Hunt2 | [118] | SVM | βBarrel | ||||
HMM: Hidden Markov Model, NN: Neural Network, RE: Regular Expression, AA: Amino Acid, SVM: Support Vector Machine
Table 2.
A summary of CoBaltDB precomputed meta-tools
| Program | Reference | Analytical method | Localizations |
|---|---|---|---|
| Subcell Specialization Server 2.5 | [119] | Multiple classifiers | 5 diderms/3 monoderms |
| SLP-Local | [120] | SVM | 3 with no distinction |
| SubLoc v1.0 | [121] | SVM | 3 with no distinction |
| Subcell (Adaboost method) | [122] | AdaBoost algorithm | 3 with no distinction |
| SOSUIGramN | [123] | Physico-chemical parameters | 5 diderms/no monoderm |
SVM: Support Vector Machine
Table 3.
A summary of CoBaltDB integrated databases and tools features.
| Databases | Reference | Features predicted | Genome(s) | Protein numbers |
|---|---|---|---|---|
| EchoLOCATION | [124] | Subcellular-location (EXP) | E. coli K-12 | 4330 (506 exp) |
| Ecce | _ | Subcellular-location | E. coli K-12 | 306 |
| LocateP DataBase | [89] | Subcellular-location | 178 MD | 542788 |
| cPSORTdb | [91] | Subcellular-location | 140 BA | 1634278 |
| ePSORTdb | [91] | Subcellular-location (EXP) | 2165 | |
| THGS | [125] | Transmembrane Helices | 689 PROK | 465411 |
| Augur | [88] | Subcellular-location | 126 MD | 111223 |
| CW-PRED | [126] | Cell-anchored (surface) | 94 MD | 954 |
| PROFtmb | [78] | Beta-barrel (OM) | 78 DD/19 MD | 2152 |
| HHomp | [127] | Beta-barrel (OM) | 12495 | |
| PRED-LIPO | [58] | Lipoprotein SPs | 179 MD | 895 |
| SPdb | [90] | Signal peptides (SPs) | 855 PROK | 7062 |
| ExProt | [128] | Signal peptides (SPs) | 23 AR/61 MD/115DD | |
| Signal Peptide Website | _ | Signal peptides (SPs) | 384 BA | 1161 (EXP) |
| PRED-SIGNAL | [129] | Signal peptides (SPs) | 48 AR | 9437 |
| TMPDB | [130] | Alpha Helices & Beta-barrel | 188 | |
| DTTSS | Shandong Univ. | Type III secretion system | 1035 | |
| TOPDB | [131] | Transmembrane Proteins | 755 BA/16 AR | |
| TMBC-Database | Andrew Garrow | Transmembrane Beta-barrel | 1219 | |
| Swissprot signal testset | [132] | Signal peptides (SPs) (EXP) | 176 SP+/122 SP- |
MD = monoderm, DD = diderm. AR (Archeae), BA (Bacteria), PROK (Prokaryotes) include both bacteria and Archaee, EXP = Experimental database
These data were organized in five "boxes" with regard to the features predicted: three boxes correspond to signal peptide detection (Lipoprotein, Tat- and Sec- dependent targeting signals); one box for the prediction of alpha-transmembrane segments (TM-Box); and one box, only available for diderms (Gram-negatives), for outer membrane localization through prediction of beta-barrels.
Data generation
There is a great diversity of web and stand-alone resources for the prediction of protein subcellular location. We retrieved and tested 99 currently (in 2009) available specialized and global tools (software resources) that use various amino acid features and diverse methods: algorithms, HMM, NN, Support Vector Machine (SVM), software suites and others), to predict protein subcellular localization (Additional file 2). All tools were evaluated: some are included in CoBaltDB, some may be launched directly from the platform (Table 4), and others were excluded because of redundancy or processing reasons or both (Table 5). Some tools are specific to Gram-negative or Gram-positive bacteria. Many prediction methods applicable to both Gram categories have different parameters for the two groups of bacteria. For these reasons, each NCBI complete bacterial and archaeal genome implemented in CoBaltDB was registered as "monoderm" or "diderm", on the basis of information in the literature and phylogeny (Additional file 3). Monoderms and diderms were considered as Gram-negative and Gram-positive, respectively. All archaea were classified as monoderm prokaryotes since their cells are bounded by a single cell membrane and possess a cell envelope [3,95]. An exception was made for Ignicoccus hospitalis as it owns an outer sheath resembling the outer membrane of gram-negative bacteria [96].
Table 4.
Tools available using CoBaltDB "post" window
| Program | Reference | Analytical method | CoBaltDB features prediction group(s) | |||
|---|---|---|---|---|---|---|
| LipPred | [133] | Naive Bayesian Network | LIPO | |||
| PRED-LIPO | [58] | HMM | LIPO | (only Monoderm) | ||
| SPEPLip | [134] | NN | LIPO | SEC | ||
| SecretomeP | [135] | Pattern & NN | ΔSEC_SP | |||
| Signal-3L | [136] | Multi-modules | SEC | |||
| Signal-CF | [137] | Multi-modules | SEC | |||
| Signal-Blast | [138] | BlastP | SEC | |||
| Sigcleave | EMBOSS | Von Heijne method | SEC | |||
| PRED-SIGNAL | [129] | HMM | SEC | (only Archae) | ||
| Flafind | [139] | AA features | T3SS Archae + T4SS Bacteria | |||
| T3SS_prediction | [110] | SVM & NN | T3SS | |||
| EffectiveT3 | [111] | Machine learning | T3SS | |||
| NtraC Signal Analysis | [140] | Pattern model | SEC (long SP) | |||
| Philius | [141] | HMM | SEC | αTMB | ||
| (SP)OCTOPUS | [142,143] | Blast Homology, NN, HMM | SEC | αTMB | ||
| MemBrain | [144] | Machine learning | SEC | αTMB | ||
| DAS | [145] | Dense Alignment Surface | αTMB | |||
| HMM-TM | [146] | HMM | αTMB | |||
| SVMtop Server 1.0 | [147] | SVM | αTMB | |||
| UMDHMM_TMHP | [148] | HMM | αTMB | |||
| waveTM | [149] | Hydropathy signals algorithm | αTMB | |||
| PRED-TMR | [150] | AA features | αTMB | |||
| TMAP | [67] | AA features | αTMB | |||
| igTM | [151] | Grammatical Inference | αTMB | |||
| TOPCONS | [152] | Tools Consensus | αTMB | |||
| TUPS | [153] | Tools Consensus | αTMB | |||
| ConPred II | [154] | Tools Consensus | αTMB | |||
| MEMSAT3 | [66,155] | NN | αTMB | |||
| SABLE | [156] | NN | αTMB | |||
| TM-Pro | [64,157] | AA features | αTMB | |||
| ProspRef | _ | Knowledge-based method | αTMB | |||
| PSIPRED | [158,159] | NN, PSSM | αTMB | |||
| NPS@ | [160] | Tools Consensus | αTMB | |||
| SAM-T08 | [161] | HMM | αTMB | |||
| PORTER | [162] | NN | αTMB | |||
| TMPred | EMBnet | Weight-matrices | αTMB | |||
| TMMOD | [163] | HMM | αTMB | |||
| TopPred II | [61] | G. von Heijne algorithm | αTMB | |||
| YASPIN | [164] | Hidden Neural Network | αTMB | |||
| MemType-2L | [165] | PseudoPSSM, classifier | Membrane Type | |||
| BOMP | [84] | AA features | βBarrel | |||
| TMBETADISC-RBF | [87] | RBF network, PSSM | βBarrel | |||
| TMBETA-NET | [117] | AA features | βBarrel | |||
| PRED-TMBB | [85] | HMM | βBarrel | |||
| ConBBPred | [76] | Tools Consensus | βBarrel | |||
| CW-PRED (submit) | [126] | HMM | Cell-Wall (only Monoderm) | |||
| ProtCompB | SoftBerry | Multi-methods | Localization | |||
| CELLO | [166] | SVM | Localization | |||
| PSL101 | [167] | SVM, structure homology | Localization | |||
| PSLpred | [168] | SVM | Localization | |||
| GPLoc-neg | [169] | Basic classifier | Localization | (only Diderm) | ||
| GPLoc-pos | [170] | Basic classifier | Localization | (only Monoderm) | ||
| LOCtree | [171] | SVM | Localization | |||
| PSORTb | [91] | Multi-modules | Localization | |||
| SLPS | [172] | Nearest Neighbor on domain | Localization | |||
| Couple-subloc v1.0 | Jian Guo | AA features | Localization | |||
| TBPRED | [173] | SVM | Localization | (only Mycobacterium) | ||
HMM: Hidden Markov Model, NN: Neural Network, AA: Amino Acid, SVM: Support Vector Machine, PSSM: Position Specific Scoring Matrix, T3SS: Type III Secretion System, RBF: Radial Basis Function
Table 5.
