Abstract
We investigated two male infant patients who were given a diagnosis of progressive mitochondrial encephalomyopathy on the basis of clinical, biochemical, and morphological features. These patients were born from monozygotic twin sisters and unrelated fathers, suggesting an X-linked trait. Fibroblasts from both showed reduction of respiratory chain (RC) cIII and cIV, but not of cI activities. We found a disease-segregating mutation in the X-linked AIFM1 gene, encoding the Apoptosis-Inducing Factor (AIF) mitochondrion-associated 1 precursor that deletes arginine 201 (R201 del). Under normal conditions, mature AIF is a FAD-dependent NADH oxidase of unknown function and is targeted to the mitochondrial intermembrane space (this form is called AIFmit). Upon apoptogenic stimuli, a soluble form (AIFsol) is released by proteolytic cleavage and migrates to the nucleus, where it induces “parthanatos,” i.e., caspase-independent fragmentation of chromosomal DNA. In vitro, the AIFR201 del mutation decreases stability of both AIFmit and AIFsol and increases the AIFsol DNA binding affinity, a prerequisite for nuclear apoptosis. In AIFR201 del fibroblasts, staurosporine-induced parthanatos was markedly increased, whereas re-expression of AIFwt induced recovery of RC activities. Numerous TUNEL-positive, caspase 3-negative nuclei were visualized in patient #1's muscle, again indicating markedly increased parthanatos in the AIFR201 del critical tissues. We conclude that AIFR201 del is an unstable mutant variant associated with increased parthanatos-linked cell death. Our data suggest a role for AIF in RC integrity and mtDNA maintenance, at least in some tissues. Interestingly, riboflavin supplementation was associated with prolonged improvement of patient #1's neurological conditions, as well as correction of RC defects in mutant fibroblasts, suggesting that stabilization of the FAD binding in AIFmit is beneficial.
Main Text
A peculiar feature of mitochondrial disorders is that only a minority are caused by mutations directly affecting the structural components of the respiratory chain (RC) complexes.1 In fact, the vast majority of mitochondrial-disease gene products are involved in ancillary roles concerning RC formation, activity, and turnover.1 Far from being diminutive, this characteristic has allowed scientists to exploit human mitochondrial disorders to dissect and characterize in detail the mitochondrial machinery at the molecular level. A further consideration is that mitochondrial RC not only catalyzes oxidative phosphorylation (OXPHOS) and thus provides most ATP, the universal energy currency of cells, but is also at the cross-point of a number of other crucial functions, including heat production, ion trafficking, nucleotide synthesis, generation of reactive oxygen species (ROS), and apoptosis.1,2 In addition, several RC-associated proteins play multiple roles and may be located in different cell compartments, besides mitochondria.3 Consequently, OXPHOS impairment determines not only various degrees of energy failure but also multiple additional effects that account, at least in part, for the intricacy and complexity of mitochondrial disorders.2,4 Primary mutations in the mtDNA or of nuclear genes encoding proteins directly involved in RC functioning can cause homeostatic alterations and perturb mitochondrial pathways that in turn contribute to the pathological mechanism.2 Is this a two-way correlation? In other terms, could defective RC and faulty OXPHOS be caused by primary abnormalities of such pathways and eventually lead to a mitochondrial disease? In support to this idea, we report here the results of genetic and molecular investigation on two male infant patients affected by severe encephalomyopathy and OXPHOS failure associated with a deleterious mutation in AIF, a master gene product of mitochondrial programmed cell death.5,6
Patient #1 was born at term after an uneventful pregnancy; an elder brother was healthy. Birth weight was 3000 g, and the Apgar score was 9-9. He was breastfed and had frequent regurgitations, but his growth remained normal until 1 year of age. However, from the first months he showed reduced spontaneous motility of the right leg and continuous involuntary movements of small amplitude in the hands and feet. At 5 months a first brain MRI was normal, but at 1 year the MRI showed bilateral abnormal signal in the neostriatum (Figures 1A and 1B). By that time, a moderate psychomotor development delay in head control had become evident: he achieved head control at as late as 5 months of age and reached the sitting position at 10 months. When he was admitted to our hospital, at 15 months of age, he could not reach and maintain the standing position by himself. Neurological examination revealed continuous fasciculations of the tongue, hypotonia, and areflexia. Verbal communication was limited to modulated babble, but no defined words, and the general quotient on the Griffith scale was 50. Lactate and pyruvate were moderately increased in plasma and cerebro-spinal fluid (CSF). Electromyography and nerve conduction velocities demonstrated axonal sensory and motor peripheral neuropathy. Because a CT scan of quadriceps muscles had shown an unexpectedly severe grade of muscular atrophy, an open muscle biopsy was performed so that the possibility of a mitochondrial disorder could be evaluated. RC activities could not be measured because the muscle tissue was largely substituted by fat, but in cultured fibroblasts complex III and IV were partially reduced (Table 1). Although mtDNA sequence analysis failed to reveal mutations, its copy number, measured by quantitative real-time PCR, was reduced to 20% of the control values in a few muscle fibers dissected out of the surrounding fat. However, mtDNA amount was normal in cultured fibroblasts. The patient was discharged from the hospital, but after a few days he developed seizures and severe respiratory distress needing assistance from the intensive care unit. The neurological conditions steadily worsened thereafter, and there was severe psychomotor regression: sitting and head control were lost, and environment contact became poor and then lost. Interestingly, riboflavin supplementation (30 mg/day) led to improvement of his clinical conditions for the following 7 months: he recovered the ability to sit unaided, grasp objects, and communicate with his parents in spite of generalized severe weakness. At two years, he suddenly developed generalized seizures, partially controlled by phenobarbital, and severe respiratory insufficiency with bronchopneumonia, which required mechanical ventilation and tracheotomy. He is now 5 years old and still alert, but he is virtually unable to communicate, is tetraplegic and wheelchair bound, and is under permanent artificial ventilation.
Figure 1.
