Abstract
Large-scale DNA databanks linked to electronic medical record (EMR) systems have been proposed as an approach for rapidly generating large, diverse cohorts for discovery and replication of genotype-phenotype associations. However, the extent to which such resources are capable of delivering on this promise is unknown. We studied whether an EMR-linked DNA biorepository can be used to detect known genotype-phenotype associations for five diseases. Twenty-one SNPs previously implicated as common variants predisposing to atrial fibrillation, Crohn disease, multiple sclerosis, rheumatoid arthritis, or type 2 diabetes were successfully genotyped in 9483 samples accrued over 4 mo into BioVU, the Vanderbilt University Medical Center DNA biobank. Previously reported odds ratios (ORPR) ranged from 1.14 to 2.36. For each phenotype, natural language processing techniques and billing-code queries were used to identify cases (n = 70–698) and controls (n = 808–3818) from deidentified health records. Each of the 21 tests of association yielded point estimates in the expected direction. Previous genotype-phenotype associations were replicated (p < 0.05) in 8/14 cases when the ORPR was > 1.25, and in 0/7 with lower ORPR. Statistically significant associations were detected in all analyses that were adequately powered. In each of the five diseases studied, at least one previously reported association was replicated. These data demonstrate that phenotypes representing clinical diagnoses can be extracted from EMR systems, and they support the use of DNA resources coupled to EMR systems as tools for rapid generation of large data sets required for replication of associations found in research cohorts and for discovery in genome science.
Introduction
The deployment of electronic medical record (EMR) systems offers the hope of improving routine care, not only by enhancing individual practitioner access to patient information but also by aggregating information for clinical research and quality improvement.1 EMRs and associated support systems can reduce medication errors, costs, and inappropriate testing and improve quality of care, physician documentation, and guideline adherence.2–8
Because EMRs contain large populations with diverse diseases, they have the potential to act as platforms for generating sets of cases and controls for clinical and translational research. Potential advantages of such an approach include rapid and inexpensive creation of large, inclusive patient sets,9 as well as support for studies of disease-disease or disease-drug interactions over time. An especially appealing, albeit complex, vision is one in which dense genomic information is accrued into EMRs, ultimately enabling discovery and incorporation into practice of new genotype-phenotype associations.10–12
Implementing such a vision requires that major obstacles be overcome, including technological, computational, ethical, and financial issues, and determining whether genomic information will meaningfully inform clinical decision making and healthcare outcomes. An important hypothesis to be tested13 is the idea that large biorepositories containing DNA samples linked to EMRs can be used to address these challenges to augment or supplement traditional research designs in which cases and controls are prospectively enrolled and phenotype data are systematically collected.
BioVU, the Vanderbilt DNA databank,14 represents such a biorepository. To test the utility of the resource in addressing these challenges, we genotyped the first ∼10,000 samples accrued (over 4 mo) for SNP sites reproducibly associated with a range of human diseases. Automated queries were then developed, validated, and deployed for the identification of cases and controls in five common diseases, and previously reported genotype-phenotype relationships were examined. Our results provide support for the concept that biorepositories linked to “real-world” EMR data represent robust tools for accelerating genome-driven diagnostics and therapeutics.
Subjects and Methods
Inclusion and Exclusion Criteria
BioVU accrues DNA samples extracted from blood remaining from routine clinical testing after the samples have been retained for 3 days and are scheduled to be discarded. A full description of the resource and its ethical protections has been published elsewhere.14 The resource contains data and tissues that are deidentified in accordance with provisions of Title 45, Code of Federal Regulations, part 46 (45 CFR 46) that define criteria for investigations that are considered “nonhuman subjects” research. Exclusion criteria are as follows: poor-quality or insufficient DNA, age < 18 yrs, absence of a signed consent-to-treatment form, a formal indication (on the consent-to-treatment form or elsewhere) that an individual wishes to opt out, or duplicate samples. In addition, a small percentage (∼2%) of patients is randomly excluded from BioVU so that it is not possible to know whether any individual's sample is or is not included in the biobank. The project has been reviewed and approved at multiple levels, including the institutional review board, internal and external ethics committees, the community advisory board and the legal department (Nashville, TN, USA), as well as the Federal Office of Human Research Protection (Washington, D.C., USA), and this oversight is ongoing. As of March 22, 2010. the resource included 80,635 samples, with an accrual rate of ∼500–700 per wk.
Phenotypes and Genotypes Analyzed
Our aim was to study SNPs previously and reproducibly associated in GWAS with susceptibility to common diseases and to determine whether those associations could be replicated with the use of only the information derived from the EMR to determine case and control status. The conditions chosen were atrial fibrillation (AF),15 Crohn disease (CD),16–18 multiple sclerosis (MS [MIM 126200]),19,20 rheumatoid arthritis (RA [MIM 180300]),16,21 and type 2 diabetes (T2D [MIM 125853]), because there were several single locus hits to replicate for each disorder and the diseases varied in level of difficulty for the EMR phenotype abstraction.16,22–24 The assayed SNPs and their primer sequences are presented in Table S1, available online. We selected 21 SNPs that had consistently shown replication for these diseases by late 2007; SNP associations studied after this date were therefore not included. The individuals studied were accrued in the first 4 mo of operation of the resource.
We selected this set of diseases and SNPs to ask the question of whether EMR-derived phenotypes can be used for human genetic-association studies. The previously reported odds ratios (ORPR) for the SNPs that we examined ranged from 1.14 to 2.36. Table S1 also presents power calculations25 performed in determining the sample size needed to replicate each ORPR at 80% power, given reported allele frequencies in cases and controls for each disease, for determining whether we were powered to detect the selected effects.
Identifying Cases and Controls
For each disease, cases and controls were identified in the Synthetic Derivative, a deidentified image of the EMR linked to BioVU by anonymous research unique identifiers.14 The Vanderbilt EMR began accumulating clinical data in the early 1990s and now includes all inpatient and outpatient billing codes, laboratory values, reports, and clinical documentation, almost all in electronic formats available for searching. It currently contains over 120 million documents on about 2 million patients.26 The synthetic derivative is refreshed monthly to add new clinical information from the EMR as it is accrued.
For each disease, content experts (listed in Acknowledgments) were consulted to develop algorithms that segmented the deidentified EMR data into four groups: definite cases (algorithm-defined); possible cases (requiring manual curation); exclusions for matching potentially overlapping diseases or symptoms or insufficient data for classifying; and controls (algorithm-defined). The selection algorithms are presented in the Appendix. In brief, cases were selected via disease-specific combinations of billing codes, patient encounters, laboratory data, and natural language processing (NLP) techniques on unstructured patient records such as medications, electrocardiograms, or past medical history. To define controls, we adopted criteria to ensure that the diagnosis had been sought and was absent. This generally included visits to primary care or internal medicine clinics with nonempty “past medical history” sections and medication lists. In addition, controls for atrial fibrillation all had electrocardiograms that did not show atrial fibrillation. We also excluded potentially overlapping conditions, such as other inflammatory arthritides from rheumatoid arthritis controls and autoimmune diseases from rheumatoid arthritis, multiple sclerosis, and Crohn disease controls.
