Abstract
Autosomal-dominant woolly hair (ADWH) is a rare disorder characterized by tightly curled hair. The molecular basis of ADWH has not previously been reported. In this study, we identified a Pakistani family with ADWH. The family showed linkage to chromosome 12q12-q14.1, containing the type II keratin gene cluster. We discovered a heterozygous mutation, p.Asn148Lys, within the helix initiation motif of the keratin 74 (KRT74) gene in all affected family members. KRT74 encodes the inner root sheath (IRS)-specific epithelial (soft) keratin 74. We demonstrate that the mutant K74 protein results in disruption of keratin intermediate filament formation in cultured cells, most likely in a dominant-negative manner. Furthermore, we sequenced the mouse Krt71-74 genes in the dominant Caracul-like 4 (Cal4) allele, which is characterized by a wavy-coat phenotype and maps to the same region of mouse chromosome 15 as the Caracul (Ca) and Reduced coat (Rco) alleles. We identified a heterozygous mutation, p.Glu440Lys, not in Krt74 but in the neighboring gene, Krt71. Krt71 was previously reported to harbor Ca and Rco mutations, as well as a coding SNP that is associated with curly-coated dogs. In this study, we define the ADWH phenotype resulting from a mutation in a hair-follicle-specific epithelial keratin in humans. Our findings not only further underscore the crucial roles of the IRS-specific epithelial keratin genes Krt71-74 in hair disorders but also open the possibility that these genes might function as genetic determinants of normal variation in hair texture across mammalian species.
Main Text
The genetic determinants of hair texture in human populations are largely undefined. One approach to identify candidate genes is to analyze hereditary hair diseases that show hair-shaft anomalies, such as woolly hair (WH). WH refers to a phenotypic variant with fine and tightly curled hair.1 Distinct from the tightly curled hair typical of African populations, WH shows hair-shaft anomalies and is sometimes associated with sparse and/or depigmented hair.1–3 WH can be classified into syndromic and nonsyndromic forms.1 Nonsyndromic WH can be inherited as either an autosomal-dominant (ADWH [MIM 194300])3 or an autosomal-recessive (ARWH [MIM 278150])2,3 trait. We and others have recently reported that mutations in the LIPH (MIM 607365) and P2RY5/LPAR6 (MIM 609239) genes underlie ARWH and/or localized autosomal-recessive hypotrichosis (LAH [MIM 604379 and 611452]).4–7 The LIPH gene encodes a phospholipase A1 family member that produces 2-acyl lysophosphatidic acid (LPA),8 and the LPAR6 gene encodes a receptor of LPA.5,9 Because both LIPH and LPAR6 are expressed in the inner root sheath (IRS) of human hair follicles (HFs),6,7 they are postulated to function in a common signaling pathway and play a critical role in hair growth in humans.
To date, no gene has been implicated in the pathogenesis of ADWH. We undertook this study to identify a gene underlying ADWH and to better understand the genetic determinants of hair texture in humans.
We recently identified a Pakistani family (ADWH1) with features consistent with dominantly inherited WH. Multiple affected family members showed clinical features at birth. The hair over the entire scalp region is coarse, lusterless, dry, and tightly curled, leading to a diffuse WH phenotype with normal hair density (Figures 1A–1C). The hair grows slowly and stops growing at a few inches. Under light microscopy, plucked hairs from affected individuals show several anomalies, such as dystrophic anagen hairs (Figure 1D), twisting (Figure 1E), knot formation (Figure 1F), and tapered distal ends (Figure 1G). These features are consistent with abnormal scalp hair growth, whereas the eyebrows, eyelashes, and beard hairs appeared normal. Affected individuals had normal teeth, nails, and sweating and did not show palmoplantar hyperkeratosis or keratosis pilaris. There was no family history of heart disease, early sudden death, neurologic abnormalities, or a high prevalence of cancers.
Figure 1.