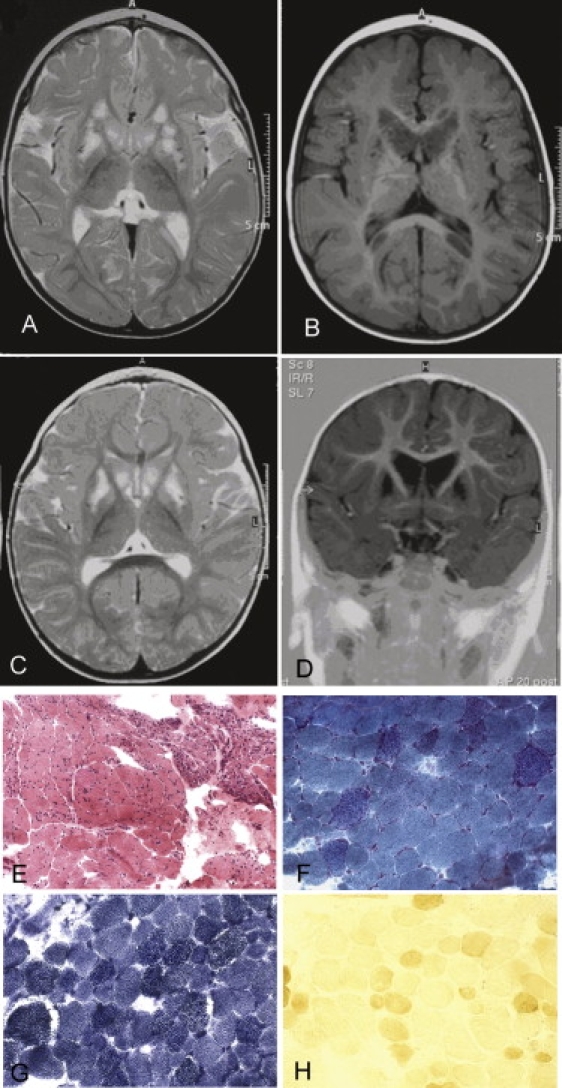
Brain Imaging and Skeletal Muscle Morphological and Histochemical Analysis
(A and B) Magnetic resonance imaging (MRI) T2-weighted (A) and T1-weighted (B) sequences on a transverse section of supratentorial brain of patient #1.
(C) MRI T2-weighted sequence on a transverse section of supratentorial brain of patient #2 at 11 months of age.
(D) MRI T1-weighted sequence on a coronal section of supratentorial brain of patient #2 at 11 months of age.
(E–H) Patient #2 muscle biopsy showing an increase of connective tissue, severe variability in fiber size, and necrotic and regenerating fibers at Hematoxylin and Eosin (H&E) (E), numerous ragged-red fibers under the modified Gomori trichrome staining (F), increased SDH (succinate dehydrogenase) histochemical staining (G), and diffuse reduction of COX histochemical reaction (H).
Table 1.
Biochemical Analysis of OXPHOS Activities
| CSa | cI | cII | SDH | cIII | cIV | cV | Treatment | |
|---|---|---|---|---|---|---|---|---|
| Muscle | ||||||||
| Ct values | 80–210 | 13–24 | 15–28 | 10.7–17.4 | 88–167 | 120–220 | 130–280 | |
| Pt#2 ms | 153 | 7.9 (43) | 13.6 (63) | 8.9 (63) | 56 (43) | 19 (11) | 71 (35) | |
| Fibroblasts | ||||||||
| Ct values | 100–200 | 10.7–26 | 8.6–18.4 | 6.5–14.3 | 70–120 | 95–150 | 65–113 | |
| Primary Fibroblasts | ||||||||
| Pt#1 fbs | 144 | 13.5 (73) | 9.4 (70) | 9.3 (89) | 63 (66) | 66 (54) | 100 (112) | no |
| Pt#2 fbs | 187 | 15.2 (82) | 15.6 (116) | 10 (96) | 66 (69) | 87,5 (71) | 205 (230) | no |
| Pt#1 fbs | 91 | 9.4 (51) | 14 (104) | 10.5 (101) | 50.4 (53) | 67.4 (55) | 88.1 (99) | 48 hr galactose |
| Pt#2 fbs | 111 | 5.9 (32) | 17 (126) | 10.7 (103) | 40.9 (43) | 66.2 (54) | 97 (109) | 48 hr galactose |
| SV40-Immortalized Fibroblasts Treated with Riboflavin | ||||||||
| Pt#2 SV40 fbs | 128 | 11,4 (62) | 11,1 (82) | 7,1 (68) | 41,8 (44) | 84,5 (69) | 107,7 (121) | Fbs immortalized with pBabe-puro SV40 |
| Pt#1 SV40 fbs | 154 | 11,6 (63) | 18,1 (134) | 8,5 (82) | 33,9 (36) | 73 (60) | 97 (109) | |
| Pt#1 SV40 fbs | 174 | n.d | n.d | n.d | 58,9 (62) | 88,2 (72) | n.d. | 1 week riboflavin |
| Immortalized Fibroblasts Transfected with an AIF-Expressing Eukaryotic Vector | ||||||||
| Pt#1 SV40 fbs mock | 147 | 11,8 (63) | 20,6 (152) | 12 (115) | 29,5 (31) | 69,9 (57) | 67,7 (76) | SV40-fbs transfected with empty pcDNA3.1 |
| Pt#1 SV40 fbs+AIF | 123 | 11,9 (65) | 16,3 (121) | 12,2 (117) | 45,2 (48) | 93,6 (77) | 68,5 (77) | SV40-fbs transfected with pcDNA3.1 + AIF |
All enzymatic activities are normalized for citrate synthase (CS) activity. The percentages of the mean control value are shown in parentheses. Values less than control values are shown in italics. Abbreviations are as follows: n.d., not determined; Ms, skeletal muscle biopsy; fbs, fibroblasts.
nmol/min mg.