Tools and Database not available in CoBaltDB
| Program | Reference | Analytical method | CoBaltDB features prediction group(s) | |||
|---|---|---|---|---|---|---|
| SpLip | [174] | Weight matrix | LIPO | (only Spirochaetal) | ||
| PROTEUS2 | [175] | Multi-Methods | SEC | αTMB | βBarrel | |
| PRED-TMR2 | [176] | NN | αTMB | |||
| PRODIV-TMHMM | [72] | Multi HMM | αTMB | |||
| S_TMHMM | [72] | HMM | αTMB | |||
| TransMem | [69] | NN | αTMB | |||
| BPROMPT | [177] | Bayesian Belief Network | αTMB | |||
| orienTM | [178] | Statistical analysis | αTMB | |||
| APSSP2 | [179] | Multi-Methods | Secondary structure | |||
| PRALINE_TM | [180] | Alignment, tools consensus | Secondary structure | |||
| OPM (DB) | [181] | Multi-Methods | Membrane orientation | |||
| MP_Topo (DB) | [182] | Experimental | TMB | |||
| PDBTM (DB) | [183] | TMDET algorithm | TMB | |||
| TMB-HMM | A.Garrow | HMM, SVM | βBarrel | |||
| TMBETA-SVM | [86] | SVM | βBarrel | |||
| TMBETA-GENOME (DB) | [184] | Multi-Methods | βBarrel | |||
| PredictProtein | [185] | Alignment, Multi-Methods | Localization | |||
| EcoProDB (DB) | [186] | Identification on 2D gels | Localization | (only E.coli) | ||
| LOCTARGET (DB) | [187] | Multi-Methods | Localization | |||
| DBMLoc (DB) | [188] | _ | Localization | |||
NN: Neural Network, HMM: Hidden Markov Model, SVM: Support Vector Machine
Currently, CoBaltDB contains pre-computed results obtained with 48 tools and databases, and additionally provides pre-filled access to 50 publicly available tools that could not be pre-computed or that provide new information (tools dedicated to a special phylum, consensus tools or tools predicting proteins secreted via other pathways). The data pre-computing process is illustrated in Figure 1; web-based and stand-alone tools were used separately. Web-based localization prediction tools were requested via a Web automat, a python automatic submission workflow using both "httplib" and "urllib" libraries. A different script was created for each tool. For web-tools with no equivalent (such as "TatP" for Tat-BOX and "LIPO" for Lipoprotein-BOX) and incompatible with automatic requests, we collected results manually. CoBaltDB also provides a platform with automatically pre-filled forms for additional submissions to a selection of fifty recent or specific web tools (Table 4). The stand-alone tools were installed on a Unix platform (unique common compatible platform) and included in a global python pipeline with the HTTP request scripts. We selected information from a up-to-date collection of 20 databases and integrated this data within CoBaltDB; these databases were retrieved by simple downloading or creating an appropriate script which navigates on the web databases to collect all protein information. The global python pipeline used multi-threading to speed up the pre-computation of the 784 proteomes.
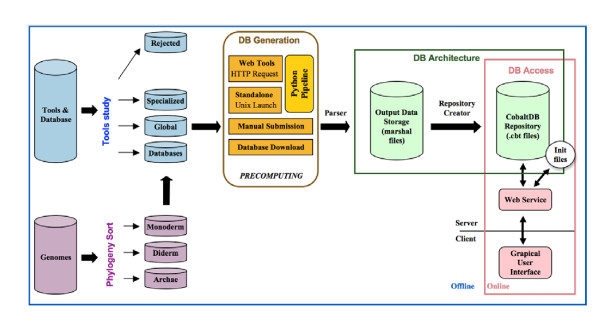
Figure 1.
A schematic view of the CoBaltDB workflow. CoBaltDB integrates the results of 43 localization predictors for 784 complete bacterial and archaeal proteomes. Each complete NCBI prokaryotic genome implemented in CoBaltDB was classified as: archaea, or monoderm or diderm bacteria. 101 protein subcellular location predictors were evaluated and few were rejected. Selected tools were classified as: feature localization tools (Specialized), localization meta-tools (Global) or databases. The data recovery process was performed manually or via a Web automat using a python automatic submission workflow for both stand-alone and web-based tools. Databases were downloaded. For each protein, ouptuts collected were parsed and selected items were stored in particular CoBaltDB formatted files (.cbt). The parsing pipeline creates one ".cbt" file per replicon to compose the final CoBaltDB repository. The client CoBaltDB Graphical User Interface communicates with the server-side repository via web services to provide graphical and tabular representations of the results.
Database Creation and Architecture
For each protein, every output collected (a HTML page for web tools and a text file for standalone applications) was parsed and selected items were stored in a particular format: binary "marshal" files. The object structure obtained by parsing tool output was directly saved into a marshal file, allowing a quick and easy opening by directly restoring the initial parsing object. Another script then creates the CoBaltDB repository, by reading and analysing all marshal files to generate a specific formatted file (".cbt") for each replicon. These files contain all the required protein information and a simplified representation of the tools' results. Some initialization files containing information about phylogeny or genome features are also used.
The repository is used by the Graphical User Interface (GUI) to display CoBaltDB information. For raw data from tools, the GUI accesses the marshal file directory.
Accessing the CoBaltDB Repository and Raw Data
The CoBaltDB platform has been developed as a client-server application. The server is installed at the Genouest Bioinformatics platform http://www.genouest.org/?lang=en. The client is a Java application that needs to be locally downloaded by the users. Queries are submitted to the server-side CoBaltDB repository using a locally installed client GUI that provides tabular and graphical representations of the data. The repository is accessed through SOAP-based web services (Simple Object Access Protocol), implemented in Java 5 using the Apache Axis 1.4 toolkit and deployed on the servlet engine Tomcat 5.5.20. CoBaltDB integrates: an initialization web service (that returns the current list of genomes supported); two repository web services that allow querying the database either by specifying a replicon or a list of locus tags; and a raw data web service that retrieves all recorded raw data generated by a given tool for the specified locus tag.
Utility
Running CoBaltDB
Our goal was to build an open-access reference database providing access to protein localization predictions. CoBaltDB was designed to centralize different types of data and to interface them so as to help researchers rapidly analyse and develop hypotheses concerning the subcellular distribution of particular protein(s) or a given proteome. This data management allows comparative evaluation of the output of each tool and database and thus straightforward identification of inaccurate or conflicting predictions.
We developed a user-friendly CoBaltDB GUI as a Java 5 client application using NetBeans 5.5.1 IDE. It presents four tabs that perform specific tasks: the "input" tab (Figure 2) allows selecting the organism whose proteome localizations will be presented, using organism name completion or through an alphabetical list. Alternatively, users may also enter a subset of proteins, specified by their locus tags. The "Specialized tools" tab (Figure 3) supplies a table showing, for each protein identified by its locus tag or protein identifier, some annotation information such as its gene name, description and links to the corresponding NCBI and KEGG web pages. Clicking on a "locus tag" opens a navigator window with the related KEGG link, and clicking on a "protein Id" opens the corresponding NCBI entry web page. The table shows, for each protein and for each feature box (Tat, Sec, Lipo, αTMB, βBarrel), a heat map (white/blue) representing the percentage of tools predicting the truth/presence of the corresponding localization feature in the protein considered. Clicking on the heat map opens a new window that shows the raw data generated by each tool of the considered feature box, thus allowing the investigator to access the tool-specific information they are used to. The predictions of related feature databases are given next to the corresponding heat-map. The proteins which are referred to by the databases implemented in CobaltDB as having an experimentally determined localization appear with a yellow background colour. This representation enables the user to observe graphically the distribution of tools predicting each type of feature. The "meta-tools" tab (Figure 4) provides the predictions given by multi-modular prediction software (meta-tools or global databases) that use various techniques to predict directly three to five subcellular protein localizations in mono- and/or diderm bacteria (Table 4). The descriptions of the localizations were standardised to ease interpretation by the investigator. Both tables may be searched for occurrences of any string of characters via the search button, facilitating retrieval of a particular locus tag, protein id, accession number or even a gene name or annotation description. Both tables may be sorted with respect to any column, i.e. in alphanumerical order for the locus tags, protein identifiers, annotation descriptions and localization predictions, or in numerical order for the percentages. This makes it straightforward to identify all proteins with particular combinations of localization features. Both tables may be saved as Excel files. Finally, the CoBaltDB "additional tools" tab (Figure 5) enables queries to be submitted to a set of 50 additional tools by pre-filling the selected forms with the selected protein sequence and Gram information whenever appropriate. For this use, the investigator might have to enter additional parameters.