Initial algorithms were developed to identify records from each of the four classifications from the samples in the synthetic derivative, and the results were reviewed by two physicians not associated with algorithm development. The results of the manual classification were then used to improve the algorithms, and the procedure was iterated until the positive predictive value (PPV) reached designated targets of ≥ 95% for definite cases and ≥ 98% for controls. In each algorithm iteration, the physicians reviewed a different set of 50 randomly selected cases and controls, and the final algorithm-performance numbers represent the performance of the algorithm on a test set not previously reviewed by either the algorithm developers or the physician reviewers. The final algorithms were then applied to the initial BioVU sample set of 9483 subjects. Populations for cases and controls for rheumatoid arthritis, Crohn disease, atrial fibrillation, and type 2 diabetes were defined exclusively by the final algorithm. Each phenotype algorithm was run on the full set of 9483 subjects such that a particular person could be a case for one condition, a control for one or more other conditions, or unknown for one or more other conditions. Because of the low count of definite and possible cases for multiple sclerosis, we tuned the multiple sclerosis algorithm for sensitivity and the physician reviewers manually reviewed the electronic records of all definite and possible cases.
Data Analysis
Genotyping was conducted by the Vanderbilt DNA Resources Core with the use of the midthroughput Sequenom genotyping platform, based on a single-base primer extension reaction coupled with mass spectrometry. Quality-control procedures included examination of marker and sample genotyping efficiency, allele-frequency calculations, and tests of Hardy-Weinberg equilibrium (HWE). As described in Results, some SNPs poorly assayed on the Sequenom platform (as indicated by deviations from HWE) were regenotyped with TaqMan assays and the ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
Ancestry was derived from administrative data recorded in the EMR; 9.2% of records did not include recorded ancestry or recorded the ancestry as “unknown.” Accordingly, the data were analyzed with respect to ancestry in two different ways. In the first, only cases and controls that had “non-Hispanic white” (European American) coded in the EMR were used, whereas in the second, records designated as either “non-Hispanic white” or “unknown” were included. Preliminary data directly examining ancestry-informative markers in the BioVU “unknown” group indicate that > 85% of cases cluster with European Americans (Ritchie et al., poster presented as part of the Illumina Technology Workshops at the ASHG 59th Annual Meeting, Honolulu, HI, USA, October 22, 2009). Thus, we expect that both the non-Hispanic white and the non-Hispanic white + unknown records are primarily European American.
Single-locus tests of association (cases versus controls) were conducted for each previously reported SNP. We calculated allelic (chi-square) and genotypic (logistic regression) ORs and 95% confidence intervals (CIs) for dominant, additive, and recessive genetic models. All statistical analyses were conducted in SAS, version 9.1, with the use of two-tailed tests.
Role of the Funding Source
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit for publication. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Identifying Cases and Controls
The positive predictive values of case and control selection algorithms were 97%–100%, except for multiple sclerosis, in which algorithm-classified possible cases were also manually reviewed because of small sample size (Table S2). For the rheumatoid arthritis and multiple sclerosis phenotypes, we identified both definite cases and definite cases with other, potentially overlapping, autoimmune diseases (“probable cases”): for rheumatoid arthritis, there were 174 definite cases of European ancestry and 346 definite + probable cases of European or unknown ancestry, and the corresponding figures for multiple sclerosis were 70 and 124, respectively.
Figure 1 shows the distribution of cases and controls across the five phenotypes analyzed. In the set of 9483 records analyzed by NLP for the five diseases, there were 1212 definite cases of European ancestry and 5114 controls. There were 4072 that served as either a case or a control in more than one analysis.
Figure 1.
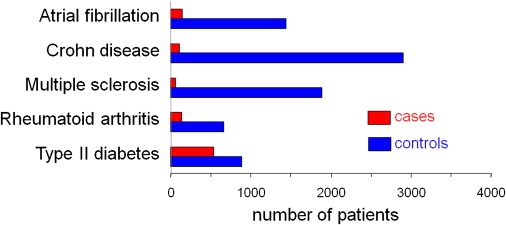
Identifying Cases and Controls
The numbers presented are for European American subjects, with a definite diagnosis. Table S2 presents the numbers for other groupings studied here.
Genotyping
A total of 9483 samples were genotyped for 23 SNPs across the five diseases with the use of the Sequenom platform. We excluded 1847 samples with genotyping efficiency < 90% from data analysis. These failures were primarily from the first several plates processed and reflected the fact that this was the first set of DNA samples genotyped with the use of the Sequenom technology in the DNA Resources Core facility. Genotyping efficiency for individual SNPs varied from 95.4%–100% (Table S3). The number of definite cases of European ancestry in which genotyping was successful ranged from 61 (multiple sclerosis) to 510–533 (type 2 diabetes; depending on the specific SNP), with 658–2884 controls (Table 1 and Table S4).
Table 1.
Allelic Odds Ratio
| SNP |
Cases |
Controls |
Cases |
Cases |
Controls |
Controls |
|||
|---|---|---|---|---|---|---|---|---|---|
| N | N | Minor Allele Frequency | Minor Allele | Minor Allele Frequency | Minor Allele | Allelic Chi-Square p Value | Odds Ratio | 95% Confidence Interval | |
| ATRIAL FIBRILLATION | |||||||||
| Ancestry: Non-Hispanic European; Case Definition: Definite | |||||||||
| rs2200733 | 147 | 1439 | 0.1599 | T | 0.1120 | T | 0.0147 | 1.5093 | (1.08–2.11) |
| rs10033464 | 143 | 1402 | 0.1084 | T | 0.0881 | T | 0.2530 | 1.2585 | (0.85–1.87) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite | |||||||||
| rs2200733 | 148 | 1467 | 0.1588 | T | 0.1115 | T | 0.0153 | 1.5048 | (1.08–2.10) |
| rs10033464 | 144 | 1432 | 0.1111 | T | 0.0887 | T | 0.2066 | 1.2844 | (0.87–1.90) |
| CROHN DISEASE | |||||||||