Patient #2 was the only child of nonconsanguineous parents. His mother was a monozygotic twin sister of the mother of patient #1. He was born by caesarean section because of fetal cardiac rhythm deceleration, but no problems were reported during the perinatal period. His psychomotor development was reported as normal until the age of 11 months, when a reduction of active movements and subsequent psychomotor regression were noticed. A brain MRI showed abnormal bilateral signal intensities in the neostriatum (Figures 1C and 1D). At 1 year of age, he presented with marked irritability, severe hypotonia, hypo-areflexia, muscle wasting, and weakness. He could sit unaided for just a few seconds and had no active movement of the lower limbs; his generalized quotient on the Griffith scale was 34. Plasma lactate was increased up to 3150 μM (normal < 2000 μM). H & E staining of a muscle biopsy showed dystrophic alterations with areas of small, regenerating fibers, next to areas with degenerating fibers (Figure 1E) but no type grouping (not shown); Gomori trichrome staining showed several ragged-red fibers (Figure 1F), which corresponded to SDH-hyperintense fibers (Figure 1G); the fibers were diffusely pale or unreactive to cytochrome-c-oxidase (COX)-specific histochemistry (Figure 1H). The activity of respiratory-chain complexes in muscle was reduced, especially that of COX (complex IV, cIV), which was as low as 11% of the normal mean. Similar to his cousin, patient #2 had complex III and IV activities that were below the normal range in fibroblasts (Table 1). Mitochondrial DNA depletion (about 35% of control value) was again present in muscle but not in fibroblasts. The patient suddenly died at 16 months of age during sleep. No autopsy was performed. The parents gave informed consent to a genetic screening according to the Ethical Committee of the Neurological Institute “Carlo Besta,” Milan, Italy.
Because our two patients were born from genetically proven (not shown) monozygotic twin sisters and from unrelated fathers (Figure 2A), we hypothesized either an mtDNA- or an X-linked trait. The former hypothesis was ruled out by sequence analysis of a normal haplogroup J1 mtDNA in all of the maternal family members, including the two propositi. Next, we carried out SNP-based haplotype analysis of the X chromosome. In the blood DNA extracted from the two patients, we found a haploidentical region between markers RS5977571 and RS5904872; this region contained 211 entries, only 75 of which corresponded to allegedly expressed protein-encoding genes. The remaining entries corresponded to pseudo-genes and micro-RNA-encoding genes. We prioritized the 75 gene sequences on the basis of known or putative mitochondrial localization of the corresponding proteins. Seven genes within the critical interval encode predicted mitochondrial proteins on the basis of combined analysis with mitochondrial-protein prediction software, including Mitprot, Target-P, Predotar, and Wolf-Psort (Table S1). However, two entries, LOC100131049 and LOC728470, were discarded because no expressed sequence tags are present in the NCBI database, suggesting that neither one corresponds to an expressed gene. Two additional genes, SLC9A6 (MIM ∗300231) and FLJ30058, encode proteins whose function is known and that are not localized in mitochondria. In particular, mutations in SLC9A6 are associated with an Angelman-like syndrome (MIM #300243), and its corresponding protein has been localized to endosomes rather than to mitochondria.7 FLJ30058 is predicted to encode a Rho-GAP, a GTPase activator protein, involved in the regulation of cell cycle and cytoskeleton formation. Sequence analysis of the exons and exon-intron boundaries of both BMCP1 (MIM ∗300242) and FAM127C failed to show mutations. However, sequence analysis of AIFM1 (Apoptosis-inducing factor mitochondrion-associated 1, MIM ∗300169), revealed in both patient #1 (III-2, Figure 2A) and patient #2 (III-3, Figure 2A) a homozygous trinucleotide deletion (c.del601-603) in exon 5 (Figure 2B), predicting the ablation of an arginine (R) residue at position 201 of the AIF polypeptide (R201 del). The two twin mothers (II-2 and II-4, Figure 2A) were both heterozygous for this change, whereas the mutation was absent in all other family members, including the maternal grandfather (I-1) and grandmother (I-2), maternal aunt (II-6), and the fathers (II-1 and II-3) of the probands, as well as the healthy brother of patient #1 (III-1). It was also absent in 210 alleles from Italian control samples. Taken together, these results suggest the occurrence of a de novo mutational event in the zygote generating the twin sisters, although germinal mosaicism of the patients' grandmother or grandfather cannot be excluded.
Figure 2.

Pedigree and AIF Features
(A) Pedigree of the family. Filled symbols indicate affected individuals (patient #1, Pt#1 and patient #2, Pt#2, also indicated by arrows). WT and ΔR201 indicate the presence of wild-type AIF sequence and the presence of the R201 deletion, respectively.
(B) Sequence analysis of exon 5 of AIFM1 showing the c.601–603 deletion in patient #1. A control sequence is shown for comparison.
(C) Schematic primary structure of the AIF protein. MTS indicates the mitochondrial targeting sequence; TM indicates a putative transmembrane domain. MPP: Mitochondrial Processing Peptidase.
The precursor, mitochondrial (AIFmit), and soluble (AIFsol) forms of the AIF protein are shown starting from the N-terminal aminoacid; the corresponding molecular weights are also indicated.
To test whether the R201 del mutation is pathogenic and responsible for the disease phenotype, we first investigated the mutant variant by in silico and in vitro analyses.