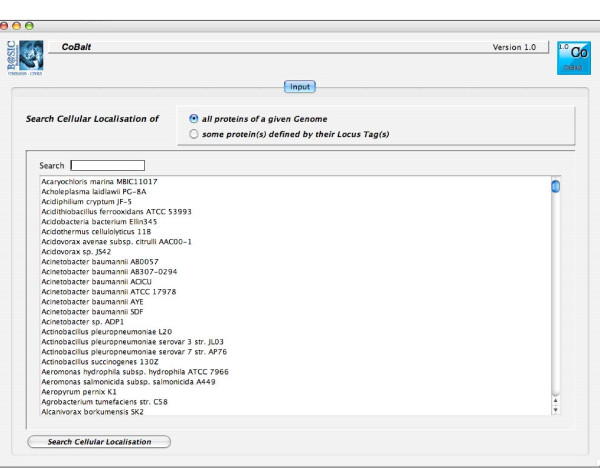
Figure 2.
A snapshot of the CoBaltDB input interface. The "input" module allows the selection of organisms, using organism name completion or through an alphabetical list. Users can also enter a subset of proteins, specified by their locus tags.
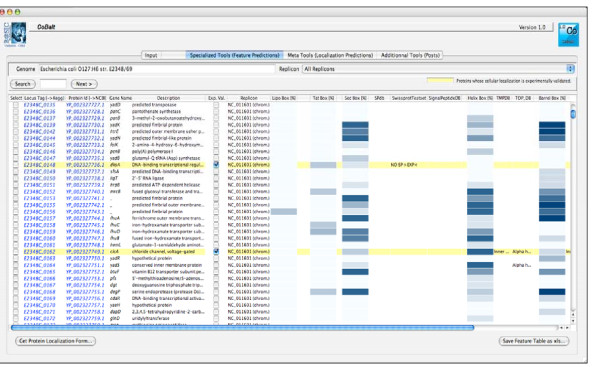
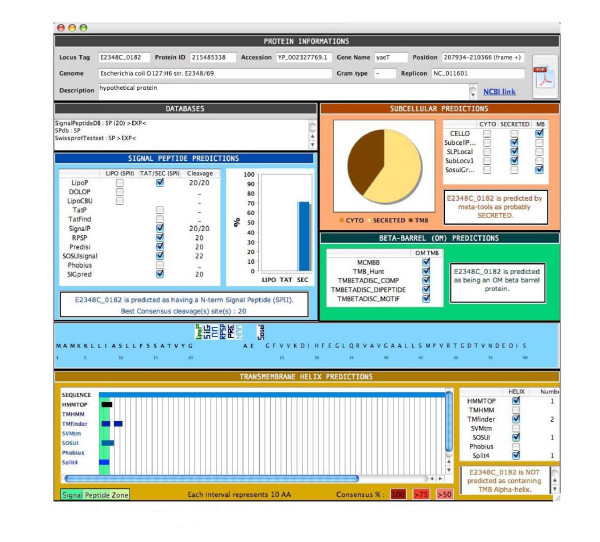
Figure 3.
The CoBaltDB Specialized Tools viewer. The "Specialized tools" browser supplies a tabular output for every protein, enriched with the protein's annotation including locus tag, protein identifier, gene name (if available) and product descriptions. Clicking on each "locus tag" opens a navigator window with related KEGG link whereas clicking on every "protein Id" opens the corresponding NCBI entry web page. Clicking on the white/blue heat map reveals the raw results of all tools corresponding to the feature box considered.
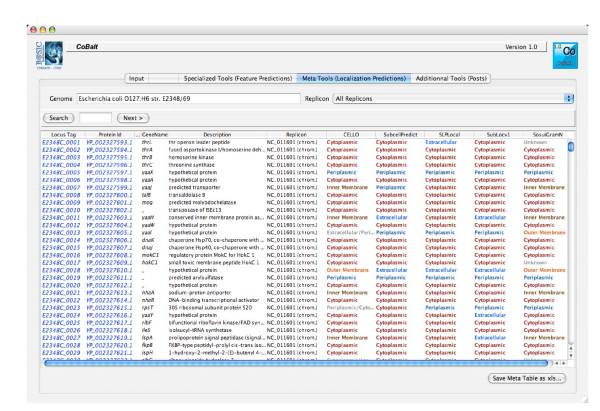
Figure 4.
The CoBaltDB Meta-Tools interface. The "meta-tools" panel presents the CoBaltDB-computed results for multi-modular prediction software that uses various techniques to directly predict 3 to 5 subcellular localizations for proteins in mono- and/or diderm bacteria.
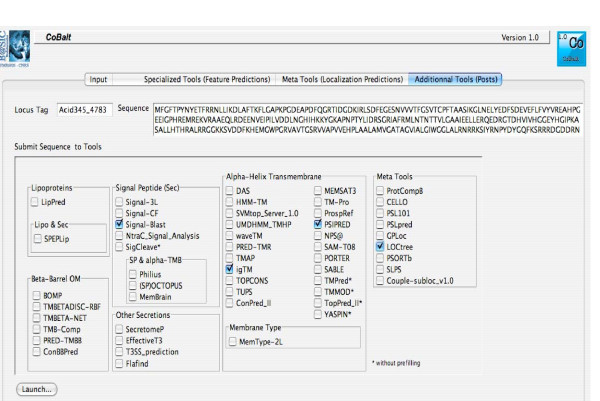
Figure 5.
The CoBaltDB Prefilled post window. The "additional tools" panel enables web page submission for a set of 50 additional tools by pre-filling selected forms with selected sequence and Gram information as appropriate.
Finally, for each protein, all results were summarized in a synopsis (Figure 6); the synopsis presents the results generated by all the tools in a unified manner, and includes a summary of all predicted cleavage sites and membrane domains. This "standardized" form thus provides all relevant information and lets the investigators establish their own hypotheses and conclusions. This form may be saved as a .pdf file (Figure 6). Examples of using the CoBaltDB synopsis are provided below in the second case study.
Figure 6.
CoBaltDB Synopsis. For any given protein, all results are summarized in a synopsis which presents, in a unified manner, a summary of all predicted cleavage sites and membrane domains. This synopsis can be stored as a .pdf file.
Selected CoBaltDB uses
We propose to illustrate briefly some possible uses of CoBaltDB.
1-Using CoBaltDB to compare subcellular prediction tools and databases
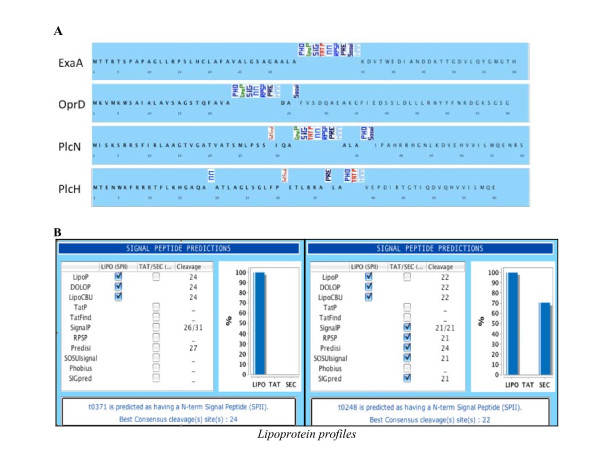
The various bioinformatic approaches developed for computational determination of protein subcellular localization exhibit differences in sensitivity and specificity; these differences are mainly the consequences of the types of sequences used as training models (diderms, monoderms, Archaea) and of the methods applied (regular expressions, machine learning or others). By interfacing the results from most of the reliable predictions tools, CoBaltDB provides immediate comparisons and constitutes an accurate and high-performance resource to identify and characterize candidate "non-cytoplasmic" proteins. As an example, using CoBaltDB to analyse the 82 proteins that compose the experimentally confirmed "Lipoproteome" of E. coli K-12 [97] shows that 72 are correctly predicted by the three precomputed tools (LipoP [59], DOLOP [57] and LIPO [56]), and that the other 10 are only identified by two of the three tools (Additional file 4A). Eight of these lipoproteins were not detected by DOLOP, because the regular expression pattern allowing detection of the lipidation sequence ([LVI] [ASTVI] [GAS] [C] lipobox) is too stringent (Additional file 4B). By comparison, the PROSITE lipobox pattern (PS00013/PDOC00013) is more permissive ([DERK](6)- [LIVMFWSTAG] (2)- [LIVMFYSTAGCQ]- [AGS]-C). This example demonstrates that using a single tool may result in errors and suggests that the best approach is to combine the various "features-based" methods available and compare their findings. This view also applies to meta-tools predictors. E. coli K12 lipoproteins can be found anchored to the inner or the outer membrane through attached lipid, but some of them are periplasmic (Additional file 4A). The comparison of in silico subcellular localization assignments with experimental findings clearly indicates that all meta-tools require significant improvements in accuracy and precision, that none should be used to the exclusion of the others. It also appears that analysis with specialized tools, organized on a "one feature at a time" basis (Lipo SPs, TAT SPs ...), most reliably gives predictions consistent with experimental data. For this purpose, CoBaltDB is a unique and innovative resource.