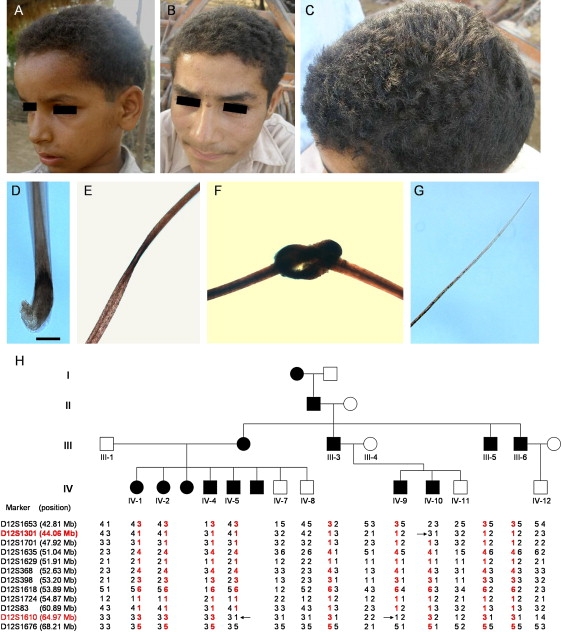
Fine Mapping of ADWH Phenotype on Chromosome 12q12-q14.1
(A–C) Clinical appearance of ADWH.
(D–G) Appearance of the hair shaft in affected individuals with ADWH. The scale bar represents 100 μm.
(H) Pedigree and haplotype analysis of a Pakistani family ADWH1. The linked haplotype is indicated in red. Critical recombination events are indicated by black arrowheads.
Informed consent was obtained from all subjects, and approval for this study was provided by the Institutional Review Board of Columbia University. The study was conducted in adherence to the Declaration of Helsinki Principles. Peripheral blood samples were collected from the family members as well as unrelated healthy control individuals of Pakistani origin. Genomic DNA was isolated from these samples with the PUREGENE DNA isolation kit (Gentra System). We initially performed genotyping by using human mapping arrays (Affymetrix 10K) on 11 members of the family. Parametric linkage analysis was performed under an autosomal-dominant model. A maximum LOD score Z = 1.56, suggestive of linkage, was identified on chromosome 12 (Figure S1A). Tests for allelic association implicated 12q13 (p = 0.005) in the region of the type II keratin gene cluster (Figure S1B). Microsatellite markers were then placed across the region, which reconfirmed linkage to the same location (Zmax = 1.57) (Figure S1C). Critical recombination events were detected between markers D12S1301 and D12S1701 in affected individual IV-10 (Figure 1H), as well as between markers D12S83 and D12S1610 in affected individuals IV-5 and IV-9 (Figure 1H). This allowed the linkage interval, flanked by markers D12S1301 and D12S1610, to be limited to 20.91 Mb. The type II keratin gene cluster resides in the candidate region, containing four inner root sheath (IRS)-specific keratin genes, KRT71-74 (MIM 608245, 608246, 608247, and 608248), previously known as K6irs1–4 (Figure 2A).10,11
Figure 2.
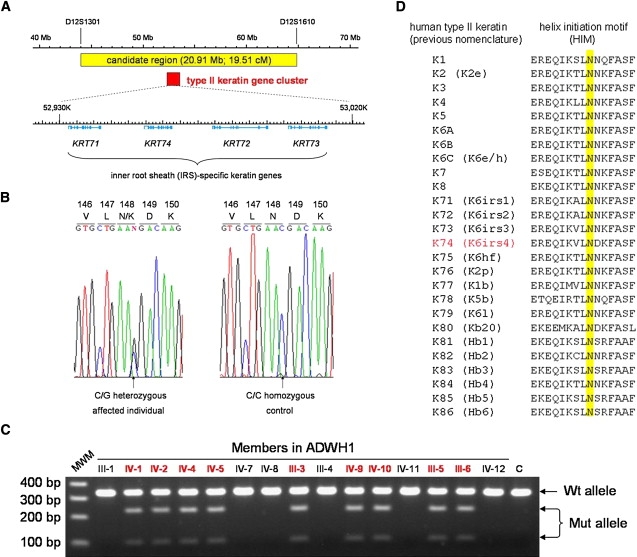
Identification of a Mutation in the KRT74 Gene
(A) Schematic representation of the candidate region harboring the ADWH gene. The candidate region and the type II keratin gene cluster are boxed in yellow and red, respectively, and localization of four IRS-specific keratin genes, KRT71–74, is shown at the bottom.