The AIF precursor protein is a 67 kDa polypeptide (Figure 2C) encoded by a gene located on chromosome Xq25-26. After mitochondrial import and cleavage of a 54-amino-acid-long mitochondrial targeting sequence, the 62 kDa, mitochondrion-specific mature AIF protein (AIFmit, Figure 2C) noncovalently binds FAD and attaches by an N-terminal trans-membrane domain to the inner mitochondrial membrane, from where it projects toward the mitochondrial inter-membrane space.8–10 Minor pools of the protein have (controversially) been proposed to occupy the outer mitochondrial membrane11 or be free in solution within the inter-membrane space. In the latter compartment, mature AIFmit homodimers function as a FAD-dependent NADH oxidase, which takes part in an unknown metabolic pathway, possibly involved in ROS control or redox sensing. Upon presentation of apoptogenic stimuli, a soluble form of AIF (AIFsol) is produced by protease-mediated cleavage of AIFmit at position 102 (Figure 2C); this cleavage is mediated by cathepsin-B, L, and S12 or calpain.13 AIFsol then translocates from mitochondria to the nucleus, where it binds to chromosomal DNA and induces chromatin condensation and DNA fragmentation by attracting and activating a set of endonucleases, possibly including endonuclease G,14 another cell-death executor normally stored in mitochondria. This caspase- and cytochrome-c-independent, peculiar form of mitochondrion-driven programmed cell death has been denominated parthanatos to distinguish it from caspase-mediated “conventional” apoptosis.15
Mammalian AIFs share >90% amino acid sequence identity and, as the X-ray structures of human and murine flavoproteins suggest,16–18 have virtually undistinguishable tertiary structures (Figure 3A). Because the crystal structure of the AIFR201 del variant is yet to be determined, we have modeled the R201 del mutation in silico. According to the X-ray data, the R201 residue is part of the AIF-specific 191–203 β-hairpin, which forms the FAD binding pouch and confers conformational stability to the flavoprotein. In oxidized AIF, the R201 residue induces the folding of the regulatory 510–560 peptide by establishing a salt bridge with E531 and thus limiting the accessibility to the FAD-containing active site (Figure 3A). In the reduced NAD-bound protein, the R201 side chain (R200 in the mouse protein) is expected to be hydrogen-bonded to the carbonyl oxygen of the neighboring F205 residue (F204 in the mouse), as observed in murine AIF (Figure 3B), spatially orienting the solvent-exposed hairpin and functionally important W196 (murine W195).18 Because R201 defines the length and positioning of the hairpin and contributes to the intricate network of polar and hydrophobic interactions between the 191–203 and 510–539 fragments, its deletion can predictably perturb the functional properties of both oxidized and reduced forms of AIF (Figure 3C).
Figure 3.

Structural and In Vitro Analyses of Recombinant AIFwt and AIF201 del
(A) Superimposed X-ray structures of oxidized (NAD-less) human and murine AIFs (PDB codes: 1M6I and 1DV416,17) show that the R201 (R200 in mouse) assists in the folding of two peptide stretches, near the FAD-binding pocket. The R201 residue, contained in the first stretch (in pink), establishes a link with the second stretch (in brown) by forming a salt bridge with the E531 residue.
(B) In the reduced NAD-bound dimer of murine AIF (PDB code 3GD418), the active form of the flavoprotein, R200 (R201 in humans), establishes a hydrogen bond with F204, helping the functionally important β-hairpin, including the W195 residue (shown in pink), to get properly oriented. F204 and W195 correspond to human F205 and W196, respectively.
(C) Computer modeling predicts that deletion of R201 in reduced human AIF can disrupt the H bond with F205 and thus shorten and distort the 191–203 β-hairpin (rendered in blue for the wild-type and in pink for the mutant).
(D) Anaerobic titration of 15 μM AIFmitR201 del with NADH (in 50 mM phosphate [pH 7.5]). The inset shows that an equimolar amount of NADH is required to fully reduce FAD.
(E) Oxidation of the NADH-reduced wild-type (wt) and R201 del mutants (mt) of AIFmit and AIFsol. A 15 μM protein solution in 50 mM phosphate (pH 7.5) was aerobically reduced with a 6-fold excess of NADH, and the FAD oxidation by air oxygen was monitored over time at 452 nm.
(F) Far-UV circular dichroism spectra of 3 μM AIFmitR201 del were recorded in 10 mM sodium phosphate (pH 7.5) in the absence or presence of a 20-fold excess of NAD(P)H. The R201 del mutation has no significant effect on the redox-dependent changes in the secondary structure of AIF.
(G) The R201 del mutation promotes the AIFsol-DNA interaction. Equal amounts of wild-type (wt) and mutant (mt) AIFsol were incubated for 15 min with 250 ng of a 100 bp DNA ladder in the absence or presence of a 20-fold excess of NAD(P)H; separation on a 2% agarose gel and visualization with ethidium bromide followed.
The R201 deletion was introduced into the AIFmit and AIFsol expression plasmids by site-directed mutagenesis with the Stratagene QuikChange kit. The mutant proteins were expressed and purified according to the procedure developed for wild-type AIF.19
To test these predictions experimentally, we performed anaerobic titration of recombinant human AIFR201 del with NADH.19 Once exposed to NADH, the AIFR201 del variant, similar to wild-type AIF (AIFwt), showed stoichiometric reduction of FAD to FADH2 and formation of a FADH2-NAD charge-transfer complex, characterized by an absorbance peak in the long-wavelength (≈700 nm) spectral region (Figure 3D). However, the lifetime of the FADH2-NAD complex formed by mutant AIFR201 del was considerably shorter than that observed with AIFwt, but there was no significant difference in t1/2 for the mature (AIFmit) and apoptogenic forms (AIFsol) (Figure 3E). Moreover, the kinetics of the redox reaction showed that AIFR201 del accepts the hydrogen atom from NADH or NADPH at a rate two orders of magnititude faster than AIFwt, and there is a lower-than-normal preference for NADH (50-fold versus 100-fold for AIFwt, Table 2). Circular dichroism spectroscopy data show nearly identical folding profiles for the recombinant mutant versus wild-type AIF (Figure 3F and Figure 2 in Churbanova et al.19), indicating that the R201 del mutation mainly affects the functions but does not drastically alter the structure of the mutant protein. However, higher sensitivity of AIFR201 del to trypsin digestion (Figure S1) suggests that subtle conformational changes caused by the R201 deletion destabilize the flavoprotein and increase its susceptibility to proteolytic cleavage. In addition, unlike AIFwt, oxidized AIFR201 del partially loses FAD during gel filtration (Figure S2), which can be another indication of structural instability of the mutant. Taken together, the in vitro results allow us to propose that AIFR201 del is an unstable variant with altered redox enzymatic properties.
Table 2.