| Ancestry: Non-Hispanic European; Case Definition: Definite | |||||||||
| rs11805303 | 107 | 2884 | 0.3271 | T | 0.3017 | T | 0.4263 | 1.1253 | (0.84–1.51) |
| rs17234657 | 106 | 2890 | 0.2028 | G | 0.1201 | G | 0.0003 | 1.8646 | (1.32–2.63) |
| rs1000113 | 107 | 2905 | 0.0935 | T | 0.0730 | T | 0.2601 | 1.3096 | (0.82–2.10) |
| rs17221417 | 107 | 2896 | 0.3785 | G | 0.2949 | G | 0.0086 | 1.4562 | (1.10–1.93) |
| rs2542151 | 107 | 2901 | 0.1542 | G | 0.1649 | G | 0.6774 | 1.0834 | (0.74–1.58) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite | |||||||||
| rs11805303 | 110 | 3175 | 0.3288 | T | 0.3054 | T | 0.4557 | 1.1145 | (0.84–1.45) |
| rs17234657 | 110 | 3182 | 0.1955 | G | 0.1204 | G | 0.0009 | 1.7756 | (1.23–2.50) |
| rs1000113 | 111 | 3199 | 0.0946 | T | 0.0769 | T | 0.3323 | 1.2542 | (0.79–1.98) |
| rs17221417 | 111 | 3188 | 0.3874 | G | 0.2917 | G | 0.0021 | 1.5353 | (1.17–2.02) |
| rs2542151 | 111 | 3195 | 0.1577 | G | 0.1635 | G | 0.8158 | 1.0446 | (0.72–1.51) |
| MULTIPLE SCLEROSIS | |||||||||
| Ancestry: Non-Hispanic European; Case Definition: Definite | |||||||||
| rs6897932 | 61 | 1861 | 0.2049 | T | 0.2515 | T | 0.2425 | 1.3036 | (0.83–2.04) |
| rs3135388 | 61 | 1892 | 0.2887 | T | 0.1427 | T | <0.0001 | 2.3210 | (1.55–3.48) |
| rs2104286 | 61 | 1888 | 0.2377 | A | 0.2582 | A | 0.6102 | 1.1163 | (0.73–1.70) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite | |||||||||
| rs6897932 | 88 | 2105 | 0.2045 | T | 0.2485 | T | 0.1855 | 1.2857 | (0.89–1.87) |
| rs3135388 | 88 | 2139 | 0.2955 | T | 0.1431 | T | <0.0001 | 2.5120 | (1.80–3.51) |
| rs2104286 | 88 | 2133 | 0.2102 | A | 0.2586 | A | 0.1503 | 1.3101 | (0.91–1.89) |
| Ancestry: Non-Hispanic European; Case Definition: Definite + Probable | |||||||||
| rs6897932 | 68 | 1861 | 0.2132 | T | 0.2515 | T | 0.3118 | 1.2396 | (0.82–1.88) |
| rs3135388 | 68 | 1892 | 0.2574 | T | 0.1427 | T | 0.0002 | 2.0818 | (1.40–3.09) |
| rs2104286 | 68 | 1888 | 0.2279 | A | 0.2582 | A | 0.4275 | 1.1790 | (0.78–1.77) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite + Probable | |||||||||
| rs6897932 | 96 | 2105 | 0.2083 | T | 0.2485 | T | 0.2072 | 1.2563 | (0.88–1.79) |
| rs3135388 | 96 | 2139 | 0.2760 | T | 0.1431 | T | <0.0001 | 2.2840 | (1.65–3.17) |
| rs2104286 | 96 | 2133 | 0.2083 | A | 0.2586 | A | 0.1190 | 1.3252 | (0.93–1.89) |
| RHEUMATOID ARTHRITIS | |||||||||
| Ancestry: Non-Hispanic European; Case Definition: Definite | |||||||||
| rs6679677 | 134 | 658 | 0.1194 | A | 0.1003 | A | 0.3496 | 1.2162 | (0.81–1.83) |
| rs2476601 | 134 | 659 | 0.1194 | A | 0.1002 | A | 0.3454 | 1.2183 | (0.81–1.84) |
| rs6457620 | 138 | 662 | 0.3370 | T | 0.4977 | T | <0.0001 | 1.9501 | (1.49–2.56) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite | |||||||||
| rs6679677 | 184 | 745 | 0.1141 | A | 0.0943 | A | 0.2609 | 1.2326 | (0.86–1.78) |
| rs2476601 | 184 | 746 | 0.1141 | A | 0.0945 | A | 0.2576 | 1.2344 | (0.86–1.88) |
| rs6457620 | 188 | 750 | 0.3601 | T | 0.4973 | T | <0.0001 | 1.6689 | (1.33–2.09) |
| Ancestry: Non-Hispanic European; Case Definition: Definite + Probable | |||||||||
| rs6679677 | 210 | 658 | 0.1286 | A | 0.1003 | A | 0.1029 | 1.3235 | (0.94–1.85) |
| rs2476601 | 210 | 659 | 0.1262 | A | 0.1002 | A | 0.1319 | 1.2975 | (0.92–1.82) |
| rs6457620 | 214 | 662 | 0.3626 | T | 0.4977 | T | <0.0001 | 1.7422 | (1.39–2.18) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite + Probable | |||||||||
| rs6679677 | 272 | 745 | 0.1250 | A | 0.0946 | A | 0.0459 | 1.3667 | (1.00–1.86) |
| rs2476601 | 272 | 746 | 0.1232 | A | 0.0945 | A | 0.0589 | 1.3459 | (0.99–1.83) |
| rs6457620 | 277 | 750 | 0.3776 | T | 0.4896 | T | <0.0001 | 1.6521 | (1.35–2.02) |
| TYPE 2 DIABETES | |||||||||
| Ancestry: Non-Hispanic European; Case Definition: Definite | |||||||||
| rs4402960 | 527 | 877 | 0.3083 | T | 0.3079 | T | 0.9787 | 1.0023 | (0.85–1.18) |
| rs10811661 | 534 | 887 | 0.1610 | C | 0.1753 | C | 0.3269 | 1.1074 | (0.90–1.36) |
| rs4506565 | 532 | 886 | 0.3524 | T | 0.3053 | T | 0.0093 | 1.2384 | (1.05–1.46) |
| rs12243326 | 520 | 876 | 0.3212 | C | 0.2785 | C | 0.0169 | 1.2253 | (1.04–1.45) |
| rs12255372 | 510 | 847 | 0.3245 | T | 0.2816 | T | 0.0178 | 1.2257 | (1.04–1.45) |
| rs5215 | 527 | 882 | 0.3672 | C | 0.3702 | C | 0.8728 | 1.0130 | (0.86–1.19) |
| rs5219 | 533 | 888 | 0.3715 | T | 0.3705 | T | 0.9580 | 1.0042 | (0.86–1.18) |
| rs8050136 | 533 | 886 | 0.4053 | A | 0.3916 | A | 0.4731 | 1.0584 | (0.91–1.24) |
| Ancestry: Non-Hispanic European + Unknown; Case Definition: Definite | |||||||||
| rs4402960 | 548 | 1089 | 0.3139 | T | 0.3159 | T | 0.9067 | 1.0094 | (0.86–1.18) |
| rs10811661 | 555 | 1103 | 0.1604 | C | 0.1727 | C | 0.3700 | 1.0931 | (0.90–1.33) |
| rs4506565 | 553 | 1100 | 0.3535 | T | 0.3100 | T | 0.0117 | 1.2172 | (1.04–1.42) |
| rs12243326 | 541 | 1088 | 0.3226 | C | 0.2845 | C | 0.0251 | 1.1976 | (1.02–1.40) |
| rs12255372 | 530 | 1048 | 0.3236 | T | 0.2863 | T | 0.0305 | 1.1928 | (1.02–1.40) |
| rs5215 | 547 | 1098 | 0.3656 | C | 0.3643 | C | 0.9404 | 1.0057 | (0.87–1.17) |
| rs5219 | 554 | 1103 | 0.3700 | T | 0.3649 | T | 0.7728 | 1.0223 | (0.88–1.19) |
| rs8050136 | 554 | 1102 | 0.4043 | A | 0.3897 | A | 0.4177 | 1.0628 | (0.92–1.23) |
Four SNPs were flagged because of significant deviations from HWE at p < 0.05. Two of these (rs7901695 and rs7903146), both in TCF7L2 (MIM 602228) and associated with type 2 diabetes, are in linkage disequilibrium with other markers studied here in the same gene (based on the HapMap) and so were eliminated from further analysis. One rheumatoid arthritis-associated SNP in the HLA cluster on chromosome 6 (MIM 142860), rs6457617, could not be successfully assayed with Sequenom or TaqMan. We therefore dropped this SNP and instead genotyped rs6457620 by using a TaqMan assay (genotyping efficiency = 99.46%; HWE p value = 0.26). The SNP rs6457620 is in complete linkage disequilibrium with rs6457617 (r2 = 1) in the International HapMap Project samples of European ancestry (CEU).27 The fourth SNP, rs2200733, an atrial-fibrillation-associated variant, had low genotyping efficiency in addition to deviation from HWE (p < 0.05). This SNP was successfully genotyped with TaqMan (genotyping efficiency = 99%; HWE p value = 0.64). The final analysis therefore included 21 SNPs.