(B) A heterozygous mutation c.444C > G (p.Asn148Lys) in the KRT74 gene in affected individuals from the family ADWH1.
(C) Segregation analysis of the mutation c.444C > G in the KRT74 gene. PCR products of 338 bp from the mutant (Mut) allele were digested into 234 bp and 104 bp fragments with the restriction enzyme AcuI, whereas those from the wild-type (Wt) allele were not digested. Affected individuals are indicated in red and show the digested bands of 234 bp and 104 bp from the Mut allele as well as the undigested band of 338 bp from the Wt allele. MWM, molecular weight markers. C, control individual.
(D) Multiple-amino-acid sequence alignment of the helix initiation motif (HIM) of human type II keratins. K74 (K6irs4) is indicated in red. The Asn residue at amino acid position 9 in the HIM, which is invariant and completely conserved among all human type II keratins, is highlighted in yellow.
Several lines of evidence led us to postulate that a mutation might lie in KRT71-74. First, mutations in the Krt71 gene have previously been identified in Caracul (Ca)12 and Reduced coat (Rco)13,14 mutant mice, which show a wavy-coat phenotype. Second, it has recently been reported that a coding SNP in the KRT71 gene is strongly associated with a curly-coat phenotype in dogs.15 Third, both LPAR6 and LIPH, the causative genes for ARWH, are abundantly expressed in the same compartment (IRS) of human HFs.6,7 We performed direct sequencing analysis of the KRT71-74 genes (Table S1) and identified a heterozygous mutation, c.444C > G, in exon 1 of the KRT74 gene in all affected individuals of the family (Figure 2B). Screening assays with the restriction enzyme AcuI showed that the mutation completely cosegregated with the disease phenotype and was excluded from 200 population-matched, unrelated, unaffected individuals (400 chromosomes) (Figure 2C; data not shown).
Keratins are a major structural component of the HF and are largely divided into two groups: type I (acidic) and type II (basic to neutral) keratins. The type I and type II keratins form keratin intermediate filaments (KIFs) in the cytoplasm through heterodimerization.16,17 All keratin proteins share a common structural organization that is composed of the N-terminal head domain, the central α-helical rod domain, and the C-terminal tail domain. The N terminus and C terminus of the central α-helical rod domain are called the helix initiation motif (HIM) and helix termination motif (HTM), respectively, which are highly conserved and critical for heterodimerization between the type I and type II keratins.16 Keratins are further classified into hair (hard) and epithelial (soft) keratins. The epithelial keratins are widely expressed in epithelia of various tissues, whereas the hair keratins are predominantly expressed in highly keratinized tissues, such as the hair shaft and nails. The hair keratins differ from the epithelial keratins by their considerably higher sulfur content in the head and tail domains. This high sulfur content is believed to be important for the extraordinarily high degree of filamentous cross-linking with hair-keratin-associated proteins.17 The mutation c.444C > G resulted in a substitution from asparagine to lysine at amino acid position 148 of the keratin 74 (K74) protein (p.Asn148Lys) (Figure 2B), and this substitution occurred within helix initiation motif (HIM) of the K74 protein. Asn148 (amino acid position 9 in the HIM) is completely conserved among all human type II keratins (Figure 2D). Furthermore, Asn-to-Lys substitutions at amino acid position 9 in the HIM of other type II keratins underlie several autosomal-dominant skin diseases in humans (Table S2).18–30 Together, these data strongly suggest the mutation p.Asn148Lys in K74 is a pathogenic mutation underlying ADWH.