Kinetic Properties of AIFWT and AIFR201 del
| AIFwta | AIFR201 del | |
|---|---|---|
| kETNADH (s−1) | 0.2 | 11 ± 1 |
| KMNADH (mM) | 0.60 | 0.35 ± 0.03 |
| kETNADH/KMNADH | 0.3 | 31 ± 2 |
| kETNADPH (s−1) | 1.0 × 10−2 | 3.3 ± 0.2 |
| KMNADPH (mM) | 3.7 | 5.0 ± 0.6 |
| kETNADPH/KMNADPH | 2.7 × 10−3 | 0.7 ± 0.1 |
Determined previously.19
Next, we tested the DNA binding ability of AIFR201 del, strictly required for induction of nuclear fragmentation.17 Our DNA retardation assays showed that the R201 deletion significantly increases AIFsol-DNA interaction (Figure 3G). Whereas AIFwt only moderately affected the electrophoretic mobility of linearized DNA, the AIFR201 del mutant protein formed a substantial amount of high-molecular weight protein-DNA complexes, indicating that AIFR201 del has higher DNA binding affinity and potential propensity to cause DNA damage.
We then investigated the consequences of the AIFR201 del mutation in cell lines. By immunovisualization on electroblotted SDS-PAGE with an anti-AIF specific antibody, we first ascertained that the amount and processing of AIFR201 del is similar to those of AIFwt in patient-derived skin fibroblasts (Figure 4A).
Figure 4.

Characterization of Patients' Fibroblasts before and after Galactose Treatment
(A) Protein-blot analysis of patient #1(Pt) and control (Ct) skin cultured fibroblasts, immunodetected with an antibody against AIF (Chemicon). Antibodies against subunit A of complex II (SDHA, Invitrogen) and actin (Sigma) were used as loading controls.
(B) Representative images of mitochondrial morphology, showing the filamentous (upper panel) or the fragmented (lower panel) mitochondrial network of fibroblasts grown in DMEM-glucose or DMEM-galactose. Cells were stained with 100 nM Mitotracker-Red CMX-Ros (Invitrogen) for 1/2 hr. The scale bar represents 10 μm.
(C) Quantification of fibroblasts with a normal or fragmented mitochondrial network in controls (Ct) and patients (Pt) after incubation in D-MEM +5 mM galactose for 48 hr.
(D) MTT cell-viability assay. Control (Ct, blue lines) and patients' (Pt#1, red lines, and Pt#2, brown lines) fibroblasts were grown in DMEM + glucose (solid lines) or DMEM + galactose (dotted lines) for the times indicated. Absorbance (Abs) is proportional to the number of viable cells.
To test whether the AIFR201 del mutant variant is directly linked to impaired OXPHOS, we analyzed mutant fibroblast cell lines grown in a galactose-rich, glucose-free medium, a condition that forces cells to rely for energy on mitochondrial respiration rather than glycolysis. After exposure to galactose (5 mM) for 48 hr, combined deficiency of cI, cIII, and cIV activities was detected in cell lines derived from both patients (Table 1). Mitotracker-based visualization of the mitochondrial network showed marked fragmentation of mitochondria in mutant cells cultured in galactose, in comparison to the normal filamentous network visualized in glucose-grown mutant cells and in glucose-grown and the majority of galactose-grown control cells (Figure 4B). We found that approximately 75% of mutant cells versus 23% control cells showed mitochondrial fragmentation under galactose treatment (Figure 4C). Fragmentation of the mitochondrial network is a well-documented morphological alteration related to reduced fusion of the mitochondrial membranes and is typically observed in faulty OXPHOS cells or in pre-apoptotic conditions.20 Finally, MTT cell viability assay showed reduced cell growth of mutant fibroblasts in glucose and showed an even greater reduction of growth in galactose medium (Figure 4D). Neither in glucose- nor in galactose-grown cells was reduction of mtDNA ever found, as opposed to the clear mtDNA depletion detected in skeletal muscle of both patients. These results indicate that AIFR201 del is directly involved in RC impairment, independently of whether mtDNA is depleted, an effect that seems to be restricted to, and aggravate the OXPHOS failure of, specific tissues, for instance skeletal muscle. To further test the pathogenic role of AIFR201 del, we expressed recombinant AIFwt in SV40-immortalized, cIII- and cIV-defective AIFR201 del mutant fibroblasts by two separate sets of transfections with a recombinant vector containing a neomycin-resistant gene as a selectable marker. After selection with the neomycin analog G-418, biochemical assays were performed in triplicate five times, two with the first transfection, three with the second one. We observed a consistent, significant (p < 0.05) recovery of biochemical activities, by approximately 55% for cIII and 35% for cIV, in comparison to mock-transfected cells (Table 1), again indicating a direct link between mutant AIFR201 del and multiple RC defects.
In order to investigate the effects of AIFR201 del on programmed cell death, we challenged AIFR201 del mutant and control fibroblasts with staurosporine (1 mM for 2 hr), a protein-kinase C inhibitor that induces cell death through mitochondrion-mediated pathways. In staurosporine-treated AIFR201 del fibroblast cells we observed a markedly higher number of morphologically altered nuclei than in identically treated control fibroblasts (Figure 5A). In cells pretreated with z-VAD-FMK (100 μM for 1/2 hr), a general inhibitor of caspases, the staurosporine-induced cell death was nearly absent in control fibroblasts, whereas as many as 34% of AIFR201 del mutant cells still displayed nuclear alterations (Figure 5B). Electron microscopy (EM) images showed a high degree of chromatin condensation, typical of parthanatos, in treated mutant fibroblasts, but hardly any cell had apoptotic bodies (Figures 5C and 5D), as are described in caspase-dependent apoptosis. Likewise, in mutant cells we found marked membrane disruption (not shown), another morphological hallmark specific to parthanatos.21 In order to establish whether staurosporine-induced parthanatos was due to the direct effect of mutant AIFR201 del or was rather a consequence of the RC defects found in mutant AIFR201 del fibroblasts, we challenged with staurosporine four fibroblast cell lines, each displaying severe, isolated defects of RC activities, namely cI (because of a pathogenic homoplasmic mutation in the mtDNA ND1 gene [MIM ∗516000]), cII (because of a pathogenic mutation in SDHAF1, a cII-specific assembly factor [MIM ∗612848]), cIII (because of pathogenic mutations in BCS1L, a cIII-specific assembly factor [MIM ∗603647]), and cIV (because of pathogenic mutations in SURF1, a cIV-specific assembly factor [MIM ∗185620]). Interestingly, although cI-, cII-, and cIII-defective fibroblasts reacted to staurosporine like the normal control fibroblasts (not shown), the cIV-defective fibroblasts displayed up to 80% more condensed nuclei than control fibroblasts. This effect was dramatically reduced (to approximately 30%), but not completely abolished, by pre-exposure to z-VAD-FMK (Figures 5A and 5B). Therefore, unlike other severe OXPHOS defects, in both AIFR201 del and SURF1 mutant cells, the higher propensity to staurosporine-induced cell death seems to occur, at least in part, through a caspase-independent mechanism, probably mediated by AIF. Further investigation will need to clarify the functional relationship that our Staurosporine (±z-VAD-FMK)-based experiments in AIFR201 del and SURF1 mutant cells suggest exists between AIF and either SURF1 or cIV, as well as the mechanism behind it.