2-Using CoBaltDB to analyse protein(s) and a proteome
One valuable property of CoBaldDB is to recapitulate all pre-computed predictions in a unique A4-formated synopsis. This summary is very helpful for assessing computational data such as the variation and frequency in the predictions of signal peptide cleavage sites: such predictions are sometimes significantly consistent, but often are not in agreement with each other (Figure 7A). However, correct identification of signal peptide cleavage sites is essential in many situations, especially for producing secreted recombinant proteins.
Figure 7.
Using CoBaltDB to analyse protein(s) and a proteome. A: Comparative analysis of SP cleavage site predictions (proteinssecreted by P. aeruginosa); B: Discriminating between SPI- and SP II cleavage sites.
The CoBaltDB synopsis could also be used to discriminate between SignalPeptidaseII- and SignalPeptidaseI-cleaved signals and between SPs and N-terminal transmembrane helices. Indeed, most localization predictors have difficulties distinguishing between type I and type II signal peptidase cleavages. CoBaltDB can be exploited in an interesting way to benchmark this prediction by displaying all cleavage site predictions in a "decreasing sensitivity" arrangement (SpII then Tat-dependant SPI then Sec-SPI). By considering lipoprotein datasets from different organisms, we evidenced two principal profiles (Figure 7B) and found that all experimentally validated lipoproteins score 100% (all tools give the same prediction) or 66% in the CoBaltDB LIPO column (see explanation in the paragraph above). In addition, in almost all of the examined cases, tools dedicated to Twin-arginine SP detection do not identify SpII-dependent SP, whereas the Sec-SP predictors detect both Sec and Tat-type I as well as type II signal-anchor sequences.
These observations allow us to propose, for our data set, thresholds for each box: as previously illustrated, lipoproteins have score > 66% in the LIPO prediction box; Tat-secreted proteins have 0% in the LIPO box and 100% for the two TAT-dedicated tools; Sec-secreted proteins have 33% in the LIPO Box (due to the fact that LipoP detects both SpI and SpII [59]), 0% in the TAT-tools, and > 80% in SEC-specialized tools. Rules of this type can be used to check entire proteomes for evaluation of the different secretomes as illustrated in the following case studies.
3-Using CoBaltDB to compare proteomes
Using CoBaltDB and the thresholds described above, we can compare the predicted lipoproteomes (Figure 8A) of the three completely sequenced substrains of E. coli K12: MG1655 and W3110 (both derived from W1485 approximately 40 years ago [98]), and DH10B which was constructed by a series of genetic manipulations [99]. Each of these three substrains encode 89 lipoproteins found in both other substrains (Additional file 4). Four additional lipoproteins are detected in DH10B (BorD, CusC, RlpA and RzoD) and are second copies lipoprotein genes, present in the 113-kb tandemly repeated region of the chromosome (Figure 8B, coordinates 514341 to 627601, [99]), and strain DH10B contains one gene encoding the Rz1 proline-rich lipoprotein from bacteriophage lambda absent from the two other substrains. Lipoprotein YghJ, that shares 64% homology with V. cholerae virulence-associated accessory colonization factor AcfD [100], is absent from the DH10B genome annotation. However, comparative genomic analysis shows that a yghJ locus could be annotated in this strain but corresponds to a pseudogene caused by a frameshift event (Figure 8C). YfbK was also overlooked in the DH10B annotation process but in this case, the gene is intact. Finally, differences between lipoprotein prediction results concerning YafY, YfiM and YmbA are due to erroneous N-terminus predictions. YafY in DH10B was predicted to be a lipoprotein due to the N-terminal 17 aa-long type II signal peptide and was published as a new inner membrane lipoprotein [101]. In substrains MG1655 and WS3110, the original annotation fused the yafY loci with its upstream pseudogene ykfK (137 N-terminal aa longer). The presumed start codons of YfiM and YmbA in MG1655 were recently changed by adding 17 (lrilfvcsllllsgcsh) and 5 (mkkwl) N-terminal amino acids, respectively (PMC1325200). These modifications substantially affect the prediction of their subcellular localization. Inspection of the genomic sequences of the two other substrains leads to equivalent changes such that YfiM and YmbA in all three substrains are now predicted to be lipoproteins. In conclusion, using CoBaltDB to compare lipoproteomes between substrains, we were able to detect genomic events as well as "annotation" errors. After correction, we can conclude that the three E. coli K12 substrains have 93 lipoproteins in common; that one locus whose function is related to virulence has been transformed into a pseudogene in DH10B; and that DH10B contains five additional lipoproteins due to duplication events and to the presence of prophages absent from the other two substrains (Figure 8D).
Figure 8.
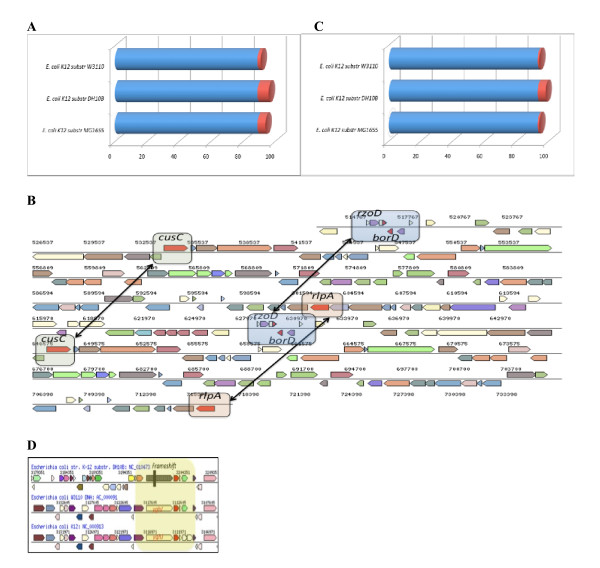
Using CoBaltDB in comparative proteomics. Example of E. coli K12 substrains lipoproteomes.
4-Using CoBaltDB to improve the classification of orthologous and paralogous proteins
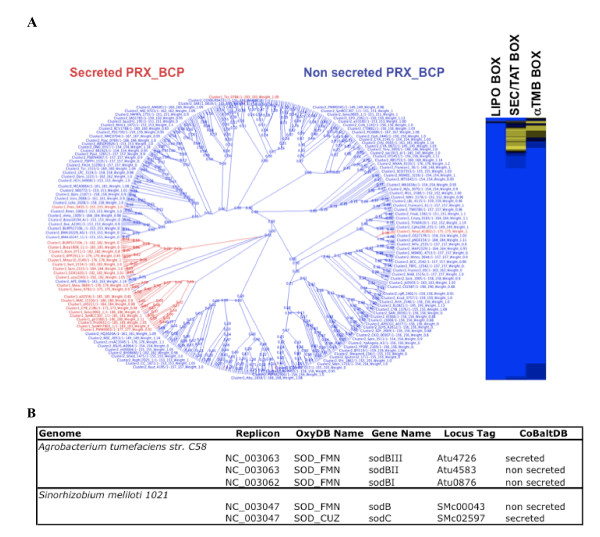
Protein function is generally related to its subcellular compartment, so orthologous proteins are expected, in most cases, to be in the same subcellular location. Consequently, inconsistencies of location predictions between orthologs potentially indicate distinct functional subclasses. Thus, CobaltDB can be used to help improve the functional annotation of orthologous proteins by adding the subcellular localization dimension. As an example, OxyGene, an anchor-based database of the ROS-RNS (Reactive Oxygen-Nitrogen species) detoxification subsystems for 664 complete bacterial and archaeal genomes, includes 37 detoxicifation enzyme subclasses [102]. Analysis of CoBaltDB subcellular localization information suggested the existence of additional subclasses. For example, 1-cystein peroxiredoxin, PRX_BCPs (bacterioferritin comigratory protein homologs), can be sub-divided into two new subclasses by distinguishing the secreted from the non-secreted forms (Figure 9a). Differences in the location between orthologous proteins are suggestive of functional diversity, and this is important for predictions of phenotype from the genotype.
Figure 9.
Using CoBalt for the analysis of orthologous and paralogous proteins. A: Phylogenetic tree of 1-cystein peroxiredoxin PRX_BCP proteins and heat map of scores in each box for each PRX_BCP protein. B: OxyGene and CoBalt predictions for SOD in Agrobacterium tumefacins str. C58 and Sinorhizobium meliloti 1021.