In order to determine the impact of p.Asn148Lys on the function of the K74 protein, we subcloned the coding sequences of wild-type (Wt) KRT74-cDNA into the mammalian expression vector pCXN2.131,32 with EcoRI and XhoI sites. The expression construct for the p.Asn148Lys mutant K74 (pCXN2.1-Mut-K74) was generated with the QuikChange II XL Site-Directed Mutagenesis Kit (QIAGEN) and the pCXN2.1-Wt-K74 construct as a template. We transfected either Wt-K74 or Mut-K74 into PtK2 (kangaroo rat kidney epithelial) and MCF-10A (human breast epithelial) cells expressing endogenous keratins such as K8/K18 and K5/K14, respectively. Double immunostainings with guinea pig polyclonal anti-K74 (diluted 1:2,000) and either mouse monoclonal anti-K18 (clone 2F8; 1:50; Sigma) or rabbit polyclonal anti-K14 (1:1,000; Covance) antibodies demonstrate that Wt-K74 protein is widely distributed throughout the cytoplasm, where it colocalizes with K18 in PtK2 cells (Figures 3A–3C) or K14 in MCF-10A cells (Figures S2A–S2C), suggesting that Wt-K74 forms a functional KIF cytoskeleton via a heterodimer with the endogenous K18 or K14 proteins. In striking contrast, Mut-K74 is mislocalized around the nucleus, where it appears collapsed and colocalizes with aggregated K18 or K14 proteins (Figures 3D–3F and Figures S2D–S2F), suggesting that the mutant K74 protein interferes with KIF formation, most likely in a dominant-negative manner.
Figure 3.
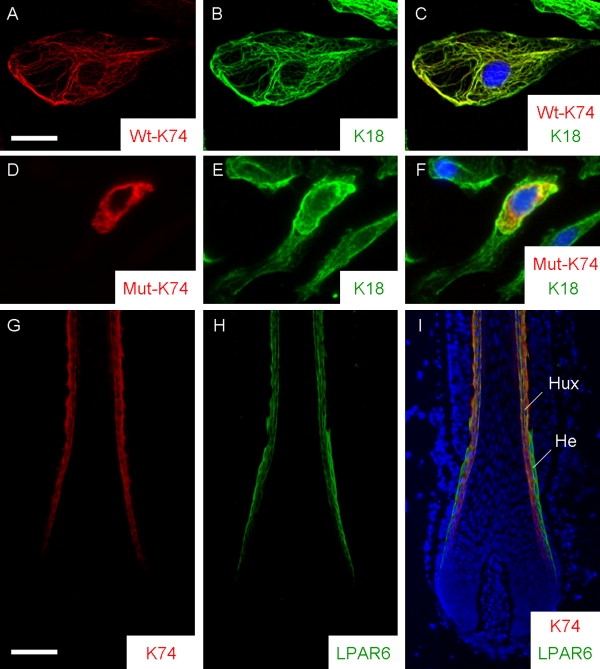
Mutant K74 Protein Disrupts Endogenous KIF Formation in PtK2 Cells, and K74 Colocalizes with LPAR6 in Huxley's Layer of Human Hair Follicles
(A–C) Ectopically expressed wild-type (Wt) K74 protein forms a KIF network via heterodimerization with endogenous K18 protein.
(D–F) The p.Asn148Lys mutant (Mut) K74 protein causes a collapse of the endogenous KIF network around the nucleus.
(G–I) Double immunostainings with guinea pig polyclonal anti-K74 (1:2,000) (G) and rabbit polyclonal anti-P2Y5 (LPAR6; 1:200; MBL International) (H) antibodies show that both K74 and LPAR6 proteins are coexpressed in Huxley's (Hux) layer of the IRS of human hair follicles from a normal control individual (I), whereas only LPAR6 is expressed in Henle's (He) layer of the IRS (I).
The right panels are merged images, and counterstaining with DAPI is shown in blue (C, F, and I). Scale bars represent 20 μm (A) and 100 μm (G).