Figure 5.
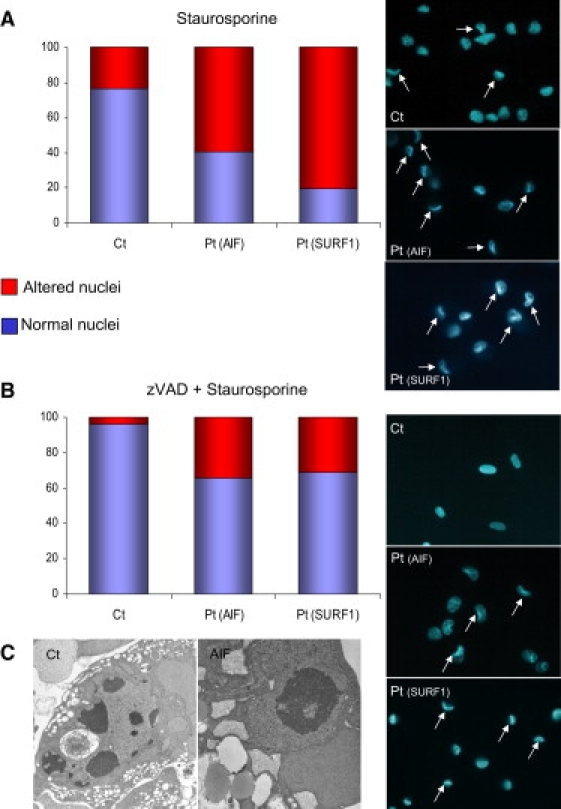
Staurosporine-Induced Cell Death
(A) Quantification of altered nuclei in control fibroblasts (Ct), in fibroblasts from patient #1, Pt(AIF) and from a patient with SURF1 mutations, Pt(SURF1) after staurosporine treatment (1 mM for 2 hr). The three panels on the right show representative fields of Hoechst-stained nuclei. White arrows indicate cells with collapsed nuclei.
(B) Samples were the same as in (A), but the treatment consisted of zVAD (100 μM for 1/2 hr) + staurosporine (1 mM for 2 hr). As in (A), the three panels on the right show representative fields of Hoechst-stained nuclei. White arrows indicate cells with collapsed nuclei.
(C) Electron microscopy (EM) image of staurosporine-treated fibroblasts. Left panel: a control fibroblast shows apoptotic bodies in the nucleus, typical of caspase-dependent apoptosis; right panel: a fibroblast from patient #1 shows chromatin condensation of the nucleus, typical of parthanatos.
Taken together, these results indicate that (i) the AIFR201 del fibroblasts are much more sensitive to apoptotic stimuli than control cells and (ii) cell death occurs predominantly via caspase-independent, AIF-mediated parthanatos. To further support this idea, we analyzed the muscle biopsy of patient #2 by using the TUNEL assay, which visualizes DNA fragmentation in situ. Numerous nuclei stained positive to TUNEL in the patient's specimen (Figure 6A), whereas hardly any staining was detected in a normal control muscle (not shown) or in three disease control muscle biopsies: (i) in a muscle affected by a nonmitochondrial muscular dystrophy, that is, Duchenne muscular dystrophy (DMD, MIM #310200) (Figure 6B); (ii) in a muscle biopsy from a patient affected by Kearns-Sayre syndrome (MIM #530000) due to a 12000 bp mtDNA deletion (Figure 6D); (iii) in a muscle from a child with multiple RC defects due to severe mtDNA depletion associated with two heterozygous allelic mutations in the TK2 gene (MIM ∗188250), encoding the mitochondrion-specific thymidine kinase isoform 2 (MIM #609560) (Figure 6F). Conversely, staining with an anti-caspase 3 antibody was virtually negative in all specimens, including the muscle biopsy of our patient (Figure 6C), again indicating that execution of programmed cell death by mutant AIFR201 del proceeds through caspase-independent parthanatos. The insets of Figures 6B, 6D, and 6F show the results of anti-Cas 3 immunostaining in the disease control muscle specimens. Only the DMD muscle showed a few Cas-3-positive fibers, whereas no anti-Cas 3-positive fibers were detected in muscle specimens carrying the mtDNA deletion or the TK2 mutation. These results were further confirmed by ultrastructural studies (Figure 6E) that showed chromatin condensation in several nuclei of the muscle biopsy of patient #2.
Figure 6.

TUNEL, Caspase 3 Immunostaining, and EM in Skeletal Muscle
(A, B, and D–F) TUNEL + DAPI staining of skeletal muscle nuclei in skeletal muscle from patient #2 and three disease controls (DMD: from a patient with Duchenne muscular dystrophy; mtDNA del: from a patient with a large heteroplasmic deletion of mtDNA; TK2: from a patient who is compound heterozygote for two pathogenic mutations in the TK2 gene). TUNEL-positive nuclei are in green, whereas TUNEL-negative nuclei are in blue, corresponding to DAPI staining. Numerous TUNEL-positive are present in patient #2; two TUNEL-positive green nuclei are present in the DMD muscle; no TUNEL-positive nuclei are detected in mtDNA del or TK muscle specimens.