Genotype-Phenotype Associations
The number of cases needed to achieve 80% power at α = 0.05 for the SNPs studied varied widely, from 75 to 3111, assuming a 1:2 ratio of cases versus controls (Table S1). This range reflects the ORPR and minor allele frequencies (MAFs) in cases and controls. Table S1 also presents the number of cases needed assuming other ratios of cases to controls, from 1:1 to 1:4. Increasing the number of controls 4-fold resulted in a modest reduction, ∼30%, of the cases needed.
Figure 2 presents the calculated allelic OR and 95% CIs, with the use of definite cases and controls and European ancestry only. In addition to the results from BioVU data, we plot the OR for the original GWAS result; subsequent publications confirming these results have very similar values (data not shown). For example, the SNP rs4506565 in TCF7L2 has previously been associated with type 2 diabetes, with an ORPR of 1.37 and MAFs of 0.32 in cases and 0.39 in controls.24 To replicate this finding assuming a similar genetic effect size, we would need to identify 503 cases and 1006 controls. Genotyping in subjects identified by the phenotype algorithms resulted in 532 definite cases (MAF 0.35) and 886 controls (MAF 0.31), resulting in a calculated allelic OR of 1.29 (95% CI: 1.09–1.53; p = 0.009; Table S4). The genotypic OR was 1.23 (1.05–1.45; p = 0.006; Table S4). Homozygotes for the minor (risk) allele had an OR of 1.7 (1.22–2.38; p = 0.0016) compared to homozygotes for the major allele or heterozygotes. The results were very similar when the 553 cases and 1100 controls of European or unknown ancestry were analyzed. Table 1 presents the results of all allelic analyses, and Table S4 presents the genotypic analyses, for both European and European + unknown ancestry, and for definite as well as definite + probable cases (rheumatoid arthritis and multiple sclerosis).
Figure 2.
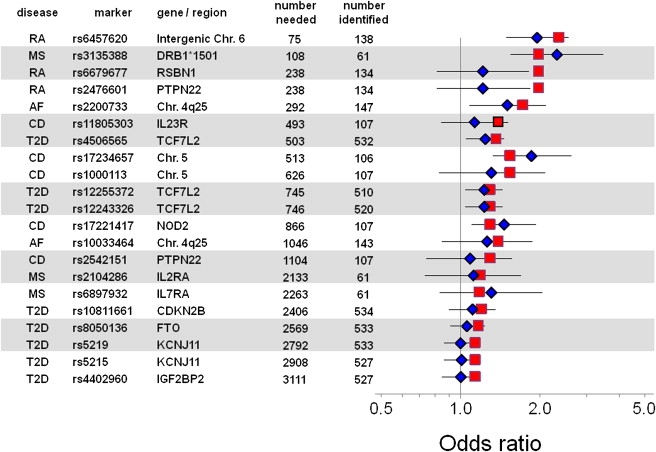
Odds Ratios for Comparisons of Cases versus Controls for Each SNP Evaluated
These are ordered by the number of cases required for replication (“number needed” column), calculated from the previously reported odds ratio (ORPR, red square; see text). The blue diamonds indicate the point estimate of the allelic OR derived from the present analysis. The error bars indicate the confidence interval of the allelic OR derived from the present analysis. This analysis used only definite cases in which European ancestry had been assigned. AF, atrial fibrillation; CD, Crohn disease; MS, multiple sclerosis; RA, rheumatoid arthritis; T2D, type 2 diabetes.
All 21 comparisons resulted in OR point estimates in the expected direction, and for each of the five diseases studied, at least one previously reported SNP association was replicated (p < 0.05 for allelic OR). Two sets of SNPs in type 2 diabetes were in LD with one another and, comfortingly, showed similar results. The SNPs rs4506565, rs1224332, and rs225537 (in or near TCF7L2) have pairwise r2 values of between 0.658 and 0.902; all three SNPs were statistically significantly associated with type 2 diabetes. Similarly, rs5215 and rs5219 (in KCNJ11 [MIM 600937]) have a pairwise r2 of 0.994, and both were nonsignificant. The analyses in Figure 2 are ordered by the number of cases required for replication, and they show that the fewer cases required, the greater the likelihood of replication with 95% CIs of the point estimate not overlapping unity. When fewer than 1000 cases were required, 8/12 associations replicated in this fashion, whereas there was no replication (0/9) of associations predicted to require more than 1000 cases. Similarly, a low ORPR implied that a large number of cases would be required for replication: indeed, 8/14 associations with an ORPR > 1.25 replicated, in contrast to 0/7 with ORPR ≤ 1.2.
Analyses that included the slightly larger numbers of subjects whose ancestry was European or unknown yielded the same results: replication of the same 8/14 associations with ORPR > 1.25. When both definite and probable cases for rheumatoid arthritis and multiple sclerosis were included in the case series, one more association was replicated in the rheumatoid arthritis set (Table 1).
Discussion
We demonstrate here that common genetic variants associated with disease can be replicated with the use of samples from a DNA databank coupled to a deidentified EMR, in effect rejecting the null hypothesis that EMR-derived phenotypes are insufficiently distinct or detailed to identify such associations that historically have been observed in carefully characterized research cohorts. We examined SNPs with a wide range of ORPR, from 1.14 to 2.36, and showed that the likelihood that we could replicate any genotype-phenotype association in this set of 9483 records varied directly with the genetic effect size of the previously reported association. These records were selected because they were the first accrued into the resource and were accrued with no knowledge of underlying disease frequencies. The time required to generate this set from which the cases and controls were identified was only 4 mo.
Accruing, defining, and accessing samples presented multiple technical challenges, so establishing appropriate quality-control checks was vital to the success of this experiment and to use of any biorepository. Before genotyping and phenotyping, we validated sample-handling algorithms by using gender testing, as previously reported.14 A critical challenge was the definition of cases and controls, both of which represent unique challenges in the EMR and require nuanced application of information-extraction techniques. It was not unusual to find cases in which a rheumatoid arthritis or multiple sclerosis code from the International Statistical Classification of Diseases and Related Health Problems, revision 9 (ICD-9 code) was associated with a clinic visit, only to have the diagnosis subsequently overturned by a specialist consultation. Thus, the case definitions each relied on both coded and unstructured, free-text data, such as diagnoses, medications, or laboratory findings. Similarly, the criteria for controls were not simply the absence of the condition, but also included the absence of clinically similar conditions and included documentation that the condition was sought as well as absence of clinically similar conditions. Assurance that controls do not have the disease is especially important for relatively common diseases such as diabetes, although it is potentially less impactful for rare diseases such as multiple sclerosis. To avoid introduction of bias in our control populations by overly restrictive criteria, these algorithms ensured that control subjects did not have the case (or similar) diseases through clinic visits with past medical history assessments, common laboratory values, and normal electrocardiograms.