To test whether mutations in the corresponding mouse Krt74 gene might underlie a new mutation, we sequenced mouse Krt71–74 genes in the Caracul-like 4 (Cal4) allele (Jackson Laboratory), which is characterized by a wavy-coat phenotype and maps to the same region of mouse chromosome 15 as Caracul (Cal)12 and Reduced coat (Rco)13,14 alleles. We identified a heterozygous mutation c.1318G > A (p.Glu440Lys) not in Krt74 but in the neighboring gene Krt71, as has been previously reported for Ca and Rco mutations (Figure S3),12–14 as well as SNPs in curly-coated dogs.15
It has recently been shown that SNPs in EDAR (MIM 604095)33 and trichohyalin (TCHH [MIM 190370])34 genes are associated with straight hair in Asian and European populations, respectively. In addition, we postulated that SNPs in the KRT71-74 genes might also be associated with a determinant of normal variations in hair texture in humans. We hypothesized that a comparison of two populations with extremely divergent distributions of hair texture would show marked differences in the allele frequencies of such SNPs. Therefore, we examined allele frequencies for SNPs across this gene cluster for an African (YRI) and a European ancestry (CEU) population from HapMap and found seven KRT71 gene SNPs that showed significant differences in allele frequencies between the two populations (Figure S4). For these seven SNPs, the reference allele frequency in YRI was 1 and ranged between 0.51 and 0.60 for CEU. Linkage disequilibrium patterns across these SNPs suggest that they share a common haplotype, and it is noteworthy that one of these, rs10783518, falls within a coding region and results in an amino acid change p.Gly464Val in the C terminus of the K71 protein, which could potentially affect KIF formation. Taken together with genetic evidence from model mouse and canine studies, these data highlight the importance of this gene cluster in hair texture across mammalian species.
A total of four type II epithelial keratins, K71–74, are differentially expressed in the IRS of human HFs. The IRS is well established as a critical structural element for supporting and molding the hair shaft.10,11 It consists of three distinct layers: IRS cuticle, Huxley's layer, and Henle's layer. The companion layer surrounds the Henle's layer. Although there are few desmosomes between the keratinized Henle's layer and the companion layer, some cells of Huxley's layer transect Henle's layer and attach directly to the companion layer, forming desmosomes.10 Therefore, Huxley's layer plays an important role in maintaining the structure of the HFs. K74 is specifically expressed in Huxley's layer of the IRS (Figure 3G).11 Disruption of K74 is predicted to result in a collapse of KIF formation in Huxley's layer and lead to perturbations of hair-shaft growth and elongation. Because expression of LIPH,7 LPAR6, and K74 overlaps in Huxley's layer (Figures 3G–3I), LIPH/LPAR6 signaling is most likely connected with KIF formation, as well as desmosomal assembly in the IRS.
Mutations in some hair keratins are known to cause hereditary hair diseases, such as monilethrix (MIM 158000).35 In addition, a coding SNP in the epithelial keratin 75 (K6hf [MIM 609025]), which is specifically expressed in the companion layer and the hair-shaft medulla, was previously associated with pseudofolliculitis barbae (PFB [MIM 612318]).36 PFB is a common condition in African populations and is characterized by ingrown hairs of the facial and submental regions. However, no pathogenic mutations have previously been found in HF-specific epithelial keratins in humans. To our knowledge, we show for the first time the phenotype that arises from a mutation in an HF-specific epithelial keratin. Because the typical hair texture in Pakistani populations is straight, Asian-type hair, we were able to easily identify ADWH. However, it might not be as obvious in populations with admixture of different hair textures. We postulate that some individuals in other populations who appear to have extremely curly hair may actually have WH and carry mutations in one of the IRS-specific keratins. Our findings indicate that disruption of K74 underlies autosomal-dominant woolly hair. More broadly, we postulate that the IRS-specific keratins KRT71-74 may be involved in the determination of hair texture in different human populations.