CoBaltDB is a very useful tool for the comparison of paralogous proteins. For example, quantitative and qualitative analysis of superoxide anion detoxification subsystems using the OxyGene platform identified three iron-manganese Superoxide dismutase (SOD_FMN) in Agrobacterium tumefaciens but only one SOD_FMN and one copper-zinc SOD (SOD_CUZ) in Sinorhizobium meliloti. The number of paralogs and the class of orthologs thus differ between these two closely related genus. However, adding the subcellular localization dimension reveals that both species have machinery to detoxify superoxide anions in both the periplasm and cytoplasm: both one of the three SOD_FMN of A. tumefaciens and the SOD_CUZ of S. meliloti are secreted (Figure 9b). CoBaltDB thus helps explain the difference suggested by OxyGene with respect to the ability of the two species to detoxify superoxide.
Discussion
CobaltDB allows biologists to improve their prediction of the subcellular localization of a protein by letting them compare the results of tools based on different methods and bringing complementary information. To facilitate the correct interpretation of the results, biologists have to keep in mind the limitations of the tools especially regarding the methodological strategies employed and the training sets used [93]. For example, most specialized tools tend to detect the presence of N-terminal signal peptides and predict cleavage sites. However the absence of an N-terminal signal peptide does not systematically indicate that the protein is not secreted. Some proteins that are translocated via the Sec system might not necessarily exhibit an N-terminal signal peptide, such as the SodA protein of M. tuberculosis, which is dependent on SecA2 for secretion and lacks a classical signal sequence for protein export [103]. Furthermore, there is no systematic cleavage of the N-terminal signal peptide as it can serve as a cytoplasmic membrane anchor [104,105]. Another example: although type II and type V secretion systems generally require the presence of an N-terminal signal peptide in order to utilise the sec pathway for translocation from cytoplasm to periplasm, type I and type III (and usually also type IV) systems can secrete a protein without any such signal [28,106]. Other proteins, such as Yop proteins exported by the Yersinia TTS system, have no classical sec-dependent signal sequences; however the information required to direct these proteins into the TTS pathway is contained within the N-terminal coding region of each gene [107-109].
Some challenges still need to be addressed in the prediction of the subcellular localization of proteins. For instance, bioinformatics has recently focussed on predicting proteins secreted via other pathways [110,111].
Conclusion
We have developed CoBaltDB, the first friendly interfaced database that compiles a large number of in silico subcellular predictions concerning whole bacterial and archaeal proteomes. Currently, CoBaltDB allows fast access to precomputed localizations for 2,548,292 proteins in 784 proteomes. It allows combined management of the predictions of 75 feature tools and 24 global tools and databases. New specialised prediction tools, algorithms and methods are continuously released, so CoBaltDB was designed to have the flexibility to facilitate inclusion of new tools or databases as required.
In general, our analysis indicates that both feature-based and general localization tools and databases have perform diversely in terms of specificity and sensitivity; the diversity arises mainly from the different sets of proteins used during the training process and from the limitations of the mathematical and statistical methodologies applied. In all our analyses with CoBaltDB, it became clear that that the combination and comparative analysis of results of heterogeneous tools improved the computational predictions, and contributed to identifying the limitations of each tool. Therefore, CoBaltDB can serve as a reference resource to facilitate interpretation of results and to provide a benchmark for accurate and effective in silico predictions of the subcellular localization of proteins. We hope that it will make a significant contribution to the exploitation of in silico subcellular localization predictions as users can easily create small datasets and determine their own thresholds for each predicted feature (type I or II SPs for example) or proteome. This is very important, as constructing an exhaustive "experimentally validated protein location" dataset is a time-consuming process --including identifying and reading all relevant papers-- and as experimental findings about some subcellular locations are very limited.
Availability and requirements
Database name: CoBaltDB
Project home page:
http://www.umr6026.univ-rennes1.fr/english/home/research/basic/software/cobalten
Operating system(s): Platform independent
Programming languages: Java, Python and BioPython
CoBaltDB package, requirements and documentations are freely available at http://www.umr6026.univ-rennes1.fr/english/home/research/basic/software/cobalten
Abbreviations
(AA): Amino acid; (aTMB): alpha-transmembrane; (CU): chaperone-usher; (ENP): extracellular nucleation-precipitation; (EXP): experimentally valitaded; (FEA): flagella export apparatus; (FPE): fimbrilin-protein exporter; (GUI): Graphical User Interface; (HMM): Hidden Markov Model; (LIPO): lipoprotein; (NN): Neural Network; (PRX_BCPs): bacterioferritin comigratory protein homologs; (PSSM): Position Specific Scoring Matrix; (RE): regular expression; (ROS-RNS): Reactive Oxygen-Nitrogen species; (Sec): Sec apparatus; (SOAP): Simple Object Access Protocol; (SOD_CUZ): one copper-zinc superoxide dismutase; (SOD_FMN): iron-manganese superoxide dismutase; (SP): signal peptide; (SVM): support vector machine; (T1-7SS): Type (1-7) Secretion System; (Tat): Twin-arginine translocation; (TM): transmembrane; (Wss): WXG100 secretion system.
Authors' contributions
DG designed and implemented the CoBaltDB database and the pre-computing pipeline for automated data retrieval. SA and DG developed the user interface. CLM and FBH tested the database for functionality, and performed bioinformatics analyses leading to valuable suggestions on utility and design. CLM and SA helped coordinate the study. FBH conceived and managed the project. All authors participated in CoBaltDB design, contributed to workflow and interface designs and helped write the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of precomputed genomes (Excel). A table of all complete procaryotic genomes and corresponding replicons available in CoBaltDB.
Procaryotic subcellular localisation tools (HTML). This page is an inventory of all tools considered during the construction of CoBaltDB. The tools and databases related to the protein localization in procaryotic genomes are sorted by type of prediction. For each tool, a short description and the corresponding web link are displayed.
Monoderm and Diderm classification of genomes (PNG). Picture showing the cellular organization type (monoderm or diderm) for phylum in CoBaltDB.
Using CoBalt in comparative proteomics (PDF). Example of the lipoproteomes of E. coli K12 substrains, experimentally confirmed by EcoGene. Table1A: Prediction results for the 89 confirmed lipoproteins in the three substrains DH10B, MG1655 et W3110. Table1B: The lipoproteins that are not recognized by DOLOP have a sequence which does not match the DOLOP lipoBox pattern [LVI] [ASTVI] [ASG] [C].
Contributor Information
David Goudenège, Email: david.goudenege@univ-rennes1.fr.
Stéphane Avner, Email: stephane.avner@univ-rennes1.fr.
Céline Lucchetti-Miganeh, Email: celine.lucchetti@univ-rennes1.fr.
Frédérique Barloy-Hubler, Email: fhubler@univ-rennes1.fr.
Acknowledgements
DG is supported by the Ministère de la Recherche. We wish to thank the bioinformatics platform of Biogenouest of Rennes for providing the hosting infrastructure.