The BioVU DNA Databank currently uses ancestry assigned in administrative (e.g., billing) databases. The 9483 records analyzed included 77% European American, 11% African American, 1% Hispanic, 1% Asian, and 1% other; an explicit statement of ancestry is absent in 9% of records. Including the samples of unknown ancestry did not alter the results, possibly because the majority of our population is non-Hispanic European American. The accuracy of ancestry information contained in administrative databases is unknown, although self-reported ancestry agrees well with genetically determined race.28 Our current evaluation of the ancestry of the “unknown” population of BioVU indicates that the majority of these individuals are European American (Ritchie et al., poster presented as part of the Illumina Technology Workshops at the ASHG 59th Annual Meeting, Honolulu, HI, USA, October 22, 2009).
Although EMR-based phenotyping can be complex, as discussed above, the approach also has potential advantages. In some instances, the case diagnosis can be firmer than that in prospective clinical trials or case-control studies, because longitudinal information from multiple, interacting, trained physicians is available. For example, differentiating Crohn disease from other conditions such as ulcerative colitis benefits from longitudinal information from multiple, interacting, trained physicians when available. Similarly, controls may also represent a more definite phenotype than those in traditional research cohorts because there is an average of 6.6 yrs of follow up in the EMR. In our study, replication of genotype-phenotype associations was slightly improved by incorporation of definite + probable phenotypes in rheumatoid arthritis, but the sample sizes are small.
In this experiment, we did not replicate any association with an ORPR < 1.25 or an estimated number of required cases > 1000. Because it was undertaken early in the growth of the biobank, the study was anticipated to be underpowered to assess many of the analyses attempted, given the prevalence of the diseases of interest in the first 10,000 samples accrued. Our failure to replicate could also have arisen because these associations could be important only in certain populations. This highlights one of the advantages of large DNA resources such as BioVU: simple extrapolation indicates that with the current size of the biobank, 18/21 tests of replication would be adequately powered. It is noteworthy that the OR point estimates for all 21 associations evaluated were in the expected direction. Larger populations may also permit discovery of new genetic associations.
However, we successfully replicated genotype-phenotype associations that were underpowered. Although this could reflect simple statistical variation for a few SNPs, it was observed in multiple analyses for multiple diseases. It is conceivable that that the phenotype criteria used here result in a more stringently defined case than in the original studies that identified the genotype-phenotype association. In addition, other studies have used controls drawn from apparently healthy populations,16 and to the extent that these include subjects who actually have disease, the EMR-based approach that we have used may be superior. It is also possible that the risk alleles are enriched in the population studied, although we observed this phenomenon with multiple diagnoses. Studies in other data sets will be required to address this issue and are a goal of the National Human Genome Research Institute's eMERGE Network.29
A long-term goal of BioVU and similar resources30 is to provide a platform for evaluating and overcoming barriers to incorporation of genomic and other high-dimensional data into clinical medicine. One challenge is to determine the added value of a particular genotype or set of genotypes in clinical care. To address this question, data sets much larger than those ordinarily accrued into clinical trials may be required.31 Thus, EMR-based biobanks like BioVU may be not only useful but in fact indispensible in advancing this field. Moreover, as this experiment demonstrates, EMR-based phenotyping can rapidly generate large numbers of subjects across multiple diseases. Thus, the approach that we describe ultimately provides a platform for assessment and validation of genotype-phenotype associations in clinical practice. In addition, the method provides an opportunity to generate a closed loop: as health care information accumulates, genotype-phenotype associations will become increasingly well defined, ultimately identifying common genotype variants that are useful in clinical medicine.
Appendix: Selection of Cases and Controls
General Approach
Phenotype-selection algorithms used a combination of queries of structured billing codes and unstructured “natural language” clinical notes. Most algorithms combined a search for disease codes taken from International Classification of Diseases, version 9-CM (ICD-9), and procedure codes from Current Procedural Terminology (CPT) billing records, with textual searches for medication names used to treat the disease. For all text searches, we included synonyms, abbreviations, acronyms, and common misspellings for all terms (including generic and brand names for medications), assisted by standardized vocabularies such as the Unified Medical Language System (UMLS; see Web Resources) and natural language processing tools such as the KnowledgeMap concept identifier.32
We required that control records have “minimum content information,” defined as an inpatient history and physical admission document or comprehensive outpatient clinic note that included nonempty medication and past medical history sections.
We removed “family medical history” sections from all clinical notes prior to processing.33
The algorithms considered inpatient admission notes, progress notes, and discharge summaries; all outpatient clinic notes (> 97% of all outpatient encounters produce notes available in electronic formats since about 2001); all problem lists (these include key diagnoses and procedures, and allergy and medication lists); and the cardiologist-generated electrocardiogram (ECG) impressions.
For Crohn disease, many records included codes for both ulcerative colitis and Crohn disease (e.g., a patient initially thought to have ulcerative colitis who later has a biopsy confirming Crohn disease). To differentiate them, the algorithm also used the ratio of billing codes for each disease.
In each case, content experts were consulted to identify potentially overlapping diseases (such as other autoimmune diseases for rheumatoid arthritis, Crohn disease, and multiple sclerosis) that we excluded from both cases and controls.
Each algorithm was validated and adjusted to achieve target positive and negative predictive values, as described in the text. The final algorithms used to select cases and controls are described in further detail below.
Atrial Fibrillation
For all cases, we considered atrial fibrillation and atrial flutter as similar entities. We excluded all patients who had received heart transplants (identified by CPT codes; Table A1).
Table A1.
ICD-9 and CPT Codes Used for Excluding Heart Transplant
| Description | ICD-9/CPT Code |
|---|---|
| Heart replaced by transplant | V42.01 |
| Anesthesia for heart transplant or heart/lung transplant | 580 |
| Complications of transplanted heart | 996.83 |
| Heart-lung transplant with recipient cardiectomy-pneumonectomy | 33935 |
| Heart transplant, with or without recipient cardiectomy | 33945 |
Definite cases: To identify definite cases, we required a cardiologist diagnosis of atrial fibrillation as identified by a natural language processing tool from the unstructured free text of the “ECG impression;” i.e., an ECG with an official interpretation of atrial fibrillation. These were identified by first taking all ECG impressions (n = 14,569) and processing them with the KnowledgeMap concept identifier. This tool34 maps unstructured free text to standardized biomedical concepts with their assertion or negation status (e.g., “no atrial fibrillation” becomes “C0004238 Atrial Fibrillation, status: negated” in which “C0004238” represents a unique identifier for the concept of “atrial fibrillation” and its synonyms). There were only two negated atrial fibrillation or flutter concepts, both in records that had prior ECGs indicating atrial fibrillation (e.g., “compared with prior ECG, atrial fibrillation has resolved”).