Acknowledgments
We are grateful to the family members for their participation in this study and to Helen Lam, Ming Zhang, Yuki Kobayashi, Claire Higgins, and Munenari Itoh (Columbia University) for technical assistance. We thank Dr. Lutz Langbein (German Cancer Research Center, Heidelberg, Germany) for generously sharing the K74 antibody and Noah A. Rosenberg (University of Michigan) and David Reich (Harvard Medical School) for helpful discussions. We thank Satoshi Ishii (Tokyo University, Tokyo, Japan) and Junichi Miyazaki (Osaka University, Osaka, Japan) for supplying the pCXN2.1 vector. This work was supported in part by United States Public Health Service National Institutes of Health grant R01AR44924 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (to A.M.C.). Y.S. is supported by a Research Career Development Award from the Dermatology Foundation.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
Ensembl Genome Browser, http://www.ensembl.org/
Human Intermediate Filament Database, http://www.interfil.org
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
References
- 1.Chien A.J., Valentine M.C., Sybert V.P. Hereditary woolly hair and keratosis pilaris. J. Am. Acad. Dermatol. 2006;54:S35–S39. doi: 10.1016/j.jaad.2005.01.092. [DOI] [PubMed] [Google Scholar]
- 2.Salamon T. Über eine familie mit recessiver Kraushaarigkeit, hypotrichose und anderen anomalien. Hautarzt. 1963;14:540–544. [PubMed] [Google Scholar]
- 3.Hutchinson P.E., Cairns R.J., Wells R.S. Woolly hair. Clinical and general aspects. Trans. St Johns Hosp. Dermatol. Soc. 1974;60:160–177. [PubMed] [Google Scholar]
- 4.Kazantseva A., Goltsov A., Zinchenko R., Grigorenko A.P., Abrukova A.V., Moliaka Y.K., Kirillov A.G., Guo Z., Lyle S., Ginter E.K. Human hair growth deficiency is linked to a genetic defect in the phospholipase gene LIPH. Science. 2006;314:982–985. doi: 10.1126/science.1133276. [DOI] [PubMed] [Google Scholar]
- 5.Pasternack S.M., von Kügelgen I., Aboud K.A., Lee Y.A., Rüschendorf F., Voss K., Hillmer A.M., Molderings G.J., Franz T., Ramirez A. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat. Genet. 2008;40:329–334. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 6.Shimomura Y., Wajid M., Ishii Y., Shapiro L., Petukhova L., Gordon D., Christiano A.M. Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nat. Genet. 2008;40:335–339. doi: 10.1038/ng.100. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura Y., Wajid M., Petukhova L., Shapiro L., Christiano A.M. Mutations in the Lipase H (LIPH) gene underlie autosomal recessive woolly hair/hypotrichosis. J. Invest. Dermatol. 2009;129:622–628. doi: 10.1038/jid.2008.290. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda H., Aoki J., Hiramatsu T., Ishida M., Bandoh K., Nagai Y., Taguchi R., Inoue K., Arai H. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J. Biol. Chem. 2002;277:34254–35263. doi: 10.1074/jbc.M201659200. [DOI] [PubMed] [Google Scholar]
- 9.Yanagida K., Masago K., Nakanishi H., Kihara Y., Hamano F., Tajima Y., Taguchi R., Shimizu T., Ishii S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J. Biol. Chem. 2009;284:17731–17741. doi: 10.1074/jbc.M808506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langbein L., Rogers M.A., Praetzel S., Aoki N., Winter H., Schweizer J. A novel epithelial keratin, hK6irs1, is expressed differentially in all layers of the inner root sheath, including specialized huxley cells (Flügelzellen) of the human hair follicle. J. Invest. Dermatol. 2002;118:789–799. doi: 10.1046/j.1523-1747.2002.01711.x. [DOI] [PubMed] [Google Scholar]