(C) Anti-Caspase 3 immunostaining of muscle from patient #2 revealed by the diamino-benzidine (DAB) method. No Cas-3-positive fibers were detected. In the insets of (B), (D), and (F), we show the same anti-Cas 3 immunostaining carried out in the corresponding muscle specimens. Only the DMD muscle shows a few Cas-3-positive, brown fibers.
(E) EM image of a nucleus with chromatin condensation in muscle tissue from patient #2.
Prolonged amelioration of clinical conditions by riboflavin supplementation was reported in patient #1. Because AIF is a FAD-dependent flavoprotein, this effect could well be explained by stabilization and partial functional correction of the mutant protein by increased availability of the FAD precursor vitamin, i.e., riboflavin. In support to this hypothesis, we found marked correction of the cIII and cIV defects in AIFR201 del immortalized mutant cells treated with riboflavin (2 mg/liter in DMEM medium) for one week (+74% and +21%, respectively, compared to pretreatment values) (Table 1). In addition, significant recovery of the filamentous network and improvement in cell viability was obtained in riboflavin-treated mutant fibroblasts under galactose medium (Figure S3), whereas hardly any difference was observed in the percentage of condensed nuclei in riboflavin-treated versus untreated mutant cells, both challenged with Staurosporine (not shown). In spite of this latter result, the data altogether offer rational, evidence-based support for riboflavin treatment in AIF mutant subjects.
Finally, we asked whether inactivation of the AIFR201 del allele in tissues of the two healthy monozygotic twin mothers had occurred randomly or under selective pressure due to the deleterious effects caused by the AIFR201 del in the hemizygous mutant offspring. An X-chromosome-inactivation test based on DNA methylation (HUMARA-PCR assay) showed that skewed inactivation22 was present in blood lymphocytes from both individuals and was associated with virtually no expression of the AIFR201 del transcript. However, the X-inactivation pattern was less skewed or not skewed at all in two other cell types (oral and urinary mucosal epithelial cells) (Table S2), suggesting variable sensitivity of different cell types to the deleterious effects of AIFR201 del expression.
In conclusion, we found and characterized a human AIF mutation clearly associated with (i) a mitochondrial early-onset encephalomyopathy, (ii) OXPHOS failure caused by multiple RC defective activities and tissue-specific mtDNA depletion, and (iii) increased susceptibility to parthanatos, the AIF-mediated programmed cell death. It is difficult for the moment to establish the quantitative contribution of these three molecular abnormalities to the clinical phenotype, but we favor the hypothesis that instability of mutant AIFR201 del ultimately causes destabilization of the inner mitochondrial membrane and damage of the respiratory chain structures and activities. The latter are possibly consequent to a combination of a direct effect of AIFR201 del on the integrity of the inner mitochondrial membrane, as well as to the reduction of the mtDNA copy number. The existence of an independent pathogenic mechanism, based on increased propensity to cell death by mutant AIFR201 del, is supported by experimental evidence from our in vitro DNA-binding assays, as well as increased caspase-independent nuclear condensation and DNA fragmentation shown in both mutant fibroblasts and muscle fibers. In addition, in silico structural prediction, in vitro characterization of the recombinant mutant protein, and ex vivo correction of RC defects by expression of the wild-type protein in mutant cell lines all converge to provide solid evidence that the AIFR201 del mutation is indeed pathogenic and responsible for the human disease. This is the first example of a human mitochondrial encephalomyopathy directly linked to impaired control of mitochondrion-driven programmed cell death. The effect of AIFR201 del on RC activities in humans was similar, but not identical, to that caused by reduced AIF expression in the Harlequin (Hq) mouse strain, a spontaneous model of AIF deficiency due to a retroviral insertion into the AIFM1 gene. The 80% reduction in the normal AIF protein expression found in the Hq model23 causes progressive degeneration of cerebellar and retinal neurons, leading to ataxia and blindness, and is associated with moderate reduction of complex I activity, restricted to affected tissues.24 Neither a defect of cI in liver and heart nor defects of cIII and cIV in brain and retina were reported in the Hq model; the mtDNA copy number was apparently normal in Hq embryonic stem cells, but no data are available for mtDNA quantification in affected tissues. The multiple RC defects, predominantly affecting cIII and cIV activities, and the mtDNA depletion detected in skeletal muscle of our patients do suggest a more complex mechanism of damage of the RC and mtDNA in the human syndrome. The differences between the Hq mouse versus human patients are possibly due, at least in part, to the different genetic lesions affecting AIF. In contrast to the Hq mouse mutation, the R201 del mutation does not reduce expression of AIF but rather determines a qualitatively abnormal behavior by affecting both redox and DNA-binding properties of the protein. Nevertheless, in both Hq mouse and human patients, the mechanism by which mutant AIF deleteriously affects the RC activity is presently unknown. In humans, the RC defects and the mtDNA reduction vary according to the cell type and OXPHOS demand, therefore favoring the idea that AIF can influence RC and mtDNA integrity by taking part in a complex, possibly tissue-specific control network that might (a) act on the stability of mitochondrial membrane structures and shape,25 (b) regulate the levels of ROS and the mitochondrial redox state, or (c) combine both mechanisms. Although the constitutive knockout of AIF is embryonic lethal in mice, an isolated partial cI defect, or combined partial cI and cIV defects, have been reported in muscle and heart, respectively, in a cardiac- and skeletal-muscle-specific conditional AIF knockout mouse model, again suggesting that the biochemical effects of defective AIF on RC activities might vary in different tissues and organisms.26 No mtDNA quantification was reported in these conditional models.