Controls: To be a control, a record had to meet all of the conditions below:
-
•
Contain at least one ECG whose impression does not mention atrial fibrillation (or synonym).
-
•
Contain no ICD-9 codes representing atrial flutter or atrial fibrillation (Table A2).
-
•
Contain no free-text matches for atrial fibrillation or synonyms.
-
•
Contain no free-text references (including synonyms) to: direct-current cardioversion, atrial tachycardia or multifocal atrial tachycardia, atrioventricular nodal ablation.
Table A2.
ICD-9 Codes for Atrial Fibrillation or Atrial Flutter
| Description | ICD-9 Code |
|---|---|
| Atrial fibrillation | 427.31 |
| Atrial fibrillation and flutter | 427.3 |
| Atrial flutter | 427.32 |
Crohn Disease
Definite cases: To be considered a definite case, a record had to contain all of the following:
-
•
At least one ICD-9 code for Crohn disease (555.∗). If also containing an ICD-9 code for ulcerative colitis (556.∗), we required that the ratio of Crohn disease ICD-9 codes to ulcerative colitis ICD-9 codes was ≥ 2.
-
•
At least one medication used to treat Crohn disease, such as: balsalazide, mesalamine, sulfasalazine, ciprofloxacin, levofloxacin, metronidazole, rifaximin, prednisone, budesonide, azathioprine, mercaptopurine, methotrexate, infliximab, adalimumab, certolizumab, natalizumab.
Controls: These were defined as a record with “minimum content information” described above, plus none of the following:
-
•
Any free-text references to key autoimmune diseases or inflammatory bowel disease: rheumatoid arthritis, Felty syndrome, juvenile arthritis, lupus, inflammatory bowel disease, Crohn disease, ulcerative colitis, reactive arthritis, sarcoidosis, ankylosing spondylitis, Hashimoto thyroiditis, polymyositis, dermatomyositis, chronic lymphocytic thyroiditis, autoimmune thyroid disease, Graves disease, Raynaud disease, multiple sclerosis.
-
•
ICD-9 codes indicating another autoimmune diseases or inflammatory bowel disease (Table A3).
Table A3.
ICD-9 Codes Indicating Another Autoimmune Disease or Inflammatory Bowel Disease
| Description | ICD-9 Code |
|---|---|
| Rheumatoid arthritis and other inflammatory polyarthropathies | 714.∗ |
| Discoid lupus erythematosus of eyelid | 373.34 |
| Lupus erythematosus | 695.4 |
| Systemic lupus erythematosus | 710.0 |
| Systemic sclerosis | 710.1 |
| Sjogren disease | 710.2 |
| Dermatomyositis | 710.3 |
| Polymyositis | 710.4 |
| Regional enteritis of small intestine | 555.∗ |
| Sarcoidosis | 135 |
| Psoriasis and similar disorders | 696 |
| Psoriatic arthropathy | 696.0 |
| Other psoriasis and similar disorders excluding psoriatic arthropathy | 696.1 |
| Other psoriasis and similar disorders | 696.8 |
| Reiter disease | 099.3 |
| Palindromic rheumatism | 719.3 |
| Ankylosing spondylitis | 720.∗ |
| Thoracic spondylosis without myelopathy | 721.2 |
| Lumbosacral spondylosis without myelopathy | 721.3 |
| Hashimoto thyroiditis | 245.2 |
| Toxic diffuse goiter | 242.0 |
| Myasthenia gravis | 358.0∗ |
| Neonatal myasthenia gravis | 775.2 |
| Raynaud syndrome | 443 |
| Reiter disease | 099.3 |
| Multiple sclerosis | 340 |
| Demyelinating disease of the central nervous system, unspecified | 341.9 |
| Irritable bowel disease | 564.1 |
| Ulcerative enterocolitis | 556.∗ |
Multiple Sclerosis
Definite cases: To be considered a definite case, a record had to meet one of the following two case definitions:
-
•
Case definition 1: Presence of an ICD-9 code for multiple sclerosis (340).
-
•Case definition 2: Any record matching all of the following:
-
○One of the following ICD-9 codes: 341.9, demyelinating disease of the central nervous system, unspecified; 323.9, transverse myelitis.
-
○Any of the following medications: interferon-β 1a, interferon-β 1b, glatiramer, natalizumab.
-
○Text match of “multiple sclerosis.”
-
○No potentially overlapping autoimmune diseases by ICD-9 code (Table A4).
-
○
Possible cases: To be considered a possible case, a record had to contain all of the following:
-
•
Text match of “multiple sclerosis” in the clinical record.
-
•
Any one of the following ICD-9 codes: other demyelinating diseases of central nervous system; demyelinating disease of the central nervous system, unspecified; optic neuritis.
-
•
No potentially overlapping autoimmune diseases by ICD-9 code (Table A4).
Because of the small number of cases of multiple sclerosis, we manually reviewed all cases and analyzed them with and without the autoimmune disease exclusions (those with potentially overlapping autoimmune diseases were referred to as “probable cases”).
Table A4.
ICD-9 Codes Indicating Another Autoimmune Disease
| Description | ICD-9 Code |
|---|---|
| Rheumatoid arthritis and other inflammatory polyarthropathies | 714.∗ |
| Discoid lupus erythematosus of eyelid | 373.34 |
| Lupus erythematosus | 695.4 |
| Systemic lupus erythematosus | 710.0 |
| Systemic sclerosis | 710.1 |
| Sjogren disease | 710.2 |
| Dermatomyositis | 710.3 |
| Polymyositis | 710.4 |
| Regional enteritis of small intestine | 555.∗ |
| Sarcoidosis | 135 |
| Psoriasis and similar disorders | 696 |
| Psoriatic arthropathy | 696.0 |
| Other psoriasis and similar disorders excluding psoriatic arthropathy | 696.1 |
| Other psoriasis and similar disorders | 696.8 |
| Reiter disease | 099.3 |
| Palindromic rheumatism | 719.3 |
| Ankylosing spondylitis | 720.∗ |
| Thoracic spondylosis without myelopathy | 721.2 |
| Lumbosacral spondylosis without myelopathy | 721.3 |
| Hashimoto thyroiditis | 245.2 |
| Toxic diffuse goiter | 242.0 |
| Myasthenia gravis | 358.0∗ |
| Neonatal myasthenia gravis | 775.2 |
| Raynaud syndrome | 443 |
Controls: To be considered a control, a record had to contain none of the following:
-
•
Text match for the string “multiple sclerosis.”
-
•
A multiple sclerosis ICD-9 code (340, 341.8, 341.9, or 377.3).
-
•
A multiple sclerosis medication.
-
•
Any free-text references to key autoimmune diseases: rheumatoid arthritis, Felty syndrome, juvenile rheumatoid arthritis, lupus, inflammatory bowel disease, Crohn disease, ulcerative colitis, sarcoidosis, ankylosing spondylitis, Hashimoto thyroiditis, reactive arthritis, polymyositis, dermatomyositis, chronic lymphocytic thyroiditis, autoimmune thyroid disease, Graves disease, Raynaud disease, multiple sclerosis.
Rheumatoid Arthritis
Definite cases: These were defined as any record that met all of the following criteria:
-
•Contains a rheumatoid arthritis ICD-9 code (Table A5).