References
- Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y. Automatic prediction of protein function. Cell Mol Life Sci. 2003;60(12):2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Hegyi H, Farkas K, Tordai H, Kozma E, Banyai L, Patthy L. Identification and correction of abnormal, incomplete and mispredicted proteins in public databases. BMC bioinformatics. 2008;9:353. doi: 10.1186/1471-2105-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends in microbiology. 2009;17(4):139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- De-la-Pena C, Lei Z, Watson BS, Sumner LW, Vivanco JM. Root-microbe communication through protein secretion. The Journal of biological chemistry. 2008;283(37):25247–25255. doi: 10.1074/jbc.M801967200. [DOI] [PubMed] [Google Scholar]
- Steward O, Pollack A, Rao A. Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: time course of appearance of recently synthesized proteins in synaptic junctions. Journal of neuroscience research. 1991;30(4):649–660. doi: 10.1002/jnr.490300408. [DOI] [PubMed] [Google Scholar]
- Russo DM, Williams A, Edwards A, Posadas DM, Finnie C, Dankert M, Downie JA, Zorreguieta A. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. Journal of bacteriology. 2006;188(12):4474–4486. doi: 10.1128/JB.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhu Z, Jing H, Zhang J, Xiong Y, Yan M, Gao S, Wu LF, Xu J, Kan B. Pleiotropic effects of the twin-arginine translocation system on biofilm formation, colonization, and virulence in Vibrio cholerae. BMC microbiology. 2009;9:114. doi: 10.1186/1471-2180-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck E, Anne J, Lammertyn E. The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology (Reading, England) 2007;153(Pt 12):3948–3953. doi: 10.1099/mic.0.2007/012039-0. [DOI] [PubMed] [Google Scholar]
- Poueymiro M, Genin S. Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Current opinion in microbiology. 2009;12(1):44–52. doi: 10.1016/j.mib.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Shrivastava R, Miller JF. Virulence factor secretion and translocation by Bordetella species. Current opinion in microbiology. 2009;12(1):88–93. doi: 10.1016/j.mib.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochimica et biophysica acta. 2008;1778(9):1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nature reviews. 2007;5(11):839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Muller M. Twin-arginine-specific protein export in Escherichia coli. Research in microbiology. 2005;156(2):131–136. doi: 10.1016/j.resmic.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annual review of microbiology. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Szabo Z, Driessen AJ. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nature reviews. 2006;4(7):537–547. doi: 10.1038/nrmicro1440. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Parham NJ, Scott-Tucker A, Henderson IR. The general secretory pathway: a general misnomer? Trends in microbiology. 2004;12(7):306–309. doi: 10.1016/j.tim.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Delepelaire P. Type I secretion in gram-negative bacteria. Biochimica et biophysica acta. 2004;1694(1-3):149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Molecular membrane biology. 2005;22(1-2):29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444(7119):567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68(4):771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Covacci A, Donati C. Protein homology network families reveal step-wise diversification of Type III and Type IV secretion systems. PLoS computational biology. 2006;2(12):e173. doi: 10.1371/journal.pcbi.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Current opinion in microbiology. 2009;12(1):11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology (Reading, England) 2008;154(Pt 6):1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M, Henderson IR, Pallen MJ. Type III secretion: what's in a name? Trends in microbiology. 2006;14(4):157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Coulthurst SJ, Palmer T. A new way out: protein localization on the bacterial cell surface via Tat and a novel Type II secretion system. Molecular microbiology. 2008;69(6):1331–1335. doi: 10.1111/j.1365-2958.2008.06367.x. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends in microbiology. 2005;13(12):581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Cornelis GR. The type III secretion system tip complex and translocon. Molecular microbiology. 2008;68(5):1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68(4):692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Parham NJ, Henderson IR. Type V protein secretion: simplicity gone awry? Current issues in molecular biology. 2004;6(2):111–124. [PubMed] [Google Scholar]
- Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71(4):551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochimica et biophysica acta. 2004;1694(1-3):259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. Mechanisms of protein export across the bacterial outer membrane. Journal of bacteriology. 2005;187(13):4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, Houben EN, Luirink J, Appelmelk BJ. Type VII secretion in mycobacteria: classification in line with cell envelope structure. Trends in microbiology. 2009;17(8):337–338. doi: 10.1016/j.tim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Khan A, Scott-Tucker A, Chaudhuri RR, Pallen MJ, Henderson IR. Genomic analysis of the protein secretion systems in Clostridium acetobutylicum ATCC 824. Biochimica et biophysica acta. 2005;1745(2):223–253. doi: 10.1016/j.bbamcr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology (Reading, England) 2003;149(Pt 11):3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Hughes KT. How and when are substrates selected for type III secretion? Trends in microbiology. 2001;9(5):209–214. doi: 10.1016/S0966-842X(01)02014-5. [DOI] [PubMed] [Google Scholar]
- Pallen MJ. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends in microbiology. 2002;10(5):209–212. doi: 10.1016/S0966-842X(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M, Talon R, Henderson IR. Outer membrane translocation: numerical protein secretion nomenclature in question in mycobacteria. Trends in microbiology. 2009;17(8):338–340. doi: 10.1016/j.tim.2009.05.008. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. European journal of biochemistry/FEBS. 1983;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic acids research. 1986;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ. On the predictive recognition of signal peptide sequences. Virus research. 1985;3(3):271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- Ladunga I, Czako F, Csabai I, Geszti T. Improving signal peptide prediction accuracy by simulated neural network. Comput Appl Biosci. 1991;7(4):485–487. doi: 10.1093/bioinformatics/7.4.485. [DOI] [PubMed] [Google Scholar]
- Schneider G, Rohlk S, Wrede P. Analysis of cleavage-site patterns in protein precursor sequences with a perceptron-type neural network. Biochemical and biophysical research communications. 1993;194(2):951–959. doi: 10.1006/bbrc.1993.1913. [DOI] [PubMed] [Google Scholar]
- Plewczynski D, Slabinski L, Ginalski K, Rychlewski L. Prediction of signal peptides in protein sequences by neural networks. Acta biochimica Polonica. 2008;55(2):261–267. [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proceedings/International Conference on Intelligent Systems for Molecular Biology; ISMB. 1998;6:122–130. [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of molecular biology. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007. pp. W429–432. [DOI] [PMC free article] [PubMed]
- Zhang Z, Henzel WJ. Signal peptide prediction based on analysis of experimentally verified cleavage sites. Protein Sci. 2004;13(10):2819–2824. doi: 10.1110/ps.04682504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC. A common export pathway for proteins binding complex redox cofactors? Molecular microbiology. 1996;22(3):393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- Rose RW, Bruser T, Kissinger JC, Pohlschroder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Molecular microbiology. 2002;45(4):943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein engineering. 1989;2(7):531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- Sankaran K, Gan K, Rash B, Qi HY, Wu HC, Rick PD. Roles of histidine-103 and tyrosine-235 in the function of the prolipoprotein diacylglyceryl transferase of Escherichia coli. Journal of bacteriology. 1997;179(9):2944–2948. doi: 10.1128/jb.179.9.2944-2948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berven FS, Karlsen OA, Straume AH, Flikka K, Murrell JC, Fjellbirkeland A, Lillehaug JR, Eidhammer I, Jensen HB. Analysing the outer membrane subproteome of Methylococcus capsulatus (Bath) using proteomics and novel biocomputing tools. Archives of microbiology. 2006;184(6):362–377. doi: 10.1007/s00203-005-0055-7. [DOI] [PubMed] [Google Scholar]
- Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. Journal of bacteriology. 2006;188(8):2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J Proteome Res. 2008;7(12):5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochimica et biophysica acta. 1985;815(3):468–476. doi: 10.1016/0005-2736(85)90375-X. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10(6):685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics (Oxford, England) 1998;14(4):378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S, Hristova K, White SH. Energetics, stability, and prediction of transmembrane helices. Journal of molecular biology. 2001;312(5):927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- Ganapathiraju M, Jursa CJ, Karimi HA, Klein-Seetharaman J. TMpro web server and web service: transmembrane helix prediction through amino acid property analysis. Bioinformatics. 2007;23(20):2795–2796. doi: 10.1093/bioinformatics/btm398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deber CM, Wang C, Liu LP, Prior AS, Agrawal S, Muskat BL, Cuticchia AJ. TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 2001;10(1):212–219. doi: 10.1110/ps.30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33(10):3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P. Prediction of membrane protein topology utilizing multiple sequence alignments. Journal of protein chemistry. 1997;16(5):453–457. doi: 10.1023/A:1026353225758. [DOI] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5(8):1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Cedano J, Oliva B, Aviles FX, Querol E. 'TransMem': a neural network implemented in Excel spreadsheets for predicting transmembrane domains of proteins. Comput Appl Biosci. 1997;13(3):231–234. doi: 10.1093/bioinformatics/13.3.231. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17(9):849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Viklund H, Elofsson A. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 2004;13(7):1908–1917. doi: 10.1110/ps.04625404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Mattick JS, Teasdale RD. SVMtm: support vector machines to predict transmembrane segments. Journal of computational chemistry. 2004;25(5):632–636. doi: 10.1002/jcc.10411. [DOI] [PubMed] [Google Scholar]