- 11.Langbein L., Rogers M.A., Praetzel S., Winter H., Schweizer J. K6irs1, K6irs2, K6irs3, and K6irs4 represent the inner-root-sheath-specific type II epithelial keratins of the human hair follicle. J. Invest. Dermatol. 2003;120:512–522. doi: 10.1046/j.1523-1747.2003.12087.x. [DOI] [PubMed] [Google Scholar]
- 12.Kikkawa Y., Oyama A., Ishii R., Miura I., Amano T., Ishii Y., Yoshikawa Y., Masuya H., Wakana S., Shiroishi T. A small deletion hotspot in the type II keratin gene mK6irs1/Krt2-6g on mouse chromosome 15, a candidate for causing the wavy hair of the caracul (Ca) mutation. Genetics. 2003;165:721–733. doi: 10.1093/genetics/165.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters T., Sedlmeier R., Büssow H., Runkel F., Lüers G.H., Korthaus D., Fuchs H., Hrabé de Angelis M., Stumm G., Russ A.P. Alopecia in a novel mouse model RCO3 is caused by mK6irs1 deficiency. J. Invest. Dermatol. 2003;121:674–680. doi: 10.1046/j.1523-1747.2003.12491.x. [DOI] [PubMed] [Google Scholar]
- 14.Runkel F., Klaften M., Koch K., Böhnert V., Büssow H., Fuchs H., Franz T., Hrabé de Angelis M. Morphologic and molecular characterization of two novel Krt71 (Krt2-6g) mutations: Krt71rco12 and Krt71rco13. Mamm. Genome. 2006;17:1172–1182. doi: 10.1007/s00335-006-0084-9. [DOI] [PubMed] [Google Scholar]
- 15.Cadieu E., Neff M.W., Quignon P., Walsh K., Chase K., Parker H.G., Vonholdt B.M., Rhue A., Boyko A., Byers A. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulombe P.A., Omary M.B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 17.Moll R., Divo M., Langbein L. The human keratins: biology and pathology. Histochem. Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J.M., Chipev C.C., DiGiovanna J.J., Bale S.J., Marekov L.N., Steinert P.M., Compton J.G. Mutations in the H1 and 1A domains in the keratin 1 gene in epidermolytic hyperkeratosis. J. Invest. Dermatol. 1994;102:17–23. doi: 10.1111/1523-1747.ep12371725. [DOI] [PubMed] [Google Scholar]
- 19.McLean W.H., Eady R.A., Dopping-Hepenstal P.J., McMillan J.R., Leigh I.M., Navsaria H.A., Higgins C., Harper J.I., Paige D.G., Morley S.M. Mutations in the rod 1A domain of keratins 1 and 10 in bullous congenital ichthyosiform erythroderma (BCIE) J. Invest. Dermatol. 1994;102:24–30. doi: 10.1111/1523-1747.ep12371726. [DOI] [PubMed] [Google Scholar]
- 20.Arin M.J., Longley M.A., Küster W., Huber M., Hohl D., Rothnagel J.A., Roop D.R. An asparagine to threonine substitution in the 1A domain of keratin 1: A novel mutation that causes epidermolytic hyperkeratosis. Exp. Dermatol. 1999;8:124–127. doi: 10.1111/j.1600-0625.1999.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee D.Y., Ahn K.S., Lee C.H., Rho N.K., Lee J.H., Lee E.S., Steinert P.M., Yang J.M. Two novel mutations in the keratin 1 gene in epidermolytic hyperkeratosis. J. Invest. Dermatol. 2002;119:976–977. doi: 10.1046/j.1523-1747.2002.00061.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith F.J., Maingi C., Covello S.P., Higgins C., Schmidt M., Lane E.B., Uitto J., Leigh I.M., McLean W.H. Genomic organization and fine mapping of the keratin 2e gene (KRT2E): K2e V1 domain polymorphism and novel mutations in ichthyosis bullosa of Siemens. J. Invest. Dermatol. 1998;111:817–821. doi: 10.1046/j.1523-1747.1998.00371.x. [DOI] [PubMed] [Google Scholar]
- 23.Whittock N.V., Ashton G.H., Griffiths W.A., Eady R.A., McGrath J.A. New mutations in keratin 1 that cause bullous congenital ichthyosiform erythroderma and keratin 2e that cause ichthyosis bullosa of Siemens. Br. J. Dermatol. 2001;145:330–335. doi: 10.1046/j.1365-2133.2001.04327.x. [DOI] [PubMed] [Google Scholar]
- 24.Stephens K., Ehrlich P., Weaver M., Le R., Spencer A., Sybert V.P. Primers for exon-specific amplification of the KRT5 gene: identification of novel and recurrent mutations in epidermolysis bullosa simplex patients. J. Invest. Dermatol. 1997;108:349–353. doi: 10.1111/1523-1747.ep12286486. [DOI] [PubMed] [Google Scholar]