Table A5.
ICD-9 Codes for Rheumatoid ArthritisDescription ICD-9 Code Rheumatoid arthritis and other inflammatory polyarthropathies 714 Rheumatoid arthritis 714.0 Felty syndrome 714.1 Other rheumatoid arthritis with visceral or systemic involvement 714.2 -
•
Contains a rheumatoid arthritis medication (any of the following): methotrexate, sulfasalazine, minocycline, hydroxychloroquine, adalimumab, etanercept, infliximab, gold, azathioprine, rituximab, anakinra, abatacept, leflunomide.
-
•
Contains a text match for “rheumatoid arthritis” in any clinical note.
-
•
Does not contain any of the following autoimmune diseases or inflammatory arthritides, by ICD-9 code (Table A6) or by text match: juvenile rheumatoid arthritis, inflammatory osteoarthritis, reactive arthritis.
As with multiple sclerosis, we analyzed rheumatoid arthritis cases with and without the autoimmune disease exclusions. Those with potentially overlapping autoimmune diseases were referred to as “probable cases.”
Table A6.
ICD-9 Codes Indicating Other Autoimmune Diseases or Inflammatory Arthritides
| Description | ICD-9 Code |
|---|---|
| Reiter disease | 099.3 |
| Sarcoidosis | 135 |
| Toxic diffuse goiter | 242.0 |
| Hashimoto thyroiditis | 245.0 |
| Gouty arthropathy | 274.0 |
| Multiple sclerosis and other demyelinating diseases | 340, 341.9, 323.9 |
| Myasthenia gravis | 358.0∗, 775.2 |
| Raynaud syndrome | 443.0 |
| Crohn disease | 555.∗ |
| Ulcerative colitis | 556.∗ |
| Irritable bowel syndrome | 564.1 |
| Lupus | 695.4,710.0, 373.34 |
| Psoriasis | 696.∗ |
| Systemic sclerosis, Sjogren disease, and polymyositis | 710.1,710.2. 710.3, 710.4 |
| Juvenile rheumatoid arthritis | 714.3∗ |
| Osteoarthrosis | 715.∗ |
| Palindromic rheumatism | 719.∗ |
| Ankylosing spondylitis and other spondylosis | 720.∗, 721.2, 721.3 |
| Rheumatism, unspecified and fibrositis | 729.0 |
Controls: These were defined as any record that did not contain any of the following:
-
•Any ICD-9 code for rheumatoid arthritis, autoimmune diseases, or other inflammatory arthritides (Table A7).
Table A7.
ICD-9 Code for Rheumatoid Arthritis, Autoimmune Diseases, or Other Inflammatory ArthritidesDescription ICD-9 Code Reiter disease 099.3 Sarcoidosis 135 Toxic diffuse goiter 242.0 Hashimoto thyroiditis 245.0 Gouty arthropathy 274.0 Multiple sclerosis and other demyelinating diseases 340, 341.9, 323.9 Myasthenia gravis 358.0∗, 775.2 Raynaud syndrome 443.0 Crohn disease 555.∗ Ulcerative colitis 556.∗ Irritable bowel syndrome 564.1 Lupus 695.4, 710.0, 373.34 Psoriasis 696.∗ Systemic sclerosis, Sjogren disease, and polymyositis 710.1, 710.2. 710.3, 710.4 Rheumatoid arthritis and other inflammatory polyarthropathies 714.∗ Osteoarthrosis 715.∗ Palindromic rheumatism 719.∗ Ankylosing spondylitis and other spondylosis 720.∗, 721.2, 721.3 Rheumatism, unspecified and fibrositis 729.0 -
•
Any of the following text in clinical notes: lupus, inflammatory bowel disease, Crohn disease, ulcerative colitis, multiple sclerosis, transverse myelitis, progressive systemic sclerosis, scleroderma, acrosclerosis, dermatomyositis, polymyositis, sarcoidosis, psoriasis / psoriatic arthritis, arthritis, osteoarthritis / degenerative joint disease, reactive arthritis, Sjogren disease, rheumatism, ankylosing spondylitis, Hashimoto thyroiditis, chronic lymphocytic thyroiditis, autoimmune thyroid disease, Graves disease, myasthenia gravis, Raynaud disease.
Type 2 Diabetes
We adapted the type 2 diabetes algorithm designed by William Lowe, Abel Kho, and Wendy Wolf at Northwestern University as part of the eMERGE Network to identify patients at Vanderbilt. Notably, ∼20% of the records identified as type 2 diabetes cases also contained diagnostic codes for type 1 diabetes. Manual review (described in the main text) of 50 of these cases revealed that all had type 2 diabetes.
Definite cases:
-
•
Any record including a type 2 diabetes ICD-9 code (Table A8).
-
•
A noninsulin hypoglycemic medication (Table A9).
Controls:
-
•
No type 2 diabetes ICD-9 codes (Table A8).
-
•
No impaired glucose ICD-9 codes (Table A9).
-
•
No family history of diabetes (including type 1, type 2, or unspecified, as identified by keyword searches of family history sections from clinical notes and problem lists), because patients with a family history of diabetes are more likely to develop diabetes in the future (and thus end up being cases).
-
•No abnormal labs: glucose < 110 mg/dl, hemoglobin A1c < 6.0%.
Table A8.
ICD-9 Codes Used for Defining Type 2 Diabetes CasesDescription ICD-9 Code Diabetes II with other coma 250.30 250.32 Diabetes II with hyperosmolarity 250.20 250.22 Diabetes II with unspecified complication 250.90 250.92 Diabetes II with other unspecified manifestation 250.80 250.82 Diabetes II with peripheral circulatory disorder 250.70 250.72 Diabetes II with neurological manifestations 250.60 250.62 Diabetes II with ophthalmic manifestations 250.50 250.52 Diabetes II with renal manifestations 250.40 250.42 Diabetes II without mention of complication 250.00 250.02 Table A9.
ICD-9 Code Exclusions for Type 2 Diabetes ControlsDescription ICD-9 Code Impaired fasting glucose 790.21 Impaired oral glucose tolerance test 790.22 Abnormal glucose, not otherwise specified 790.29 Glycosuria 791.5 Gestational diabetes 648.∗ Dysmetabolic syndrome 277.7 Diabetes—asymptomatic 790.29 Diabetes (all) 250.∗ Renal glycosuria 271.4
Supplemental Data
Supplemental Data include four tables and can be found with this article online at http://www.ajhg.org.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Electronic Medical Records and Genomics (eMERGE) Network, http://www.gwas.net
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Unified Medical Language System, http://www.nlm.nih.gov/pubs/factsheets/umls.html
Acknowledgments
Development of each phenotype definition required input from clinicians who were expert in that disease: C. Michael Stein, Subramaniam Sriram, David Schwartz, Hiroshi Watanabe, Dawood Darbar, and Prince Kannankeril (all from Vanderbilt University School of Medicine), as well as William Lowe and Abel Kho (from Northwestern University). The Vanderbilt DNA Core Resource that houses all of the samples also performed the genotyping under the supervision of Cara Sutcliffe. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and analytical support for this work. Deede Wang and Sunny Wang assisted with the phenotype algorithms. This work was supported in part by U01 HG04603, a site of the National Human Genome Research Institute's eMERGE Network. The data sets used for the analyses described were obtained from Vanderbilt University Medical Center's BioVU biobank, supported in part by the Vanderbilt Institute for Clinical and Translational Research (VICTR), as well as a a Clinical and Translational Science Award (CTSA) grant (1UL1 RR024975-01 from the National Center for Research Resources and the National Institutes of Health [NIRR/NIH]).