- Garrow AG, Agnew A, Westhead DR. TMB-Hunt: an amino acid composition based method to screen proteomes for beta-barrel transmembrane proteins. BMC bioinformatics. 2005;6:56. doi: 10.1186/1471-2105-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrow AG, Westhead DR. A consensus algorithm to screen genomes for novel families of transmembrane beta barrel proteins. Proteins. 2007;69(1):8–18. doi: 10.1002/prot.21439. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Hamodrakas SJ. Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method. BMC bioinformatics. 2005;6:7. doi: 10.1186/1471-2105-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli PL, Fariselli P, Krogh A, Casadio R. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics (Oxford, England) 2002;18(Suppl 1):S46–53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic acids research. 2004;32(8):2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall A, Cheng J, Sweredoski M, Baldi P. TMBpro: secondary structure, beta-contact and tertiary structure prediction of transmembrane beta-barrel proteins. Bioinformatics (Oxford, England) 2008;24(4):513–520. doi: 10.1093/bioinformatics/btm548. [DOI] [PubMed] [Google Scholar]
- Bigelow H, Rost B. PROFtmb: a web server for predicting bacterial transmembrane beta barrel proteins. Nucleic acids research. 2006. pp. W186–188. [DOI] [PMC free article] [PubMed]
- Hu J, Yan C. A method for discovering transmembrane beta-barrel proteins in Gram-negative bacterial proteomes. Computational biology and chemistry. 2008;32(4):298–301. doi: 10.1016/j.compbiolchem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Waldispuhl J, Berger B, Clote P, Steyaert JM. transFold: a web server for predicting the structure and residue contacts of transmembrane beta-barrels. Nucleic acids research. 2006. pp. W189–193. [DOI] [PMC free article] [PubMed]
- Zhai Y, Saier MH Jr. The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci. 2002;11(9):2196–2207. doi: 10.1110/ps.0209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004. pp. W394–399. [DOI] [PMC free article] [PubMed]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004. pp. W400–404. [DOI] [PMC free article] [PubMed]
- Park KJ, Gromiha MM, Horton P, Suwa M. Discrimination of outer membrane proteins using support vector machines. Bioinformatics. 2005;21(23):4223–4229. doi: 10.1093/bioinformatics/bti697. [DOI] [PubMed] [Google Scholar]
- Ou YY, Gromiha MM, Chen SA, Suwa M. TMBETADISC-RBF: Discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Computational biology and chemistry. 2008;32(3):227–231. doi: 10.1016/j.compbiolchem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Billion A, Ghai R, Chakraborty T, Hain T. Augur--a computational pipeline for whole genome microbial surface protein prediction and classification. Bioinformatics. 2006;22(22):2819–2820. doi: 10.1093/bioinformatics/btl466. [DOI] [PubMed] [Google Scholar]
- Zhou M, Boekhorst J, Francke C, Siezen RJ. LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC bioinformatics. 2008;9:173. doi: 10.1186/1471-2105-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH, Tan TW, Ranganathan S. SPdb--a signal peptide database. BMC bioinformatics. 2005;6:249. doi: 10.1186/1471-2105-6-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FS. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005. pp. D164–168. [DOI] [PMC free article] [PubMed]
- Park S, Yang JS, Jang SK, Kim S. Construction of Functional Interaction Networks through Consensus Localization Predictions of the Human Proteome. J Proteome Res. 2009;8(7):3367–3376. doi: 10.1021/pr900018z. [DOI] [PubMed] [Google Scholar]
- Restrepo-Montoya D, Vizcaino C, Nino LF, Ocampo M, Patarroyo ME, Patarroyo MA. Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinformatics. 2009;10:134. doi: 10.1186/1471-2105-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YQ, Burger G. 'Unite and conquer': enhanced prediction of protein subcellular localization by integrating multiple specialized tools. BMC Bioinformatics. 2007;8:420. doi: 10.1186/1471-2105-8-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. The natural evolutionary relationships among prokaryotes. Critical reviews in microbiology. 2000;26(2):111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- Rachel R, Wyschkony I, Riehl S, Huber H. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea (Vancouver, BC) 2002;1(1):9–18. doi: 10.1155/2002/307480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28(1):60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Okayama T, Hashimoto H, Takeda J, Davis RW, Mori H, Gojobori T. A low rate of nucleotide changes in Escherichia coli K-12 estimated from a comparison of the genome sequences between two different substrains. FEBS letters. 1999;450(1-2):72–76. doi: 10.1016/S0014-5793(99)00481-0. [DOI] [PubMed] [Google Scholar]
- Durfee T, Nelson R, Baldwin S, Plunkett G, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190(7):2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Mekalanos JJ. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infection and immunity. 1988;56(11):2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. The Journal of biological chemistry. 2004;279(38):39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- Thybert D, Avner S, Lucchetti-Miganeh C, Cheron A, Barloy-Hubler F. OxyGene: an innovative platform for investigating oxidative-response genes in whole prokaryotic genomes. BMC genomics. 2008;9:637. doi: 10.1186/1471-2164-9-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR Jr. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Molecular microbiology. 2003;48(2):453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- Goder V, Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS letters. 2001;504(3):87–93. doi: 10.1016/S0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends in cell biology. 1998;8(10):410–415. doi: 10.1016/S0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Current opinion in microbiology. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science (New York, NY) 1997;278(5340):1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Schneewind O. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Molecular microbiology. 1999;31(4):1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G. Secretion of Yop proteins by Yersiniae. Infection and immunity. 1990;58(9):2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower M, Schneider G. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One. 2009;4(6):e5917. doi: 10.1371/journal.pone.0005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes HW, Horn M, Rattei T. Sequence-based prediction of type III secreted proteins. PLoS pathogens. 2009;5(4):e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K, Grote A, Scheer M, Munch R, Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004. pp. W375–379. [DOI] [PMC free article] [PubMed]
- Gomi M, Sonoyama M, Mitaku S. High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio Informatics Journal. 2004;4(4):142–147. doi: 10.1273/cbij.4.142. [DOI] [Google Scholar]
- Mitaku S, Hirokawa T, Tsuji T. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics. 2002;18(4):608–616. doi: 10.1093/bioinformatics/18.4.608. [DOI] [PubMed] [Google Scholar]
- Juretic D, Zoranic L, Zucic D. Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci. 2002;42(3):620–632. doi: 10.1021/ci010263s. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Hamodrakas SJ. Finding beta-barrel outer membrane proteins with a Markov Chain Model. WSEAS Transactions on Biology and Biomedecine. 2004;1(2):186–189. [Google Scholar]
- Gromiha MM, Ahmad S, Suwa M. TMBETA-NET: discrimination and prediction of membrane spanning beta-strands in outer membrane proteins. Nucleic Acids Res. 2005. pp. W164–167. [DOI] [PMC free article] [PubMed]
- Garrow AG, Agnew A, Westhead DR. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005. pp. W188–192. [DOI] [PMC free article] [PubMed]
- Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20(4):547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Vert JP, Saigo H, Ueda N, Toh H, Akutsu T. A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci. 2005;14(11):2804–2813. doi: 10.1110/ps.051597405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001;17(8):721–728. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- Niu B, Jin YH, Feng KY, Lu WC, Cai YD, Li GZ. Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins. Molecular diversity. 2008;12(1):41–45. doi: 10.1007/s11030-008-9073-0. [DOI] [PubMed] [Google Scholar]
- Imai K, Asakawa N, Tsuji T, Akazawa F, Ino A, Sonoyama M, Mitaku S. SOSUI-GramN: high performance prediction for sub-cellular localization of proteins in Gram-negative bacteria. Bioinformation. 2008;2(9):417–421. doi: 10.6026/97320630002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horler RS, Butcher A, Papangelopoulos N, Ashton PD, Thomas GH. EchoLOCATION: an in silico analysis of the subcellular locations of Escherichia coli proteins and comparison with experimentally derived locations. Bioinformatics. 2009;25(2):163–166. doi: 10.1093/bioinformatics/btn596. [DOI] [PubMed] [Google Scholar]
- Fernando SA, Selvarani P, Das S, Kumar Ch K, Mondal S, Ramakumar S, Sekar K. THGS: a web-based database of Transmembrane Helices in Genome Sequences. Nucleic Acids Res. 2004. pp. D125–128. [DOI] [PMC free article] [PubMed]
- Litou ZI, Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of cell wall sorting signals in gram-positive bacteria with a hidden markov model: application to complete genomes. Journal of bioinformatics and computational biology. 2008;6(2):387–401. doi: 10.1142/S0219720008003382. [DOI] [PubMed] [Google Scholar]
- Remmert M, Linke D, Lupas AN, Soding J. HHomp--prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009. pp. W446–451. [DOI] [PMC free article] [PubMed]