- 25.Liao H., Sayers J.M., Wilson N.J., Irvine A.D., Mellerio J.E., Baselga E., Bayliss S.J., Uliana V., Fimiani M., Lane E.B. A spectrum of mutations in keratins K6a, K16 and K17 causing pachyonychia congenita. J. Dermatol. Sci. 2007;48:199–205. doi: 10.1016/j.jdermsci.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Smith F.J., Liao H., Cassidy A.J., Stewart A., Hamill K.J., Wood P., Joval I., van Steensel M.A., Björck E., Callif-Daley F. The genetic basis of pachyonychia congenita. J. Investig. Dermatol. Symp. Proc. 2005;10:21–30. doi: 10.1111/j.1087-0024.2005.10204.x. [DOI] [PubMed] [Google Scholar]
- 27.Smith F.J., McKenna K.E., Irvine A.D., Bingham E.A., Coleman C.M., Uitto J., McLean W.H. A mutation detection strategy for the human keratin 6A gene and novel missense mutations in two cases of pachyonychia congenita type 1. Exp. Dermatol. 1999;8:109–114. doi: 10.1111/j.1600-0625.1999.tb00356.x. [DOI] [PubMed] [Google Scholar]
- 28.Lin M.T., Levy M.L., Bowden P.E., Magro C., Baden L., Baden H.P., Roop D.R. Identification of sporadic mutations in the helix initiation motif of keratin 6 in two pachyonychia congenita patients: further evidence for a mutational hot spot. Exp. Dermatol. 1999;8:115–119. doi: 10.1111/j.1600-0625.1999.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 29.Winter H., Clark R.D., Tarras-Wahlberg C., Rogers M.A., Schweizer J. Monilethrix: a novel mutation (Glu402Lys) in the helix termination motif and the first causative mutation (Asn114Asp) in the helix initiation motif of the type II hair keratin hHb6. J. Invest. Dermatol. 1999;113:263–266. doi: 10.1046/j.1523-1747.1999.00685.x. [DOI] [PubMed] [Google Scholar]
- 30.Korge B.P., Hamm H., Jury C.S., Traupe H., Irvine A.D., Healy E., Birch-MacHin M., Rees J.L., Messenger A.G., Holmes S.C. Identification of novel mutations in basic hair keratins hHb1 and hHb6 in monilethrix: Implications for protein structure and clinical phenotype. J. Invest. Dermatol. 1999;113:607–612. doi: 10.1046/j.1523-1747.1999.00722.x. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 32.Noguchi K., Ishii S., Shimizu T. Identification of p2y9/GPR23 as a novel G protein-coupled receptor for lysophosphatidic acid, structurally distant from the Edg family. J. Biol. Chem. 2003;278:25600–25606. doi: 10.1074/jbc.M302648200. [DOI] [PubMed] [Google Scholar]
- 33.Mou C., Thomason H.A., Willan P.M., Clowes C., Harris W.E., Drew C.F., Dixon J., Dixon M.J., Headon D.J. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum. Mutat. 2008;29:1405–1411. doi: 10.1002/humu.20795. [DOI] [PubMed] [Google Scholar]
- 34.Medland S.E., Nyholt D.R., Painter J.N., McEvoy B.P., McRae A.F., Zhu G., Gordon S.D., Ferreira M.A., Wright M.J., Henders A.K. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter H., Rogers M.A., Langbein L., Stevens H.P., Leigh I.M., Labrèze C., Roul S., Taieb A., Krieg T., Schweizer J. Mutations in the hair cortex keratin hHb6 cause the inherited hair disease monilethrix. Nat. Genet. 1997;16:372–374. doi: 10.1038/ng0897-372. [DOI] [PubMed] [Google Scholar]
- 36.Winter H., Schissel D., Parry D.A., Smith T.A., Liovic M., Birgitte Lane E., Edler L., Langbein L., Jave-Suarez L.F., Rogers M.A. An unusual Ala12Thr polymorphism in the 1A α-helical segment of the companion layer-specific keratin K6hf: Evidence for a risk factor in the etiology of the common hair disorder pseudofolliculitis barbae. J. Invest. Dermatol. 2004;122:652–657. doi: 10.1111/j.0022-202X.2004.22309.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