Physical interactions between AIF and RC complexes and between AIF and OPA1, a morphogenetic and apoptosis controller of mitochondria, have been proposed to participate in the pathogenesis of DOA (dominant optic atrophy, MIM #165500), a condition associated with OPA1 mutations.27 Although we failed to replicate these data in our conditions (not shown) and found no obvious abnormalities in the composition of OPA1 isoforms in AIFR201 del mutant fibroblasts (Figure S4), the possible role of AIF in morphogenesis of mitochondria and RC stabilization is suggested by disruption of the mitochondrial network in our mutant cells grown in galactose and by the partial mtDNA depletion that suggests instability of the mitochondrial nucleoid structure. In addition, although the role of AIFsol in parthanatos is well documented and firmly established, the role of AIFmit as a NADH oxidase flavoprotein is completely unknown, as is the correlation, if any, between these two functions. There is clearly more work to be done on the multiple homeostatic and metabolic roles of this intriguing factor so that we can fully understand the molecular and cellular pathogenesis associated with its deficiency or defect. The relation between AIF and RC activity, established by investigation of both mouse and human AIF-related disorders, offers to future research an interesting and further exploitable clue. Finally, we observed in patients, and showed quantitatively in mutant cell lines, that riboflavin supplementation can partially correct increased cell death and severe respiratory-chain deficiency. This result, which has relevant implication for therapy, is most likely due to the fact that AIF is a FAD-containing protein whose redox activity is strictly required for normal mitochondrial functioning.28 Because the flavin binding is partially impaired in AIFR201 del, the mutant cells and organisms could benefit from higher availability of FAD, which can be achieved by a simple increase in the supply of its vitamin precursor.
Acknowledgments
We are grateful to Flavia Blasevich for technical support in electron microscopy studies. This work was supported by the Pierfranco and Luisa Mariani Foundation Italy, Fondazione Telethon-Italy grant number GGP07019, and grant RF-INN-2007-634163 of the Italian Ministry of Health.
Supplemental Data
Web Resources
The URL for data presented herein is as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
References
- 1.Schapira A.H. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 2.Smeitink J.A., Zeviani M., Turnbull D.M., Jacobs H.T. Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Regev-Rudzki N., Pines O. Eclipsed distribution: A phenomenon of dual targeting of protein and its significance. Bioessays. 2007;29:772–782. doi: 10.1002/bies.20609. [DOI] [PubMed] [Google Scholar]
- 4.McFarland R., Turnbull D.M. Batteries not included: Diagnosis and management of mitochondrial disease. J. Intern. Med. 2009;265:210–228. doi: 10.1111/j.1365-2796.2008.02066.x. [DOI] [PubMed] [Google Scholar]
- 5.Modjtahedi N., Giordanetto F., Madeo F., Kroemer G. Apoptosis-inducing factor: Vital and lethal. Trends Cell Biol. 2006;16:264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Joza N., Pospisilik J.A., Hangen E., Hanada T., Modjtahedi N., Penninger J.M., Kroemer G. AIF: Not just an apoptosis-inducing factor. Ann. N. Y. Acad. Sci. 2009;117:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- 7.Brett C.L., Wei Y., Donowitz M., Rao R. Human Na+/H+ exchanger isoform 6 is found in recycling endosomes of cells, not in mitochondria. Am. J. Physiol. Cell Physiol. 2002;282:C1031–C1041. doi: 10.1152/ajpcell.00420.2001. [DOI] [PubMed] [Google Scholar]
- 8.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Constantini P., Loeffler M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 9.Arnoult D., Parone P., Martinou J.C., Antonsson B., Estaquier J., Ameisen J.C. Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J. Cell Biol. 2002;159:923–929. doi: 10.1083/jcb.200207071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otera H., Ohsakaya S., Nagaura Z., Ishihara N., Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu S.W., Wang Y., Frydenlund D.S., Ottersen O.P., Dawson V.L., Dawson T.M. Outer mitochondrial membrane localization of apoptosis-inducing factor: Mechanistic implications for release. ASN Neuro. 2009;1 doi: 10.1042/AN20090046. Published online November 18, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuste V.J., Moubarak R.S., Delettre C., Bras M., Sancho P., Robert N., d'Alayer J., Susin S.A. Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 2005;12:1445–1448. doi: 10.1038/sj.cdd.4401687. [DOI] [PubMed] [Google Scholar]
- 13.Polster B.M., Basañez G., Etxebarria A., Hardwick J.M., Nicholls D.G. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 2005;280:6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Yang C., Chai J., Shi Y., Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 15.Andrabi S.A., Dawson T.M., Dawson V.L. Mitochondrial and nuclear cross talk in cell death: Parthanatos. Ann. N Y Acad. Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maté M.J., Ortiz-Lombardia M., Boitel B., Haouz A., Tello D., Susin S.A., Penninger J., Kroemer G., Alzari P.M. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat. Struct. Biol. 2002;9:442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 17.Ye H., Candé C., Stephanou N.C., Jiang S., Gurbuxani S., Larochette N., Dauglas E., Garrido C., Kroemer G., Wu H. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat. Struct. Biol. 2002;9:680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 18.Sevrioukova I.F. Redox-linked conformational dynamics in apoptosis-inducing factor. J. Mol. Biol. 2009;390:924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Churbanova I.Y., Sevrioukova I.F. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2008;283:5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 20.Youle R.J., Karbowski M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Dawson V.L., Dawson T.M. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp. Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen R.C., Zoghbi H.Y., Moseley A.B., Rosenblatt H.M., Belmont J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 23.Klein J.A., Longo-Guess C.M., Rossmann M.P., Seburn K.L., Hurd R.E., Frankel W.N., Bronson R.T., Ackerman S.L. The harlequin mouse mutation down-regulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 24.Vahsen N., Cande C., Briere J.J., Benit P., Joza N., Larochette N., Mastroberardino P.G., Pequignot M.O., Casares N., Lazar V. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung E.C., Joza N., Steenaart N.A., McClellan K.A., Neuspiel M., McNamara S., MacLaurin J.G., Rippstein P., Park D.S., Shore G.C. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joza N., Oudit G.Y., Brown D., Benit P., Kassiri Z., Vahsen N., Benoit L., Patel M.M., Nowikovsky K., Vassault A. Muscle-specific loss of Apoptosis-Inducing Factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol. Cell. Biol. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanna C., Ghelli A., Porcelli A.M., Karbowski M., Youle R.J., Schimpf S., Wissinger B., Pinti M., Cossarizza A., Vidoni S. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–367. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 28.Urbano A., Lakshmanan U., Choo P.H., Kwan J.C., Ng P.Y., Guo K., Dhakshinamoorthy S., Porter A. AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. EMBO J. 2005;24:2815–2826. doi: 10.1038/sj.emboj.7600746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