- Saleh MT, Fillon M, Brennan PJ, Belisle JT. Identification of putative exported/secreted proteins in prokaryotic proteomes. Gene. 2001;269(1-2):195–204. doi: 10.1016/S0378-1119(01)00436-X. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. Prediction of signal peptides in archaea. Protein Eng Des Sel. 2009;22(1):27–35. doi: 10.1093/protein/gzn064. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Arai M, Okuno T, Shimizu T. TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res. 2003;31(1):406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Kalmar L, Simon I. TOPDB: topology data bank of transmembrane proteins. Nucleic Acids Res. 2008. pp. D234–239. [DOI] [PMC free article] [PubMed]
- Menne KM, Hermjakob H, Apweiler R. A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics. 2000;16(8):741–742. doi: 10.1093/bioinformatics/16.8.741. [DOI] [PubMed] [Google Scholar]
- Taylor PD, Toseland CP, Attwood TK, Flower DR. LIPPRED: A web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation. 2006;1(5):176–179. doi: 10.6026/97320630001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariselli P, Finocchiaro G, Casadio R. SPEPlip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics. 2003;19(18):2498–2499. doi: 10.1093/bioinformatics/btg360. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Signal-3L: A 3-layer approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;363(2):297–303. doi: 10.1016/j.bbrc.2007.08.140. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;357(3):633–640. doi: 10.1016/j.bbrc.2007.03.162. [DOI] [PubMed] [Google Scholar]
- Frank K, Sippl MJ. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics. 2008;24(19):2172–2176. doi: 10.1093/bioinformatics/btn422. [DOI] [PubMed] [Google Scholar]
- Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschroder M. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol. 2007;189(3):772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiss JA, Resch E, Schreiner A, Meissner M, Starzinski-Powitz A, Schneider G. Domain organization of long signal peptides of single-pass integral membrane proteins reveals multiple functional capacity. PLoS One. 2008;3(7):e2767. doi: 10.1371/journal.pone.0002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SM, Kall L, Riffle ME, Bilmes JA, Noble WS. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol. 2008;4(11):e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H, Elofsson A. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 2008;24(15):1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- Viklund H, Bernsel A, Skwark M, Elofsson A. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24(24):2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- Shen H, Chou JJ. MemBrain: improving the accuracy of predicting transmembrane helices. PLoS One. 2008;3(6):e2399. doi: 10.1371/journal.pone.0002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10(6):673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Hamodrakas SJ. Algorithms for incorporating prior topological information in HMMs: application to transmembrane proteins. BMC Bioinformatics. 2006;7:189. doi: 10.1186/1471-2105-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A, Chiu HS, Sung TY, Lyu PC, Hsu WL. Enhanced membrane protein topology prediction using a hierarchical classification method and a new scoring function. J Proteome Res. 2008;7(2):487–496. doi: 10.1021/pr0702058. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. Predicting the topology of transmembrane helical proteins using mean burial propensity and a hidden-Markov-model-based method. Protein Sci. 2003;12(7):1547–1555. doi: 10.1110/ps.0305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashou EE, Litou ZI, Liakopoulos TD, Hamodrakas SJ. waveTM: wavelet-based transmembrane segment prediction. Silico Biol. 2004;4(2):127–131. [PubMed] [Google Scholar]
- Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS, Hamodrakas SJ. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 1999;12(5):381–385. doi: 10.1093/protein/12.5.381. [DOI] [PubMed] [Google Scholar]
- Peris P, Lopez D, Campos M. IgTM: an algorithm to predict transmembrane domains and topology in proteins. BMC Bioinformatics. 2008;9:367. doi: 10.1186/1471-2105-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009. pp. W465–468. [DOI] [PMC free article] [PubMed]
- Zhou H, Zhang C, Liu S, Zhou Y. Web-based toolkits for topology prediction of transmembrane helical proteins, fold recognition, structure and binding scoring, folding-kinetics analysis and comparative analysis of domain combinations. Nucleic Acids Res. 2005. pp. W193–197. [DOI] [PMC free article] [PubMed]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004. pp. W390–393. [DOI] [PMC free article] [PubMed]
- Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23(5):538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59(3):467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- Ganapathiraju M, Balakrishnan N, Reddy R, Klein-Seetharaman J. Transmembrane helix prediction using amino acid property features and latent semantic analysis. BMC Bioinformatics. 2008;9(Suppl 1):S4. doi: 10.1186/1471-2105-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005. pp. W36–38. [DOI] [PMC free article] [PubMed]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25(3):147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Karplus K. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 2009. pp. W492–497. [DOI] [PMC free article] [PubMed]
- Pollastri G, McLysaght A. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics. 2005;21(8):1719–1720. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- Kahsay RY, Gao G, Liao L. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics. 2005;21(9):1853–1858. doi: 10.1093/bioinformatics/bti303. [DOI] [PubMed] [Google Scholar]
- Lin K, Simossis VA, Taylor WR, Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics. 2005;21(2):152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. MemType-2L: a web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biochem Biophys Res Commun. 2007;360(2):339–345. doi: 10.1016/j.bbrc.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64(3):643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Su EC, Chiu HS, Lo A, Hwang JK, Sung TY, Hsu WL. Protein subcellular localization prediction based on compartment-specific features and structure conservation. BMC Bioinformatics. 2007;8:330. doi: 10.1186/1471-2105-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin M, Garg A, Raghava GP. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 2005;21(10):2522–2524. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Large-scale predictions of gram-negative bacterial protein subcellular locations. J Proteome Res. 2006;5(12):3420–3428. doi: 10.1021/pr060404b. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Gpos-PLoc: an ensemble classifier for predicting subcellular localization of Gram-positive bacterial proteins. Protein Eng Des Sel. 2007;20(1):39–46. doi: 10.1093/protein/gzl053. [DOI] [PubMed] [Google Scholar]
- Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol. 2005;348(1):85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Jia P, Qian Z, Zeng Z, Cai Y, Li Y. Prediction of subcellular protein localization based on functional domain composition. Biochem Biophys Res Commun. 2007;357(2):366–370. doi: 10.1016/j.bbrc.2007.03.139. [DOI] [PubMed] [Google Scholar]
- Rashid M, Saha S, Raghava GP. Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinformatics. 2007;8:337. doi: 10.1186/1471-2105-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Reis M, Matsunaga J, Haake DA. Lipoprotein computational prediction in spirochaetal genomes. Microbiology. 2006;152(Pt 1):113–121. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomerie S, Cruz JA, Shrivastava S, Arndt D, Berjanskii M, Wishart DS. PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res. 2008. pp. W202–209. [DOI] [PMC free article] [PubMed]
- Pasquier C, Hamodrakas SJ. An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng. 1999;12(8):631–634. doi: 10.1093/protein/12.8.631. [DOI] [PubMed] [Google Scholar]
- Taylor PD, Attwood TK, Flower DR. BPROMPT: A consensus server for membrane protein prediction. Nucleic Acids Res. 2003;31(13):3698–3700. doi: 10.1093/nar/gkg554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos TD, Pasquier C, Hamodrakas SJ. A novel tool for the prediction of transmembrane protein topology based on a statistical analysis of the SwissProt database: the OrienTM algorithm. Protein Eng. 2001;14(6):387–390. doi: 10.1093/protein/14.6.387. [DOI] [PubMed] [Google Scholar]
- Raghava GP. APSSP2: A combination method for protein secondary structure prediction based on neural network and example based learning. CASP5. 2002;A-132 [Google Scholar]
- Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005. pp. W289–294. [DOI] [PMC free article] [PubMed]
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S, Hristova K, White SH. MPtopo: A database of membrane protein topology. Protein Sci. 2001;10(2):455–458. doi: 10.1110/ps.43501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005. pp. D275–278. [DOI] [PMC free article] [PubMed]
- Gromiha MM, Yabuki Y, Kundu S, Suharnan S, Suwa M. TMBETA-GENOME: database for annotated beta-barrel membrane proteins in genomic sequences. Nucleic Acids Res. 2007. pp. D314–316. [DOI] [PMC free article] [PubMed]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004. pp. W321–326. [DOI] [PMC free article] [PubMed]
- Yun H, Lee JW, Jeong J, Chung J, Park JM, Myoung HN, Lee SY. EcoProDB: the Escherichia coli protein database. Bioinformatics. 2007;23(18):2501–2503. doi: 10.1093/bioinformatics/btm351. [DOI] [PubMed] [Google Scholar]
- Nair R, Rost B. LOCnet and LOCtarget: sub-cellular localization for structural genomics targets. Nucleic Acids Res. 2004. pp. W517–521. [DOI] [PMC free article] [PubMed]
- Zhang S, Xia X, Shen J, Zhou Y, Sun Z. DBMLoc: a Database of proteins with multiple subcellular localizations. BMC Bioinformatics. 2008;9:127. doi: 10.1186/1471-2105-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of precomputed genomes (Excel). A table of all complete procaryotic genomes and corresponding replicons available in CoBaltDB.
Procaryotic subcellular localisation tools (HTML). This page is an inventory of all tools considered during the construction of CoBaltDB. The tools and databases related to the protein localization in procaryotic genomes are sorted by type of prediction. For each tool, a short description and the corresponding web link are displayed.
Monoderm and Diderm classification of genomes (PNG). Picture showing the cellular organization type (monoderm or diderm) for phylum in CoBaltDB.
Using CoBalt in comparative proteomics (PDF). Example of the lipoproteomes of E. coli K12 substrains, experimentally confirmed by EcoGene. Table1A: Prediction results for the 89 confirmed lipoproteins in the three substrains DH10B, MG1655 et W3110. Table1B: The lipoproteins that are not recognized by DOLOP have a sequence which does not match the DOLOP lipoBox pattern [LVI] [ASTVI] [ASG] [C].