References
- 1.Stead W.W. Rethinking electronic health records to better achieve quality and safety goals. Annu. Rev. Med. 2007;58:35–47. doi: 10.1146/annurev.med.58.061705.144942. [DOI] [PubMed] [Google Scholar]
- 2.Mahoney C.D., Berard-Collins C.M., Coleman R., Amaral J.F., Cotter C.M. Effects of an integrated clinical information system on medication safety in a multi-hospital setting. Am. J. Health Syst. Pharm. 2007;64:1969–1977. doi: 10.2146/ajhp060617. [DOI] [PubMed] [Google Scholar]
- 3.Walsh K.E., Landrigan C.P., Adams W.G., Vinci R.J., Chessare J.B., Cooper M.R., Hebert P.M., Schainker E.G., McLaughlin T.J., Bauchner H. Effect of computer order entry on prevention of serious medication errors in hospitalized children. Pediatrics. 2008;121:e421–e427. doi: 10.1542/peds.2007-0220. [DOI] [PubMed] [Google Scholar]
- 4.Galanter W.L., Hier D.B., Jao C., Sarne D. Computerized physician order entry of medications and clinical decision support can improve problem list documentation compliance. Int. J. Med. Inform. 2008 doi: 10.1016/j.ijmedinf.2008.05.005. Published online July 1, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Kazley A.S., Ozcan Y.A. Do hospitals with electronic medical records (EMRs) provide higher quality care?: an examination of three clinical conditions. Med. Care Res. Rev. 2008;65:496–513. doi: 10.1177/1077558707313437. [DOI] [PubMed] [Google Scholar]
- 6.Ammenwerth E., Schnell-Inderst P., Machan C., Siebert U. The effect of electronic prescribing on medication errors and adverse drug events: a systematic review. J. Am. Med. Inform. Assoc. 2008;15:585–600. doi: 10.1197/jamia.M2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holdsworth M.T., Fichtl R.E., Raisch D.W., Hewryk A., Behta M., Mendez-Rico E., Wong C.L., Cohen J., Bostwick S., Greenwald B.M. Impact of computerized prescriber order entry on the incidence of adverse drug events in pediatric inpatients. Pediatrics. 2007;120:1058–1066. doi: 10.1542/peds.2006-3160. [DOI] [PubMed] [Google Scholar]
- 8.Kaushal R., Jha A.K., Franz C., Glaser J., Shetty K.D., Jaggi T., Middleton B., Kuperman G.J., Khorasani R., Tanasijevic M., Bates D.W., Brigham and Women's Hospital CPOE Working Group Return on investment for a computerized physician order entry system. J. Am. Med. Inform. Assoc. 2006;13:261–266. doi: 10.1197/jamia.M1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood G.C., Still C.D., Chu X., Susek M., Erdman R., Hartman C., Yeager S., Blosky M.A., Krum W., Carey D.J. Association of chromosome 9p21 SNPs with cardiovascular phenotypes in morbid obesity using electronic health record data. Genomic Med. 2008;2:33–43. doi: 10.1007/s11568-008-9023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell D.R., Mitchell J.A. Status of clinical gene sequencing data reporting and associated risks for information loss. J. Biomed. Inform. 2007;40:47–54. doi: 10.1016/j.jbi.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman M.A. The genome-enabled electronic medical record. J. Biomed. Inform. 2007;40:44–46. doi: 10.1016/j.jbi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Sax U., Schmidt S. Integration of genomic data in Electronic Health Records—opportunities and dilemmas. Methods Inf. Med. 2005;44:546–550. [PubMed] [Google Scholar]
- 13.Ginsburg G.S., Burke T.W., Febbo P. Centralized biorepositories for genetic and genomic research. JAMA. 2008;299:1359–1361. doi: 10.1001/jama.299.11.1359. [DOI] [PubMed] [Google Scholar]
- 14.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudbjartsson D.F., Arnar D.O., Helgadottir A., Gretarsdottir S., Holm H., Sigurdsson A., Jonasdottir A., Baker A., Thorleifsson G., Kristjansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 16.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., Roberts R.G., Nimmo E.R., Cummings F.R., Soars D., Wellcome Trust Case Control Consortium Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., NIDDK IBD Genetics Consortium, Belgian-French IBD Consortium, Wellcome Trust Case Control Consortium Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., Ivinson A.J., International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 20.Gregory S.G., Schmidt S., Seth P., Oksenberg J.R., Hart J., Prokop A., Caillier S.J., Ban M., Goris A., Barcellos L.F., Multiple Sclerosis Genetics Group Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 21.Julià A., Ballina J., Cañete J.D., Balsa A., Tornero-Molina J., Naranjo A., Alperi-López M., Erra A., Pascual-Salcedo D., Barceló P. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58:2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 22.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 23.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groves C.J., Zeggini E., Minton J., Frayling T.M., Weedon M.N., Rayner N.W., Hitman G.A., Walker M., Wiltshire S., Hattersley A.T., McCarthy M.I. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55:2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 25.Dupont W.D., Plummer W.D. Power and sample size calculations. A review and computer program. Control. Clin. Trials. 1990;11:116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 26.Giuse D.A. Supporting communication in an integrated patient record system. AMIA Annu. Symp. Proc. 2003:1065. [PMC free article] [PubMed] [Google Scholar]
- 27.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaeger R., Avila-Bront A., Abdul K., Nolan P.C., Grann V.R., Birchette M.G., Choudhry S., Burchard E.G., Beckman K.B., Gorroochurn P. Comparing genetic ancestry and self-described race in african americans born in the United States and in Africa. Cancer Epidemiol. Biomarkers Prev. 2008;17:1329–1338. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolio T.A. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics. 2009;10:235–241. doi: 10.2217/14622416.10.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilke R.A., Berg R.L., Peissig P.L., Kitchner T.E., Sijercic B., McCarty C.A., McCarty D.J. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin. Med. Res. 2007;5:1–7. doi: 10.3121/cmr.2007.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis J.P., Trikalinos T.A., Khoury M.J. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am. J. Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 32.Denny J.C., Smithers J.D., Miller R.A., Spickard A. “Understanding” medical school curriculum content using KnowledgeMap. J. Am. Med. Inform. Assoc. 2003;10:351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denny J.C., Miller R.A., Johnson K.B., Spickard A. Development and evaluation of a clinical note section header terminology. AMIA. Annu. Symp. Proc. 2008:156–160. [PMC free article] [PubMed] [Google Scholar]
- 34.Denny J.C., Miller R.A., Waitman L.R., Arrieta M.A., Peterson J.F. Identifying QT prolongation from ECG impressions using a general-urpose Natural Language Processor. Int. J. Med. Inform. 2008;78(Suppl 1):S34–S42. doi: 10.1016/j.ijmedinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


